Significance
Endocytosis plays a fundamental role in Wnt signaling, leading to the sequestration of cytosolic GSK3 kinase inside multivesicular endosomes. Here we show an unexpected role for the enzyme protein arginine methyltransferase 1 (PRMT1) during Wnt signaling. In the case of the soluble tumor suppressor transcription factor Smad4, modification of a particular arginine was required before this protein could be phosphorylated by GSK3. Wnt3a addition caused rapid endocytosis that strikingly expanded the cellular liquid-phase compartment within 5–20 min. Wnt-induced vesicles contained arginine-methylated proteins, GSK3 and PRMT1 itself, within membrane-bound organelles. The translocation of these proteins into multivesicular endosomes via microautophagy was essential for canonical Wnt signaling.
Keywords: endocytosis, methyl Arginine, microautophagy, lysosomes, multivesicular bodies
Abstract
Arginine methylation has emerged as a widespread and reversible protein modification with the potential to regulate a multitude of cellular processes, but its function is poorly understood. Endolysosomes play an important role in Wnt signaling, in which glycogen synthase kinase 3 (GSK3) becomes sequestered inside multivesicular bodies (MVBs) by the process known as microautophagy, causing the stabilization of many proteins. Up to 20% of cellular proteins contain three or more consecutive putative GSK3 sites, and of these 33% also contain methylarginine (meArg) modifications. Intriguingly, a cytoskeletal protein was previously known to have meArg modifications that enhanced subsequent phosphorylations by GSK3. Here, we report the unexpected finding that protein arginine methyltransferase 1 (PRMT1) is required for canonical Wnt signaling. Treatment of cultured cells for 5–30 min with Wnt3a induced a large increase in total endocytic vesicles which were also positive for asymmetric dimethylarginine modifications. Protease protection studies, both biochemical and in situ in cultured cells, showed that many meArg-modified cytosolic proteins became rapidly translocated into MVBs together with GSK3 and Lys48-polyubiquitinated proteins by ESCRT-driven microautophagy. In the case of the transcription factor Smad4, we showed that a unique arginine methylation site was required for GSK3 phosphorylation and Wnt regulation. The enzyme PRMT1 was found to be essential for Wnt-stimulated arginine methylation, GSK3 sequestration, and canonical Wnt signaling. The results reveal a cell biological role for PRMT1 arginine methylation at the crossroads of growth factor signaling, protein phosphorylation, membrane trafficking, cytosolic proteolysis, and Wnt-regulated microautophagy.
Arginine methylation is emerging as a fundamental protein modification as it is reversible and as prevalent as phosphorylation in both the nucleus and cytoplasm (1–3). The significance of methylarginine (meArg) in cell signaling and metabolism is incompletely understood. Protein arginine methyltransferase 1 (PRMT1) functions in cell signaling, playing an essential role in initiating bone morphogenetic protein (BMP)-induced Smad1 activation (4). In addition, Albrecht et al. (5) had previously reported that meArg modification by PRMT1 is required for initiating the subsequent phosphorylation of desmoplakin by GSK3, raising the intriguing possibility of an association between arginine methylation and Wnt signaling.
The canonical Wnt pathway is essential in cell physiology and signals through the regulation of GSK3 activity (6). Wnt induces GSK3 to become sequestered together with the Wnt coreceptors Frizzled and low-density lipoprotein receptor-related protein 6 (LRP6) and other proteins inside late endosomes/multivesicular bodies (MVBs) through microautophagy (7, 8). Microautophagy is an underappreciated cell biological process by which cytosolic proteins become incorporated into late endosomes/lysosomes through the invagination of small intraluminal vesicles (9–12). In the absence of Wnt, GSK3 phosphorylation marks proteins for Lys48 polyubiquitination (polyUb) and degradation in proteasomes. Wnt-induced GSK3 sequestration into MVBs causes the stabilization of many cytosolic proteins, concurrently with microautophagy of a subset of GSK3-phosphorylated and Lys48–polyUb-modified cytosolic substrates into endolysosomes (13, 14). GSK3 may phosphorylate up to 20% of the human proteome, and we observed that 33% of meArg proteins (15) were also putative GSK3 substrates (7).
These threads of evidence prompted us to test whether meArg was involved in the Wnt/GSK3 signaling pathway. Here we report that Wnt treatment triggers arginine methylation of protein substrates, which become engulfed inside late endosomes/MVBs together with GSK3 within 5–30 min. This is accompanied by a striking increase in endocytic vesicles. Translocation of cytosolic GSK3 substrates such as Smad4 and Lys48-polyubiquitinated proteins (14) into endolysosomes required arginine methylation. PRMT1 was sequestered together with GSK3 inside MVBs and its activity was essential for Wnt signaling. The results uncover unexpected connections between arginine methylation, protein phosphorylation, Wnt signaling, cytosolic proteolysis, membrane trafficking, and regulated microautophagy.
Results
Wnt3 Triggers Rapid Accumulation of meArg Proteins Inside Membrane-Bound Organelles.
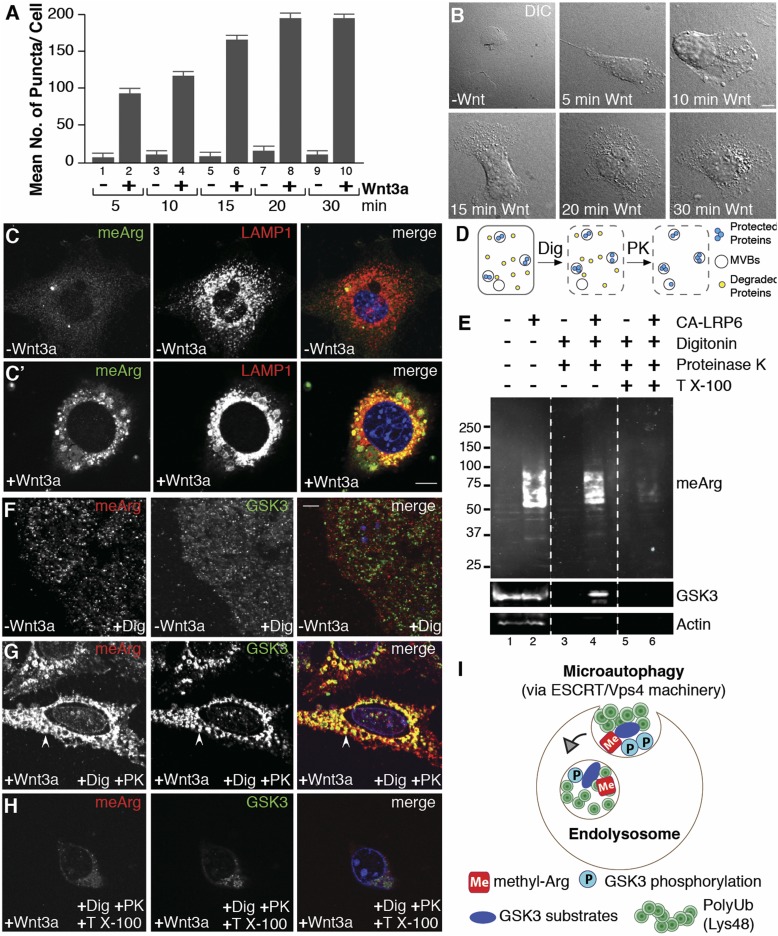
We first noticed a striking increase in puncta or vesicles visible by phase contrast microscopy after only 5–15 min of Wnt treatment (Fig. 1 A and B), both with purified Wnt3a protein or conditioned medium (SI Appendix, Fig. S1 A–F). Large vesicles positive for asymmetric dimethylarginine monoclonal antibody were observed in 3T3 cells, and their limiting membranes were positive for LAMP1, indicating that they corresponded to late endosomes/lysosomes (Fig. 1C). To determine whether meArg-modified proteins were indeed within vesicles, we used a protease protection assay, which is the gold standard to analyze membrane-bound organelles (16). Digitonin dissolves cholesterol patches on the plasma membrane, making soluble cytosolic proteins accessible to proteinase K degradation while protecting those inside intracellular membrane-bound organelles (Fig. 1D). HeLa cells transfected with constitutively active LRP6 (CA-LRP6), which generates a strong Wnt signal (7), showed an increase in arginine-methylated proteins that was resistant to proteinase K, but only in the absence of Triton X-100 (which dissolves all membranes) (Fig. 1E, lanes 1–6).
Fig. 1.
Wnt3a treatment triggers the accumulation of arginine-methylated proteins inside endolysosomes. (A) Wnt signaling rapidly increased the number of puncta visualized by phase microscopy in HeLa cells (ImageJ). (B) Wnt-induced vesicles accumulated within 5–30 min. (C and C′) Wnt signaling by Wnt3a triggered relocalization of arginine-methylated proteins (meArg) (green) into LAMP1 vesicles (red) in 3T3 cells. Panel C′ was treated with Wnt3a for 20 min. (D) Diagram of protease protection assay. (E) meArg proteins were increased by Wnt activation following CA-LRP6 transfection (lanes 1–2) and became protected from proteinase K treatment (lanes 3–4), but only in the absence of Triton X-100 (lanes 3–6). GSK3 and actin served as MVBs and cytosolic protein controls, respectively. (F–H) In situ protease protection assay showing that Wnt-induced meArg proteins localized inside the same vesicles as GSK3 (arrowhead). (I) During Wnt signaling, cytosolic substrates modified by meArg, GSK3 phosphorylation, and K48-polyUb are sequestered into endolysosomes through microautophagy via the ESCRT/Vps4 machinery. (Scale bars, 10 μm.)
These protease-protection results were confirmed in situ at the cellular level by permeabilizing cells with digitonin on coverslips, followed by proteinase K digestion with or without Triton. Using this method, which to our knowledge is unique, we found that meArg was strongly protected within the same Wnt-induced organelles that sequestered GSK3 (Fig. 1 F–H). These organelles were indeed endolysosomes/MVBs because the intralysosomal portion of LAMP1 was also protected from proteinase K digestion (SI Appendix, Fig. S1 G–I). The results suggested that many cytosolic proteins undergo meArg modification under the influence of Wnt signaling and become sequestered by microautophagy inside endolysosomes (Fig. 1I). The proteins with increased methylation fell in a range of 37–100 kDa and formed a smear that may be consistent with polyubiquitination by Lys48-linked ubiquitin chains which would be expected in Wnt signaling MVBs after proteins are phosphorylated by GSK3 (14).
Wnt Increases Overall Endocytosis and meArg Microautophagy.
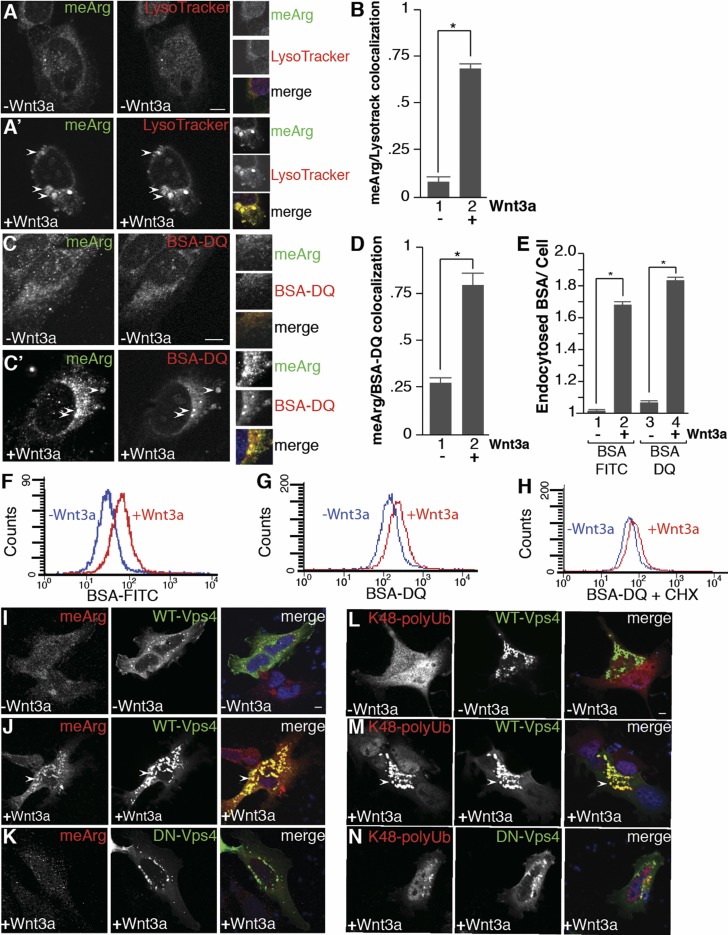
Wnt-induced meArg accumulated within LysoTracker (which accumulates inside acidic organelles) and BSA-dequenched (DQ)-positive (which fluoresces only after BSA is degraded in endolysosomes) vesicles within 15 min of Wnt treatment (Fig. 2 A–D). The increases in colocalization between meArg and lysosomal markers were over threefold. The rapid entry of BSA into the cell after Wnt treatment could be visualized in real time through the accumulation and degradation of BSA-DQ 20 min after the addition of Wnt3a to 3T3 fibroblasts (SI Appendix, compare Movies S1 and S2). Remarkably, the degradation of BSA-DQ took place in the presence of 10% serum; since the endocytosis of BSA is nonreceptor mediated, this suggests that Wnt induces a large increase in endolysosomal trafficking. In agreement with this, total endocytosis into the liquid-phase compartment of HeLa cells was strongly increased by Wnt3a after 15 min of addition of either BSA-FITC or BSA-DQ (Fig. 2E). Quantification by flow cytometry confirmed this increase in overall endocytosis, even in the presence of cycloheximide (Fig. 2 F–H). Thus, canonical Wnt signaling emerges as a major regulator of the endocytic capacity of the cell.
Fig. 2.
Wnt signaling increases endocytosis, and arginine-methylated proteins are sequestered by microautophagy. (A and A′) meArg colocalized with LysoTracker. Panel A′ was treated with Wnt3a for 15 min. (B) Wnt increased colocalization of meArg puncta with LysoTracker endolysosomes by Pearson’s correlation coefficient. (C and C′) Methylarginine and BSA-DQ lysosomal marker (arrowheads) colocalize with Wnt3a. Panel C′ was treated with Wnt3a for 15 min. (D) Wnt increased colocalization of meArg puncta with BSA-DQ endolysosomes by Pearson’s correlation coefficient. (E) Wnt signaling (15 min) increased endocytosis of BSA-FITC and BSA-DQ into the cell liquid phase by quantification of fluorescence per cell; n > 20 cells per sample. (F and G) Wnt signaling increased BSA-FITC and BSA-DQ uptake by flow cytometry (4-h treatment) (36). (H) Blocking protein synthesis with cycloheximide did not prevent Wnt-induced BSA-DQ endocytosis. (I–K) Wnt-induced meArg sequestration into MVBs marked by Vps4-GFP (arrowheads) was blocked by DN-Vps4. (L–N) Lys48-polyUb proteins colocalized with Vps4-GFP MVBs (arrowheads) 15 min after Wnt3a treatment and this required active Vps4. (Scale bars, 10 μm.) *P < 0.05.
To investigate the cellular mechanism by which meArg proteins are relocalized during Wnt signaling, vacuolar protein sorting-associated protein 4 (Vps4) was transfected into HeLa cells with or without Wnt treatment. Vps4, a component of the ESCRT machinery required for the final stages of intraluminal vesicle formation in MVBs (17), colocalized strongly with meArg proteins following Wnt activation (Fig. 2 I and J). A dominant-negative Vps4 (DN-Vps4-GFP) containing a point mutation within the ATPase domain inhibits microautophagy (18). In the presence of DN-Vps4-GFP, meArg did not accumulate in vesicles even in the presence of Wnt3a (Fig. 2K). Many cytosolic proteins are Lys48-polyubiquitinated and degraded in proteasomes. In the presence of Wnt, however, GSK3-phosphorylated proteins modified by Lys48-polyUb accumulate in MVBs by microautophagy (14). As in the case of meArg, Lys48-polyUb colocalized with Vps4-GFP after Wnt3a addition, but not in the presence of a DN-Vps4-GFP (Fig. 2 L–N). These results indicate that the microautophagy ESCRT machinery is required for Wnt-induced meArg and Lys48-polyUb protein sequestration in MVBs.
Methylation Is Required for GSK3 Sequestration.
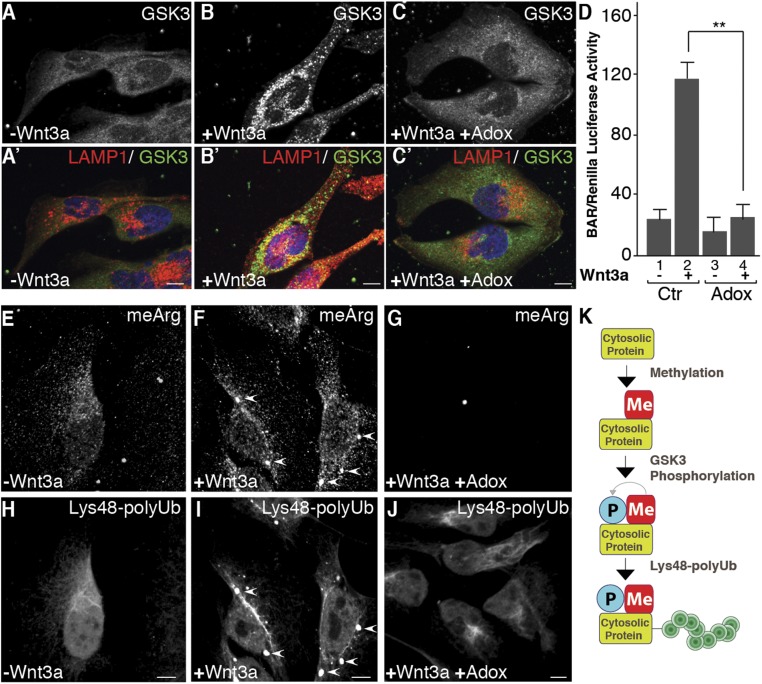
To test whether methylation is required for Wnt-induced GSK3 sequestration, we used the competitive methylation inhibitor adenosine-2′,3′-dialdehyde (Adox). Preincubation of HeLa cells with 20 μM Adox prevented the relocalization of GSK3 into LAMP1-positive vesicles after treatment with Wnt3a for 15 min (Fig. 3 A–C′). In signaling assays, the β-catenin-activated luciferase reporter (BAR) activation by Wnt3a was significantly inhibited in the presence of Adox (Fig. 3D). Adox inhibited arginine methylation (Fig. 3 E–G) and prevented the accumulation of Lys48-polyUb in membrane vesicles (Fig. 3 H–J). Thus, methylation is required for the sequestration of GSK3 and cytosolic substrates during Wnt signaling.
Fig. 3.
Arginine-methylation inhibition blocks GSK3 sequestration and Wnt signaling. (A–C′) GSK3 sequestration into endolysosomes by Wnt signaling was inhibited by the methylation inhibitor Adox (24-h preincubation, 15 min of Wnt3a). Panels indicated with prime were treated with Wnt3a. (D) Adox blocks Wnt signaling (BAR/Renilla assay) (39). (E–J) Wnt-induced meArg and Lys48-polyUb puncta colocalize (arrowheads) and are inhibited by Adox. (K) Model in which soluble cytosolic proteins are first modified by meArg, which facilitates GSK3 phosphorylation and polyubiquitination (see also Fig. 4). (Scale bars, 10 μm.) **P < 0.01.
An Arginine Methylation Site in Smad4 Is Required for Endolysosomal Localization and Wnt Regulation.
The results thus far suggested the translocation of a large number of cellular proteins modified by meArg into lysosomes under the influence of Wnt. To provide a specific example of a cytosolic Wnt-regulated protein that becomes sequestered in MVBs, we chose Smad4 (19, 20), a GSK3 substrate that our laboratory has studied in detail (21) (Fig. 4A). The choice of Smad4, also called deleted in pancreatic carcinoma 4 (DPC4), offered many advantages because it is not only an important tumor suppressor protein that is frequently inactivated in prostate, colorectal, and pancreatic carcinomas, but also a signaling protein for which sensitive luciferase reporter functional assays exist (19, 20). In addition, we had available custom-made antibodies that recognize specifically Smad4 phosphorylated by GSK3 (pSMAD4GSK3) (21) that allow one to follow its intracellular localization.
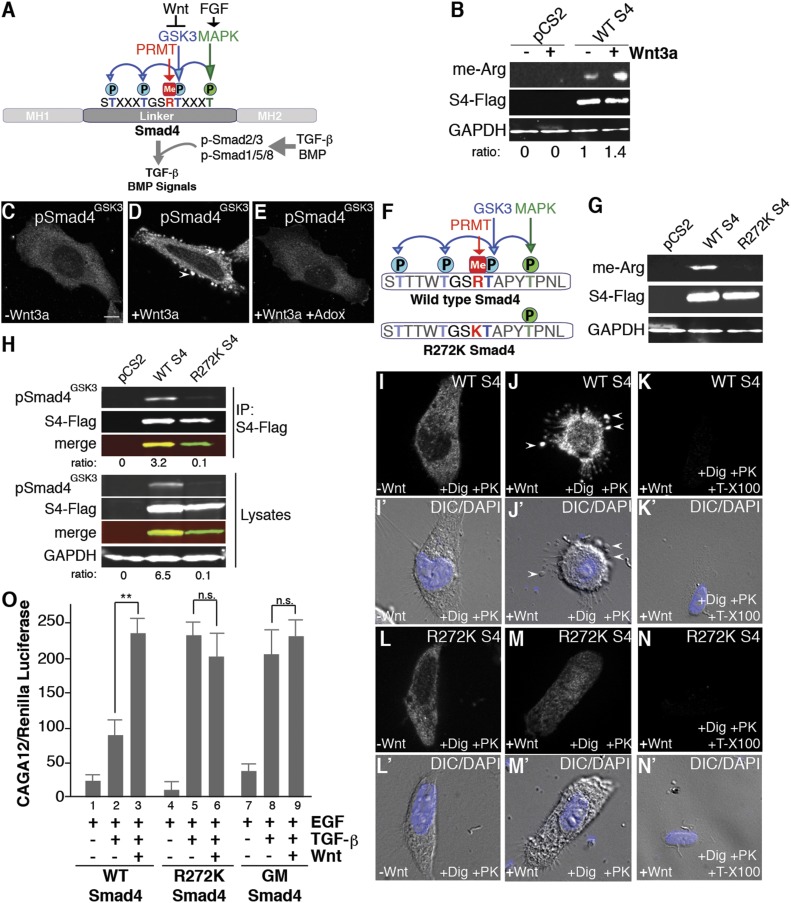
Fig. 4.
Smad4 provides an example of a protein that must be methylated before it can be phosphorylated by GSK3 and translocated into MVBs by Wnt signaling. (A) Diagram of how FGF/EGF, Wnt, and TGF-β/BMP signaling cross-talk at the level of Smad4. MAPK/FGF (green) primes phosphorylation by GSK3 (blue) at three sites; the meArg site discovered in this study is shown in red. (B) Wnt addition for 20 min increased Smad4 methylation in transfected HEK-293T cells. S4-Flag and GAPDH serve as loading controls. (C–E) Phospho-Smad4-Flag relocalized to vesicular structures after 15 min of Wnt3a addition, but only in the absence of the competitive methylation inhibitor Adox. (F) A potential Smad4 arginine-methylation site was mutated (R272K) to prevent arginine methylation with minimal effect on the protein. (G) Smad4-Flag-WT immunoprecipitated from transfected HEK-293T lysates was recognized by asymmetric dimethyl-Arg antibody while the Smad4-Flag-R272K mutant was not. Thus, Smad4 contains a single meArg site. (H) Smad4 phosphorylation by GSK3 requires arginine methylation. Ratios under each lane and the merge panels indicate GSK3 phosphorylated Smad4/total Smad4-Flag. (I–N) In situ protease protection assay using digitonin and proteinase K showing that wild-type Smad4-Flag was translocated inside membrane-bound organelles when Wnt was added for 15 min (J) but digested when Triton X-100 was added (K). Smad4-R272K-Flag was not translocated into membrane vesicles and was degraded by proteinase K (L and M). Panels I′–N′ show DAPI staining and differential interference contrast microscopy to visualize cellular contours in the corresponding cells shown above. (O) Smad4 wild type (WT), Smad4-R272K, and Smad4 mutated at the three GSK3 sites (phosphorylation-resistant Smad4-GM) were tested in TGF-β signaling assays. HaCaT cells permanently transfected with the CAGA12-luciferase reporter and constitutive CMV-Renilla (in which MAPK activation was primed by addition of EGF) were used. This indicates that in the context of TGF-β signaling arginine methylation is required for the integration of FGF, Wnt, and TGF-β signals. **P < 0.01; n.s., not significant statistically.
The transcription factor Smad4 functions as a co-Smad that binds to receptor-phosphorylated Smads (R-Smads) and is an essential component of the transforming growth factor β (TGF-β) and BMP signaling pathways (22) (Fig. 4A). Smad4 is a soluble protein located in the cytoplasm or in the nucleus depending on TGF-β/BMP signaling, which causes Smad4 binding to R-Smads phosphorylated by TGF-β or BMP receptors (Fig. 4A). Until recently, Smad4 was considered a constitutive and unregulated signaling component, but work from our laboratory has shown that the linker (middle) region of Smad4 contains a site phosphorylated by mitogen-activated protein kinase (MAPK) activated by FGF or EGF receptors and three GSK3 phosphorylation sites inhibited by Wnt (21) (Fig. 4A). MAPK phosphorylation promotes maximal activity by activating a Smad transcriptional activation domain and at the same time serves as a priming site for three subsequent inhibitory phosphorylations by GSK3. The GSK3 phosphorylations down-regulate transcriptional activity and recruit the E3 polyubiquitin ligase βTRCP leading to its degradation (21). TGF-β activity signaling is maximal when the FGF/EGF pathway and the Wnt pathway (which inhibits GSK3 phosphorylation) are activated. In this way, Wnt3a increases TGF-β/BMP signaling but only in the presence of FGF/EGF (21). This regulatory mechanism at the level of Smad4 is important for germ layer specification in the embryo and probably also in the function of Smad4 as a barrier against tumor progression (21, 23).
We discovered a putative meArg site Gly-Ser-Arg (GSR) sequence within the GSK3 phosphorylation sites of Smad4 (Fig. 1A, indicated in red). Importantly, this is the same sequence previously found by Albrecht et al. (5) to be arginine-methylated within GSK3 phosphorylation sites in desmoplakin, indicating that Smad4 was an excellent candidate for studying the role of arginine methylation at a key regulatory node at the intersection of important signaling pathways such as TGF-β, FGF/EGF, and Wnt. In Western blots, Smad4 was found to be arginine methylated and this modification was increased by Wnt3a treatment (Fig. 4B). Using a custom-made antibody for phospho-SmadGSK3, we observed that phosphorylated Smad4 was translocated into vesicular structures after 15 min of Wnt3a addition, and that this relocalization was inhibited by preventing methylation with Adox (Fig. 4 C–E). Mutation of Arg-272 into Lys in the GSR region revealed that this was the only meArg site in Smad4 (Fig. 4 F and G). Phosphorylation by GSK3 was greatly decreased or eliminated in the methylation-resistant Smad4R272K mutant (Fig. 4H). Further, in situ proteinase K protection assays showed that while Flag-tagged Smad4WT was translocated inside membrane-bound organelles, the methylation-resistant mutant was not sequestered (Fig. 4 I–N). In TGF-β signaling assays using the CAGA12-luciferase reporter (21), Wnt3a increased TGF-β signaling in the presence of EGF with wild-type transfected Smad4 (Fig. 4O, lanes 2 and 3) as expected, while both methylation- and GSK3 phosphorylation-resistant mutants (Smad4-GM) had maximal activity but were insensitive to Wnt stimulation (Fig. 4O, lanes 4–9). The superactivity of Smad4-GM in which the three GSK3 sites are mutated into alanines had been documented previously (21); in this mutant MAPK phosphorylation is still required for maximal activity. The fact that Smad4-R272K is just as transcriptionally active as Smad4-GM or Smad4-WT treated with Wnt supports the view that the sole arginine-methylation site in Smad4 is required for subsequent phosphorylation by GSK3.
These experiments identify Smad4 as a GSK3 and arginine methylase soluble substrate that undergoes methylation-dependent translocation into protease-protected organelles after the addition of Wnt. Methylation-resistant Smad4 was maximally active but Wnt insensitive, suggesting indirectly that MAPK phosphorylation takes place, even though this construct is not phosphorylated by GSK3. The results are consistent with a biochemical pathway in which meArg modification of proteins facilitates GSK3 phosphorylation which, in turn, facilitates protein degradation in lysosomes (see diagram in Fig. 3K).
PRMT1 Is Required for Canonical Wnt Signaling.
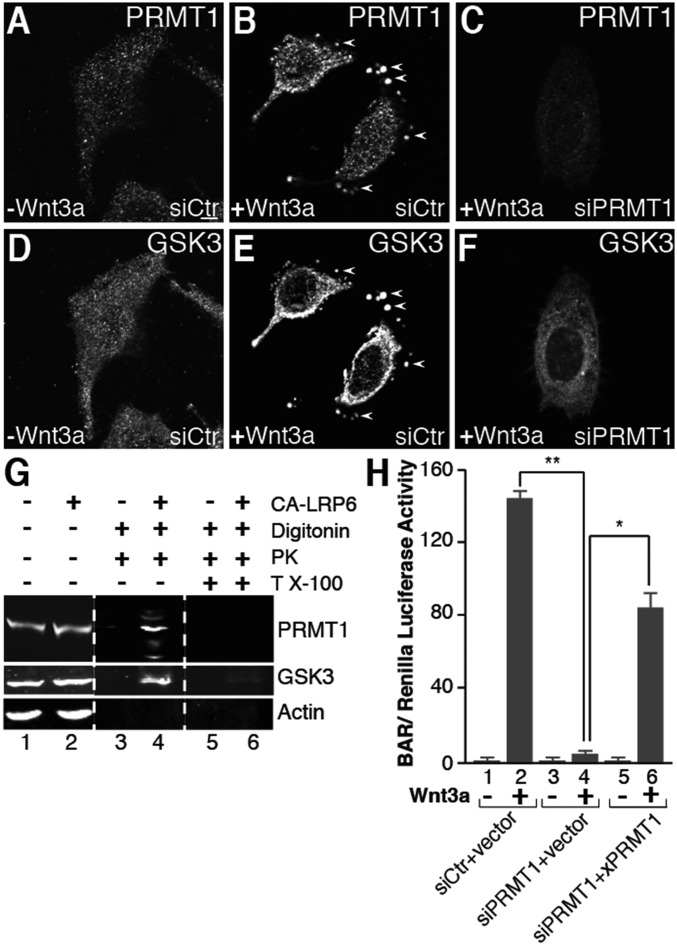
Which arginine methyltransferase is responsible for promoting Wnt signal transduction? There are nine PRMTs in mammals, of which six produce asymmetric dimethylarginine modifications recognized by the antibody used in this study (2, 3). PRMT1 accounts for 85% of cellular meArg (24). In Xenopus, zebrafish, and mouse, PRMT1 is expressed at high levels in intestinal crypt stem cells which express Lrg5, a marker of high Wnt signaling (25). We therefore examined the role of PRMT1 in the context of Wnt signaling. A PRMT1 antibody showed that Wnt caused the relocalization of PRMT1 inside vesicular structures in digitonin-permeabilized cells treated with proteinase K (Fig. 5, compare A and B). These vesicles were the same ones into which GSK3 became sequestered during Wnt signaling (Fig. 5, compare B and E). PRMT1 siRNA depleted PRMT1 levels (Fig. 5C) and prevented the sequestration of GSK3 during Wnt signaling (Fig. 5F). A similar relocalization of PRMT1 was also observed, even in the absence of digitonin, for endogenous or epitope-tagged transfected PRMT1 (SI Appendix, Fig. S2 A–F). In biochemical protease protection assays, PRMT1 became protected from proteinase K digestion together with GSK3 when Wnt signaling was activated (Fig. 5G, compare lanes 4 and 6).
Fig. 5.
Protein arginine methyltransferase 1 (PRMT1) is sequestered in MVBs by Wnt3a and is required for Wnt signaling. (A and B) Endogenous PRMT1 was sequestered in vesicles by Wnt signaling in in situ protease protection assays (arrowheads). (C) PRMT1 was depleted by siRNA. (D and E) GSK3 was sequestered by Wnt3a into the same vesicles as PRMT1 (arrowheads). (F) PRMT1 was required for the Wnt-induced endolysosomal sequestration of GSK3. siRNA targeting PRMT1 ablated GSK3 relocalization into vesicles following treatment with Wnt3a. (G) PRMT1 was protected by proteinase K inside membrane organelles during Wnt signaling. Actin serves as a negative control. (H) PRMT1 was required for Wnt signaling (lanes 2 and 4, bracket). Signaling was partially rescued by expression of a Xenopus PRMT1 (26) in siPRMT1-treated cells (lane 6, bracket). (Scale bars, 10 μm.) *P < 0.05; **P < 0.01.
Importantly, in BAR assays PRMT1 siRNA strongly inhibited Wnt signaling compared with siRNA scrambled control, both in HEK-293T (Fig. 5H, lanes 2 and 4) and in HeLa cells (SI Appendix, Fig. S2G). Wnt inhibition by PRMT1 siRNA was partially rescued by cotransfection of PRMT1 from Xenopus (26) which differs in the siRNA-targeted sequences (Fig. 5H, lanes 4 and 6). These results indicate that PRMT1 is a required enzyme for GSK3 sequestration and canonical Wnt signaling.
Discussion
Endocytosis and Wnt Signaling.
A key finding, which made possible the present study, was that Wnt greatly increased endolysosomes within 5–15 min of signaling. These vesicles were less evident several hours after Wnt treatment, which could explain why this striking phenomenon eluded previous studies (7). LRP6 receptor is thought to be present in small amounts at the cell surface (27), yet the extensive endocytosis suggests an abundance of available Wnt receptors. Wnt stabilizes many proteins in addition to β-catenin (7, 13). Stabilization of GSK3 substrates is so extensive that Wnt increases cell size (13). Wnt signaling is maximal during the G2/M phase of the cell cycle (28) and it has been proposed that this Wnt-stabilization of proteins (Wnt/STOP) serves to slow down GSK3-dependent protein degradation in proteasomes to accumulate materials in preparation for cell division (13, 29). Wnt/STOP is accompanied by a shift in the degradation of cytosolic proteins already phosphorylated by GSK3 from proteasomes into endolysosomes via microautophagy, resulting in the accumulation of Lys48-polyUb in MVBs (14). The present results indicate that meArg greatly facilitates microautophagy during Wnt signaling.
In the case of Smad4, we were able to identify a specific meArg site that is required for its subsequent phosphorylation by GSK3 and endolysosomal translocation. Since Smad4 is a key tumor suppressor at the intersection of TGF-β/BMP, FGF/EGF, and Wnt signaling, the results also suggest a hypothetical role for PRMT1 inhibition in opposing cancer progression via Smad4 (19, 23). In addition, PRMT1 gain of function might also increase Wnt oncogenic signals (30).
The results indicate that PRMT1 is an essential enzyme for GSK3 microautophagy and canonical Wnt signaling. In Xenopus and zebrafish, it has been shown that PRMT1 is highly expressed in the crypt Lrg5-positive intestinal stem cells (25). Importantly, gain- and loss-of-function experiments in Xenopus showed that PRMT1 is essential for intestinal stem cell development (25). Lrg5 is the receptor for R-spondin, which when activated, down-regulates a transmembrane ubiquitin ligase that normally degrades Frizzled receptor, resulting in the maximal Wnt signaling required for development of many stem cell types (6). Our present results suggest that the high expression of PRMT1 in intestinal crypt stem cells may function to facilitate maximal Wnt pathway activity.
Microautophagy-Regulated Protein Degradation.
The Wnt-induced switch from cytosolic proteasomes to the endolysosomal pathway (14) may help conserve energy to promote cell growth as the proteasome consumes large amounts of ATP (31), while microautophagy recycles membrane and cytosolic components via lysosomal hydrolases that do not consume ATP. This switch could provide building blocks for cell growth by degrading in endolysosomes a subset of the proteome composed of PRMT1 and GSK3 Wnt-regulated substrates. There has been a recent renaissance in our understanding of lysosomes and the key role of their limiting membrane in integrating cellular energy metabolism (32–34). Our study extends the central role of endolysosomes to the regulation of intraluminal vesicle formation and microautophagy by Wnt signaling. The results suggest an unforeseen general role for arginine methylation in facilitating GSK3 phosphorylation that may go well beyond the individual cases of desmoplakin (5) and Smad4 (Fig. 4). The mechanism by which PRMT1 and GSK3 are translocated into MVBs is unclear. Axin is a protein that plays a central scaffolding role in the β-catenin destruction complex. Upon addition of Wnt, axin binds to the Wnt receptor complex (6) and is translocated inside MVBs together with GSK3 and the Wnt coreceptors (8). Interestingly, it has been reported that axin is a PRMT1 substrate, and that methylation of Arg-378 in axin increases its binding to GSK3 (35). Thus, axin could provide a possible mechanism for PRMT1 sequestration. However, since many proteins containing GSK3 consensus sites have also been found in mass spectrometry screens to be arginine-methylated proteins, the translocation of PRMT1 and GSK3 into late endosomes could be mediated by a variety of other proteins. Upon Wnt signaling, many proteins methylated by PRMT1 and phosphorylated by GSK3 are rapidly translocated inside endolysosomes by microautophagy. PRMT1 is emerging as a key regulator of growth factor signaling, at least in the case of BMP (4) and Wnt signaling (this study).
Materials and Methods
Antibodies and Reagents.
Antibodies were obtained from the following sources: mouse monoclonal antibody against PRMT1 (Santa Cruz Biotechnology, sc-59648; 1:1,000 for immunostaining and Western blots); antibody against Lamp1 (Cell Signaling, 3243; 1:1,000 for immunostaining); antibodies against asymmetric dimethylarginine modifications (MilliporeSigma, ASYM24; 1:500 for immunostaining or Western blots); and (Abcam 21C7; 1:500 for immunostaining or Western blots); antibody against GSK3 (BD Biosciences, 610201; 1:500 for immunostaining and 1:1,000 for Western blots); antibody against K48-polyubiquitin (MilliporeSigma, 05–1307; 1:1,000 for immunostaining); antibody against Flag (MilliporeSigma, F1804; 1:5,000 for immunostaining or Western blots); antibody against GAPDH (Cell Signaling Technology, 2118; 1:1,000 for Western blots); and actin (MilliporeSigma, A2066; 1:1,000 for Western blots) were used as loading controls for Western blots. Secondary antibodies used were infrared dyes (IRDs) 800CW donkey anti-rabbit IgG (IgG) (LI-COR Biosciences, 926–32213; 1:5,000) and IRDye 680RD donkey anti-mouse IgG (LI-COR Biosciences, 926–68072, 1:5,000). A custom pSmad4GSK3 antibody (1:5,000) was generated previously in our laboratory (21) using the synthetic peptide ([H]-CKK-Acp-NSTTTWT(PO3)GSRT(PO3)APY-[NH2]) to immunize rabbits (Covance).
LysoTracker (Invitrogen, L7528), BSA-DQ (Thermo Fisher, D-12051) and fluorescein BSA conjugate (Thermo Fisher, A23015) were used to monitor lysosomes and fluid-phase endocytosis (36). The BioT transfection reagent (Bioland Scientific) was used for overexpression and siRNA knockdown in cultured cells. Growth factor ligands EGF (MilliporeSigma, E9644) and TGF-β (Sigma-Aldrich, T7039) were used for luciferase assays in CAGA12-HaCaT cells (Fig. 4O). Reagents for protease protection assays were digitonin (Abcam), proteinase K (Invitrogen), and Triton X-100 (MilliporeSigma). Protein A/G sepharose (Abcam, ab193262) was used for immunoprecipitations. Mounting medium for coverslips on slides was from Life Technologies, P36931. Other chemicals used in this study included cycloheximide (Sigma-Aldrich, C-7698; 20 mg/mL) and arginine methyl-inhibitor adenosine-2′,3′-dialdehyde (Adox) (MilliporeSigma, A7154; 20 μM). The QuikChange site mutagenesis kit was used to generate point mutations in Smad4 (Stratagene).
For quantitatively analyzing LysoTracker, BSA-DQ, and BSA-FITC, fluorescence was quantified in control and Wnt3a-treated cells using ImageJ software analyses (n > 20 cells per condition). Briefly, this includes normalizing fluorescence in images and measuring fluorescence in individual cells. Results from three or more independent experiments were presented as the mean ± SEM.
Cell Lines and Tissue Culture.
CAGA12-HaCaT, HeLa, HEK-293T, L cells (ATCC, CRL-2648), as well as L-Wnt3a cells (ATCC, CRL-2647) (37), or NIH 3T3 permanently transfected with a LAMP1-RFP lentivector (38), were cultured in DMEM supplemented with 10% FBS (GIBCO), 1% penicillin/streptomycin, and 1% glutamine. The cells were maintained at 37 °C and 5% CO2.
Plasmids.
Plasmids used in this study were wild-type Vps4-GFP and dominant-negative DN-Vps4-GFP (18); constitutively active LRP6 (CA-LRP6) (7); BAR-Luc and Renilla reporters, a generous gift from Randall Moon, University of Washington, Seattle (39); wild-type Smad4-Flag and GM Smad4-Flag (21), and R272K Smad4-Flag generated in this study by PCR-based site-directed mutagenesis (QuikChange II Site-Directed Mutagenesis, Stratagene). The primers employed to generate the arginine methylation-resistant Smad4-R272K were: 5′-ctaccacctggactggaagtaagactgcaccatac-3′ and 5′-gtatggtgcagtcttacttccagtccaggtggtag-3′. Mutations were confirmed by sequencing. Human PRMT1-pCMV6 was purchased from OriGene (RC224239) and subcloned into pCS2+ using ClaI and StuI restriction enzymes; Xenopus PRMT1 cDNA (26) was a generous gift from Yun-Bo Shi, NIH. A pCS2+ xPRMT1 was generated by PCR amplification followed by an intermediate step of cloning xPRMT1 in the pGEM-4Z vector and excision with BamHI and ClaI restriction enzymes. The human PRMT1 siPRMT1 oligos (Santa Cruz Biotechnology, sc-41069) targeted three regions with the following sequences: 1: 5′-gcaactccatgtttcataa-3′; 2: 5′-gttccagtatctctgatta-3′; and 3: 5′-cgacatgtcttgcatcaaa-3′, all of which were not conserved in the Xenopus sequence which was used to rescue Wnt signaling inhibition by PRMT1 siRNA. The negative control was Scrambled siRNA (Thermo Fisher, D-001810).
Wnt Treatments and Endocytosis.
Recombinant murine Wnt3a protein (315–20) from PeproTech was used for Wnt signaling studies (80 ng/mL). Similar results were obtained with Wnt conditioned medium obtained from L cells stably expressing mouse Wnt3a (ATCC, CRL-2647) or control L cells were used to collect Wnt medium as described (37). A major difference between this study and our previous studies is that for immunoblotting, protease protection, and immunofluorescence studies, Wnt3a was added to cells for short time periods before fixation, ranging from 5 to 20 min at 37 °C. For luciferase assays, Wnt3a was added for 16 h. To measure endocytosis following Wnt activation, we assessed LysoTracker and BSA-DQ or BSA-FITC. Cells were plated onto 12-well dishes (60% confluency) and incubated with 5 μg/mL of BSA-DQ or BSA-FITC in the presence or absence of Wnt3a. For flow cytometry, following 4 h of Wnt activation, cells were washed twice with PBS, fixed in suspension with 4% (wt/vol) paraformaldehyde (PFA) for 15 min at room temperature, and washed with PBS at room temperature. Endocytosis was measured as the intensity of fluorescence from BSA-DQ and BSA-FITC by flow cytometry. Quantification of fluorescence by flow cytometry was performed in an LSRII flow cytometer (Becton Dickinson) as described previously (36). Ten thousand events per sample were collected by FACSDivav6.0 and analyzed by CellQuest software.
For immunofluorescence analyses, cells plated on 12-well dishes were incubated with LysoTracker (1:1,000), BSA-DQ, or BSA-FITC for 15 min in the presence or absence of Wnt3a. Cells were subsequently washed with PBS and subjected to standard immunofluorescence processing which included fixation with 4% PFA for 15 min at room temperature in the dark, washing with PBS, and mounting with ProLong Gold antifade reagent with DAPI (Life Technologies).
Protease Protection Assays.
Protease protection assays were performed to assess whether proteins were contained within membrane-bound organelles (7, 8, 16). For in situ protease protection assays, which to our knowledge are unique, cells were incubated with Wnt3a for 5- to 20-min short time points, placed on ice, washed twice with PBS, incubated with 6.5 μg/mL digitonin in PBS for 15 min on ice, washed twice with ice-cold PBS to remove digitonin, and subsequently incubated with proteinase K (1 μg/mL) for 10 min. As a control to show that proteinase K was active, Triton X-100 (0.1%), which dissolves all membranes, was added together with the protease. The reaction was stopped by fixing with PFA, followed by immunofluorescence analyses as described below. To examine protease-protected proteins by immunoblotting analyses, 2 million cells were plated on 10-cm dishes and transfected with 5 μg of CA-LRP6 to activate Wnt signaling. Then, 24 h after transfection, cells still attached to the culture dish were incubated with a 6.5 μg/mL digitonin solution containing 100 mM potassium phosphate pH 6.7, 5 mM MgCl2, 250 mM sucrose (15 min on ice), washed with PBS, and incubated with proteinase K (1 μg/mL, 10 min), with or without Triton X-100 (0.1%). Finally, the reaction was stopped by adding cell lysis buffer 2× Laemmli sample buffer and boiling samples at 95 °C, before immunoblot analyses. Actin and GAPDH served as cytosolic protein controls.
Immunofluorescence Assays.
Immunofluorescence analyses were performed as described previously (7, 14). In short, cells were grown in 10-cm dishes and replated onto 12-well dishes containing glass coverslips. Cells were allowed to attach to coverslips for 24 h before Wnt treatment. Cells were washed twice with PBS to remove medium, fixed in 4% (wt/vol) PFA (Sigma, P6148) in PBS for 20 min, and permeabilized with 0.2% (vol/vol) Triton X-100 in PBS for 10 min on ice. Next, coverslips were washed with PBS, blocked for 1 h in BSA blocking buffer composed of 5% (vol/vol) goat serum plus 0.5% (wt/vol) BSA at room temperature, and incubated with primary antibodies in blocking buffer overnight at 4 °C (1:100–1:1,000, diluted in blocking buffer). Finally, cells were washed three times with PBS, incubated with secondary antibodies produced in donkey for 45 min (1:1,000, diluted in blocking buffer), and mounted onto glass slides in ProLong Gold antifade reagent with DAPI (Life Technologies) to stain cell nuclei. Images were acquired on a Zeiss Imager Z.1 microscope with Apotome using a 63× oil immersion objective. Zeiss and ImageJ (NIH) imaging software were used for image analyses. The excitation lasers used to capture the images were 488 nm, 568 nm, and 405 nm (DAPI) using Alexa 568- and Alexa 488-conjugated secondary antibodies. To detect asymmetric dimethylarginine in endolysosomes, two antibodies were used. All results shown here were with mouse monoclonal Abcam 21C7; however, rabbit polyclonal Sigma-Aldrich ASYM24 resulted in similar results with respect to Wnt-induced endolysosomes, but had more nuclear staining (presumably because it also reacts with monomethylarginine modifications).
Statistical Analyses and Image Quantification.
Two-tailed t tests were used for two-sided comparisons, and *P < 0.05, **P < 0.01, and ***P < 0.001 were considered to be statistically significant. Wnt-induced vesicles were quantified from light microscopy [differential interference contrast microscopy (DIC)] images using computer-assisted particle analysis in ImageJ software. Individual channels were first given thresholds using MaxEntropy, available in the FIJI ImageJ software package. Next, individual vesicles were separated using the binary watershed function and vesicles were counted using the analyze particles function at particle size 0.2–5 μm, circularity –1. More than 25 cells were counted per experiment, numbered, and outlined copied to a spreadsheet. Pearson’s correlation coefficients were calculated using ImageJ software to assess the degree of colocalization between two different channels.
Additional methods for transfections, immunoprecipitations, Western blots, and luciferase reporter assays can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Yun-Bo Shi (NIH) for the Xenopus PRMT1 constructs; Roberto Zoncu (University of California, Berkeley) for Lamp1-RFP-lentivirus; Caroline Hill (Francis Crick Institute) for CAGA-HaCaT; Randy Moon (University of Washington) for reporter genes; Richard L. Watson (Cedars-Sinai Medical Center), Duane G. Albrecht (University of Texas at Austin), undergraduate students Jane Kwong and Maggie Bui, and other members of the E.M.D.R. laboratory for comments on the manuscript. L.V.A. is supported by an NIH postdoctoral fellowship (NIH F32 GM123622) and N.T.-M., by a University of California Institute for Mexico and the United States (UC MEXUS) and El Consejo Nacional de Ciencia y Tecnologia (CONACYT) postdoctoral fellowship (FE-17-65). This work was supported by the Norman Sprague Endowment and the Howard Hughes Medical Institute, of which E.M.D.R. is an investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804091115/-/DCSupplemental.
References
- 1.Murn J, Shi Y. The winding path of protein methylation research: Milestones and new frontiers. Nat Rev Mol Cell Biol. 2017;18:517–527. doi: 10.1038/nrm.2017.35. [DOI] [PubMed] [Google Scholar]
- 2.Bedford MTM, Clarke SGS. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, et al. Arginine methylation initiates BMP-induced Smad signaling. Mol Cell. 2013;51:5–19. doi: 10.1016/j.molcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht LV, et al. GSK3- and PRMT-1-dependent modifications of desmoplakin control desmoplakin-cytoskeleton dynamics. J Cell Biol. 2015;208:597–612. doi: 10.1083/jcb.201406020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Taelman VF, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinyoles M, et al. Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Mol Cell. 2014;53:444–457. doi: 10.1016/j.molcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 9.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 10.Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem. 1988;263:2506–2512. [PubMed] [Google Scholar]
- 11.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 12.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acebron SP, Karaulanov E, Berger BS, Huang Y-L, Niehrs C. Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol Cell. 2014;54:663–674. doi: 10.1016/j.molcel.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Vick P, Hedtke J, Ploper D, De Robertis EM. Wnt signaling translocates Lys48-linked polyubiquitinated proteins to the lysosomal pathway. Cell Rep. 2015;11:1151–1159. doi: 10.1016/j.celrep.2015.04.048. [DOI] [PubMed] [Google Scholar]
- 15.Larsen SC, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 16.Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 18.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Ding Z, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demagny H, Araki T, De Robertis EM. The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-β signaling. Cell Rep. 2014;9:688–700. doi: 10.1016/j.celrep.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demagny H, De Robertis EM. Smad4/DPC4: A barrier against tumor progression driven by RTK/Ras/Erk and Wnt/GSK3 signaling. Mol Cell Oncol. 2015;3:e989133. doi: 10.4161/23723556.2014.989133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda H, Shi Y-B. An essential and evolutionarily conserved role of protein arginine methyltransferase 1 for adult intestinal stem cells during postembryonic development. Stem Cells. 2010;28:2073–2083. doi: 10.1002/stem.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda H, Paul BD, Choi CY, Hasebe T, Shi Y-B. Novel functions of protein arginine methyltransferase 1 in thyroid hormone receptor-mediated transcription and in the regulation of metamorphic rate in Xenopus laevis. Mol Cell Biol. 2009;29:745–757. doi: 10.1128/MCB.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan Z, Vijayakumar S, de la Torre TV, Rotolo S, Bafico A. Analysis of endogenous LRP6 function reveals a novel feedback mechanism by which Wnt negatively regulates its receptor. Mol Cell Biol. 2007;27:7291–7301. doi: 10.1128/MCB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson G, et al. Cell cycle control of wnt receptor activation. Dev Cell. 2009;17:788–799. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell. 2015;163:1225–1236. doi: 10.1016/j.cell.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 31.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castellano BM, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu X, et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358:813–818. doi: 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abu-Remaileh M, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha B, et al. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 2011;30:2379–2389. doi: 10.1038/onc.2010.610. [DOI] [PubMed] [Google Scholar]
- 36.Ploper D, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 38.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.