Significance
The delta glutamate receptors (GluD1 and GluD2) shared high homology with ionotropic glutamate receptors but, surprisingly, are not gated by glutamate, or any other known ligand. GluD2 is only expressed in cerebellar Purkinje cells, where it forms a scaffolding complex with Cbln1 and presynaptic neurexin 1β (+S4). Might other synapses in the brain use a similar mechanism? We have found that GluD1, a long-neglected receptor subtype, powerfully and specifically regulates synapses in the hippocampus. We show that Cbln2 interacts with GluD1 and presynaptic neurexin 1β (+S4). This tripartite complex plays critical roles in both synaptogenesis and maintenance. Finally, we provide evidence that our results likely apply to synapses throughout the forebrain where GluD1 is widely expressed.
Keywords: GluD1, Cbln2, neurexin, hippocampus, synapse
Abstract
The δ1 glutamate receptor (GluD1) was cloned decades ago and is widely expressed in many regions of the brain. However, its functional roles in these brain circuits remain unclear. Here, we find that GluD1 is required for both excitatory synapse formation and maintenance in the hippocampus. The action of GluD1 is absent in the Cbln2 knockout mouse. Furthermore, the GluD1 actions require the presence of presynaptic neurexin 1β carrying the splice site 4 insert (+S4). Together, our findings demonstrate that hippocampal synapse assembly and maintenance require a tripartite molecular complex in which the ligand Cbln2 binds with presynaptic neurexin 1β (+S4) and postsynaptic GluD1. We provide evidence that this mechanism may apply to other forebrain synapses, where GluD1 is widely expressed.
Ionotropic glutamate receptors are divided into three families: AMPA receptors (AMPARs), NMDA receptors (NMDARs), and kainate receptors (KARs). However, there is a fourth family of receptors that shares high homology with these ionotropic receptors, referred to as the delta family of glutamate receptors (GluD1 and GluD2) (1, 2). Despite their high homology, these receptors are not gated by glutamate and, in fact, no ligand has been found to gate these receptors. Most research has focused on the GluD2 receptor, which is highly and specifically expressed in cerebellar Purkinje cells. GluD2 is selectively localized to parallel fiber (PF)-Purkinje cell synapses and its deletion results in a severe loss of PF synapses (3) and AMPAR trafficking/long-term depression (4). In addition, a soluble glycoprotein, called Cbln1, binds to postsynaptic GluD2 as well as to presynaptic neurexins (5–7), and is required for synaptogenesis of cerebellar PF synapses.
On the other hand, the GluD1 receptor, which shares 50% amino acid identity with GluD2, is expressed throughout the forebrain (8, 9). However, remarkably few studies have been carried out on GluD1 and its role, if any, has remained a mystery. Anatomical studies show that it is present in the postsynaptic density (9). GluD1 knockout (KO) mice are reported to have a number of behavioral abnormalities (10), but effects at the cellular level are confusing. In the medial prefrontal cortex and hippocampus dendritic spine density and the frequency of miniature excitatory postsynaptic currents (mEPSCs) are increased (11), suggesting an increase in the number of excitatory synapses in the absence of GluD1. In contrast, KO of GluD1 had no effect on synaptic transmission and plasticity in the hippocampus (12). Finally, the number of PF synapses onto interneurons in the cerebellum is reduced (8). To summarize, except for the cerebellum, the KO studies do not support a widespread synaptogenic role for GluD1, similar to that shown for GluD2 in the cerebellum. Expression of GluD1 in HEK293 cells cocultured with cortical neurons either induces inhibitory synapses preferentially (13), or both excitatory and inhibitory synapses (14). Taken together, the literature on the possible synaptic role of GluD1 is confusing and, in some cases, seemingly conflicting.
In the present study we find that expressed WT GluD1 localizes to synapses in CA1 hippocampal pyramidal cells but, as expected, generates no current. However, it enhances both AMPA and NMDA synaptic currents. GluD1 also increases the density of dendritic spines, indicating a synaptogenic effect. On the other hand knockdown of GluD1 decreases AMPA and NMDA currents, as well as spine density, indicating that GluD1 is required for normal synapse function. Furthermore, knockdown of GluD1 in the adult hippocampus decreases synaptic transmission and spine density, suggesting that GluD1 is required for synaptic transmission and maintenance of synapses in the adult hippocampus. The functional role of GluD1 requires its amino-terminal domain (ATD). More importantly, Cbln2 is the endogenous ligand for GluD1 and is required for the actions of GluD1. Finally, the action of GluD1 requires presynaptic neurexin 1β carrying the splice site 4 insert (+S4). Our findings demonstrate that Cbln2 bridges presynaptic neurexin 1β (+S4) and postsynaptic GluD1 to mediate hippocampal synapse formation and maintenance.
Results
Synaptic GluD1 Enhances Excitatory Synaptic Transmission by Increasing Synapse Number.
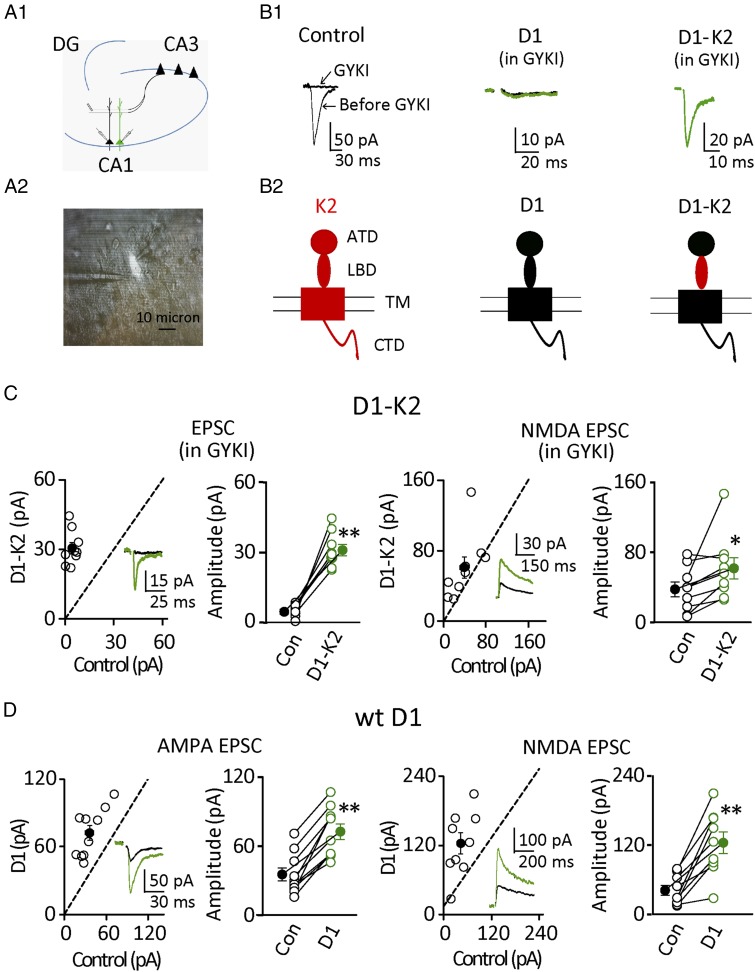
For these experiments we used slice culture from 6- to 8-d-old rats and biolistically expressed various constructs. We then did paired recordings from D1 biolistic-transfected cells (shown in green in Fig. 1A1 and the bright cell in left side of Fig. 1A2) and neighboring control cells in CA1, and evoked response by electrical stimulation of the Schaffer collateral pathway (Fig. 1A1). After 6 d of expression, we simultaneously recorded from a transfected cell and a neighboring control cell. In the presence of the selective AMPAR antagonist GYKI53655 (GYKI), synaptic responses recorded at −70 mV from CA1 neurons were fully blocked (Fig. 1B1). Expression of GluD1 in pyramidal cells in the presence of GYKI failed to generate synaptic current (Fig. 1B1), as expected because heterologously expressed GluD1 fails to generate currents (1). However, there are a number of other possibilities that could account for this negative result, such as a failure to express the receptor, inability of GluD1 to target to the synapse, and so forth. To overcome these uncertainties, we used a chimeric receptor in which the ligand-binding domain (LBD) of the GluK2 subtype of the KAR was swapped into the GluD1 receptor (D1-K2), as shown in the cartoon models (Fig. 1B2). This modified GluD1 receptor is gated by glutamate and has the pharmacological properties of a KAR (15, 16). Thus, we expressed this receptor in the presence of GYKI and found that this modified receptor was clearly capable of targeting to synapses (Fig. 1 B and C). However, much to our surprise, the expression of this receptor consistently enhanced the NMDA EPSC (Fig. 1C). This finding raises the possibility that GluD1 is having a more general effect on excitatory synapses. We therefore expressed the WT GluD1 and found that it also caused a robust enhancement of both AMPA and NMDA EPSCs (Fig. 1D) (AMPA, n = 10, P < 0.01; NMDA, n = 9, P < 0.01).
Fig. 1.
Synaptic GluD1 receptors enhanced excitatory synaptic transmission. (A1) Cartoon diagram of paired recordings from D1 biolistic transfected cell (shown in green) and neighboring control cell in CA1, evoked by electrical stimulation of Schaffer collateral pathway. (A2) IR-DIC image of culture slice and paired recordings from D1 transfected cell (bright cell with black gold particle dot) and control cell. (B1) Basic characterizations of D1 receptors: (Left) GYKI (30 µM) completely inhibited AMPA current in control cells; (Center) GYKI (30 µM) completely inhibited AMPA currents in both control and D1 overexpression cells; (Right) D1-K2 overexpression cells generated a GYKI (30 µM) -resistant current. Black traces are control, green are transfected. (B2) Cartoon models of GluK2 (K2), GluD1 (D1), and GluD1-K2 (D1-K2). (C) Summary data of D1-K2 mediated synaptic current in the presence of GYKI [30 µM, control cell (Con): 4.0 ± 0.9 pA; D1-K2: 30.8 ± 2.4 pA; n = 9, P < 0.01]; Overexpression of D1-K2 increased NMDA current (Con: 37.7 ± 8.4 pA; D1-K2: 61.8 ± 12.2 pA; n = 9, P < 0.05). Open circles are individual pairs, filled circle is mean ± SEM. (D) Overexpression of WT D1 increased AMPA (Left, Con: 35.5 ± 5.5 pA; D1: 72.6 ± 6.6 pA; n = 10, P < 0.01) and NMDA currents (Right, Con: 41.8 ± 8.4 pA; D1: 124.2 ± 18.5 pA; n = 9, P < 0.01). **P < 0.01, *P < 0.05.
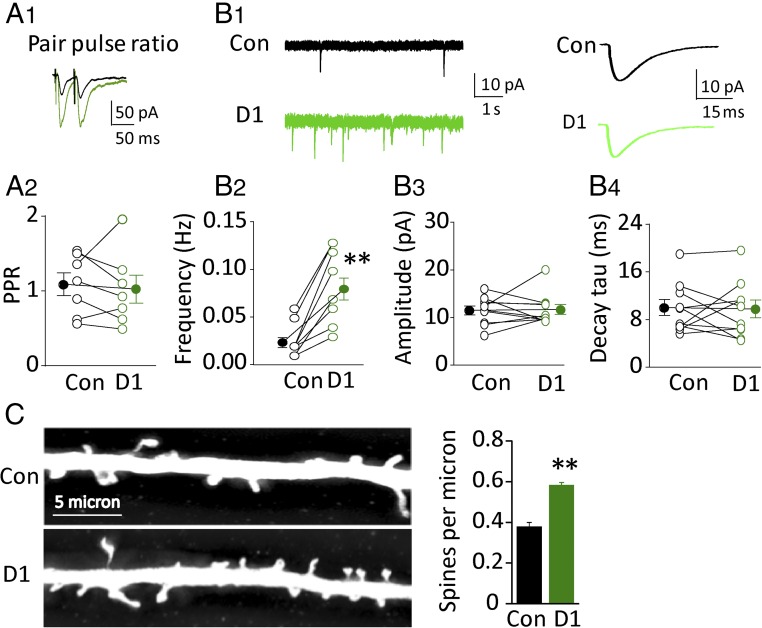
The GluD1-induced enhancement was not accompanied with a change in the paired pulse ratio (PPR) (Fig. 2 A1 and A2), suggesting no change in the probability of release. There was a large increase in the frequency of mEPSCs (Fig. 2 B1 and B2), without a change in amplitude (Fig. 2B3). Furthermore, there was no change in the decay of mEPSCs (Fig. 2B4). These findings, along with the increase in the NMDAR EPSCs, are consistent with an increase in the number of functional synapses. To address this possibility more directly, we examined the density of dendritic spines, a proxy for the number of excitatory synapses, which preferentially contact spines. GluD1-expressing neurons had a robust increase in the density of dendritic spines (Fig. 2C) (n = 15, P < 0.01) but had no effect on spine head diameter, spine neck diameter, or spine length (Fig. S1A). We also found that GluD1 overexpression increased inhibitory postsynaptic current (IPSC) (Fig. S1B).
Fig. 2.
Mechanism of synaptic enhancement by GluD1 overexpression. (A1) Representative traces of PPR in control cell (black trace) and D1 overexpression cell (green trace); PPR, two consecutive stimulations separated by 40 ms. (A2) Summary data of PPR (second EPSC normalized to first EPSC) in control cells and D1 overexpression cells [control cell (Con): 1.1 ± 0.1; D1: 1.0 ± 0.2; n = 7, P > 0.05]. Open circles are individual pairs, filled circle is mean ± SEM. (B1, Left) Representative traces of mEPSC in control cell and D1 overexpression cell; (Right) average trace of 100 individual mEPSC events in control cell and D1 overexpression cell. (B2–B4) D1 overexpression increased frequency of mEPSC (Con: 0.13 ± 0.06 Hz; D1: 0.54 ± 0.19 Hz; n = 10, P < 0.01), but had no effect on amplitude (Con: 11.49 ± 0.92 pA; D1: 11.7 ± 1.03 pA; n = 10, P > 0.05) and decay time constant (Con: 9.7 ± 1.3 ms; D1: 9.5 ± 1.5 ms; n = 10, P > 0.05) of mEPSC. (C, Left) Representative pictures of a dendrite in Con and D1 overexpression cell (D1); (Right) summary data showing D1 overexpression increased spine density (Con: 0.38 ± 0.03; D1: 0.58 ± 0.01; n = 15, P < 0.01). Spine density expressed as spines per micrometer ± SEM **P < 0.01.
Knockdown of GluD1 Decreases Excitatory Synaptic Transmission.
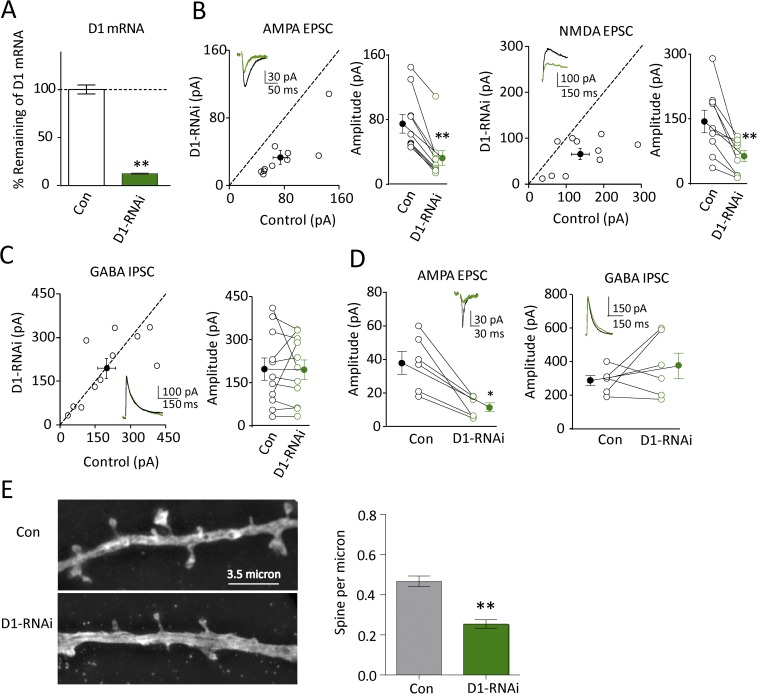
The above results indicate that GluD1 has a profound effect on excitatory synapses. However, the results do not address whether GluD1 normally plays a role at these synapses. To address this we expressed a microRNA directed against GluD1. This construct reduced GluD1 mRNA by 88% (Fig. 3A). Both the AMPAR and NMDAR EPSCs were substantially and similarly reduced in cells expressing the microRNA (Fig. 3B) (n = 10, P < 0.01). This effect was not due to off-target effects of the GluD1-RNAi because overexpression of RNAi-resistant GluD1 rescued GluD1-RNAi effects (Fig. S2A). In contrast, there was no effect on the IPSC (Fig. 3 C and D). These results indicate that endogenous GluD1 plays an important role in excitatory, but not inhibitory synapse function. The reduction in synaptic responses was accompanied with a similar reduction in the density of spines (Fig. 3E), consistent with a synaptogenic role. To confirm GluD1 functional role in the hippocampus, we also tested the effects of GluD1 knockdown on mEPSC and synaptic plasticity. We found GluD1 knockdown reduced frequency of mEPSC (Fig. S2B), but had no effect on long-term potentiation (LTP) (Fig. S2C).
Fig. 3.
Knockdown of GluD1 reduced excitatory synaptic transmission but had no effect on inhibitory synaptic transmission. (A) RT-PCR analysis of GluD1 mRNA levels by GluD1 [control cell (Con): 100 ± 4%; D1-RNAi: 12 ± 1%; n = 3, P < 0.01]. (B) Knockdown of D1 reduced AMPA current (Left, Con: 74.8 ± 11.4 pA; D1-RNAi: 32.5 ± 8.9 pA; n = 10, P < 0.01) and NMDA current (Right, Con: 144 ± 26.1 pA; D1-RNAi: 63.3 ± 12.8 pA; n = 10, P < 0.01). Black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. (C) Knockdown of D1 did not change GABA IPSC (Con: 197.4 ± 39.0 pA; D1-RNAi: 195.0 ± 34.2 pA; n = 11, P > 0.05). (D) Simultaneous recordings of AMPA eEPSC (Con: 35.2 ± 8.2 pA; D1-RNAi: 12.7 ± 2.7 pA; n = 6, P < 0.05) and GABA eIPSC (Con: 284 ± 30 pA; D1-RNAi: 373 ± 75 pA; n = 6, P > 0.05) in the same cell. eEPSC and eIPSC were recorded when membrane potential was held at −70 mV and 0 mV, respectively. (E, Left) Representative pictures of a dendrite in Con and D1 knockdown cells (D1-RNAi) of day 6 biolistic-transfected hippocampus culture slice; (Right) summary data showing knockdown of D1 decreased spine density of day 6 transfected hippocampal CA1 pyramidal neuron (Con: 0.46 ± 0.02, n = 11; D1-RNAi: 0.25 ± 0.02, n = 14; P < 0.01). Spine density expressed as spines per micrometer ± SEM *P < 0.05, **P < 0.01.
GluD1 Maintains Excitatory Synapses in the Adult Hippocampus.
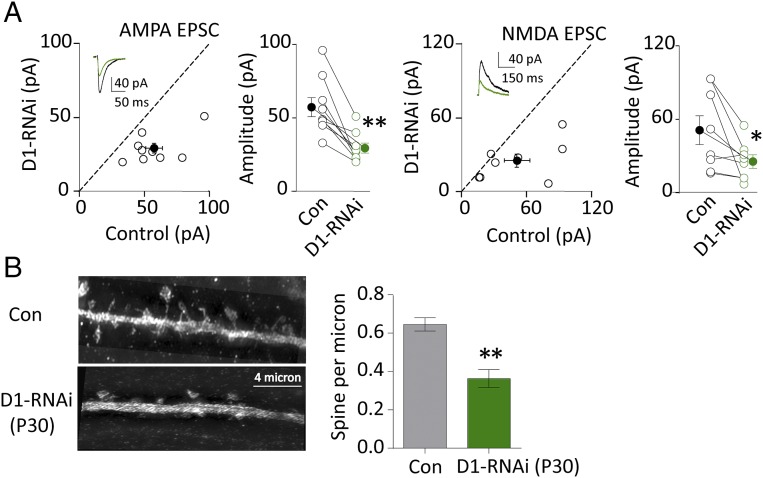
Developmentally, the synapse connectivity in the adult becomes dynamically stable. Thus, we wonder whether GluD1 was required for maintaining synapses in the adult hippocampus. We found that lentivirus injection of GluD1-microRNA into the adult hippocampus decreased excitatory synaptic transmission to the same extents as at the juvenile stage (Fig. 4A). Consistent with these physiological results, spine density of CA1 pyramidal neurons was reduced by ∼50% (Fig. 4B). Thus, these results indicate that GluD1 is essential for maintaining excitatory synaptic transmission in the adult hippocampus.
Fig. 4.
GluD1 maintains excitatory synaptic transmission in the adult hippocampus. (A) Knockdown of D1 decreased both AMPA [Left, control cell (Con): 57.3 ± 6.4 pA; D1-RNAi: 29.4 ± 3.3 pA; n = 9, P < 0.01] and NMDA currents (Right, Con: 51.1 ± 11.7 pA; D1-RNAi: 25.5 ± 5.5pA; n = 8, P < 0.05) in P30 virus infected hippocampus CA1 pyramidal neuron; black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. (B, Left) Representative pictures of a dendrite in Con and D1 knockdown cells (D1-RNAi) of postnatal day (P) 30 virus-infected acute slices; (Right) summary data showing knockdown of D1 decreased spine density of P30 virus infected hippocampal CA1 pyramidal neuron (Con, 0.64 ± 0.03, n = 14; D1-RNAi, 0.36 ± 0.04, n = 10; P < 0.01). Spine density expressed as spines per micrometer ± SEM. Note that the spine images shown here are a montage from maximum intensity projection, which processes all images captured at different “z” axes. *P < 0.05, **P < 0.01.
GluD1 Requires the ATD for Its Function.
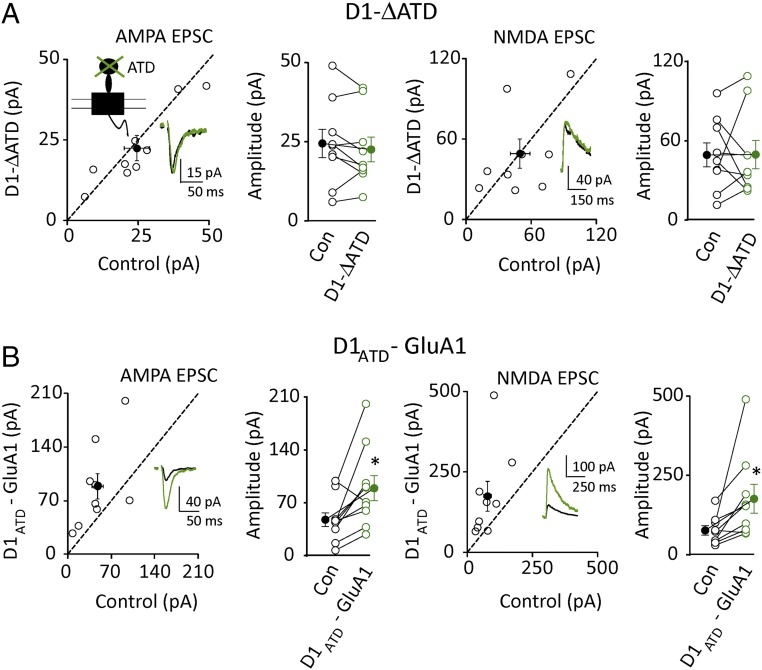
To determine what domains in GluD1 are responsible for the functional effects, we deleted the ATD, referred to as ΔATD GluD1 (Fig. 5A, Inset). ΔATD GluD1 had no effect on excitatory synapses (Fig. 5A). However, this negative result could be explained by a number of possibilities, such as failure to express this truncated receptor. We therefore expressed ΔATD GluD1 with the KAR ligand binding site. In this case, the chimeric receptor generated GYKI-resistant currents (Fig. S3, Left), indicating that it is functional and targets to the synapse. However, it had no effect on the NMDAR response, indicating that the ATD is essential for the functional effect. Finally we made a chimeric receptor of GluA1, in which the ATD of GluA1 was swapped for the ATD of GluD1. Overexpression of WT GluA1 had no effect on the size of the AMPA or NMDA EPSC (Fig. S4 A1 and B). To ensure that GluA1 is actually targeted to the synapse in these experiments we measured rectification, because overexpressed GluA1 in CA1 pyramidal cells forms homomeric rectifying receptors that are constitutively trafficked to the synapse (17). The presence of rectification in cells overexpressing GluA1 indicates that the expressed homomeric receptors are present at synapses (Fig. S4A2). These results with the WT GluA1 indicate that, while it is expressed at the synapse, it fails to enhance synaptic transmission. However, expression of a GluA1 receptor containing the ATD of GluD1, caused a dramatic enhancement of both AMPAR and NMDAR currents (Fig. 5B), further establishing the role for the ATD of GluD1 in enhancing synaptic function. These results are consistent with recent findings about the roles of ATD in GluD2 and GluA4 (5, 6, 18, 19).
Fig. 5.
GluD1 required the ATD for its function. (A) Deletion of ATD of D1 blocked the effects of D1 on both AMPA [Left, control cell (Con): 24.4 ± 4.4 pA; D1-ΔATD: 22.5 ± 3.9 pA; n = 9, P > 0.05] and NMDA currents (Right, Con: 49.3 ± 9.0 pA; D1-ΔATD: 49.6 ± 10.7 pA; n = 9, P > 0.05). (Inset) Schematic topology of a GluD1 receptor showing the following domains: ATD, LBD, transmembrane (TM), C-terminal domain (CTD). Black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. (B) Replacing GluA1 ATD with D1 ATD increased both AMPA (Left, Con: 47.5 ± 9.0 pA; D1ATD- GluA1: 89.2 ± 16.4 pA; n = 10, P < 0.05) and NMDA currents (Right, Con: 76.4 ± 15.0 pA; D1ATD- GluA1: 176.2 ± 45.5 pA; n = 9, P < 0.05). *P < 0.05.
Cbln2 Is the Endogenous Ligand for GluD1 in the Hippocampus.
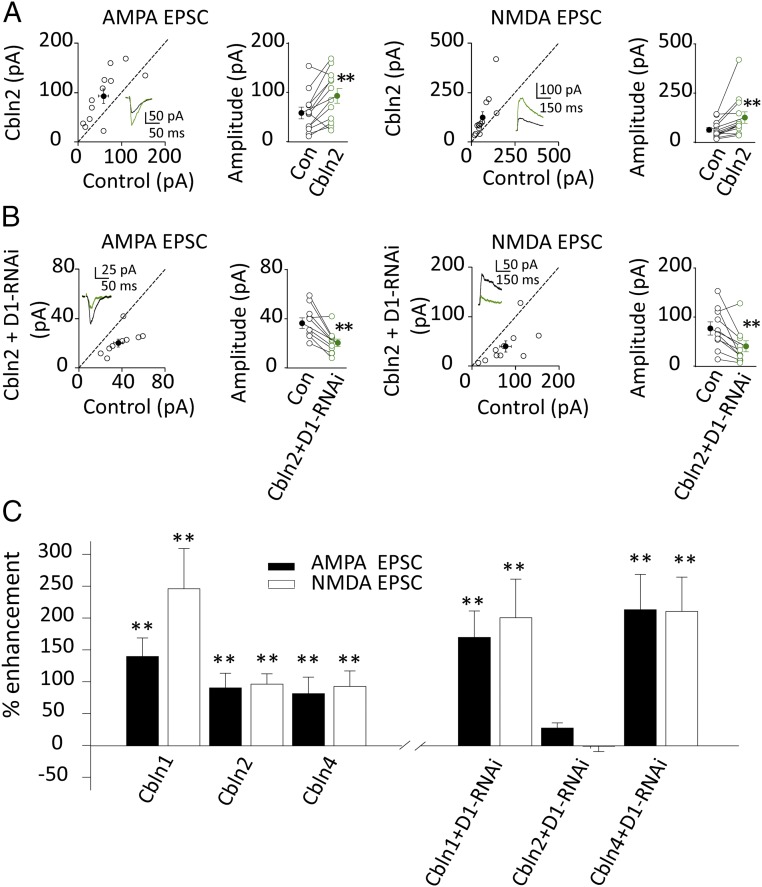
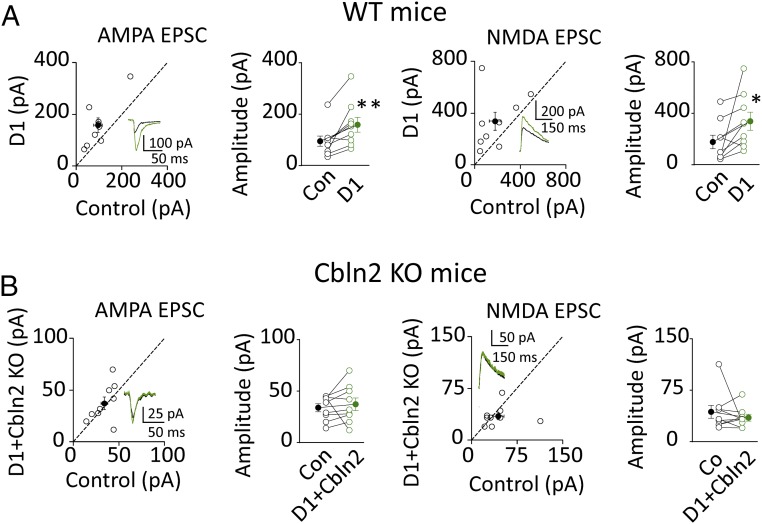
How might GluD1 exert its transsynaptic effects? At PF-Purkinje cell synapses the soluble glycoprotein, referred to as Cbln1, binds to the ATD of GluD2. Cbln1 belongs to the C1q and tumor necrosis factor superfamily (20). There are four Cblns and three of them are expressed in the hippocampus: Cbln1, Cbln2, and Cbln4 (21–24). Thus, we tested whether these three members of the Cbln family are involved in the action of GluD1. A simple experiment was to express these soluble glycoproteins in CA1 pyramidal neurons to determine if they could mimic the action of GluD1, keeping in mind that, analogous to the PF-Purkinje cell synapse, the origin of the endogenous source of glycoprotein would presumably come from the presynaptic terminals. We found that Cbln2 (Fig. 6 A and C), Cbln1 (Fig. 6C and Fig. S5A), and Cbln4 (Fig. 6C and Fig. S5C) all enhanced excitatory synaptic transmission. Does this enhancement require the presence of GluD1? To answer this question we repeated the overexpression experiments but in the presence of the microRNA for GluD1. We found that the enhancing effect of both Cbln1 (Fig. 6C and Fig. S5B) and Cbln4 (Fig. 6C and Fig. S5D) remained intact after knocking down GluD1. However, in striking contrast, the enhancing effects of Cbln2 were absent after knocking down GluD1 (Fig. 6 B and C). The results of all these experiments are summarized in the bar graph of Fig. 6C and suggest that Cbln2 mediates the transsynaptic effects of GluD1 at CA1 excitatory synapses and Cbln2 is the endogenous ligand for GluD1. To further confirm this conclusion, we tested whether the actions of GluD1 are absent in the Cbln2 KO mouse. Indeed, the enhancing effects of GluD1 observed in WT mice (Fig. 7A) were absent in Cbln2 KO mice (Fig. 7B), indicating that the actions of GluD1 require Cbln2. To exclude off-target effects of the Cbln2 KO, we also found that overexpression of GluD1 and Cbln2 rescued the Cbln2 KO phenotypes (Fig. S6).
Fig. 6.
GluD1 actions require its ligand Cbln2. (A) Overexpression of Cbln2 increased both AMPA [Left, control cell (Con): 58.4 ± 11.9 pA; Cbln2: 93.0 ± 14.7 pA; n = 12, P < 0.01] and NMDA currents (Right, Con: 63 ± 11.4 pA; Cbln2: 125.4 ± 29.1 pA; n = 13, P < 0.01); black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. (B) Cbln2 did not rescue the depression of GluD1-RNAi on excitatory synaptic transmission (Left, AMPA current, Con: 36.4 ± 4.3 pA; Cbln2+D1-RNAi: 20.4 ± 3.0 pA, n = 10, P < 0.01; Right, NMDA currents, Con: 76.9 ± 13.4 pA; Cbln2+D1-RNAi: 40.5 ± 11.2 pA, n = 10, P < 0.01). (C) Summary of Cblns and GluD1-RNAi on excitatory synaptic transmission, all data were normalized to their own control. **P < 0.01.
Fig. 7.
GluD1 actions are absent in Cbln2 knockout mice. (A) Overexpression of D1 in WT mouse increased both AMPA current [Left, control cell (Con): 95.1 ± 19.7 pA; D1: 158.8 ± 28.9 pA, n = 9, P < 0.01] and NMDA current (Right, Con: 177.3 ± 52.9 pA; D1: 338.5 ± 70.0 pA, n = 9, P < 0.05). Black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. (B) Overexpression of D1 in Cbln2 KO mice did not increase AMPA current (Left, Con: 33.8 ± 3.9 pA; D1: 37.2 ± 6.1 pA, n = 9, P > 0.05) and NMDA current (Right, Con: 43.5 ± 9.3 pA; D1: 35 ± 4.9 pA, n = 9, P > 0.05). *P < 0.05, **P < 0.01.
GluD1 Actions Require Presynaptic Neurexin 1β (+S4).
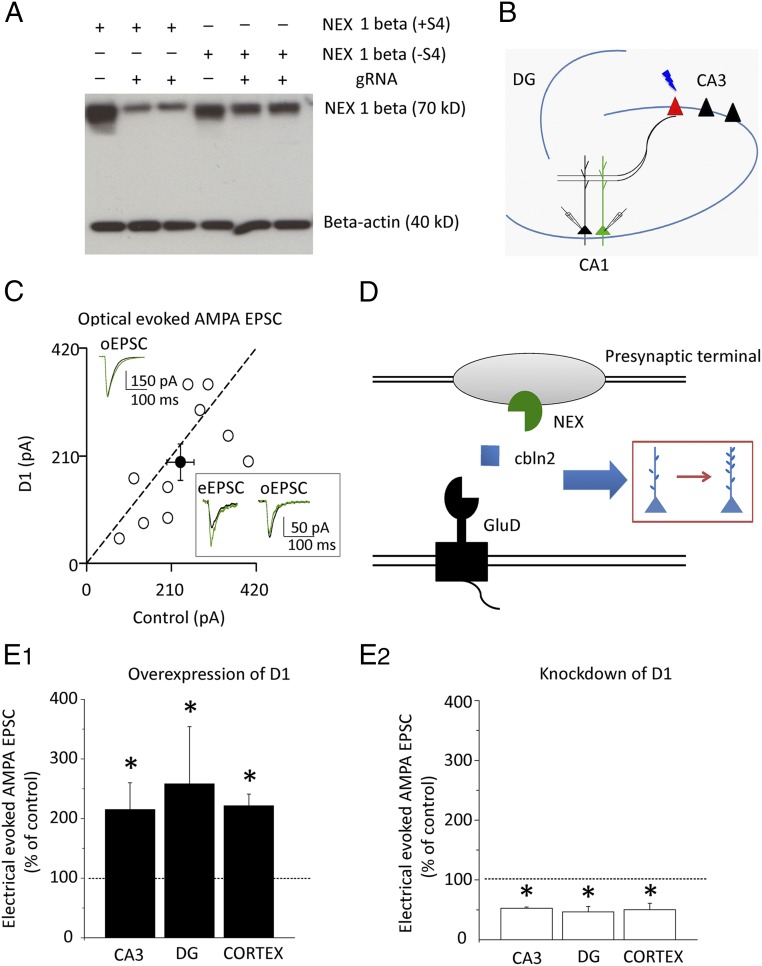
What might be the presynaptic partner to which Cbln2 binds? Previous studies have shown that presynaptic neurexin 1β (+S4) binds Cbln2 in the hippocampus (25). We therefore examined whether neurexin 1β (+S4) might carry out the action of GluD1 in the hippocampus. To test this, we deleted neurexin 1β (+S4) in CA3 neurons, which synapse onto CA1 neurons, by injecting AAV virus containing a guide RNA (gRNA) targeting neurexin 1β (+S4) (Fig. 8A), together with channelrhodopsin2 (ChR2) into Cas-9 knockin mice (Fig. 8B). This allowed us to activate only those synapses in which neurexin had been deleted. We found that in synapses lacking presynaptic neurexin 1β (+S4) there was no difference in the size of AMPA EPSCs between control cells and cells expressing GluD1 (Fig. 8C) (n = 10, P > 0.05), indicating that neurexin 1β (+S4) is required for the action of GluD1. This effect was specific to neurexin 1β (+S4), because GluD1 effects on excitatory transmission were restored by coexpression of CRISPR-resistant neurexin 1β (+S4) and gRNA targeting neurexin 1β (+S4) in Cas-9 mouse slices (Fig. S7D). As control, the enhancing effects of GluD1 on excitatory synaptic transmission (optogenetically evoked EPSCs) were still present when neurexin 1β (+S4) was intact (Fig. S7 A and B). Interestingly, when the AAV infection was sparse, we found in the same cell that GluD1 actions were intact when examined with electrical stimulation (eEPSC), but absent when examined with photostimulation (optogenetically evoked EPSC, oEPSC) (Fig. 8C, Inset).
Fig. 8.
GluD1 actions require presynaptic neurexin 1β (+S4). (A) CRISPR targeting at neurexin 1β (+S4) reduced the protein expression of neurexin 1β (+S4). HA-tagged neurexin 1β (+S4), HA-tagged neurexin 1β (−S4), and gRNA targeting at neurexin 1β (+S4) were transfected in HEK293 cells according to experiment designs. px458 DNA which contains Cas-9 gene was included in all conditions. (B) Experimental designs showing paired recordings from optical stimulation-evoked responses. Green is biolisitc expression of D1 in CA1, while red is the AAV transfection of CRISPR targeting neurexin 1β (+S4) in CA3. (C) AAV injection of CRISPR construct targeting neurexin 1β (+S4) blocked GluD1 actions on AMPA current evoked by optical stimulus [control cell (Con): 231.6 ± 32.7 pA; D1: 198.7 ± 34.9 pA, n = 10, P > 0.05]. Black traces are control, green are transfected. Open circles are individual pairs, filled circle is mean ± SEM. Note, Inset box, a group of representative traces in the same cell showing that AAV blocked GluD1 actions on AMPA current evoked by optical stimulus, but not the effect on AMPA current evoked by electrical stimulus. (D) Cartoon model showing the tripartite molecular complex in which the ligand Cbln2 binds with presynaptic neurexin 1β (+S4) and postsynaptic GluD1 that assemble hippocampus synapses. (E1) D1 overexpression increased AMPA current in CA3 (normalized, 2.2 ± 0.4, n = 6, P < 0.05), DG (normalized, 2.6 ± 0.9, n = 6, P < 0.05), and cortical layer 2/3 pyramidal neurons (normalized, 2.2 ± 0.2, n = 7, P < 0.05). (E2) D1-RNAi reduced AMPA current in CA3 (normalized, 0.52 ± 0.02, n = 6, P < 0.05), DG (normalized, 0.46 ± 0.08, n = 5, P < 0.05), and cortical layer 2/3 pyramidal neurons (normalized, 0.50 ± 0.1, n = 6, P < 0.05), indicating that D1 actions are widely spread in forebrain synapses. *P < 0.05.
A Role for GluD1 at Other Cortical Synapses.
How widespread might a role for GluD1 be? To begin to address this question we examined the effect of GluD1 overexpression and knockdown on other excitatory synapses. We found that overexpression of GluD1 enhanced excitatory synaptic transmission in CA3 pyramidal cells, dentate granule cells, and layer 2/3 pyramidal cells of the somatosensory cortex (Fig. 8E1 and Fig. S7C). Finally, we provide evidence that endogenous GluD1 plays a synaptogenic role at these synapses by expressing the microRNA for GluD1. In all cases we observed a strong depression of excitatory synaptic transmission (Fig. 8E2). Based on this limited sampling of forebrain synapses, we suggest that the mechanism we describe in CA1 pyramidal cells may be widespread.
Discussion
This study addresses the role of a long-neglected glutamate receptor subtype present at excitatory synapses in the hippocampus, the GluD1 receptor. More specifically, we identify the endogenous ligand for GluD1 and its synaptic function in the hippocampus. GluD1, along with the GluD2, have been referred to as “orphan receptors” because they are not gated by glutamate or any other known ligand. Although localized at excitatory synapses on CA1 pyramidal cells, GluD1 receptors, as expected, did not contribute to excitatory synaptic currents. However, overexpression of GluD1 caused a large increase in both AMPA and NMDA EPSCs. The density of dendritic spines was also increased by GluD1 overexpression. Knockdown of GluD1 caused a large reduction in AMPA EPSCs, NMDA EPSCs, and spine density in both juvenile and adult rats. The functional effect of GluD1 required its extracellular ATD. Additionally, Cbln2 mimicked the effect of GluD1 and this action was prevented by GluD1 knockdown, indicating that Cbln2 is the ligand for GluD1. Finally, GluD1 actions require its ligand Cbln2 and presynaptic neurexin 1β (+S4). Taking these data together, we find that this tripartite transsynaptic molecular complex plays a critical role in the assembly and maintenance of hippocampal synapses (Fig. 8D). Given that neurexin–Cbln2–GluD1 is widely expressed in the forebrain, it seems likely that the mechanism we describe here is widespread (Fig. 8E).
Because GluD1 generates no ionic current, it has been difficult to study its functional implications in the context of a specific neuronal circuit. To circumvent this issue, we used a chimeric GluD1, which contained the KAR LBD and is gated by glutamate (15, 16). This construct generated synaptic currents, indicating that the GluD1 receptor is targeted to synapses. Expression of the WT GluD1 caused a large increase in AMPA and NMDA EPSCs and in the density of dendritic spines. Our results indicate that overexpression of GluD1 has a synaptogenic effect on excitatory synapses. To determine if endogenous GluD1 plays a role, we knocked down the protein with microRNA. This resulted in a large decrease in both AMPA and NMDA EPSCs, as well as spine density, indicating that GluD1 plays an essential role in synaptogenesis of excitatory synapses. These results contrast with the results from germline deletion of GluD1, which found no impairment in excitatory synaptic transmission (12), suggesting that compensation is occurring in the germline deletion of GluD1. In contrast to the robust depression of EPSCs with knockdown of GluD1, there was no effect on IPSCs, indicating that synaptogenesis of inhibitory synapses is independent of GluD1. This conclusion appears to be at odds with studies in which GluD1 expressing HEK cells are cocultured with neurons (13, 14). However, we also observed an enhancement in IPSCs when overexpressing GluD1, suggesting that it can engage a preexisting synaptogenic machinery that is present at inhibitory synapses.
We examined a number of soluble glycoproteins that might mediate the transsynaptic effects of GluD1. Although Cbln1 and Cbln4 enhanced synaptic transmission, their actions remained intact after the knockdown of GluD1, ruling out their involvement in the action of GluD1. However, the enhancing effect of Cbln2, which is widely expressed in the forebrain, was absent when it was coexpressed with GluD1 microRNA, suggesting that Cbln2 mediates the effects of GluD1 and thus Cbln2 is the endogenous ligand for GluD1. This conclusion was confirmed by the observation that the actions of GluD1 are absent in the germline KO of Cbln2. At the PF-Purkinje cell synapse Cbln1 binds to presynaptic neurexin β (+S4). Similarly, we found that Cbln2 also bridges neurexin 1β (+S4). This in vivo bridging of the neurexin 1β (+S4) –Cbln2–GluD1 complex is further supported by in vitro biochemistry data: Cbln2 binds to GluD1 (7, 13, 22) and Cbln2 binds to neurexin 1β (+S4) (7, 13, 22, 26).
Previous studies have elegantly demonstrated the critical role of the GluD2/Cbln1/neurexins transsynaptic complex in PF-Purkinje cell synaptogenesis (5, 27). GluD2 is only expressed in Purkinje cells, raising the question as to whether a similar mechanism occurs at other excitatory synapses. Here, we report that another member of the delta receptor family, GluD1, has a dramatic synaptogenic role at hippocampus synapses and is essential for maintaining excitatory synaptic transmission in the adult. Although there are functional similarities between GluD1 and GluD2, their molecular mechanisms are different: the endogenous ligand for GluD2 in the cerebellum is Cbln1, while for GluD1 in the hippocampus, it is Cbln2. The finding that synaptic transmission in the CA3, dentate gyrus, and cerebral cortex are also dependent of GluD1 suggests that the model we describe in the CA1 region of the hippocampus may be widespread. However, it is clear that other signaling pathways contribute to synaptogenesis and maintenance. For example, deletion of GluD1, Cbln2, or neurexin does not abolish synaptic transmission in the CA1 region. In addition, although Cbln1 and Cbln4 are not involved in GluD1-mediated effects, their enhancement of synaptic transmission suggests that they may be tapping into another mechanism. Although GluD1 plays no role at inhibitory synapses, its overexpression engages a preexisting synaptogenic machine. Furthermore, different synapses may favor one mechanism over another. Recent RNA-seq data (from the Hippocampus RNA-seq Atlas, hipposeq.janelia.org/) shows that GluD1 is expressed at relatively higher amounts in CA1, while Cbln2 is more highly expression in CA3. This evidence also suggests that different brain regions may utilize different signaling pathways by controlling the expression of different bridge molecules. In summary, our results showing dramatic roles in synaptogenesis and synapse maintenance for neurexin–Cbln2–GluD1 in the hippocampus suggest that this mechanism may well be widespread, given that neurexin–Cbln2–GluD1 are widely expressed in the forebrain.
Materials and Methods
Animals.
Cbln2 KO mice were obtained from James I. Morgan’s laboratory at St. Jude Children’s Hospital, Memphis, TN, and were bred at the University of California, San Francisco. Cas-9 knockin mice were purchased from The Jackson Laboratory (donated by Feng Zhang, McGovern Institute for Brain Research at MIT, Cambridge, MA; Stock no. 024858 |Rosa26-Cas9 knockin). All of the experimental procedures on animals were approved by the University of California, San Francisco Animal Care and Use Committee.
Slice Culture and Biolistic Transfection.
Hippocampal cultured slices are obtained from 6- to 8-d-old rats or mice. Biolistic transfection was done 1 d after sectioning, by using a Helio Gene Gun with 1-μm DNA-coated gold particles (Bio-Rad). Slices were maintained at 34 °C and the medium was changed every 2 d.
Electrophysiological Recording.
Dual whole-cell voltage clamp recordings were obtained from a fluorescent transfected cell and a neighboring control pyramidal cell in CA1 region of hippocampus. Pyramidal neurons were identified by location and morphology. All recordings were made at 20–25 °C. More experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Michael Hollmann for the GluD1 and GluD1-K2 plasmids; Dr. Peter Scheiffele for the neurexin 1β constructs; Dr. Karl Deisseroth (from Addgene) for pAAV-CaMKцa-hChR2 (H134R)-mCherry; Dr. James I. Morgan for Cbln2 knockout mice; Dr. Massimo Scanziani and members of the R.A.N. laboratory for comments on the manuscript; and Dan Qin for technical assistance. This research was supported by grants from the National Institute of Mental Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802737115/-/DCSupplemental.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid SM, Hollmann M. Bridging the synaptic cleft: Lessons from orphan glutamate receptors. Sci Signal. 2010;3:pe28. doi: 10.1126/scisignal.3136pe28. [DOI] [PubMed] [Google Scholar]
- 3.Kashiwabuchi N, et al. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- 4.Hirai H, et al. New role of delta2-glutamate receptors in AMPA receptor trafficking and cerebellar function. Nat Neurosci. 2003;6:869–876. doi: 10.1038/nn1086. [DOI] [PubMed] [Google Scholar]
- 5.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 6.Matsuda K, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- 8.Konno K, et al. Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. J Neurosci. 2014;34:7412–7424. doi: 10.1523/JNEUROSCI.0628-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hepp R, et al. Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Struct Funct. 2015;220:2797–2815. doi: 10.1007/s00429-014-0827-4. [DOI] [PubMed] [Google Scholar]
- 10.Yadav R, et al. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta SC, et al. Essential role of GluD1 in dendritic spine development and GluN2B to GluN2A NMDAR subunit switch in the cortex and hippocampus reveals ability of GluN2B inhibition in correcting hyperconnectivity. Neuropharmacology. 2015;93:274–284. doi: 10.1016/j.neuropharm.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, et al. Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol Cell Biol. 2007;27:4500–4512. doi: 10.1128/MCB.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasumura M, et al. Glutamate receptor δ1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J Neurochem. 2012;121:705–716. doi: 10.1111/j.1471-4159.2011.07631.x. [DOI] [PubMed] [Google Scholar]
- 14.Ryu K, Yokoyama M, Yamashita M, Hirano T. Induction of excitatory and inhibitory presynaptic differentiation by GluD1. Biochem Biophys Res Commun. 2012;417:157–161. doi: 10.1016/j.bbrc.2011.11.075. [DOI] [PubMed] [Google Scholar]
- 15.Schmid SM, Kott S, Sager C, Huelsken T, Hollmann M. The glutamate receptor subunit delta2 is capable of gating its intrinsic ion channel as revealed by ligand binding domain transplantation. Proc Natl Acad Sci USA. 2009;106:10320–10325. doi: 10.1073/pnas.0900329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orth A, Tapken D, Hollmann M. The delta subfamily of glutamate receptors: Characterization of receptor chimeras and mutants. Eur J Neurosci. 2013;37:1620–1630. doi: 10.1111/ejn.12193. [DOI] [PubMed] [Google Scholar]
- 17.Granger AJ, Shi Y, Lu W, Cerpas M, Nicoll RA. LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature. 2013;493:495–500. doi: 10.1038/nature11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sia GM, et al. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Pelkey KA, et al. Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron. 2015;85:1257–1272. doi: 10.1016/j.neuron.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishore U, Reid KB. C1q: Structure, function, and receptors. Immunopharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 21.Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–760. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- 22.Wei P, et al. The Cbln family of proteins interact with multiple signaling pathways. J Neurochem. 2012;121:717–729. doi: 10.1111/j.1471-4159.2012.07648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otsuka S, et al. Roles of Cbln1 in non-motor functions of mice. J Neurosci. 2016;36:11801–11816. doi: 10.1523/JNEUROSCI.0322-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seigneur E, Südhof TC. Cerebellins are differentially expressed in selective subsets of neurons throughout the brain. J Comp Neurol. 2017;525:3286–3311. doi: 10.1002/cne.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traunmüller L, Gomez AM, Nguyen TM, Scheiffele P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- 26.Rong Y, et al. Comparison of Cbln1 and Cbln2 functions using transgenic and knockout mice. J Neurochem. 2012;120:528–540. doi: 10.1111/j.1471-4159.2011.07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito-Ishida A, Okabe S, Yuzaki M. The role of Cbln1 on Purkinje cell synapse formation. Neurosci Res. 2014;83:64–68. doi: 10.1016/j.neures.2014.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.