Significance
Nucleic acids mediate storage and expression of genetic information. Extracellular DNA (exDNA) and exRNA are traces of nucleic acids released from cells into the extracellular environment. Their use as disease biomarkers has been limited by technical challenges in their isolation caused by abundant RNA- and DNA-degrading enzymes in biofluids. Using isolation protocols developed especially for biofluids, we generated plasma and serum exRNA reference profiles from 13 healthy volunteers over time and determined the effect of critical clinical parameters such as gender and fasting. Surprisingly, we encountered one participant with dramatically increased endocrine-origin exRNA contributions stable over 1 year and detectable in all of his samples, thereby demonstrating the robustness of this approach and the clinical potential of circulating RNAs as biomarkers.
Keywords: extracellular nucleic acids, exRNA reference profiling, exRNA biomarker, biofluid RNA isolation, biofluid DNA isolation
Abstract
Circulating extracellular RNAs (exRNAs) have the potential to serve as biomarkers for a wide range of medical conditions. However, limitations in existing exRNA isolation methods and a lack of knowledge on parameters affecting exRNA variability in human samples may hinder their successful discovery and clinical implementation. Using combinations of denaturants, reducing agents, proteolysis, and revised organic extraction, we developed an automated, high-throughput approach for recovery of exRNAs and exDNA from the same biofluid sample. We applied this method to characterize exRNAs from 312 plasma and serum samples collected from 13 healthy volunteers at 12 time points over a 2-month period. Small RNA cDNA library sequencing identified nearly twofold increased epithelial-, muscle-, and neuroendocrine-cell–specific miRNAs in females, while fasting and hormonal cycle showed little effect. External standardization helped to detect quantitative differences in erythrocyte and platelet-specific miRNA contributions and in miRNA concentrations between biofluids. It also helped to identify a study participant with a unique exRNA phenotype featuring a miRNA signature of up to 20-fold elevated endocrine-cell–specific miRNAs and twofold elevated total miRNA concentrations stable for over 1 year. Collectively, these results demonstrate an efficient and quantitative method to discern exRNA phenotypes and suggest that plasma and serum RNA profiles are stable over months and can be routinely monitored in long-term clinical studies.
The presence of extracellular RNA (exRNA) and exDNA in circulation is typically considered a consequence of cellular turnover or formation of microvesicles from blood cells, platelets, and other phagocytic and secretory cells (1) residing in tissues such as liver (2), placenta (3), endocrine organs (4, 5), or tumors (6), especially under conditions of tissue injury. While the earliest observations date back almost five decades (7–10), systematic study of extracellular nucleic acids became increasingly prominent with the introduction of deep-sequencing technologies (11). Protected by vesicle enclosure and/or associated RNA- and DNA-binding proteins, extracellular nucleic acids persist in cell-free environments despite the presence of secreted and highly active nucleases (12–14). Considering the distinct cellular origins of cell-free nucleic acids, variations in its extracellular concentration and composition may reflect unique metabolic states and disease processes, and determining their sequence composition and abundance has been considered valuable for the discovery of new biomarkers (1, 2, 4, 5, 15–20). In pregnant women, fetal exDNA released from shedding vesicles of the placental syncytiotrophoblast cells contribute 10–50% of total exDNA in maternal circulation, and polymorphic variation specific to the fetus can reveal chromosomal aberrations (21, 22).
Progress in the exRNA field has been hampered by an inability to efficiently and reproducibly recover small quantities of cell-free nucleic acids present in biofluids, nucleic acid size biases intrinsic to existing isolation methods, and rapid degradation of exRNAs by nucleases ubiquitously found in biofluids (23–26) during or following isolation. Varying yield and quality encountered during exRNA isolation prompted us to modify nucleic acid isolation protocols until they became suitable for application to cell-free serum and plasma (27, 28) to recover nanogram quantities typically present in 0.5-mL-sized clinical samples at high reproducibility and purity required for enzymatic cDNA library preparation and Illumina deep sequencing.
To generate reference data of healthy individuals for future biomarker studies, we applied this protocol to determine the extracellular small RNA composition of 312 serum and plasma samples collected over a 2-mo period from 13 healthy volunteers. In the case of one individual with a unique and strongly deviating exRNA signature, an additional time point 1 y later was collected. The sampling intervals and timescale of this study extend beyond existing studies and enable detailed examination of exRNA variation across primary clinical variables such as male vs. female, fasting vs. nonfasting morning and afternoon, time and individual variability, critical for advancement of exRNA composition analysis in biomarker discovery, which has been limited by a lack of knowledge concerning the influence of these basic factors.
Results
Development of an Extracellular Nucleic Acid Isolation Method Suitable for Automation.
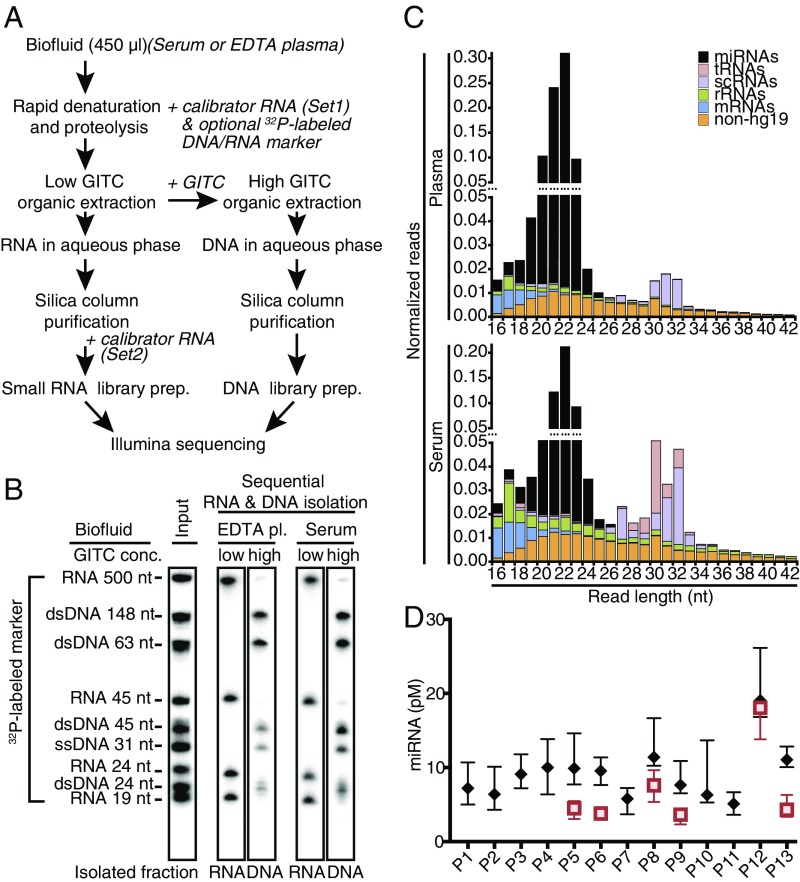
Isolating nucleic acids from cells or biofluids is challenging due to omnipresent nucleases involved in nucleic acid turnover, while physicochemical parameters during purification also strongly impact their recovery. To monitor nucleic acid integrity, partitioning, and recovery during isolation, tracer mixes of 5′-32P–labeled single-stranded 19- to 45-nt RNAs and a 31-nt ssDNA, 5-32P–labeled dsDNA of 24–148 bp, and an internally labeled 500-nt ssRNA in vitro transcript were spiked in with the denaturant or extracting agent before the biofluid was first contacted. This assay enabled us to rectify limitations in commercial extraction-based RNA isolation and column purification approaches, in particular carryover of nucleases or denaturants and recovery and separation of exRNA and exDNA (SI Text: Method Optimization of exRNA and exDNA Isolation and Fig. S1). The resulting protocol consists of (i) hot sample denaturation and proteolytic digestion, (ii) interphase-deprived organic extraction of digested proteins and DNA while keeping RNA in the aqueous phase, (iii) reextraction of DNA from the organic phase, and (iv) individual column purifications of each nucleic acid type using silica matrix columns and vacuum manifolds or centrifugation (Fig. 1 A and B) in small volumes free of residual denaturants and other inhibitors of downstream enzymatic processing steps.
Fig. 1.
exRNA isolation from serum and plasma. (A) Schematic overview of extracellular RNA and DNA isolation procedure detailing addition of calibrator or radiolabeled DNA and RNA size markers. (B) Examples of RNA and DNA isolations from plasma and serum samples visualized by phosphorimaging of 32P-labeled spike-in DNA and RNA tracers. Less than 0.1 pmol of labeled tracer nucleic acids (input lane) were added with the denaturant to individual 450-μL biofluid samples at the first step of the procedure. (C) Average composite read length distributions of 5′P/3′OH-containing exRNAs in platelet-depleted EDTA plasma and serum samples. (D) Individual miRNA concentration ranges in EDTA plasma and serum. miRNA concentrations were calculated using read abundance data of Set1 calibrators added with the denaturant at the beginning of the RNA isolation. Boxplots show first to third interquartile ranges and their medians for plasma (black diamonds) and serum samples (red squares) of all 13 study participants.
EDTA Plasma and Serum exRNA Isolation and Small RNA cDNA Library Preparation.
Platelet-depleted EDTA plasma and serum samples were collected from seven male and six female healthy adults at 12 time points on 4 d evenly spaced over 2 mo (Table S1). On collection days, time points reflected three prandial (food intake) states: AM fasting, 1 h postprandial, and 4 h postprandial. Cell-free exRNA was isolated in batches of up to 96 samples from 216 EDTA plasma samples including 60 technical replicates of all study participants and 96 serum samples including 24 replicates of six study participants (Dataset S1). cDNA libraries were generated by processing 24 samples per batch using barcoded 3′ adapters, and subjecting each batch to single-read 50-bp Illumina HiSeq sequencing (29). To monitor the recovery of exRNAs from each sample and batch, we spiked the denaturant solution at the start of RNA isolation with a defined amount of a mixture of 10 synthetic 5′-phosphorylated nonradioactive 22-nt calibrator RNAs (Set1), which served as an external standard in subsequent analysis. Before preparation of the small RNA cDNA libraries, another mixture of 10 synthetic 5′-phosphorylated nonradioactive 22-nt calibrator RNAs (Set2) was added to the sample. All calibrator RNA sequences were distinct from human genome assembly 19 (hg19) genome or transcriptome by at least two mismatches (30).
We obtained an average of 169 million reads per cDNA library batch. Of these reads, an average of 53% comprised sequence inserts within the targeted size range, while 28% were insertless or comprised repeated adapter sequences. Following barcode extraction, the average serum and plasma sample-derived cDNA library yielded 4.5 and 3.2 million reads, respectively (summarized in Table 1; detailed metadata and read statistics in Dataset S1). Sequence reads were mapped to annotated reference RNAs (31). Of these reads, 3.0 and 2.3 million, respectively, mapped to hg19 including the RNA annotation classes miRNAs, rRNAs, tRNAs, small cytoplasmic RNAs (scRNAs), and mRNAs. For comparison across samples, read counts were normalized and reported as relative read frequencies (31).
Table 1.
Summary table of sequencing reads and annotated read statistics
| Calibrators | RNA category, % | ||||||||||||
| Biofluid | Total | Set 1 | Set 2 | Sample | miRNA | rRNA | tRNA | scRNA | Other cRNA | mRNA | non-hg19 | Technical | |
| Serum, n = 96 | Average | 4,568,021 | 12,090 | 18,909 | 2,984,332 | 53.4 | 8.5 | 6.6 | 9.7 | 1.6 | 5.2 | 14.8 | 1,552,689 |
| Median | 2,190,478 | 6,384 | 10,202 | 1,473,132 | 54.5 | 7.8 | 5.6 | 7.9 | 1.6 | 4.6 | 12.3 | 602,986 | |
| Min | 91,780 | 214 | 723 | 51,795 | 17.7 | 1.0 | 0.3 | 2.1 | 0.7 | 0.8 | 2.5 | 25,798 | |
| Max | 31,993,329 | 124,488 | 193,438 | 23,141,839 | 81.6 | 19.8 | 20.2 | 39.1 | 2.4 | 21.8 | 62.2 | 12,056,077 | |
| Plasma, n = 216 | Average | 3,242,576 | 7,784 | 11,448 | 2,255,037 | 77.8 | 2.8 | 0.6 | 3.1 | 1.2 | 4.7 | 9.8 | 968,308 |
| Median | 2,041,473 | 5,883 | 8,524 | 1,411,639 | 80.9 | 2.5 | 0.4 | 2.5 | 0.9 | 3.7 | 7.0 | 429,009 | |
| Min | 21,955 | 47 | 143 | 13,300 | 26.6 | 0.7 | 0.2 | 0.2 | 0.3 | 0.8 | 1.1 | 2,011 | |
| Max | 19,671,378 | 56,088 | 81,127 | 15,970,087 | 94.4 | 9.2 | 6.5 | 17.5 | 6.6 | 29.5 | 58.7 | 15,767,594 | |
Listed are median, minimal (min), and maximal (max) mapped reads on a per-sample basis for plasma and serum. The technical read category includes reads mapped against plasmid and genome sequences of the expression system used in production of RNA ligase 1 and 2, which are used for the 5′- and 3′-adapter ligations in cDNA library preparation, as well as marker sequences and reaction side products. Individual sample read reports are detailed in Dataset S1.
The relative read frequencies for calibrators varied between 0.1% and 7% per sample, with average read frequencies of 0.7% for Set1 and 1.2% for Set2. Based on relative read frequency for Set1 compared with Set2, an exRNA recovery efficiency of ∼42% was achieved. Rare instances of less than 2% recovery were traced to obstructed microtiter plate wells (production errors) during plate-based nucleic acid purification.
ExRNA Classes and Their Size Distribution in Serum and Plasma of Healthy Volunteers.
The overall composition of plasma and serum by class of RNA was distinct. In plasma, miRNA was captured with a median of 80.9% of the sample-derived nontechnical reads (total reads – technical reads), with less than 4% each of rRNA, scRNA, or mRNA, and 0.4% of tRNA (Table 1). In serum, the miRNA content was lower with a median of 54.5%, while rRNA, scRNA, and tRNA were each near 10%. The relative contribution of reads annotated as mRNA was similar in plasma and serum at a median close to 4%. The read size distribution of each class of RNA was similar in plasma and serum and peaked at 22 nt for miRNAs, 30 nt for tRNAs, and 32 nt for scRNA, while mRNA, rRNA, and nonannotated reads were more broadly distributed between 17 and 40 nt (Fig. 1C and Fig. S2).
The read depth for sample-derived nontechnical reads was ≥100,000 and ≥2,000,000 in 96.2% and 39.7% of all samples, respectively. Thirteen samples with less than 100,000 sample reads were excluded from further analysis.
Extracellular miRNA Content and Composition of Plasma and Serum.
Concentrations of extracellular miRNA in biofluids were calculated based on the ratios of miRNA read to calibrator-Set1 read counts, the molar amount of Set1 spike, and the sample volume. Total miRNA concentration medians in repeat samples of study subjects (excluding subject P12; see below) ranged from 4.4 to 10.9 pM for plasma and 3.8 to 7.7 pM for serum (Fig. 1D).
Relative read frequencies for miRNA and calibrator were reported in separate heatmaps subjected to unsupervised clustering (Fig. 2). Considering samples with ≥2,000,000 reads, calibrator profiles did not cluster by batch, subject, or type of biofluid (Fig. 2A); however, subsets of samples clustered by 3′ adapter reflecting small, yet noticeable sequence biases of enzymatic adapter ligation to a small subset of input RNA sequences (32). Similar results were obtained when calibrators of all samples with ≥100,000 sample reads were included (Fig. S3A), although, in some samples with less than 2,000,000 total reads, calibrator clustering was driven by the absence of infrequent sequences.
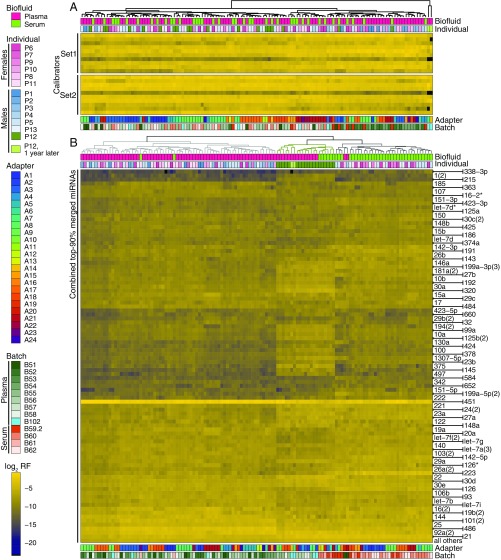
Fig. 2.
Unsupervised clustering of miRNAs from human biofluid samples reveal distinct miRNA signatures for serum and plasma and segregate subject P12 from otherwise indistinguishable study participants. For each sample, individual calibrator and miRNA read frequencies from Dataset S2 are log2-transformed and color-coded. Study subject metadata and library preparation details are color-coded and indicated as annotation. This figure features samples with ≥2,000,000 hg19-mapped reads; an equivalent panel of samples with a lower cutoff ≥100,000 reads is included as Fig. S3. (A) The calibrator heatmap reports unsupervised clustering of two sets of 10 synthetic 22-nt 5′P/3′OH-containing oligoribonucleotides spiked into samples at the beginning of RNA isolation (Set1) and before small RNA cDNA library preparation (Set2). (B) The miRNA heatmap is based on the union of the top 90% of miRNAs present in any sample. Sample order (columns) and miRNA arrangement (rows) were determined by hierarchical clustering. All biofluid samples from study subject P12 (green) clustered together yet were different from the rest of samples.
For unsupervised clustering of miRNA profiles, we selected the union of miRNAs represented within the top 90% of cumulative miRNA-annotated reads of any sample. Clustering separated samples according to biofluid type but not by individual or other metadata (Fig. 2B and Fig. S3B). Surprisingly, all samples of P12 characterized by elevated extracellular RNA content clustered distinctly from other study subjects, although they still showed some separation by biofluid type. Batch effects were unnoticeable, although some samples subclustered by 3′ adapter within each of the biofluid branches.
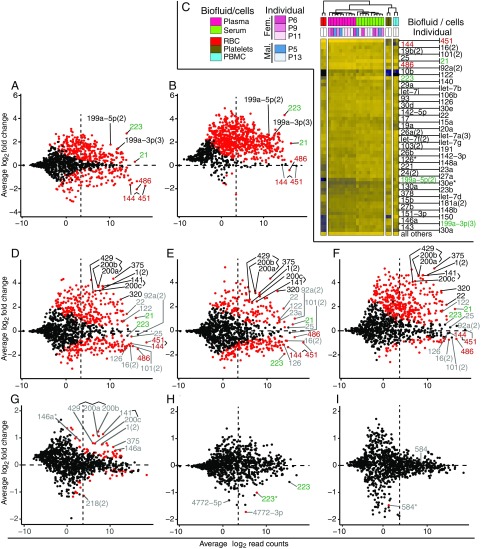
For differential expression analyses (DEAs), miRNA size factor estimation (SFE) for count normalization typically considered all miRNA counts in a sample unless stated otherwise; considering miRNAs with base mean counts of at least 10, DEA of serum (n = 84) vs. plasma (n = 194) samples excluding P12 identified 98 and 15 miRNAs, which showed absolute log2 fold changes (lfcs) ≥1 to <2 and ≥2, respectively (Padj ≤ 0.05, Fig. 3A and Dataset S4, A). The abundant red blood cell (RBC) enriched miR-144, 451, and 486 (Fig. 3C) (33) were prevalent in platelet-depleted EDTA plasma (lfc ≈ −2) used in this study. In contrast, the platelet (PLT) and peripheral blood mononuclear cell (PBMC)-enriched miR-223 (lfc = 2.7) and 199a-5p (lfc = 1.7) (33) were prevalent in serum. Reevaluation of serum-to-plasma comparisons ratios utilizing Set1 calibrator reads as external normalization standards confirmed the approximately threefold reduced abundance of many miRNAs including RBC miRNAs in serum (Dataset S4, A_Set1), which is likely a consequence of increased exRNA degradation or their adsorption to the blood clot during coagulation to obtain serum. The resulting twofold reduction in total individual miRNA concentrations of serum vs. plasma (Fig. 1D) is likely caused by miRNA release from PLTs during coagulation (34). Cellular miRNA profiles from RBC, PBMCs, and PLTs, and 10 representative serum and plasma samples, excluding study subject P12, subjected to unsupervised clustering (Fig. 3C) support this interpretation.
Fig. 3.
Differences in miRNA abundance based on type of biofluid, individual P12, gender, female hormonal cycle, and prandial state. MA plots are based on merged miRNA read counts from Dataset S3 and tabulated values describing lfc values, and their significance can be found in Dataset S4. Significant differences in abundance are indicated by red dots for Padj < 0.05. Blood-based miRNAs are labeled in red (RBCs) and green (platelets/PBMCs). Select examples of additional miRNAs discussed in the text are labeled black when their abundance variance is greater than fourfold (lfcs ≤ −2 or ≥2), while smaller magnitudes are shown in gray. A count of 10 normalized reads, which has been chosen as a lower threshold for data analyses to eliminate noise, is indicated by a dashed vertical line. Count normalization in this analysis considers all miRNA sample reads, unless stated otherwise. (A and B) Serum vs. plasma, excluding individual P12, considering all miRNA reads (A) or utilizing miR-451 and 144 as standards (B) for SFE/miRNA read count normalization. (C) Unsupervised clustering separating miRNA signatures of biofluid representatives and those of RBC, PBMC, and platelets; only a few abundant miRNAs show enrichment in some of these major cellular or cell-derived blood constituents (red, green: higher relative abundance in RBC and platelets/PBMCs, respectively). (D–F) Study subject P12 vs. other male study participants, plasma (D and F) and serum (E). In F, we used Set1 calibrator reads for SFE/read count normalization. (G) Effect of gender, comparing female vs. male (plasma). For the corresponding serum comparison, see Fig. S4A. (H) Effect of menstrual cycle, comparing follicular vs. luteal state, in plasma. For the corresponding serum comparisons, see Fig. S4E. (I) Effect of fasting: preprandial vs. 4-h postprandial state, in plasma. For the 1-h preprandial vs. postprandial state comparison, see Fig. S4F.
Assuming that overall miR-451 und 144 abundances in the biofluid do not notably change during serum formation and may be utilized as internal standards for SFE, miRNA contributions from platelets may increase 20-fold during coagulation compared with platelet-depleted plasma, as suggested by abundance changes of platelet specific miR-223 (Fig. 3B and Dataset S4, A_451,144).
Study Subject P12 Shows a Unique Extracellular miRNA Composition Stable over 1 y.
The total miRNA concentrations of subject P12 corresponded to 19.0 and 18.5 pM for plasma and serum samples, respectively, and were at least twofold higher compared with other healthy controls (Fig. 1D). The difference was highly significant (Padj ≤ 0.0002, Tables S2 and S3), while the other interindividual miRNA concentration comparisons showed no significant systematic differences or sporadic significant differences conserved across biofluid types. Considering an average read count of at least 10 DESeq2-normalized reads, comparison of biofluid samples from study subject P12 to other male healthy volunteers’ differences observed in sera highly correlated with differences found in plasma samples (RPearson = 0.92 for 166 common lfc pairs with Padj ≤ 0.05 in both comparisons). Totals of 115 and 57 significant abundance differences in plasma with absolute lfcs ≥1–2 and ≥2, respectively (Fig. 3D and Dataset S4, B1), were observed, and 93 and 40 in serum (Fig. 3E and Dataset S4, B2) (Padj ≤ 0.05; n = 12 and 22 for P12; n = 36 and 92 for other males in serum and plasma, respectively). Negative lfcs including the most abundant miRNAs in plasma and serum of subject P12 compared with other males as well as across different biofluid types invoked substantial changes in abundance, which were also supported by the doubled miRNA concentrations in P12 (Fig. 1D); in this situation, default miRNA read normalization for DEA may be compromised, because it assumes minimal abundance changes for the majority of miRNAs across all samples to reliably estimate sample size factors, which are based on the medians of individual size factors of each miRNA present in a sample. Reevaluating the DEA of P12 vs. other males utilizing Set1 calibrator counts as external normalization standards returned the most abundant RBC-derived miRNA lfcs close to zero and left only few miRNAs with significant negative lfcs (Fig. 3F and Dataset S4, B1_Set1 and B2_Set1), suggesting a similar miRNA background in P12’s circulation as in the other participants. This result was confirmed utilizing RBC miR-451 and 144 as internal standards for SFE (Dataset S4, B1_451,144 and B2_451,144). Consequently, many more miRNA members including neuroendocrine miR-375, endothelial miR-141 and 200a-c, muscle-specific miR-1, and liver-specific miR-122 (33, 35, 36) now showed greater positive lfcs, indicating an increased contribution from these cell types to miRNAs in the circulation of P12. miRNA abundance differences of similar magnitude were observed, when P12 was compared with all other healthy volunteers. A thorough medical, surgical, and social evaluation both at the time of the initial blood draw and 1 y later, as well as a complete blood count, blood chemistries, liver function tests, and an abdominal MRI were normal (Fig. S5 and Table S4) despite the fact that his signature prevailed (Fig. 2B, samples “P12, 1 y later”).
Sex-Dependent Differences in Serum and Plasma miRNA.
Considering miRNAs with an average abundance ≥10 DESeq2-normalized reads, comparison of samples of female vs. male healthy volunteers showed intermediate correlation between plasma and serum (RPearson = 0.68 for 15 common lfc pairs with Padj ≤ 0.05 in both comparisons). Disregarding study participant P12, 15 and 49 miRNAs in women’s plasma (n = 102) and serum (n = 48), respectively, showed at least 1.5-fold altered abundance relative to men’s plasma (n = 96) and serum (n = 32) (lfcs ≤ −0.6 or ≥0.6, Padj < 0.05; Fig. 3G, Fig. S4A, and Dataset S4, C1 and C2). The maximum miRNA changes in plasma and serum were 2.4- and 4.4-fold, respectively. Among those elevated in women in both biofluids were epithelial cell-type–enriched cistronically organized miR-141, 200c, 429, and 200a, 200b, muscle-specific miR-1, and neuroendocrine-specific miR-375 (33, 35, 36).
To evaluate the extent by which sex-dependent differences are impacted by personal baseline miRNA contributions of volunteers, we performed two approaches using the larger plasma dataset. We first compared two groups composed of three male and three female volunteers each (P1, P3, P5, P9, P10, P14 vs. P2, P4, P6, P7, P11, P13), and observed five miRNAs with at least 1.5-fold altered abundance (lfcs ≤ −0.6 or ≥0.6; Padj < 0.05; Fig. S4D and Dataset S4, C5), including some of the 15 sex-dependent miRNAs observed in plasma above. We also omitted one individual from the male or female groups above and recalculated abundance differences; in particular, omission of female P11 reduced the contributions of epithelial cell-type miRNAs from average 2.0- to average 1.6-fold differences (Padj values ranging from 1.13E-02 to 1.90E-07; Fig. S4 B and C and Dataset S4, C3 and C4). These controls indicate that apparent gender differences are small and influenced by personal miRNA baseline variation.
Comparison of miRNA abundance by follicular (n = 28) and luteal (n = 52) state of the female menstrual cycle identified no significant changes in plasma (Fig. S4E and Dataset S4, D2). In serum (n = 12, 24 for follicular and luteal state, respectively), seemingly significant changes (Padj ≤ 0.05) were observed for two low-abundance passenger miRNAs also referred to as miRNA* (37), for which, however, the corresponding mature miRNA showed lesser and not significant lfcs (Fig. 3H and Dataset S4, D2). We conclude that menstrual cycle states do not affect extracellular miRNA profiles.
Food Intake Does Not Impact Plasma and Serum miRNA Composition.
Comparison of miRNA abundance between fasted (n = 61, 26) and postprandial states identified no significant changes 1 h after food intake (n = 67, 28) in serum and plasma, respectively (Fig. S4F and Dataset S4, E1 and E2). Four hours after food intake (n = 66, 30), one seemingly significant change (Padj ≤ 0.05) was detected in plasma, but not in serum (Fig. 3I and Dataset S4, E2 and E4). As stated above, the variation was observed for one low-abundance passenger strand miRNAs, for which, however, the corresponding much more abundant mature miRNA was unchanged. We therefore conclude that prandial state does not affect plasma or serum miRNA profiles.
Discussion
Our exRNA isolation method facilitated automated, high-throughput sequential recovery of exRNA and exDNA from biofluids with improved speed, yield, and intactness, overcoming size biases and limitations caused by low-input nucleic acid concentrations and high nuclease activities in conventional TRIzol-based extraction and column-based purification methods. Revision of extraction conditions enabled doubling of sample volumes compared with current commercial RNA extraction solutions, allowing the processing of up to 450 µL of biofluid in standard 1-mL deep-well plates compatible with automated liquid handling, as recommended by the latest guidelines for total exRNA recovery from biofluids (https://exrna.org/resources/protocols/). Elimination of interphases, a common limitation in existing extraction methods, which notably affects RNA recovery and increases processing times, allowed full automation of the method, enabling processing of hundreds of samples per day using a standard 96-well liquid handling system. This methodology is already feasible for detailed and cost-effective characterization of large sample collections, and in combination with small RNA cDNA sequencing, it allows screening of large sample collections for exRNA biomarker discovery. Combining our exRNA isolation protocol with RT-qPCR assays for validating a limited set of exRNAs may be a practical and even faster alternative, and adaptable to routine screening applications in the clinic.
For RNA quantification and quality control throughout the entire process of sample handling, we introduced two distinctive sets of 10 22-nt calibrator RNAs each, which do not map against the human genome. The sum of calibrator-annotated reads and their defined molar amounts spiked-in during RNA isolation were utilized to determine miRNA concentrations in biofluids. Calibrator Set1 allows for determination of miRNA concentration before RNA isolation, while calibrator Set2, spiked after RNA isolation, provides information about the RNA recovery yield. Furthermore, the use of calibrators also proved critical to the unanticipated discovery of global twofold elevated extracellular miRNA concentration in plasma and serum of study participant P12, stable over an observation period of more than 1 y. Combining absolute miRNA quantification and calibrator-guided scale factor estimation in DEA helped to overcome normalization biases caused by differences in global sample composition changes and allowed to discover and dissect the origins and background contributions of extracellular miRNAs. Because external standardization using calibrators appears less prone to changes in global composition than internal standardization approaches, we expect it to be beneficial to acquire composition changes more accurately in samples, which differ considerably in their exRNA compositions such as different biofluid types.
Biofluid type, serum or plasma, was a biological variable that impacted the miRNA profile and therefore must be taken into account when collecting and analyzing extracellular miRNA profiles. Differences between serum and plasma miRNA composition are substantial, and additional platelet contributions before and after coagulation are significant. Consequently, the extent of platelet depletion in plasma samples is a critical variable, as well as losses of exRNA by surface adsorption on the blood clot or additional RNA degradation paired with the release of platelet-contained RNAs upon coagulation. Platelet-depleted plasma generated immediately after blood collection therefore constitutes the preferred sample collection process for exRNA biomarker discovery.
Gender-associated abundance variations are small and characterized by 1.5- to 2-fold differences in abundance of cistronically expressed, epithelial cell-specific miR-200a, 200b, 429, and miR-141, 200c, as well as muscle-specific miR-1. While these miRNAs show abundance differences, the personal variability is high, and individually they provide sensitivity and specificity values of up to 73% (miR-200b) and therefore do not allow to accurately discern gender. Two other studies (38, 39) reported similar effect sizes but their gender-discriminating miRNAs agreed neither with each nor with the miRNAs identified in our study, indicating that gender differences are small and variation by individual and experimental noise are of almost similar magnitude. Other clinical parameters such as fasting and the menstrual cycle did not measurably affect miRNA profiles and need not be controlled for.
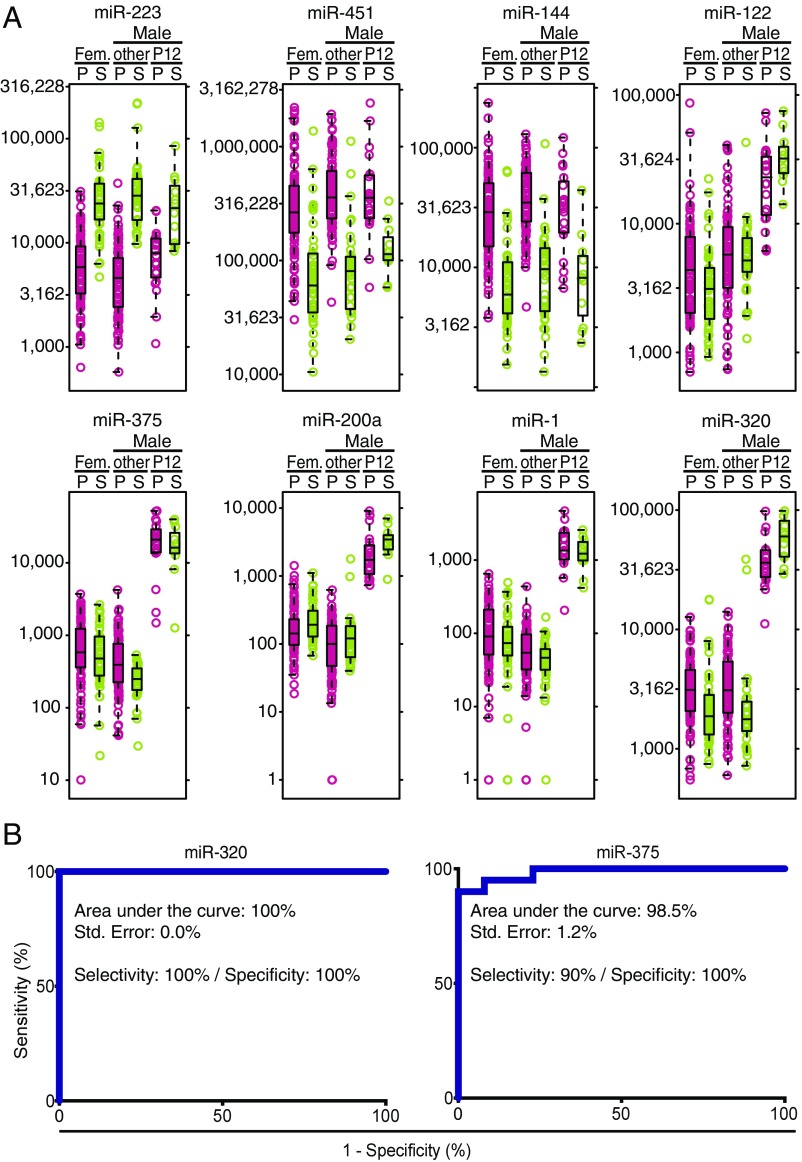
Considering that the blood-derived background of abundant miRNAs in the circulation of subject P12 is similar to that of other individuals, despite his twofold-increased total miRNA concentration in circulation, >130 up-regulated miRNAs may allow identification of additional contributing tissue sources (Figs. 3 D and E and 4A). Among them are approximately twofold relative increases of the prevalent miR-21, abundant in many proliferating cells, and liver-specific miR-122 and 22. miR-375, specific to neuroendocrine cells, such as those present in the pancreas and pituitary gland (33, 35, 36), showed an even greater relative 20-fold increase compared with all other males. miR-320, which is more broadly expressed including neuroendocrine cells (35), showed a relative 6.2-fold increase. Displaying median read frequencies of about 1% in P12 and less than 0.1% in all individuals, both miR-375 and 320 were of low abundance, yet their differential expressions were highly significant (Padj < 10−20), allowing us to discern this phenotype in virtually every sample originating from subject P12 (Fig. 4A), and could therefore serve as diagnostic markers with ≥90% sensitivity and specificity (Fig. 4B).
Fig. 4.
miRNA abundance variability in individual P12 compared with other individuals. (A) Boxplots featuring Set1-calibrator normalized read counts of select miRNAs, which are either prevalent in plasma (pink) and serum (sage) and show no significant difference (miR-451, 144, 223); or which are tissue specifically expressed and are significantly more abundant in individual P12 (miR-1, 122, 200a, 375, 320). miRNA abundance was computed using Set1-normalized count data. (B) Receiver operating characteristic curves based on miRNA 375 and 320 read counts.
Despite this remarkably different exRNA phenotype, to date we have not identified an etiology that accounts for this profile. Further longitudinal studies of this individual and a larger pool of study subjects including family members will provide insight into whether this phenotype predicts a genetic condition and predisposition to disease or represents a benign variant of cellular RNA metabolism and/or vesicular sorting and secretion, contributing to RNA release into biofluids, or exRNA turnover and secretion.
Finally, the discovery of an atypical extracellular miRNA phenotype in the course of this study, albeit without thus far identified clinical implications, indicates that our isolation, processing, and analyses are sufficiently sensitive and reproducible to capture even a singular abundance change of lower-prevalence miRNAs, independent of time, biofluid type, and gender. The implemented automation and adaptation to routine clinically collected biofluid volumes will facilitate larger clinical studies and accelerate RNA biomarker discovery.
Materials and Methods
All materials and methods for optimization of extracellular nucleic acid isolations from biofluids are listed in SI Text: Method Optimization of exRNA and exDNA Isolation. All oligonucleotide sequences used in exRNA/exDNA isolation and sRNA-derived cDNA library preparation are also listed in this text.
Human Subjects.
All clinical investigation has been conducted according to Declaration of Helsinki principles. All aspects of this study were reviewed and approved by the institutional review boards at Albert Einstein College of Medicine (IRB #2013–2248) and The Rockefeller University (under protocol number TTU0707). Signed informed consent was received from participants before inclusion in the study.
Plasma and Serum Collection.
Thirteen healthy subjects, seven males and six females, were recruited from Bronx, New York, to participate in this study; their demographic and additional metadata are listed in Table S1. Inclusion criteria were absence of any chronic or active diseases, not using any medications, and having a normal body mass index (18.5–29.9 kg/m2). Study subjects had to be on normal day schedules. Female study participants were required to have regular menstrual cycles of 26–32 d and could not be using any hormonal contraceptives. For each study subject, blood samples were collected on 4 d within a 2-wk interval. On each collection day, a standard 12-h clinical biochemistry fasting protocol was executed; the fasting blood sample was collected at 8 AM, a postprandial sample at 9 AM, and an afternoon sample at 4 PM. Records were kept of the activity of the study participants including food and drink ingested, physical activity, and sleep/wake cycles. Menstrual cycle information was recorded for female participants. Considering that coagulation releases platelet-containing miRNAs and thereby diminishes the signal of exRNAs originating from other cell types (16), we collected and sequenced less serum than plasma samples.
Sample collection was performed over a period of 4 mo, from November 19, 2014, to March 24, 2015. For each blood collection, the first 1 mL of peripheral blood after venipuncture was collected into a 3-mL plastic additive-free tube (BD; 366703) and discarded. Then, blood was collected into three 10-mL K2EDTA Vacutainer tubes (BD; 366643) and one 7-mL serum tube (BD; 366431). The K2EDTA tubes were immediately inverted 12 times to mix anticoagulant additive with blood. Blood in the serum tube was kept at room temperature for 45 min to allow clot formation. To obtain plasma/serum, blood samples were fractionated by centrifugation at 2,500 × g for 15 min at room temperature in a swinging-bucket centrifuge (Eppendorf; 5810R). Plasma/serum was aspirated into 50- and 15-mL tubes using disposable transfer pipettes (VWR; 414004-047) and centrifuged again using the same parameter. The supernatant plasma/serum supernatant was carefully transferred into new 50- and 15-mL tubes, mixed by inverting three times, and then aliquoted into 2-mL DNA LoBind tubes (Eppendorf; 0224310210). Samples were snap-frozen in liquid nitrogen and stored at −80 °C.
Optimized RNA Isolation, cDNA Library Preparation, Deep Sequencing, and Bioinformatic Analysis.
Detailed step-by-step protocols for RNA isolation and sRNA-derived cDNA library preparation are available in the SI Appendix. Before processing, all samples were randomized and organized in batches of 24 samples, and each 450-µL biofluid sample was quickly mixed with 105 µL of 60 °C-preheated detergent buffer P [30% (wt/vol) SDS, 66 mM Tris⋅HCl, pH 7.5, 19.8 mM EDTA, 265 mM 2-mercaptoethanol, pH 8.0] containing Set1 calibrators, a mixture of 10 unique non-hg19–mappable 22-nt 5′P/3′OH RNA spike-ins at a concentration of 10 pM each, so that the absolute total amount of Set1 calibrators added to each sample was 20 amol. Denaturation was followed by addition of 28 µL of proteinase K solution (5.4% glycerol, 3.2 mM CaCl2, 5.4 mM Tris⋅HCl, pH 8, 2.1 mg/mL proteinase K) and proteolytic digestion for 10 min at 60 °C, then organic extraction with 513 µL of buffer ED2 [37.4 mM citric acid/sodium citrate, pH 4.3, 0.4% sarcosyl, 1.6 M guanidinium isothiocyanate (GITC) and 80% phenol, 50 mM 2-mercaptoethanol], cooling to 10 °C, and phase separation with 103 µL of chloroform and centrifugation at 3,700 × g for 5 min. Column binding was performed by mixing 650 µL of the aqueous phase with 1,200 µL of buffer VB2G [98.2% isopropanol, 7.2 mM MgCl2, 2.4 mM CaCl2, 1 M GITC, and 5.0 mM tris(2-carboxyethyl)phosphine (TCEP)] and vacuum-applying mixtures of each sample to a Zymo-I 96-well plate (Zymo Research Corporation), followed by vacuum-applied washes with 970 µL of solution EWL (18 mM NaCl, 2.7 mM MgCl2, 0.9 mM CaCl2, 0.5% Triton X-100, 360 mM GITC, 5 mM TCEP) twice, 970 µL of 100% ethanol once, and 500 µL of 80% ethanol twice. RNA was eluted in 21 µL of 10 mM Tris⋅HCl, pH 7.4, by centrifugation at 3,700 × g for 5 min. If optional DNA extraction was desired, organic phases from organic extraction were reextracted using 500 µL of buffer EA3 (35.5 mM citric acid/sodium citrate, pH 4.3, 4 mM GITC), followed by centrifugation, aqueous phase aspiration, binding, washes, and elution from columns as described above for the purification of RNA. Nine microliters of each eluate, representing ∼50% of the isolated RNA extracted from one 450-µL biofluid sample, was used for cDNA library generation. Calibrator Set2 was added to each 9-µL aliquot, using absolute amounts of 1 amol each, so that the resulting total amount of Set2 calibrators added was 10 amol. Each RNA sample was individually 3′-adapter ligated. Up to 24 reaction products were then pooled, size-selected by denaturing 15% PAGE from 19 to 45 nt, 5′-adapter ligated, reverse transcribed, PCR amplified, and sequenced in a single Illumina HiSeq lane. Reads were demultiplexed, mapped against a curated hg19-based miRNA reference transcriptome, sorted, and tabulated into different RNA categories (summarized in Table 1, individually listed with complete metadata annotations in Dataset S1).
Raw sequencing data were processed (Illumina software suite), followed by read extraction using an in-house RNA-sequencing data analysis platform (RSDAP) (40) specifying a size range of 16–45 nt and default parameters. Demultiplexed RNA sequencing data were mapped against our curated human reference transcriptome to obtain miRNA raw read and read frequency profiles as well as abundance of fragments of other RNA classes, such as tRNAs, snRNAs, scRNAs, and rRNAs. Mapped data were used to generate RNA summary tables, as well as detailed miRNA raw read and read frequency tables, which were used for DEA and unsupervised clustering, respectively. Sequencing data reported in this paper have been deposited in NCBI’s Gene Expression Omnibus (41) and are accessible through GEO Series accession no. GSE113994. Per-sample fastq files have also been deposited in the Extracellular RNA Communication Consortium (42) exRNA atlas (43) and are accessible through accession no. EXRTTUSC1gCrGDHAN.
Data Processing.
Reads annotated as calibrator, expression system (plasmid and Escherichia coli), marker, and adapter were considered as reads of technical origin or “technical reads”; those remaining were considered as reads derived from the sample and referred to as “sample reads.” A total of 299 biofluid samples (95.8%) of a total of 312 samples were considered for data evaluation, and 13 samples with less than 100,000 sample reads were excluded. Reduced read coverage in these samples appears to be caused by column obstructions, RNA-ligation biases, low RNAseq efficiency, and combinations thereof: five samples were derived of batch 58 and 102 showing reduced sequencing depths of 110 and 56 million total reads, respectively (Dataset S1, column V); six samples showed notably reduced RNA isolation efficiencies (ratio of Set1 and Set2 calibrator counts; Dataset S1, column W and X); nine samples were barcoded with adapters 6, 11, 13, and 19, which tend to result in lesser than average read counts in multiplexed setups (29).
Estimation and comparison of miRNA concentrations using Set1 calibrators as external standards.
miRNA concentrations were calculated from the sum of Set1 calibrator read counts, considering their added molar amounts and volumes of biofluid samples. miRNA concentrations of study subjects were compared by one-way ANOVA assuming no matching or pairing, followed by Tukey’s multiple-comparisons test (44) using GraphPad Prism (version 7.00 for Mac; GraphPad Software, www.graphpad.com). Only comparisons where the level of difference was found to be of high significance (Padj < 0.01) were reported.
Calculation of read length distributions.
Read length distributions were based on length-dependent fractional values considering a range of 19–45 nt computed for major exRNA classes (miRNA, tRNA, rRNA, mRNA, ncRNA) using collapsed exRNA-seq data extracted from RSDAP master tables: (i) For each RNA class in each sample, individual length distribution fractions were generated for each length l in the range, by dividing the sum of sequences featuring the length l by the total number of sequences; in both cases, RNA class-specific sequences were considered. (ii) For each sample class (serum, plasma), averaged length distribution profile, fractions, and their SDs were calculated for each length l of each major exRNA class from individual read fraction values (see i) of relevant sample within that class. (iii) Composite sRNA length distribution fractions were calculated for each length l of each major RNA class of each sample class, by multiplying averaged fractional values (see ii) with averaged total read fraction values for that combination of RNA class and sample class, extracted from merged stat summary tables.
Unsupervised hierarchical clustering.
Tabulated shared read fractions of merged miRNAs and calibrators reported by RSDAP (Dataset S2) were used to generate log2-transformed heatmaps using the heatmap package (Raivo Kolde, 2015; pheatmap: Pretty Heatmaps; R package, version 1.0.8, CRAN.R-project.org/package=pheatmap). Unsupervised clustering of fractional miRNA abundance values was performed using the Complete clustering method and Manhattan clustering distances for both rows (miRNAs) and columns (samples) considering the combined top 85% most abundant miRNAs across all samples, unless stated otherwise. Calibrator heatmaps were generated accordingly by considering fractional calibrator read abundances of all members in calibrator Set1 and Set2, except that only the sample order was subjected to clustering while the order of calibrators was kept constant. Selected metadata (individual, gender, biofluid type, prandial state, female reproductive cycle, processing batch, and adapter detailed in Fig. 2 and Fig. S3) were included as metadata annotation where required.
DEAs.
For miRNA count data, we used the package DESeq2 (45), which provides methods to test for differential expression by use of negative binomial generalized linear models. This approach has been used to analyze and quantitate exRNA abundance differences (46), while also providing statistical significance with multiple testing corrections of all differences found. Tabulated shared raw reads of merged miRNAs reported by RSDAP (Dataset S3) at a level of at least five counts across all samples were used to investigate abundance differences associated with biofluid (serum vs. plasma), gender (female vs. male), female reproductive cycle (follicular vs. luteal state), prandial state (postprandial vs. preprandial state), and individual miRNA profiles. Single case profiles (e.g., P12 vs. other male subjects) required a comparison of data from a single individual with that from a group of controls, to ascertain whether the patient’s biofluid miRNA profile can be viewed as significantly different from that of controls. Various methods have been used in classical statistical inference to deal with such data, including repeated-measures ANOVA (47). Use of external standards based on read counts of all 10 oligonucleotides in the Set1 calibrator mixture for the size factor optimization proved to be critical for the correct description of a phenotype with a notably deviating exRNA composition. Although this phenotype has only been observed in one study participant, it is readily detectable in his plasma (n = 22) and serum (n = 12) samples and was consistent for over a year. Log2-transformed abundance differences were tabulated (Dataset S4) and plotted as scatterplots (Fig. 3) against log2-transformed mean abundance values (MA plots) and only considered significant for Padj < 0.05 using ggplot2 (48) (ggplot2.org). Boxplots were generated using the ReportingTools package (49).
Calculation of correlation coefficients.
Pearson product moment correlation coefficients were calculated for sample pairs using Corrplot, and pairwise comparisons were performed for groups of technical replicates as well as groups of all independent samples. Sample classes were taken into consideration where specified. To describe overall correlations within the groups, their medians and first and third quartile values are reported.
Supplementary Material
Acknowledgments
We thank William Thistlethwaite for help with the deposition of exRNA sequencing data and metadata in the Extracellular RNA Communication Consortium (ERCC) exRNA atlas and in the National Center for Biotechnology Information’s Gene Expression Omnibus. We thank Viktoriya Paroder for performing magnetic resonance cholangiopancreatography on study subject P12, providing documentation and helping to interpret the results. This work was funded through the NIH ERCC Grants NIH U19CA179564 and R01HD086327.
Footnotes
Conflict of interest statement: T.T. is cofounder of Alnylam Pharmaceuticals and is on the scientific advisory board of Regulus Therapeutics. K.E.A.M., K.B., K.M.A., K.A.B., J.L., P.M., I.Z.B.-D., X.L., Z.R.W., A.A., A.S., T.G.D., C.A., and Z.W. declare no competing financial interests.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data reported in this paper have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession no. GSE113994. Per-sample fastq files have also been deposited in the Extracellular RNA Communication Consortium exRNA atlas and are accessible through accession no. EXRTTUSC1gCrGDHAN.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714397115/-/DCSupplemental.
References
- 1.Weiland M, Gao X-H, Zhou L, Mi Q-S. Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 2.Momen-Heravi F, et al. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams Z, et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA. 2013;110:4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brase JC, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 5.Latreille M, et al. miR-375 gene dosage in pancreatic β-cells: Implications for regulation of β-cell mass and biomarker development. J Mol Med (Berl) 2015;93:1159–1169. doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 7.Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest. 1966;45:1732–1740. doi: 10.1172/JCI105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes GR, Cohen SA, Lightfoot RW, Jr, Meltzer JI, Christian CL. The release of DNA into serum and synovial fluid. Arthritis Rheum. 1971;14:259–266. doi: 10.1002/art.1780140211. [DOI] [PubMed] [Google Scholar]
- 9.Kamm RC, Smith AG. Nucleic acid concentrations in normal human plasma. Clin Chem. 1972;18:519–522. [PubMed] [Google Scholar]
- 10.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 11.Swaminathan R, Butt A, Gahan P. Circulating nucleic acids in plasma and serum IV. Proceedings of the Fourth International Conference on Circulating Nucleic Acids in Plasma/Serum. September 4–6, 2005. London, United Kingdom. Ann N Y Acad Sci. 2006;1075:1–353. [PubMed] [Google Scholar]
- 12.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: A collision of two paradigms. Trends Biochem Sci. 2016;41:883–892. doi: 10.1016/j.tibs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward J, et al. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci USA. 2014;111:12169–12174. doi: 10.1073/pnas.1412608111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akat KM, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA. 2014;111:11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2015;12:135–142. doi: 10.1038/nrcardio.2014.207. [DOI] [PubMed] [Google Scholar]
- 19.Matse JH, et al. Discovery and prevalidation of salivary extracellular microRNA biomarkers panel for the noninvasive detection of benign and malignant parotid gland tumors. Clin Cancer Res. 2013;19:3032–3038. doi: 10.1158/1078-0432.CCR-12-3505. [DOI] [PubMed] [Google Scholar]
- 20.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Lo YMD, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat Med. 2007;13:218–223. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 22.Robinson C, van den Boom D, Bombard AT. Noninvasive prenatal detection of aneuploidy. Clin Obstet Gynecol. 2014;57:210–225. doi: 10.1097/GRF.0000000000000016. [DOI] [PubMed] [Google Scholar]
- 23.Lee I, Baxter D, Lee MY, Scherler K, Wang K. The importance of standardization on analyzing circulating RNA. Mol Diagn Ther. 2017;21:259–268. doi: 10.1007/s40291-016-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent LC, et al. Meeting report: Discussions and preliminary findings on extracellular RNA measurement methods from laboratories in the NIH Extracellular RNA Communication Consortium. J Extracell Vesicles. 2015;4:26533. doi: 10.3402/jev.v4.26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Mauro M, Williams Z. Comparison of plasma extracellular RNA isolation kits reveals kit-dependent biases. Biotechniques. 2015;59:13–17. doi: 10.2144/000114306. [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–534, 536–537. [PubMed] [Google Scholar]
- 28.Chomczynski P, Mackey K, Drews R, Wilfinger W. DNAzol: A reagent for the rapid isolation of genomic DNA. Biotechniques. 1997;22:550–553. doi: 10.2144/97223pf01. [DOI] [PubMed] [Google Scholar]
- 29.Hafner M, et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58:164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bissels U, et al. Absolute quantification of microRNAs by using a universal reference. RNA. 2009;15:2375–2384. doi: 10.1261/rna.1754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farazi TA, et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods. 2012;58:171–187. doi: 10.1016/j.ymeth.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laffont B, et al. Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–261. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 35.Renwick N, et al. Multicolor microRNA FISH effectively differentiates tumor types. J Clin Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 37.Ambros V, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan T, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi: 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freedman JE, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 2016;7:11106. doi: 10.1038/ncomms11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown M, Suryawanshi H, Hafner M, Farazi TA, Tuschl T. Mammalian miRNA curation through next-generation sequencing. Front Genet. 2013;4:145. doi: 10.3389/fgene.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ainsztein AM, et al. The NIH Extracellular RNA Communication Consortium. J Extracell Vesicles. 2015;4:27493. doi: 10.3402/jev.v4.27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian SL, et al. Integration of extracellular RNA profiling data using metadata, biomedical ontologies and Linked Data technologies. J Extracell Vesicles. 2015;4:27497. doi: 10.3402/jev.v4.27497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tukey JW. Comparing individual means in the analysis of variance. Biometrics. 1949;5:99–114. [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeri A, et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep. 2017;7:44061. doi: 10.1038/srep44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mycroft RH, Mitchell DC, Kay J. An evaluation of statistical procedures for comparing an individual’s performance with that of a group of controls. Cogn Neuropsychol. 2002;19:291–299. doi: 10.1080/02643290143000150. [DOI] [PubMed] [Google Scholar]
- 48.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 49.Huntley MA, et al. ReportingTools: An automated result processing and presentation toolkit for high-throughput genomic analyses. Bioinformatics. 2013;29:3220–3221. doi: 10.1093/bioinformatics/btt551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.