Significance
The overproduction and purification of membrane proteins are intrinsically difficult, making their analysis challenging. We purified the TraE membrane protein from a bacterial conjugation system that is involved in plasmid transfer. Our results suggest that this protein forms hexamers with a central pore, and we also show that it binds to the TraD protein. The structure of TraE is completely different from that of the previously characterized periplasmic domain. This has intriguing implications for the role of TraE and of its interaction partner TraD in substrate translocation across the bacterial cell envelope. This work makes an important contribution to understanding of the mechanism of plasmid transfer, contributing to the design of approaches to inhibit the spread of antibiotic resistance genes.
Keywords: type IV secretion, VirB8-like, VirB6-like, conjugation, plasmid
Abstract
Type IV secretion systems (T4SSs) are multiprotein assemblies that translocate macromolecules across the cell envelope of bacteria. X-ray crystallographic and electron microscopy (EM) analyses have increasingly provided structural information on individual T4SS components and on the entire complex. As of now, relatively little information has been available on the exact localization of the inner membrane-bound T4SS components, notably the mostly periplasmic VirB8 protein and the very hydrophobic VirB6 protein. We show here that the membrane-bound, full-length version of the VirB8 homolog TraE from the plasmid pKM101 secretion system forms a high-molecular-mass complex that is distinct from the previously characterized periplasmic portion of the protein that forms dimers. Full-length TraE was extracted from the membranes with detergents, and analysis by size-exclusion chromatography, cross-linking, and size exclusion chromatography (SEC) multiangle light scattering (MALS) shows that it forms a high-molecular-mass complex. EM and small-angle X-ray scattering (SAXS) analysis demonstrate that full-length TraE forms a hexameric complex with a central pore. We also overproduced and purified the VirB6 homolog TraD and show by cross-linking, SEC, and EM that it binds to TraE. Our results suggest that TraE and TraD interact at the substrate translocation pore of the secretion system.
Secretion systems mediate the passage of macromolecules across cellular membranes. In bacteria, secretion systems are essential for survival and they also play an important role for bacterial virulence. The best-studied examples of bacterial secretion systems that are virulence factors are the type III secretion system (T3SS) and type IV secretion system (T4SS) (1–3). The bacterial T4SS family can be divided into three functional groups. First, as in Brucella species, T4SSs deliver effector proteins into eukaryotic cells during the course of an infection. Second, in pathogens such as Helicobacter pylori, T4SSs mediate DNA uptake from and release of DNA into the extracellular environment. Finally, as in the incompatibility group N plasmid pKM101 studied here, T4SSs mediate the conjugative transfer of plasmids into other bacteria (4).
The well-characterized Agrobacterium tumefaciens system serves as an example of the typical composition of T4SSs, which are generally composed of 12 proteins (1, 2). T4SSs include three ATPases (VirB4, VirB11, and VirD4) that power complex assembly and substrate translocation. The extracellular pilus consists of a minor subunit (VirB5) and a major subunit (VirB2). The central periplasmic core complex comprises seven proteins (VirB1, VirB3, VirB6, VirB7, VirB8, VirB9, and VirB10), including an inner membrane channel believed to be composed of VirB6, VirB8, and VirB10. Here, we focus on VirB8- and VirB6-like proteins that are essential for the function of all T4SSs in which they have been studied. Recent results using high-resolution, negative-stain electron microscopy (EM) have provided groundbreaking insights into the T4SS structure, but the exact localization of VirB8 and VirB6 proteins is currently unknown (5–7). VirB8 homologs are thought to be assembly factors that interact with many other T4SS components in a mostly transient fashion (8–16). They are membrane proteins of typically 25 kDa comprising a short N-terminal cytoplasmic region of about 40 amino acids, one transmembrane α-helix, and a periplasmic region of 18 kDa. Since work with membrane proteins is intrinsically challenging, only the structures of the periplasmic portions of VirB8 homologs have been solved until now, but it is quite likely that the N terminus and the transmembrane α-helix make important contributions to protein structure and function (17–20). So far, structural information is not available for VirB6 proteins, which are very hydrophobic inner membrane proteins containing five or more transmembrane helices that are believed to locate at or close to the translocation pore (13, 21–23). VirB6 and VirB8 can be cross-linked to the translocated DNA substrate in A. tumefaciens, which led to the notion that they may participate directly in substrate translocation (24). These data suggest that they act at the same or subsequent steps during substrate translocation, but an interaction between these proteins has not been directly shown until now.
Membrane proteins constitute ∼30% of the proteome in each living organism and play crucial roles in many fundamental cell processes. They are the targets of the majority of available drugs, but due to technical challenges for structural studies, the structures of relatively few membranes proteins are known (25, 26). To address this lack of information in the case of VirB8 and VirB6 homologs, we studied the full-length version of the VirB8 homolog TraE and the VirB6 homolog TraD from plasmid pKM101. Using a combination of cross-linking experiments, multiangle light scattering (MALS), single-particle EM, and small-angle X-ray scattering (SAXS), we show that the quaternary structure of TraE is very different from that of the previously characterized periplasmic domains of VirB8 homologs. Our data demonstrate that TraE forms a hexamer with a central pore and that it interacts directly with TraD.
Results
Expression and Purification of Full-Length VirB8 Homologs.
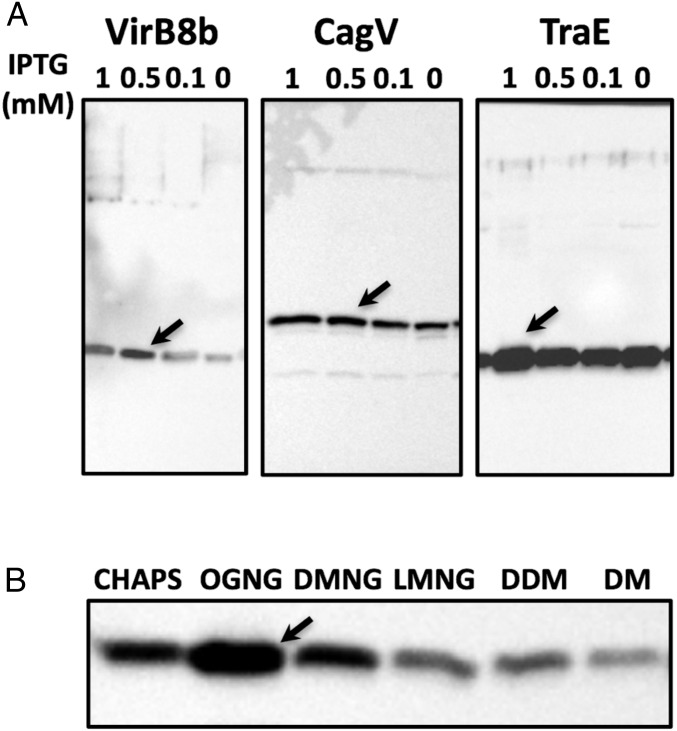
Since expression of membrane proteins can be a considerable challenge, we first optimized the conditions for overproduction of N-terminally His-tagged, full-length VirB8 homologs from Brucella suis (VirB8b), H. pylori (CagV), and plasmid pKM101 (TraE) (Fig. 1A). Since expression of traE resulted in the highest amount of overproduction, we pursued the analysis of this protein and analyzed the efficacy of extraction from the membrane with six different detergents (Fig. 1B). Extraction with octyl glucose neopentyl glycol (OGNG) gave the highest yield and was used for membrane extraction, followed by purification via immobilized metal affinity chromatography (IMAC) and size exclusion chromatography (SEC) (27).
Fig. 1.
Overexpression and detergent solubilization of VirB8-like full-length proteins. (A) Western blot analysis with a His-tag–specific antiserum to test the overexpression of VirB8-like proteins using the indicated concentrations of the expression inducer IPTG: Brucella (VirB8b; 30 °C, 6 h), Helicobacter (CagV; 30 °C, 6 h), and pKM101 (TraE; 18 °C, 16 h). (B) Western blot analysis with a His-tag–specific antiserum to test the solubilization of TraE in several detergents (CHAPS, DM, DMNG, LMNG, and OGNG). CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; DM, decyl maltoside; DMNG, decyl maltose neopentyl glycol; LMNG, lauryl maltose neopentyl glycol; OGNG, octyl glucose neopentyl glycol. Arrows indicate optimal conditions.
Cross-Linking and SEC-MALS Show That Full-Length TraE Forms a High-Molecular-Mass Complex.
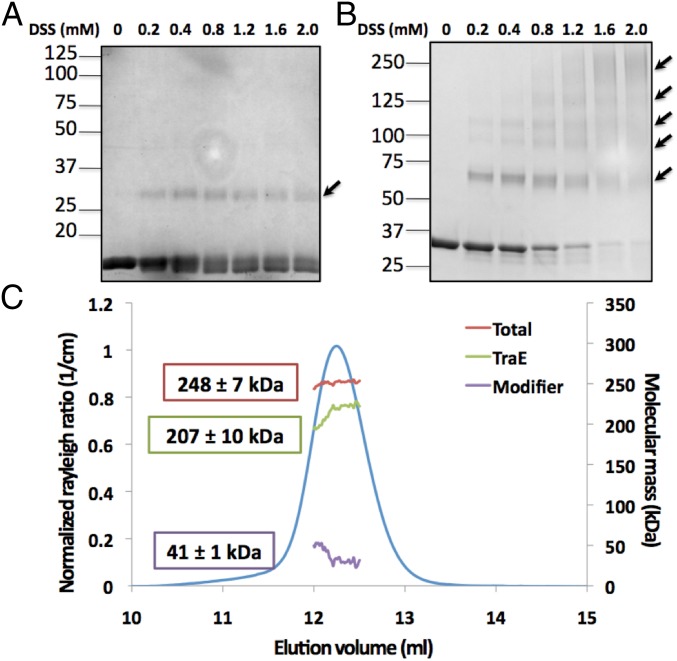
In our previous work, we have shown that the periplasmic domains of VirB8b and TraE form concentration-dependent dimers, and multimer formation can also be monitored by cross-linking (8, 10). To compare the multimeric state of full-length TraE relative to that of its periplasmic domain, we subjected equal amounts of the proteins to cross-linking with varying concentrations of the homo-bifunctional cross-linking agent disuccinimidyl suberate (DSS). The periplasmic domain primarily forms dimers (Fig. 2A), and low amounts of higher molecular-mass multimers are observed at higher protein concentrations (8). In contrast, when the full-length protein is incubated with DSS, dimers, as well as higher molecular-mass multimers, successively form and the monomeric protein diminishes, indicating high cross-linking efficacy (Fig. 2B). This result suggests that the quaternary structure of the full-length protein is quite distinct from that of the periplasmic portion. To obtain quantitative information, we further characterized TraE by SEC-MALS analysis, enabling us to calculate the absolute molecular masses of the TraE oligomer (207 kDa), of OGNG micelles (41 kDa) (SI Appendix, Fig. S1), and of the protein–detergent complex (248 kDa) (Fig. 2C). According to the molecular mass derived from MALS analysis, TraE may form hexamers or heptamers, and we next obtained direct insights into the structure of these complexes.
Fig. 2.
Analysis of the oligomerization state of TraE. SDS/PAGE analysis of the purified periplasmic domain of TraE at 1 mg/mL (A) and of purified full-length TraE at 1 mg/mL (B) in the absence (0 mM) and presence of increasing concentrations (0.2–2.0 mM) of the cross-linking agent DSS. Proteins in the gels were stained with Coomassie blue dye, and arrows indicate higher molecular-weight complexes formed after cross-linking. (C) Elution profile of the TraE oligomer is shown with the molecular weight estimated by MALS. The molar masses corresponding to the total complex, the TraE oligomer, and the modifier (detergent micelle) throughout the elution peaks are shown.
EM and SAXS Analyses Suggest That TraE Forms Hexamers.
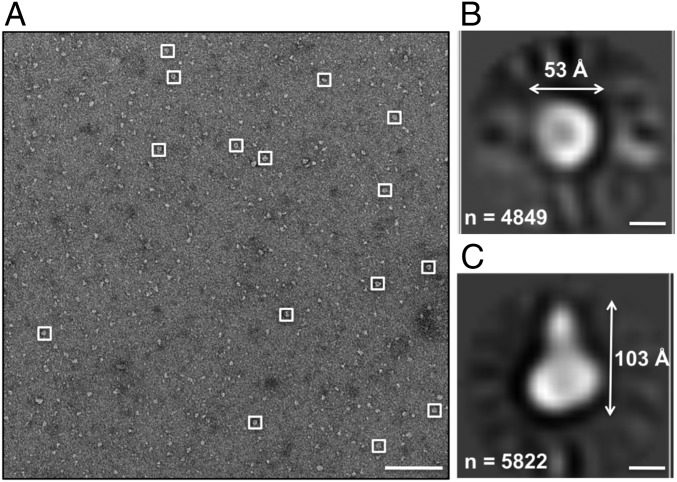
To obtain independent evidence for the quaternary structure of TraE, we analyzed purified detergent-solubilized TraE by negative-stain EM. We observe uniform ring-like particles of ∼130 Å diameter, and the particles apparently have similar orientations (Fig. 3A). Particles were manually picked and extracted, and 2D image analysis was performed. Approximately half of the class averages show featureless rings with varying pore sizes (SI Appendix, Fig. S2). The remaining class averages contain regions of density within the rings, and we observe six regions of density (Fig. 3 B and C). These results suggest that TraE forms a hexameric complex, which is consistent with the MALS data. In parallel, detergent-solubilized TraE complexes were characterized by SAXS, using inline SEC to ensure the homogeneity of the analyzed sample (Fig. 4A). The molecular weight estimated from the Porod volume (MW = Vp/1.6) and the volume of correlation are in the range of 200–225 kDa, which is in accord with the molecular weight calculated by SEC-MALS. The radius of gyration (Rg) of TraE in solution is 44 Å, and the normalized pairwise distance distribution of the curve [P(R)] (Fig. 4B) shows a maximum distance (Dmax) of 137 Å. To interpret the SAXS results at the molecular level, an ab initio model was calculated and we were able to fit the theoretical scattering profile for a rigid body model with the experimental SAXS data (Fig. 4C), with a χ2 value of 1.16. Based on the results of cross-linking, MALS, and EM analyses, we assumed that TraE forms hexamers and this symmetry was applied to reconstruct an ab initio protein structure shown in Fig. 4 D, E, and F, respectively. Interestingly, the SAXS-based model suggests that a pore exists at the center of the hexameric complex. This result is consistent with observations made by EM, and it has interesting implications for the functional interpretation of these data.
Fig. 3.
EM analysis of the TraE structure. (A) Typical negative-stain micrograph of the TraE complex showing uniform particles of ∼130 Å. (Scale bar, 500 Å.) (B) Representative 2D class averages following alignment, reference-free clustering, and multireference alignment of 1,061 particles. Numbers of particles used for generating each average are shown in the upper left portion of the panels. (Scale bars, 50 Å.) (C) Projections of TraE structures after a low-pass filter at 15 Å and the approximate dimensions are illustrated. (Scale bars, 50 Å.)
Fig. 4.
SEC-SAXS analysis of TraE. (A) SEC profile of the TraE sample used for the inline SAXS experiment. mAU, arbitrary units. (B) Normalized pair distribution functions [P(R)] calculated automatically with AutoGNOM. (C) Fit of the theoretical scattering profile for the rigid body model (gray plot) with the experimental SAXS data (black line). A top view (D), side view (E), and down view (F) of the average molecular envelope calculated for TraE (Small Angle Scattering Biological Databank SASDB75) are shown. The approximate envelope dimensions are illustrated.
TraE and TraD Interact and Form a High-Molecular-Mass Complex.
Previous data suggest that VirB6 and VirB8 interact at or close to the translocation pore, but no direct biochemical evidence was available for this notion. To address this question, we overexpressed N-terminally His-tagged TraD (His6-TraD) and we also coexpressed His6-TraD and nontagged TraE in Escherichia coli, after which the proteins were extracted from the membrane with detergents and purified by IMAC and SEC. After coexpression and detergent extraction, TraE coeluted with His6-TraD from the IMAC column, suggesting that these proteins interact, and analysis by SEC shows that they form a complex of ∼200 kDa (Fig. 5A), while nontagged TraE alone did not bind to the column. To assess whether His6-TraD and the His6-TraD–TraE complex form multimers similar to TraE, we subjected the proteins to cross-linking with varying concentrations of DSS. We observed that similar to TraE alone (Fig. 2B), His6-TraD (Fig. 5B) and His6-TraD–TraE form higher molecular-mass complexes in the presence of DSS (Fig. 5 C and D). These results show that TraD and TraE interact and form multimers, but analysis by SDS/PAGE and Western blotting did not enable us to identify distinct high-molecular-mass complexes that can be assigned to distinct TraD or TraD–TraE complexes.
Fig. 5.
SEC and cross-linking analysis of TraD and the TraD–TraE complex. (A) SEC profile of the TraD–TraE complex showing an apparent molecular mass of 200 kDa. SDS/PAGE and Western blot analysis of the SEC peak fraction with TraE-specific antiserum and His-tag–specific antiserum to detect N-terminally His-tagged TraD (His6-TraD), SDS/PAGE, and Coomassie blue staining shows the purity of the complex. mAU, arbitrary units. (B) Purified His6-TraD was incubated with varying concentrations of DSS, and cross-linking products were detected after SDS/PAGE and Western blotting with His-tag–specific antiserum. (C) Purified His6-TraD–TraE complex was incubated with varying concentrations of DSS, and cross-linking products were detected after SDS/PAGE and Western blotting with antiserum specific for His6-TraD. (D) Purified His6-Tra–TraE complex was incubated with varying concentrations of DSS, and cross-linking products were detected after SDS/PAGE and Western blotting with TraE-specific antiserum. Arrows indicate higher molecular-weight complexes formed after cross-linking.
EM Confirms That TraD and TraE Form a High-Molecular-Mass Complex.
To obtain additional insights into the structure of the TraD–TraE complex, we conducted negative-staining EM analyses revealing that these proteins form a complex of smaller dimensions compared with the TraE complex (Fig. 6 and SI Appendix, Fig. S3). This result is consistent with the analysis by SEC and the overall asymmetrical shape of this complex (53 Å × 103 Å), comprising a core body with an elongated extension that is quite distinct from the symmetrical hexamer, with a diameter of 130 Å formed by TraE alone (Fig. 3A). To corroborate the presence of TraD and TraE in the complex, we conducted immuno-EM analysis with TraE-specific antisera, His6-specific beads, and gold bead-labeled secondary antibodies; negative-staining EM confirms the presence of both proteins in the complex (SI Appendix, Fig. S4).
Fig. 6.
EM analysis of the TraD–TraE complex structure. (A) Typical negative-stain micrograph of the TraD–TraE complex showing uniform particles with dimensions of ∼60 Å and ∼100 Å, respectively. (Scale bar, 500 Å.) (B and C) Representative 2D class averages following alignment, reference-free clustering, and multireference alignment of 84,236 particles. Numbers of particles used for generating each average are shown in the bottom left portion of the panels, and the approximate dimensions are illustrated. (Scale bars, 30 Å.)
Discussion
The results presented here provide insights into the structures of integral inner-membrane T4SS components. Until now, VirB8-like proteins were primarily considered as assembly factors, and this notion was based on the fact that they interact with many other T4SS components, mostly in a relatively transient fashion. Biochemical work showing these interactions was performed primarily with the purified periplasmic domains of the protein (8–10, 13, 28), but interactions of full-length VirB8 homologs were also shown using the bacterial and yeast two-hybrid systems (12, 14, 29–31). The fact that the quaternary structure of full-length TraE is very different from that of the periplasmic portion underlines a critical role of the N-terminal transmembrane helix and of the cytoplasmic domain for protein assembly and function. The role of these parts of the protein attracted relatively little attention until now, but studies with the bacterial two-hybrid system showed that they contribute to dimerization (29–31). The fact that purified full-length TraE assembles into a hexamer with a possible pore at the center was unexpected and suggests that the protein also plays a structural role in the T4SS. This notion is consistent with previous observations suggesting that the protein binds the translocated DNA substrate in Agrobacterium (24).
The exact localization of VirB8-like proteins in T4SS complexes is currently unknown, but the SAXS model proposed here enables us to dock the protein into the available structures obtained by X-ray crystallography and EM (6, 7, 32). The dimensions of the SAXS model, combined with knowledge on the localization of TraE at the inner membrane, enable docking into the T4SS structure to the top of the “arch” observed in the structure of the plasmid R388 T4SS3–10 (7) (SI Appendix, Fig. S5). Considering the available data on VirB8/TraE protein interactions with VirB4 and VirB6 homologs, this localization would be biologically significant and is consistent with the previously observed VirB8-VirB6-VirB4 cofractionation (9). The hexameric TraE structure may localize on top of VirB4-like proteins that were shown to be present in two copies of hexamers (7) (SI Appendix, Fig. S5A). The observation of VirB8 hexamers and the docking of two copies on top of VirB4 complexes are also consistent with the experimentally determined stoichiometry of ∼12.6 VirB8 molecules in the T4SS subcomplex from plasmid R388 T4SS3–10 (7). Previous work in the Agrobacterium system showed cross-linking of VirB8 to translocated DNA substrates, which suggested that VirB8 and VirB6 form a functional subcomplex that possibly constitutes the pore of the T4SS (24). We here present direct evidence for a complex between VirB8- and VirB6-like proteins, supporting the notion that they act together in substrate translocation. The TraE multimer is strikingly similar to the higher molecular-mass complex observed in the case of TraM; its homolog from the plasmid R64 conjugation system and complexes of similar dimensions were also observed in the case of the homolog DotI from the Legionella pneumophila T4SS (33). The SAXS and EM results presented here demonstrate that this protein forms a hexamer with a central pore, and this discovery has important implications for the mechanism of type IV secretion. In addition, docking of the TraE structure into the center of the structure of a T3SS (34) shows that the hexamer would fit the dimensions of these macromolecular transporters in a similar fashion, even if T3SSs are not evolutionarily related and do not contain VirB8 homologs (SI Appendix, Fig. S5B).
Based on the available data, we propose a model for the assembly of VirB8/TraE-like proteins at the inner membrane of gram-negative bacteria (SI Appendix, Fig. S6). VirB8-like proteins integrate into the inner membrane following the standard protein insertion pathway, followed by dimerization. The available X-ray structures of VirB8 from Brucella (17) and Agrobacterium (18) and of TraE from pKM101 (8) show alternative modes of dimerization. These may reflect alternative conformational states of the protein that assembles into a hexamer, possibly exposing different interfaces for interactions with other T4SS components (e.g., with the pilus components VirB2 and VirB5), and this process may be coordinated via its interaction with VirB4 (9). The interactions with structural components such as VirB10 (11, 14) may link VirB8 complexes to the central substrate translocation pore that may be formed by VirB6 (13, 24). The TraD–TraE complex characterized here with biochemical methods and by EM substantiates the notion that these proteins interact, but it is smaller than the TraE hexamer alone. Therefore, the TraD–TraE complex likely represents an intermediate state for the assembly of these proteins at the T4SS core. In the future, higher resolution structural studies of VirB8/TraE and of its complexes with VirB6 homologs and with other proteins (e.g., by X-ray crystallography or cryo-EM) are required to test this model and to establish the role of the versatile VirB8-like proteins and their complexes with VirB6 in T4SS assembly and function.
Experimental Procedures
Strains, Plasmids, and DNA Manipulation.
The strains and plasmids used are described in SI Appendix, Table S1. The E. coli strain XL-1 Blue or DH5α was used as a host for cloning, and the E. coli strain BL21(DE3)star was used for VirB8b, CagV, TraD, TraE, and TraD–TraE complex protein overproduction. Miniprep kits (Qiagen) were used to isolate plasmid DNA. Standard techniques were employed for the cloning, transformation, preparation, and restriction analysis of plasmid DNA from E. coli (35).
Small-Scale Membrane Protein Expression and Solubilization Tests.
For protein overproduction, the E. coli strain BL21star (λDE3) carrying expression plasmids was grown under aerobic conditions at 37 °C in LB to exponential phase (OD600 of 0.4–0.8). For VirB8b, CagV, and TraE, expression was induced by the addition of 0, 0.1, 0.5, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG); temperatures at 18 °C, 25 °C, 30 °C, and 37 °C; and cultures left shaking for 1–16 h at 220 rpm. Cells were collected by centrifugation at 32,500 × g for 10 min at 4 °C. Bacterial pellets were suspended in lysis buffer [50 mM Hepes (pH 8.0), 25% sucrose] containing 5 mM MgCl2, 1% Triton X-100, 1 mg/mL lysozyme, and one tablet of cOmplete Mini Protease Inhibitor mixture (Roche) and kept on ice for 1 h. To separate soluble and insoluble fractions, centrifugation at 32,500 × g for 30 min at 4 °C was done, and the supernatant was used for isolation of the membranes. Proteins in cell lysates were detected by Western blotting with an anti–His-tag antiserum (1:5,000 dilution, AM1010a; Abgent). Following ultracentrifugation at 250,000 × g for 1 h at 4 °C, total membranes were collected and solubilized for 1.5 h at 4 °C in 50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, 40 mM imidazole, and 1% (wt/vol) detergents [decyl maltoside, dodecyl maltoside (DDM), lauryl maltose neopentyl glycol, decyl maltose neopentyl glycol (DMNG), OGNG, and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)] with cOmplete Mini Protease Inhibitor mixture. This material was then centrifuged for 45 min at 34,500 × g, and detergent-solubilized proteins in the supernatant were detected by Western blotting with an anti–His-tag antiserum (1:5,000 dilution).
TraE and TraD-TraE Membrane Protein Expression and Purification.
E. coli strain BL21star (λDE3) harboring pHTTraE, pHTTraD, and pETDuetTraDTraE was grown in LB supplemented with 50 μg/mL kanamycin or 100 μg/mL ampicillin only for pETDuetTraDTraE. Overnight precultures in LB were used to inoculate 1-L cultures (37 °C) until they reached an OD600 of 0.4–0.8. Expression was induced by addition of 1 mM IPTG at 18 °C, and cultures were left incubated for 16 h. For purification, bacterial cells were harvested, resuspended in binding buffer [50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, 40 mM imidazole] with cOmplete Mini Protease Inhibitor mixture and DNase I at 100 μg/mL, and lysed twice using a One Shot cell disrupter (Constant Systems, Inc.) at 27 kpsi and 4 °C. Debris was removed by centrifugation twice at 36,700 × g for 30 min at 4 °C, and the supernatant was retained. Pursuing ultracentrifugation at 250,000 × g for 1 h at 4 °C, total membranes were collected and solubilized for 1.5–2 h at 4 °C in 50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, 40 mM imidazole, and 1% (wt/vol) detergent OGNG with cOmplete Mini Protease Inhibitor mixture. This material was then centrifuged for 45 min at 34,500 × g to collect OGNG-solubilized TraE or DDM-solubilized TraD and TraD–TraE complex for purification over a HisTrap Ni-chelate column (GE Healthcare) and eluted using a linear 50-mL gradient of 40–500 mM imidazole. For SAXS, EM, and biochemical analysis, TraE, TraD, and TraD–TraE complex were further purified by SEC (27) using a Superdex 200 column (GE Healthcare). TraE and TraD–TraE complex protein concentrations were determined using molar extinction coefficients at 280 nm of 35,870 M−1⋅cm−1 (with 6xHis-tag) and 89,270 M−1⋅cm−1 (with 6xHis-tag), respectively.
Protein–Detergent Complex Analysis.
The TraE–OGNG complex was analyzed by SEC-MALS with the use of an ÄKTAmicro system (GE Healthcare) coupled to a Dawn HELEOS II MALS detector and an OptiLab T-rEX online refractive index detector (Wyatt Technology). The absolute molecular mass was calculated by analyzing the scattering data using the ASTRA analysis software package, version 6.1.6.5 (Wyatt Technology). Protein samples were separated on a Superdex 200 10/300 analytical SEC column (GE Healthcare) with a flow rate of 0.3 mL⋅min−1. BSA was used for calibration. A 0.1-mL sample of TraE at a concentration of 1 mg/mL was injected and eluted in 50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, 40 mM imidazole, and 0.15% OGNG. The molecular masses of TraE and OGNG were determined by the dual detection method implemented in the conjugated analysis mode of the ASTRA analysis software. The refractive index increment of OGNG was calculated using the dn/dc determination method developed by Wyatt Technology (SI Appendix, Fig. S1). The refractive index increments of TraE and OGNG used were 0.185 mL⋅g−1 and 0.118 mL⋅g−1, respectively. The extinction coefficient of TraE for UV detection at 280 nm was calculated from the amino acid sequence.
Analysis of Homo-Oligomerization of TraE, Homo-Oligomerization of TraD, and Hetero-Oligomerization of TraD–TraE Complex by Cross-Linking.
Chemical cross-linking with DSS (Pierce) was performed as described elsewhere (9). The cross-linking product formation for homo-oligomerization of TraE was monitored by SDS/PAGE and staining with Coomassie blue dye. For homo-oligomerization of TraD and hetero-oligomerization of TraD–TraE complex, the formation of cross-linking products was detected by Western blotting with an anti–His-tag antiserum (1:5,000 dilution) to detect His6-TraD and with an anti-TraE antiserum (1:3,000 dilution), respectively.
Negative-Stain EM, Image Analysis, and Gold Labeling.
Parlodion-supported and carbon-coated copper grids (SPI Supplies) were negatively glow-discharged (Leica Microsystems) before adsorbing 5 μL of SEC-purified sample at 2 ng/μL for 1 min and stained with 5 μL of freshly prepared 1.5% uranyl formate (Electron Microscopy Sciences) for 1 min. Samples were imaged at room temperature using an FEI Tecnai T12 transmission electron microscope (TEM) equipped with a tungsten filament and operated at 80 kV. Images were collected at defocus between 2 and 4 μm on an FEI Eagle 4k × 4k CCD camera at a magnification of ∼67,000×, with a pixel size of 1.64 Å. For TraE, image processing was performed using the EMAN2 package (36). A total of 1,061 particles were picked manually and extracted with a 160 × 160-pixel box size. The SPARX software suite was used for particle alignment, K means clustering (reference-free), and multireference alignment (37). No symmetry was applied at any point. For analysis of the TraD–TraE complex, image processing and 2D classification were performed using Scipion and XMIPP software. A total of 84,236 particles were picked automatically and extracted with 60 × 60-pixel box size. For colloidal gold labeling of the TraD–TraE complex and negative controls (buffer only, BSA, and ovalbumin), samples were applied for 1 min onto glow-discharged, carbon-coated copper grids. Excess liquid was blotted, and grids were incubated for 30 min at 4 °C on a drop of 5 nm of Ni-NTA-Nanogold beads solution (Nanoprobes). Following incubation, grids were washed three times on a drop of ice-cold purification buffer containing 30 nM imidazole and two times on a drop of ice-cold H2O. For labeling of TraD, samples were stained for 1 min with 1.5% uranyl formate. For dual labeling, grids were blocked for 15 min with ovalbumin before a 1-h incubation with anti-TraE rabbit antiserum (1:4,000 dilution), washed three times with PBS-Triton, and finally blocked a second time with ovalbumin before a 30-min incubation with 10 nm protein A-coupled gold beads (1:50 dilution). Samples were then negatively stained with 1.5% uranyl formate. Samples were imaged using an FEI Tecnai T12 TEM operated at an acceleration voltage of 120 kV. Images were collected at defocus between 2 and 4 μm on an FEI Eagle 4k × 4k CCD camera at a range of magnification between ∼67,000× and ∼1,000,000×.
SAXS Data Collection and Analysis.
An inline SEC-SAXS configuration was used for data collection (SI Appendix, Table S2). Protein samples were separated on a Superdex 200 analytical SEC column (GE Healthcare) with a flow rate of 0.3 mL⋅min−1. A sample of 0.5 mL of TraE at a concentration of 1 mg/mL was injected and eluted in 50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl, 40 mM imidazole, and 0.15% OGNG. Scattering data were measured at beamline G1 of the Biological Small-Angle X-Ray Solution Scattering source at the Macromolecular Diffraction at the Cornell High Energy Synchrotron Source. Data were collected on a dual Pilatus 100K-S SAXS/wide-angle X-ray scattering detector at a wavelength of 1.245 Å, covering a scattering vector range (q = 4πsinθ/λ) from 0.008 to 0.8 Å−1. Data were analyzed, integrated, and averaged with RAW software package, version 1.0.1 (38). Buffer blanks were averaged and subtracted from the data. A linear Guinier fit plot was calculated using the RAW software. Guinier analysis and Rg estimation were performed in PRIMUS and confirmed by automatic analysis using AutoRG (39). The largest dimension of the molecule, Dmax, and Vp were calculated using GNOM (40). The pair distribution function P(R) and forward scattering I(0) were computed with AutoGNOM (41) and compared with those determined in Primusqt (39). GASBOR (42) was used to reconstruct an ab initio protein structure by a chain-like ensemble of dummy residues, and a P6 symmetry was applied. Structural figures were prepared using PyMOL (43) and UCSF Chimera (44). To fit the SAXS envelope with X-ray and EM structures, the MultiFit add-on from UCSF Chimera was used (45), and, finally, a manual orientation of the X-ray structure was done. Data have been deposited in the Small Angle Scattering Biological Databank (www.sasbdb.org; SASDB75).
Supplementary Material
Acknowledgments
This work was supported by Canadian Institutes of Health Research Grants MOP-84239 (to C.B.) and MOP-110972 (to A.N.), the Natural Sciences and Engineering Research Council Collaborative Research And Training Experience (CREATE) program on the Cellular Dynamics of Macromolecular Complexes, the Groupe d’études des proteines membranaires, the Canada Foundation for Innovation, and the Fonds de recherche du Québec-Santé. We thank Jurgen Sygusch (Université de Montréal) for discussions on SAXS analysis, Jesse Hopkins and Richard Gillian [Cornell High Energy Synchrotron Source (CHESS)] for assistance with the SAXS measurements and Joaquin Ortega (McGill University) for advice on single-particle EM analysis. Synchrotron SAXS data were collected at the Macromolecular Diffraction at the Cornell High Energy Synchrotron Source (MacCHESS) beamline G1. The CHESS is supported by the National Science Foundation (NSF) and NIH/National Institute of General Medical Sciences (NIGMS) via NSF Award DMR-1332208, and the MacCHESS resource is supported by NIH/NIGMS Award GM-103485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Small Angle Scattering Biological Databank, www.sasbdb.org (accession no. SASDB75).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802501115/-/DCSupplemental.
References
- 1.Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol. 2014;27:16–23. doi: 10.1016/j.sbi.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandran Darbari V, Waksman G. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem. 2015;84:603–629. doi: 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 3.Burkinshaw BJ, Strynadka NC. Assembly and structure of the T3SS. Biochim Biophys Acta. 2014;1843:1649–1663. doi: 10.1016/j.bbamcr.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 4.Winans SC, Walker GC. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low HH, et al. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casu B, et al. Structural analysis and inhibition of TraE from the pKM101 type IV secretion system. J Biol Chem. 2016;291:23817–23829. doi: 10.1074/jbc.M116.753327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Q, et al. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem. 2005;280:26349–26359. doi: 10.1074/jbc.M502347200. [DOI] [PubMed] [Google Scholar]
- 10.Paschos A, et al. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc Natl Acad Sci USA. 2006;103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivanesan D, Hancock MA, Villamil Giraldo AM, Baron C. Quantitative analysis of VirB8-VirB9-VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly. Biochemistry. 2010;49:4483–4493. doi: 10.1021/bi902201y. [DOI] [PubMed] [Google Scholar]
- 12.Sivanesan D, Baron C. The dimer interface of Agrobacterium tumefaciens VirB8 is important for type IV secretion system function, stability, and association of VirB2 with the core complex. J Bacteriol. 2011;193:2097–2106. doi: 10.1128/JB.00907-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villamil Giraldo AM, et al. Type IV secretion system core component VirB8 from Brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry. 2012;51:3881–3890. doi: 10.1021/bi300298v. [DOI] [PubMed] [Google Scholar]
- 14.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilar J, Zupan J, Cameron TA, Zambryski PC. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc Natl Acad Sci USA. 2010;107:3758–3763. doi: 10.1073/pnas.0914940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilar J, Cameron TA, Zupan J, Zambryski P. Membrane and core periplasmic Agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. MBio. 2011;2:e00218-11. doi: 10.1128/mBio.00218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terradot L, et al. Crystal structures of the periplasmic domains of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc Natl Acad Sci USA. 2006;103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie JJ, et al. Structural insight into how bacteria prevent interference between multiple divergent type IV secretion systems. MBio. 2015;6:e01867-15. doi: 10.1128/mBio.01867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goessweiner-Mohr N, et al. The 2.5 Å structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem. 2013;288:2018–2028. doi: 10.1074/jbc.M112.428847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judd PK, Kumar RB, Das A. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol Microbiol. 2005;55:115–124. doi: 10.1111/j.1365-2958.2004.04378.x. [DOI] [PubMed] [Google Scholar]
- 23.Villamil Giraldo AM, Mary C, Sivanesan D, Baron C. VirB6 and VirB10 from the Brucella type IV secretion system interact via the N-terminal periplasmic domain of VirB6. FEBS Lett. 2015;589:1883–1889. doi: 10.1016/j.febslet.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 24.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22:326–332. doi: 10.1016/j.sbi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 27.Ndongo S, et al. Autoimmune cholangitis or seronegative biliary cirrhosis: Report of one case. Dakar Méd. 2008;53:127–130. [PubMed] [Google Scholar]
- 28.Fercher C, et al. VirB8-like protein TraH is crucial for DNA transfer in Enterococcus faecalis. Sci Rep. 2016;6:24643. doi: 10.1038/srep24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patey G, Qi Z, Bourg G, Baron C, O’Callaghan D. Swapping of periplasmic domains between Brucella suis VirB8 and a pSB102 VirB8 homologue allows heterologous complementation. Infect Immun. 2006;74:4945–4949. doi: 10.1128/IAI.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourg G, Sube R, O’Callaghan D, Patey G. Interactions between Brucella suis VirB8 and its homolog TraJ from the plasmid pSB102 underline the dynamic nature of type IV secretion systems. J Bacteriol. 2009;191:2985–2992. doi: 10.1128/JB.01426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrieux L, Bourg G, Pirone A, O’Callaghan D, Patey G. A single amino acid change in the transmembrane domain of the VirB8 protein affects dimerization, interaction with VirB10 and Brucella suis virulence. FEBS Lett. 2011;585:2431–2436. doi: 10.1016/j.febslet.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Chandran V, et al. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda T, et al. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci Rep. 2015;5:10912. doi: 10.1038/srep10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen SS, et al. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Cryst. 2009;42:959–964. [Google Scholar]
- 39.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 40.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 41.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1–Towards automated and web-supported small-angle scattering data analysis. J Appl Cryst. 2007;40:S223–S228. [Google Scholar]
- 42.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. PyMOL Molecular Graphics System (2010), Version 1.3r1 (Schrödinger, LLC, New York)
- 44.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Lasker K, Topf M, Sali A, Wolfson HJ. Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J Mol Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.