Significance
Rett syndrome (RTT) is a severe neurological disease affecting girls in their early childhood. The underlying cause of most RTT cases is mutations in the gene methyl-CpG-binding protein 2 (MECP2). How the loss of MeCP2 function in the brain due to these mutations causes such severe neurological symptoms remains a mystery. Here, we show widespread aberrations in gene expression and anomalous patterns of alternative splicing, specifically when neurons of RTT mice are stimulated. Furthermore, these aberrations occur in conjunction with higher seizure susceptibility in response to neuronal stimulation in these RTT mice. Our findings suggest that MeCP2 is required for adjusting the robust changes in gene transcription and for proper regulation of alternative splicing during neuronal stimulation.
Keywords: Rett syndrome, MeCP2, neuronal activity, gene expression, alternative splicing
Abstract
Rett syndrome (RTT) is a severe neurodevelopmental disorder that affects about 1 in 10,000 female live births. The underlying cause of RTT is mutations in the X-linked gene, methyl-CpG-binding protein 2 (MECP2); however, the molecular mechanism by which these mutations mediate the RTT neuropathology remains enigmatic. Specifically, although MeCP2 is known to act as a transcriptional repressor, analyses of the RTT brain at steady-state conditions detected numerous differentially expressed genes, while the changes in transcript levels were mostly subtle. Here we reveal an aberrant global pattern of gene expression, characterized predominantly by higher levels of expression of activity-dependent genes, and anomalous alternative splicing events, specifically in response to neuronal activity in a mouse model for RTT. Notably, the specific splicing modalities of intron retention and exon skipping displayed a significant bias toward increased retained introns and skipped exons, respectively, in the RTT brain compared with the WT brain. Furthermore, these aberrations occur in conjunction with higher seizure susceptibility in response to neuronal activity in RTT mice. Our findings advance the concept that normal MeCP2 functioning is required for fine-tuning the robust and immediate changes in gene transcription and for proper regulation of alternative splicing induced in response to neuronal stimulation.
Rett syndrome (RTT) is a severe postnatal neurodevelopmental disorder characterized by normal development up to 12–18 mo of age, followed by regression in already acquired milestones, such as speech and motor skills, as well as by the development of a myriad of other neurological and behavioral abnormalities, including seizures, learning disabilities, and autism (1). The underlying cause of over 95% of RTT cases is mutations in the X-linked gene methyl-CpG-binding protein 2 (MECP2) (1, 2), yet how the loss of MeCP2 function in the brain mediates RTT remains poorly understood. Specifically, although MeCP2 was characterized as a transcriptional repressor, transcriptional profiling studies on the whole RTT mouse brain and even specific areas of the brain revealed numerous genes that are differentially expressed, while the changes in transcript levels were mostly subtle (3–5). This conundrum has led to other studies, some of which have suggested that MeCP2 is not a gene-specific transcriptional repressor but rather a global repressor that abundantly binds methylated DNA and dampens transcriptional noise genomewide (6), that MeCP2 acts as both a repressor and an activator (5, 7, 8), that MeCP2 functions in a cell-type-specific manner (9, 10), and that MeCP2 is a long-gene-specific repressor (4). Other studies have suggested that MeCP2 functions posttranscriptionally as a regulator of alternative splicing (11, 12), or as a regulator of microRNA expression (13–15). Although these studies advanced our understanding of the function of MeCP2, they also clearly emphasize that the molecular mechanism which mediates RTT is likely complex and has yet to be deciphered.
Several recent studies, including our own, support the notion that MeCP2 function is required for maintaining the proper structure/function of the neuronal networks in the maturing as well as in the fully matured brain (16–20). Specifically, these studies have shown that reactivation of MeCP2 in symptomatic adult RTT mice reverses the RTT phenotype (19), while depletion of MeCP2 even at the adult stage results in a severe RTT phenotype (16–18, 20). In addition, other studies have shown that there is an excitatory/inhibitory imbalance in the RTT brain, resulting specifically in hyperexcitability, which likely leads to specific RTT symptoms, including seizures and breathing abnormalities (21–25). Together, these studies suggest that MeCP2 might have a key role, specifically during neuronal network activity.
In this study, we tested the hypothesis that MeCP2 function is required for proper regulation of the activity-dependent transcriptome during neuronal stimulation. We found that neuronal activity, elicited either in vitro by potassium chloride (KCl) or in vivo by kainic acid (KA), results in a significantly higher expression of many activity-dependent genes in RTT cortical neurons and in the hippocampi of RTT mice, respectively, compared with their WT counterparts. Furthermore, hundreds of genes were differentially alternatively spliced specifically in response to neuronal activity, of which many of them were alternatively spliced in opposite directions (inclusion vs. exclusion) in the hippocampi of RTT mice. Thus, we propose that MeCP2 is required to regulate the robust changes in the transcriptome that occur in response to neuronal activity.
Results
KCl-Induced Membrane Depolarization Results in Differential Expression of Many Activity-Dependent Genes in Mecp2-Null Compared with WT Neuron-Enriched Cortical Cultures.
Because RTT is associated with hyperexcitability of the cortex and hippocampus (21–23), we reasoned that analysis of the gene expression profile in response to neuronal activity, where the immediate and robust changes in gene expression should be tightly regulated, could provide a new perspective and important insight into the function of MeCP2. This approach also allowed us to examine mainly the primary, rather than the secondary, effect of the loss of MeCP2 on the induced gene expression program.
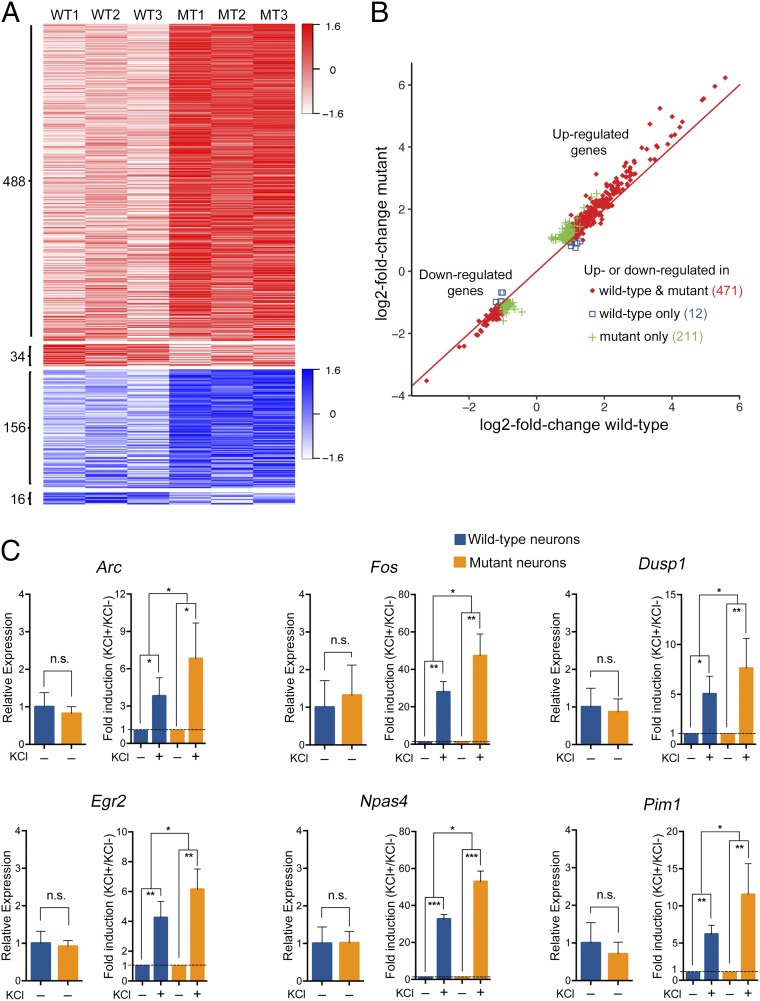
We first isolated cortical neurons from WT and Mecp2-null littermate male mice at postnatal day 0–1 and cultured them for 10 d. We analyzed by immunostaining the composition of the cultures, using several cell-type-specific markers, and verified the enrichment of neurons (65%) compared with glia (∼33% astrocytes, ∼2% oligodendrocytes, and <0.2% microglia), and the lack of bias for specific cell types between the WT and Mecp2-null cultures (SI Appendix, Fig. S1 A and B). We further validated by quantitative RT-PCR that the WT and the Mecp2-null cortical cultures expressed similar levels of neuronal and glial specific markers (SI Appendix, Fig. S1C). We induced membrane depolarization in neurons by treating the cultures with 30 mM KCl for 3 h (without presilencing of the spontaneous neuronal activity), performed RNA sequencing (RNA-Seq) analysis, and compared the mRNA profiles of untreated and KCl-treated, WT and mutant neuron-enriched cortical cultures. Our analysis shows that at steady-state conditions (untreated), only a small number of genes displayed differential gene expression between mutant and WT cultures. Specifically, of the ∼14,000 expressed genes, only 19 genes were down-regulated and eight genes were up-regulated in mutant compared with the WT (Dataset S1). In contrast, analysis of the KCl-induced activity-dependent genes, as defined by up- or down-regulation of at least twofold (q-value < 0.05) in the WT and/or mutant, showed that hundreds of genes were differentially expressed between the mutant and WT neuron-enriched cultures (Fig. 1 A and B and Datasets S2 and S3). Interestingly, most of the up-regulated genes showed a propensity toward a higher increase in gene expression in mutant compared with WT cultures (Fig. 1 A and B and Dataset S2). Specifically, of the 522 activity-dependent genes that were up-regulated, 488 genes were up-regulated to a higher level in mutant than in WT, while only 34 genes were up-regulated to a higher level in WT than in mutant cultures (Fig. 1A and Dataset S2). Furthermore, of the 172 genes that were down-regulated, 156 showed a propensity toward a greater decrease in expression in mutant than in WT, while only 16 genes showed a greater decrease in the WT cultures (Fig. 1A and Dataset S3). Detailed analysis of the activity-dependent genes showed that 471 genes were up- or down-regulated in both mutant and WT (fold change >2, q-value < 0.05), with the majority showing a higher increase in expression levels in mutant cultures (Fig. 1B, red). Notably, 211 genes were significantly up- or down- regulated in mutant (fold change >2, q-value < 0.05), but not in WT (Fig. 1B, green), while only 12 genes were significantly up- or down-regulated in WT but not in mutant cultures (Fig. 1B, blue). We validated, by quantitative RT-PCR, several of the differentially expressed activity-dependent immediate-early genes identified by RNA-Seq, including Arc, Egr2, Dusp1, Pim1, Npas4, and Fos. While there was no significant difference between untreated WT and mutant in the expression level of these genes, these genes were up-regulated to significantly higher levels in the mutant than in the WT neuron-enriched cortical cultures, upon KCl treatment (Fig. 1C).
Fig. 1.
Membrane depolarization induced by KCl results in greater changes in expression level of activity-dependent genes in Mecp2-null cortical cultures. (A) Heat map showing the relative mRNA expression level of activity-dependent genes (based on |log2-fold change| > 1.0 and q-value < 0.05) between KCl-treated and untreated WT or mutant (MT) (Mecp2-null) cortical cultures (n = 3 pairs). Red–white color range indicates the relative log2-fold changes (scaled by SDs) for up-regulated genes. Blue–white color range indicates the relative log2-fold changes (scaled by SDs) for down-regulated genes. The darker shade of blue or red corresponds to a greater level of down-regulation or up-regulation, respectively. (B) Scatter plot showing the changes in the expression level of activity-dependent genes upon KCl treatment in mutant and WT cortical cultures. Red diamonds indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in both mutant and WT cultures; green pluses indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in mutant but not in WT cultures; blue squares indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in WT but not in mutant cultures. Note that KCl treatment resulted in greater |log2-fold changes| in mutant samples in most genes shown. (C) Quantitative RT-PCR for the indicated immediate-early genes comparing their relative expression between untreated (KCl –) WT and untreated mutant cortical cultures (left for each gene) and their fold induction [treated (KCl +) vs. untreated (KCl −)] between WT and mutant cultures upon depolarization with KCl (right for each gene). n = 4 biological replicates. Bar graphs display the mean relative RNA expression level + SEM. Statistical significance was determined by paired two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Together, these data suggest that the loss of MeCP2 generates significant aberrations in the level of expression of many activity-dependent genes during neuronal stimulation.
Neuronal Activity Induced by KA in Vivo Results in Differential Expression of Many Activity-Dependent Genes in the Hippocampi of Mecp2-Null Mice Compared with WT Mice.
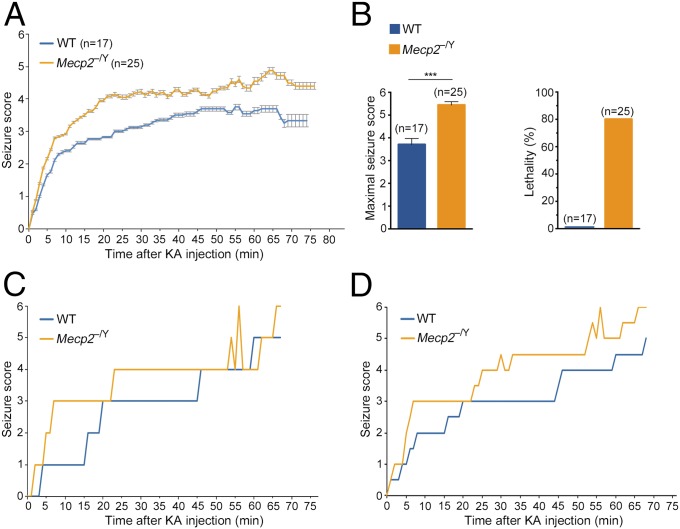
We next examined the effect of the loss of MeCP2 on the gene expression profile induced in response to neuronal activity in vivo. Because seizure is one of the overt characteristics of the RTT phenotype, we sought to induce neuronal activity in mutant RTT mice using the chemoconvulsant KA and analyze the transcription profile of their hippocampi. For this, we used WT and Mecp2-null (Mecp2−/y) littermate male mice at 7 wk of age, a time when the mutant mice show overt but not severe RTT phenotype, reaching a median score of 4–6, based on the observational phenotypic scoring system (0–10) we previously described (16). We injected the mice with 23 mg/kg body weight of KA, monitored them closely for the development of seizures, and recorded the seizure intensity stage on a scale from 1 to 6 as previously described (26). Kinetics of seizure development showed that while there was no significant difference between WT and mutant mice in the latency of seizure initiation after KA injection, there was a significant difference in the intensity of seizure development (Fig. 2A). Specifically, mutant mice reached a higher seizure score in shorter time than WT mice (Fig. 2A), and while the average maximal seizure score of the WT mice was between 3 and 4, the average maximal seizure score of mutant mice was between 5 and 6 (Fig. 2B, Left). In addition, 80% of the mutant mice did not survive following KA treatment, whereas none of the WT mice died throughout the duration of the experiment, even those that reached a high seizure score (Fig. 2B, Right). In fact, none of the mutant mice survived beyond 68 min if they reached a seizure score of 5–6. These findings support a previous study showing a differential seizure score between mutant RTT and WT mice (27).
Fig. 2.
Neuronal activity induced by KA results in a higher seizure score followed by lethality in Mecp2-null mice. (A) Kinetics of seizure development in 7-wk-old WT and mutant (Mecp2−/y) male mice injected with KA. Seizure score represents the seizure stage (1–6). Error bars display the ± SEM. (B) Bar graphs showing the mean + SEM of the maximal seizure score (Left) and percent lethality (Right) in WT and mutant mice. Statistical significance was determined by unpaired two-tailed Student’s t test. ***P < 0.001. (C and D) Kinetics of seizure development during 68 min of KA treatment of two pairs of 7-wk-old WT and mutant littermate male mice used for RNA-Seq analysis.
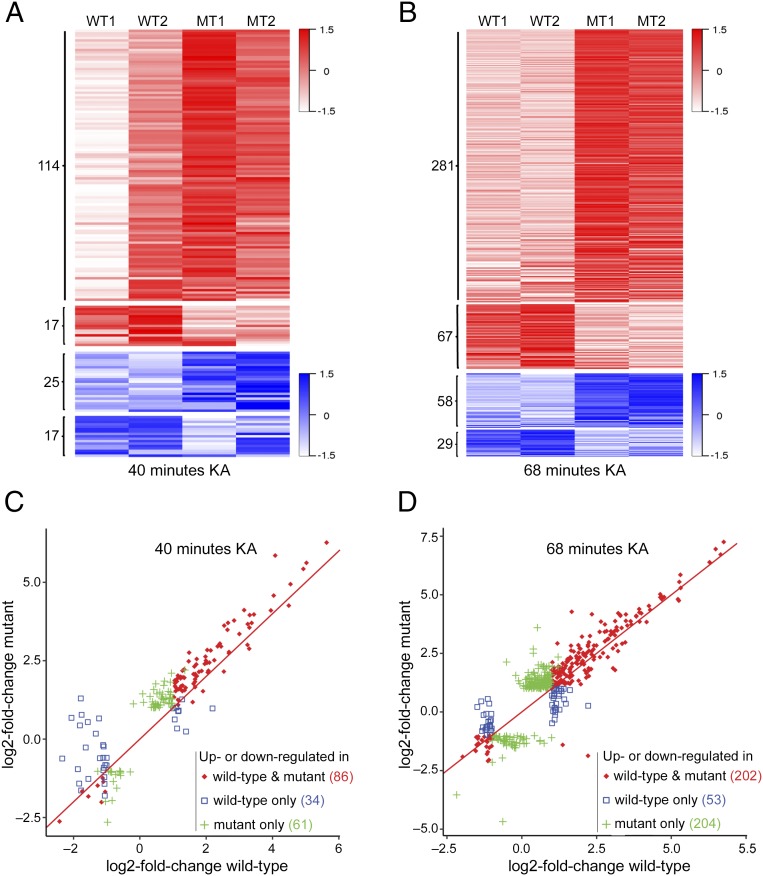
Next, we analyzed the activity-dependent gene expression profiles in the hippocampi of WT and mutant mice, at 40 and 68 min after administration of KA, compared with their gene expression profiles at steady-state conditions (untreated). To eliminate bias of the level of seizure on gene expression, we used WT and mutant littermate mice that both reached a comparable seizure score of 5–6 (Fig. 2 C and D). Our RNA-Seq analysis detected ∼14,000 expressed genes in the hippocampi of untreated WT and mutant mice. Of these expressed genes, 80 were differentially expressed between the mutant and WT mice (Dataset S4). Among them, 48 genes were up-regulated and 32 were down-regulated in the mutant mice compared with their WT counterparts (Dataset S4). Importantly, similar to the in vitro data, RNA-Seq analysis of the mutant and WT hippocampi, at 40 and 68 min of KA treatment, revealed widespread differential expression of hundreds of activity-dependent genes (fold change >2, q-value < 0.05 in WT and/or mutant) between the mutant and WT mice (Fig. 3). Specifically, of the 131 (40 min) and 348 (68 min) genes that were up-regulated, 114 (40 min) and 281 (68 min) genes were up-regulated to a higher level in the mutant mice compared with WT mice, while only 17 (40 min) and 67 genes (68 min) were up-regulated to a higher level in WT mice compared with mutant mice (Fig. 3 A and B and Datasets S5 and S7). In addition, 25 (40 min) and 58 (68 min) genes were down-regulated to a lower level in mutant than in WT mice, while only 17 (40 min) and 29 (68 min) genes were down-regulated to a lower level in WT than in mutant mice (Fig. 3 A and B and Datasets S6 and S8). Of the activity-dependent genes that were up- or down-regulated in both mutant and WT hippocampi (86 at 40 min and 202 at 68 min), the vast majority were up-regulated upon KA treatment and to a higher level in mutant than in WT mice (Fig. 3 C and D, red). Furthermore, 61 genes (40 min) and 204 genes (68 min) were significantly up- or down- regulated in mutant mice (fold change >2, q-value < 0.05), but not in WT mice (Fig. 3 C and D, green), while only 34 (40 min) and 53 (68 min) genes were significantly up- or down-regulated in WT mice, but not in mutant mice (Fig. 3 C and D, blue).
Fig. 3.
KA-induced neuronal activity in vivo results in greater changes in expression level of activity-dependent genes in the hippocampi of mutant mice. (A and B) Heat maps showing the changes in mRNA expression level of activity-dependent genes in the hippocampi of WT and mutant (MT) mice at 40 min (A) and 68 min (B) following KA treatment compared with their untreated counterparts (|log2-fold change| > 1.0 and q-value < 0.05 in mutant and/or WT samples). Red–white color range indicates the relative log2-fold changes (scaled by SDs) for up-regulated genes. Blue–white color range indicates the relative log2-fold changes (scaled by SDs) for down-regulated genes. The darker shade of blue or red corresponds to a greater level of down-regulation or up-regulation, respectively. (C and D) Scatter plots showing the log2-fold changes in expression of activity-dependent genes in the hippocampi of WT and mutant mice at 40 min (C) and 68 min (D) after KA treatment. Red diamonds indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in both mutant and WT mice; green pluses indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in mutant but not in WT mice; blue squares indicate genes that were up- or down-regulated (|log2-fold change| > 1.0 and q-value < 0.05) in WT but not in mutant mice. Note that KA treatment resulted in greater changes in gene expression in mutant samples in most activity-dependent genes.
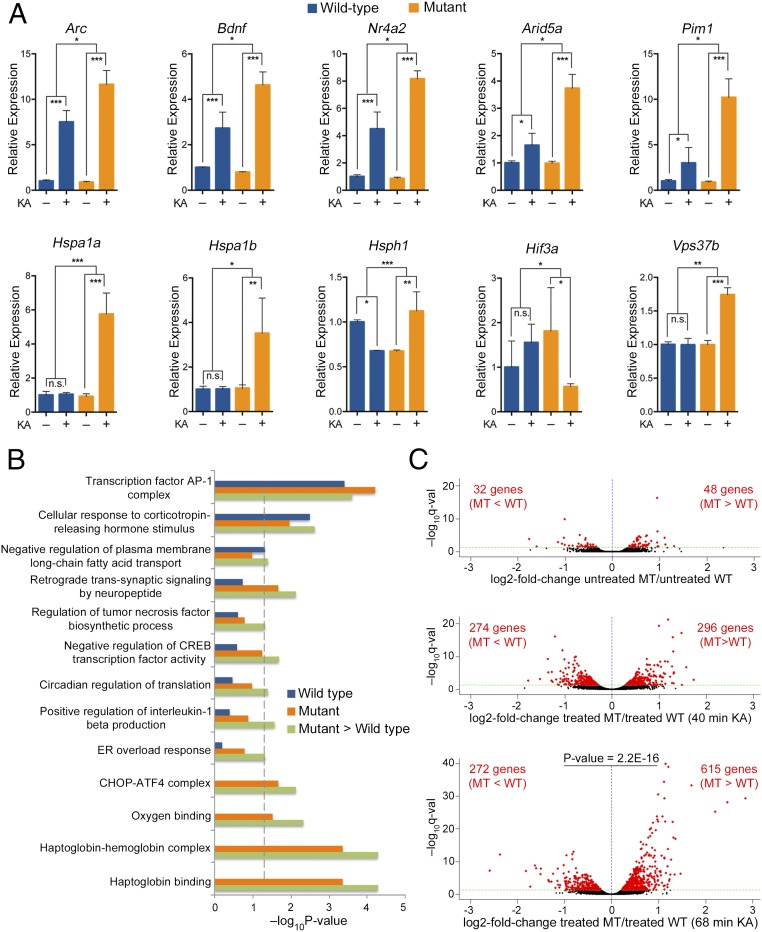
We validated, by quantitative RT-PCR, the differential expression of several known activity-dependent genes, identified by our RNA-Seq analysis, including Arc, Bdnf, Nr4a2, Arid5a, and Pim1. This analysis showed that, while there was no significant difference in the expression level of these genes between the hippocampi of untreated WT and mutant mice, these genes were up-regulated to a significantly higher level in the hippocampi of mutant mice compared with the WT mice, upon neuronal activity induced by KA (Fig. 4A, Top). Interestingly, stress-response genes, such as those encoding heat shock proteins (HSP) (Hspa1a, Hspa1b, and Hsph1) and Vps37b were significantly up-regulated only in mutant mice upon KA treatment, while the hypoxia-induced factor gene, Hif3a, was down-regulated only in the mutant mice (Fig. 4A, Bottom).
Fig. 4.
Differential gene expression in the hippocampi of mutant mice compared with WT mice upon KA-induced neuronal activity. (A) Quantitative RT-PCR for the indicated genes comparing the fold change in their expression level, upon 68 min of KA treatment (KA+/KA−), between WT and mutant mice. Bar graphs display the mean relative mRNA expression level + SEM. n = 4 biological replicates. Statistical significance was determined by two-way ANOVA with multiple pairwise comparisons (LSD method). *P < 0.05, **P < 0.01, ***P < 0.001. (B) GO analysis for up-regulated activity-dependent genes (log2-fold change > 1, q-value < 0.05) upon 68 min of KA treatment in the WT group (blue bars), mutant group (orange bars), and in mutant > WT group (green bars). Enriched GO terms in mutant > WT group were based on activity-dependent genes (log2-fold change > 1, q-value < 0.05) in mutant mice that showed a greater level of expression than in WT mice after 68 min of KA treatment (log2-fold change in mutant minus log2-fold change in WT > 0, q-value < 0.05). Statistically significant GO terms are based on a Fisher’s exact test P < 0.05. The vertical dashed line serves as a marker of the −log10(0.05). Absence of a bar indicates that the genes of that GO term were not enriched in this specific category. (C) Volcano plots showing the relative log2-fold change in the expression level of genes between mutant (MT) and WT hippocampi at steady state (untreated) (Top), after 40 min KA treatment (Middle), and after 68 min KA treatment (Bottom). Red dots represent all genes with statistically significant fold change in expression between untreated mutant and WT hippocampi or between KA-treated mutant and WT hippocampi (q-value < 0.05). Black dots represent genes with q-value > 0.05. Note the significant increase in the number of differentially expressed genes as well as in their fold change and -log10(q-value), between mutant and WT mice treated with KA, compared with untreated. Note also the significant bias (two-tailed exact binominal test) toward a higher expression level in the mutant compared with the WT for the 68-min group.
To obtain further insight into the cellular and molecular processes that might be affected by the differential expression of activity-dependent genes between mutant and WT mice, we performed a Gene Ontology (GO) analysis. Because our data suggest that the most prominent effect of the loss of MeCP2 is increased expression of activity-dependent genes in mutant mice compared with the WT mice (Figs. 1, 3, and 4), we performed the GO analysis on this group of genes. Specifically, we examined the enrichment of the GO terms for three groups of activity-dependent genes, upon 68 min of KA treatment: up-regulated in WT mice, up-regulated in mutant mice, and up-regulated to a significantly higher level in mutant mice than in the WT mice (mutant > WT after KA treatment, q-value < 0.05) (Fig. 4B). This analysis showed that several GO terms with significant enrichment in the mutant greater than WT group were related to synaptic plasticity, stress response, and regulation of gene expression. Moreover, most of these GO terms were enriched in the mutant group but not in the WT group. This suggests that those biological processes were significantly stimulated during neuronal activity in mutant hippocampi but not in WT hippocampi. Other GO terms such as transcription factor AP1 complex, which consists of immediate-early genes, were enriched in all three groups (Fig. 4B), indicating that while enriched in both WT and mutant they were expressed to higher levels in mutant mice.
Notably, analysis of the global gene expression pattern (including activity-dependent and non-activity-dependent genes), following KA-induced neuronal activity, showed a robust increase in the total number of differentially expressed genes between mutant and WT mice at 40 and 68 min after the KA treatment compared with steady-state (untreated) conditions (Fig. 4C). Additionally, at 68 min after KA-treatment there was a significant increase in the number of genes that were up-regulated to a higher level in the mutant mice compared with the WT mice (615 genes) relative to the number of genes that were down-regulated to a lower level in the mutant mice compared with the WT mice (272) (P = 2.2E-16) (Fig. 4C).
These data together suggest that while the loss of MeCP2 affects both the levels of the up-regulated and the down-regulated genes in the activity-dependent transcriptome, the most prominent effect of its loss is increased levels of gene expression.
Neuronal Activity Induced by KA in Vivo Results in Dramatic Alterations in Alternative Splicing Events in the Hippocampi of Mutant Mice Compared with WT Mice.
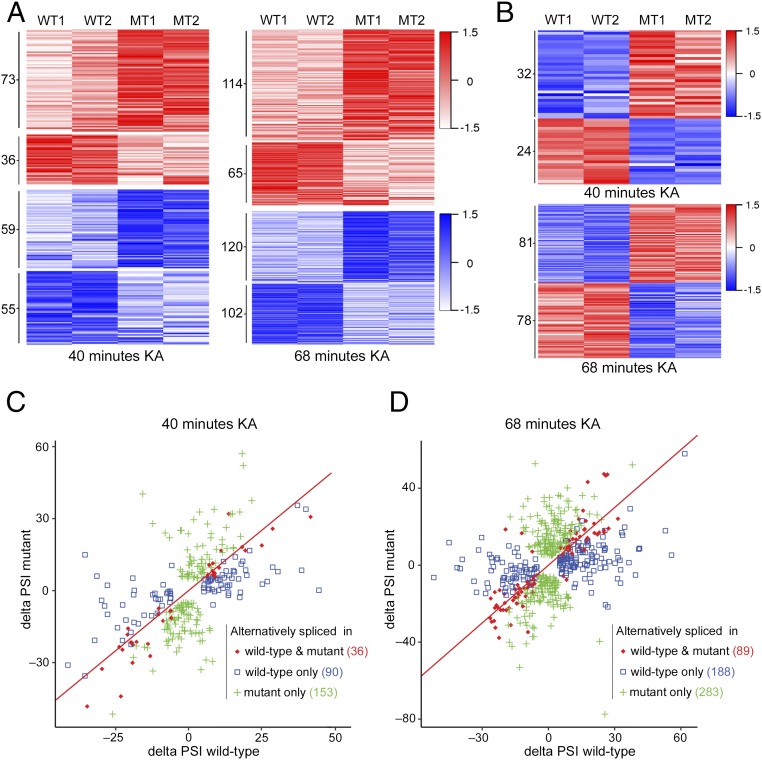
Neuronal activity induces not only changes in gene expression but also in alternative splicing (28). Therefore, we sought to analyze how the loss of MeCP2 affects the alternative splicing program during neuronal activity. We first asked whether there are differential alternative splicing events in the hippocampi of mutant mice compared with the WT mice at steady-state conditions. We detected only 27 genes with increased exon or intron inclusion and 19 genes with increased exon or intron exclusion in mutant mice compared with the WT mice, with no bias to any specific type of alternative splicing (Dataset S9). We then analyzed the splicing events occurring during 40 and 68 min of KA-induced neuronal activity. We identified a total of 279 and 560 differential alternative splicing events between WT and mutant mice that occurred upon 40 and 68 min of KA treatment, respectively (Fig. 5 A and B). Of these events, while 223 (40 min) and 401 (68 min) generally occurred in the same direction, that is, inclusion (red) or exclusion (blue) in both mutant and WT hippocampi, the change in the fraction of the included or excluded isoforms [delta “percent spliced in” (ΔPSI)] differed (Fig. 5A). Specifically, we detected 73 (40 min) and 114 (68 min) alternative splicing events with increased inclusion in mutant mice relative to the WT mice and 36 (40 min) and 65 (68 min) alternative splicing events with increased inclusion in the WT mice compared with mutant mice (Fig. 5A and Datasets S10 and S12). In addition, we identified 59 (40 min) and 120 (68 min) alternative splicing events with increased exclusion in the mutant mice compared with WT mice and 55 (40 min) and 102 (68 min) alternative splicing events with increased exclusion in the WT mice compared with mutant mice (Fig. 5A and Datasets S11 and S13). Interestingly, we also identified a group of alternative splicing events that occurred in opposite directions, that is, that underwent increased inclusion in mutant mice and increased exclusion in WT mice, and vice versa, during both 40 min (56 events) and 68 min (159 events) of KA treatment (Fig. 5B and Dataset S14). Of these events, 32 (40 min) and 81 (68 min) underwent increased inclusion in mutant mice vs. increased exclusion in WT mice, whereas 24 (40 min) and 78 (68 min) events underwent increased exclusion in mutant mice vs. increased inclusion in WT mice (Fig. 5B and Dataset S14). Detailed analysis of all the alternative splicing events showed that 36 (40 min) and 89 (68 min) events occurred in both mutant and WT mice (absolute ΔPSI > 5%, q-value < 0.05) during KA treatment (Fig. 5 C and D, red diamonds), with many of them being differentially spliced between mutant and WT mice. Importantly, 153 (40 min) and 283 (68 min) splicing events occurred in mutant (based on absolute ΔPSI > 5%, q-value < 0.05) but not in WT mice (Fig. 5 C and D, green pluses), while 90 (40 min) and 188 (68 min) alternative splicing events occurred in WT mice but not in mutant mice (Fig. 5 C and D, blue squares).
Fig. 5.
Differential alternative splicing events in the hippocampi of mutant mice compared with WT mice upon KA-induced neuronal activity. (A) Heat maps showing the changes in percentage of isoforms that include the alternative exon/intron (ΔPSI) in WT and mutant (MT) mice (based on |ΔPSI| > 5%, q-value < 0.05 in WT and/or mutant) at 40 min (Left) and 68 min (Right) of KA treatment compared with the untreated state. Red–white color range indicates high–low ΔPSIs (scaled by SDs) of alternative splicing events that have positive ΔPSIs (inclusion events). Blue–white color range indicates low–high ΔPSIs (scaled by SDs) of alternative splicing events that have negative ΔPSIs (exclusion events). The darker shade of blue or red corresponds to a greater level of exclusion or inclusion, respectively. (B) Heat maps showing alternative splicing events with opposite ΔPSIs, either positive in WT and negative in mutant or negative in WT and positive in mutant (based on |ΔPSI| > 5% and q-value < 0.05 in WT and/or mutant), at 40 min or 68 min following KA treatment compared with no treatment. Red–white color range indicates relative positive ΔPSIs (inclusion events); blue–white color range indicates relative negative ΔPSIs (exclusion events) as described in A. (C and D) Scatter plots showing the for the alternative splicing events in mutant and WT hippocampi at 40 min (C) and 68 min (D) following KA treatment (based on |ΔPSI| > 5%, q-value < 0.05 in WT and/or mutant). Red diamonds indicate genes that underwent alternative splicing events in both mutant and WT mice; Green pluses indicate genes that underwent alternative splicing events only in mutant mice; blue squares indicate genes that underwent alternative splicing events only in WT mice.
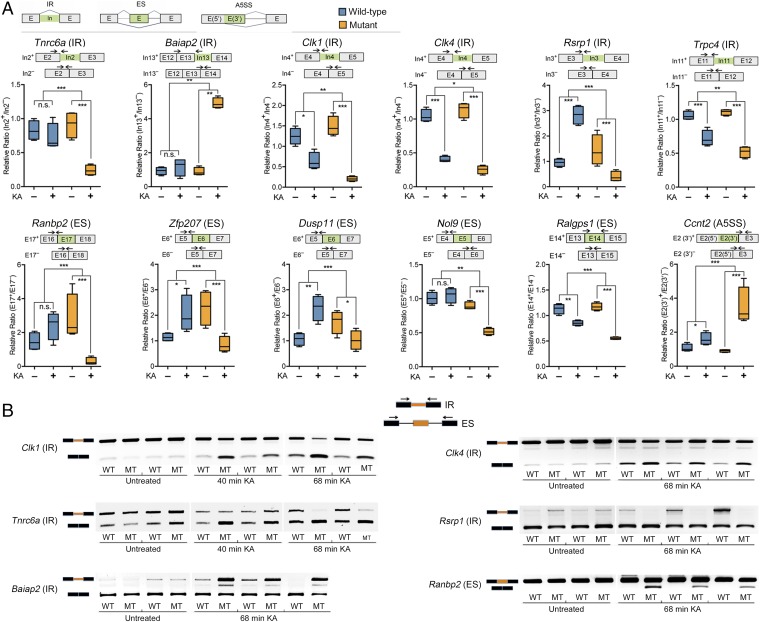
We validated several of the differentially alternative spliced genes between mutant and WT mice, using quantitative RT-PCR and semiquantitative RT-PCR (Fig. 6 and SI Appendix, Fig. S2). Our data confirmed several genes that underwent altered alternative splicing events, including altered intron retention (IR), exon skipping (ES), and alternative 5′ splice sites (A5SS), in mutant mice compared with WT mice, upon the KA treatment. Notably, and in agreement with the RNA-Seq analysis, these data show that while some of the differentially spliced RNAs, such as Tnrc6a (IR), Baiap2 (IR), Ranbp2 (ES), and Nol9 (ES), remained unchanged during neuronal activity in the hippocampi of the WT mice, they underwent significant alternative splicing changes in the mutant mice. Other genes, such as Clk1 (IR) and Clk4 (IR), recently shown to undergo intron excision during neuronal activity (29), Trpc4 (IR), and Ralgps1 (ES), underwent alternative splicing in both, mutant and WT mice but showed a greater degree of excision in mutant mice, whereas Ccnt2 (A5SS) exhibited a greater degree of inclusion in the mutant mice (Fig. 6 and SI Appendix, Fig. S2). Importantly, and in agreement with the RNA-Seq analysis, several genes, such as Rsrp1 (IR), Zfp207 (ES), and Dusp11 (ES), were alternatively spliced in opposite directions (increased inclusion vs. increased exclusion) in the mutant compared with WT mice (Fig. 6 and SI Appendix, Fig. S2).
Fig. 6.
Validations for genes that underwent differential alternative splicing in the hippocampi of mutant mice compared with WT mice upon KA treatment. (A) Quantitative RT-PCR for genes that underwent differential splicing in WT compared with mutant mice, upon 68 min of KA treatment. Top row shows diagrams of IR, ES, and A5SS. Box plots show the relative ratio of retained vs. excised introns (IR) or included vs. excluded exons (ES, A5SS) between KA-treated (+) and untreated (−) conditions. The diagram above each individual gene shows the two possible isoforms (included exon or intron in green, or excluded) with arrows indicating the location of the primers used for the 5′ junction of the included intron or exon isoform and the primers for the excluded intron or exon isoform. n = 4 biological replicates. Statistical significance for the quantitative PCR analyses was determined by two-way ANOVA with multiple pairwise comparisons (LSD method). *P < 0.05, **P < 0.01, ***P < 0.001. (B) Semiquantitative RT-PCR for the indicated genes that underwent differential alternative splicing in mutant mice compared with WT mice. Gel images show the retained vs. spliced isoform intensity. Diagram on top shows the location of the primers for the IR and ES splicing events.
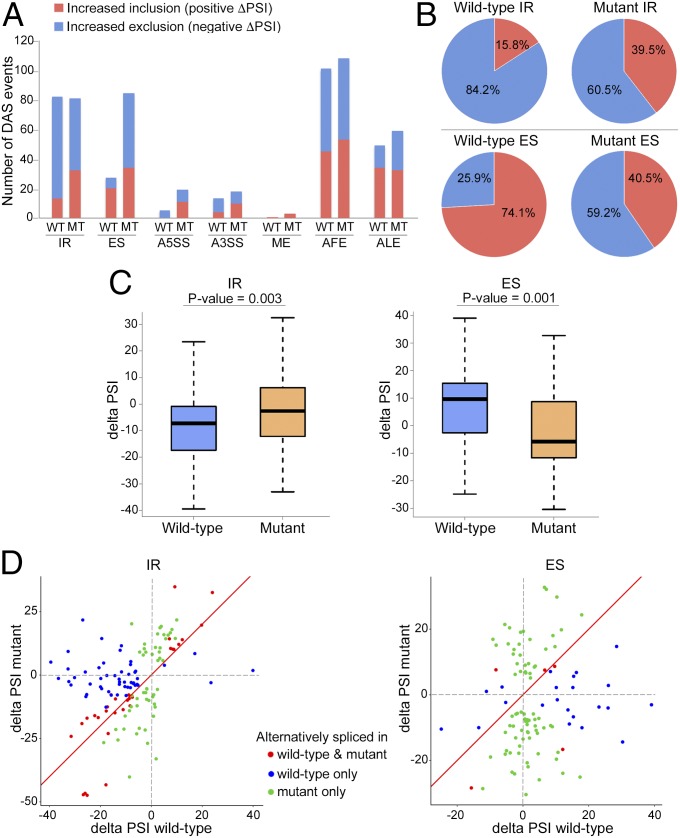
Because many more alternative splicing events were detected in the 68 min of KA treatment group than in the 40-min group, we pursued further analyses on the 68-min group. Analysis of the specific types of alternative splicing events showed a differential pattern of alternative splicing modalities employed during neuronal activity (Fig. 7A). In the WT mice, the most commonly detected typical splicing events upon 68 min of KA-induced neuronal activity were IR with a total of 82 events, comprised of 84% excised intron events and 16% retained intron events (Fig. 7 A and B). These data are consistent with a recent study showing an increase in intron excision during neuronal activity (29). Notably, while a similar number of IR events occurred in mutant mice, there was a clear shift toward an increase in retained intron events (60% excised vs. 40% retained) (Fig. 7 A and B). Furthermore, while only 27 ES events occurred in the WT mice during 68 min of KA treatment, they show a strong preference toward increased exon inclusion events (74% increased inclusion events vs. 26% increased exclusion events) (Fig. 7 A and B). Remarkably, in the mutant mice, there was a threefold increase in the total number of ES events (84 events) relative to WT mice (27 events), and these events showed a significant bias toward exon exclusion (41% increased inclusion vs. 59% increased exclusion) (Fig. 7 A and B). Statistical analysis of the IR and ES events showed a significant increase in retained introns and a significant increase in skipped (excised) exons in mutant mice compared with the WT mice, in response to neuronal activity (Fig. 7C). Furthermore, a scatter plot of the individual IR and ES events not only shows the bias in exclusion/inclusion but also further highlights the finding that many events occurred in opposite directions (positive/negative ΔPSI) in mutant vs. WT mice (Fig. 7D). Although differential alternative splicing between mutant and WT mice was also evident for other types of alternative splicing, including A5SS, alternative 3′ splice sites (A3SS), and mutually exclusive exons (ME) (Datasets S10–S14), the number of these events induced by neuronal activity was relatively low in both mutant and WT mice (Fig. 7A). In addition, we found many alternative first exon [(AFE), generated by a combination of alternative promoter usage and alternative splicing] and alternative last exon [(ALE), generated by a combination of alternative polyadenylation and alternative splicing] events, which underwent differential exon usage between mutant and WT mice (Datasets S10–S14). However, in contrast to IR and ES, there was no significant bias in included/excluded exons in the AFE and ALE events between WT and mutant mice (Fig. 7A).
Fig. 7.
Differential alternative splicing patterns in the hippocampi of mutant mice compared with WT mice upon 68 min of KA-induced neuronal activity. (A) Bar graph showing the number of inclusion (positive ΔPSI, red) and exclusion (spliced) (negative ΔPSI, blue) events for each type of alternative splicing modality in mutant (MT) and WT mice (based on with q-value < 0.05). Note the difference in the ratio of intron inclusion and exclusion events (IR), and the stark disparity in the total number of ES events, particularly in the excision events, between WT and mutant mice. (B) Pie charts showing the relative percentage of intron inclusion (positive ΔPSI, red) and exclusion (negative ΔPSI, blue) events (Top), and exon inclusion (positive ΔPSI, red) and exclusion (negative ΔPSI, blue) events (Bottom) for the WT and mutant groups. (C) Box plot displaying the ΔPSI distribution of IR and ES events in WT and mutant hippocampi during KA activation. Note that mutant mice have a significant bias toward more retained introns (less negative ΔPSI) compared with WT mice (Left) and that mutant mice have a significant bias toward exon exclusion (negative ΔPSI) compared with WT mice (positive ΔPSI) (Right). The P values are based on Mann–Whitney–Wilcoxon test and are indicated at the top of each panel. (D) Scatter plots showing changes in IR and ES events in mutant and WT mice upon KA treatment. Red dots represent alternative splicing events that occurred in both WT and mutant mice ( and q-value < 0.05). Blue and green dots represent alternative splicing events occurred in either WT or mutant mice (based on and q-value < 0.05), respectively. Note the shifts in positive and negative ΔPSIs in IR and ES events between mutant and WT mice and the significantly higher number of ES events that occurred in mutant mice, many of which are in the opposite direction to those in WT mice.
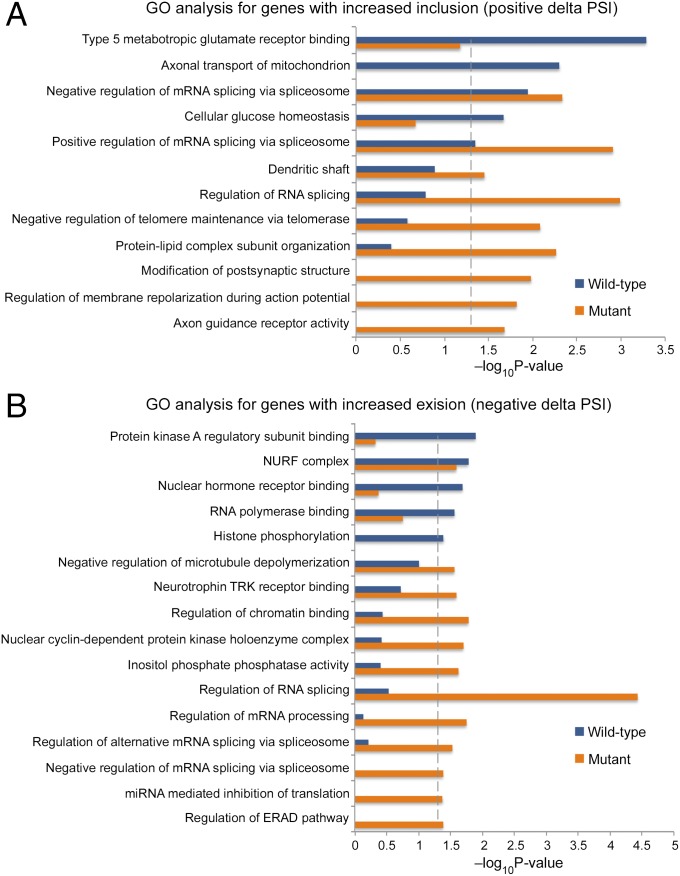
Finally, we performed GO analysis to gain insight into the potential functional impacts of the genes that underwent splicing in mutant and WT mice upon KA treatment. Alternative splicing events were separated into increased inclusion (Fig. 8A) and increased exclusion (Fig. 8B) cohorts. Our analysis showed that GO terms pertaining to neuronal structure and plasticity were enriched predominantly in the inclusion cohort, whereas terms pertaining to chromatin regulation and gene expression were enriched in the exclusion cohort. Interestingly, many processes related to the regulation of RNA splicing were enriched in both the inclusion and exclusion cohorts. Other processes pertaining to neuronal signaling and cellular stress were also enriched in both cohorts. Importantly, very few processes, in the inclusion and exclusion cohorts, showed enrichment in both WT and mutant mice. In fact, the majority of the GO terms showed a significant enrichment only in the mutant mice, while only a few GO terms showed enrichment only in the WT mice. This suggests that aberrations in alternative splicing during neuronal activity in the mutant mice affect many fundamental cellular and molecular processes.
Fig. 8.
GO analysis for genes that underwent differential alternative splicing in the hippocampi of WT and mutant mice upon KA treatment. (A and B) Enriched GO terms (Fisher’s exact test P < 0.05) for genes that underwent inclusion (ΔPSI > 5%, q-value < 0.05) (A) or exclusion (ΔPSI < −5% q-value < 0.05) (B) events, upon 68 min of KA treatment. The blue and orange bars represent −log10 (P value) of the GO enrichment for the WT and mutant mice, respectively. The vertical dashed line serves as a marker for P value = 0.05. Absence of a bar indicates that genes of that GO term were not enriched in this specific category.
Discussion
In this study, we analyzed the impact of the loss of MeCP2 on the gene expression programs, induced specifically in response to neuronal activity, and revealed myriad aberrations in the activity-dependent transcriptome of the mutant RTT mice, which are otherwise not detected at steady-state conditions. This analysis highlights aberrations not only in the gene transcription program but also in the posttranscriptional program, specifically, alternative splicing, which together dramatically alter the activity-dependent transcriptome landscape in mutant RTT mice.
Our study revealed that—following both in vitro KCl-induced membrane depolarization of cortical neurons and in vivo KA-induced neuronal excitation in the hippocampus—the loss of MeCP2 in mutant mice results in significantly higher expression levels of the majority of the activity-dependent up-regulated genes, compared with WT mice. Aberrations in the expression levels of the activity-dependent down-regulated genes, due to the loss of MeCP2, were also evident; however, this group of genes was relatively small compared with the dysregulated group of activity-dependent up-regulated genes. These down-regulated genes are likely downstream to the primary wave of the up-regulated genes, although the possibility that they are down-regulated directly in response to neuronal activity cannot be ruled out.
The predominance of increased levels of up-regulated activity-dependent genes in mutant mice suggests that MeCP2 functions mostly as a repressor during neuronal activity. Further, it suggests that MeCP2 is likely not a gene-specific repressor but rather a global repressor that fine-tunes the immediate and robust increase in gene transcription in response to neuronal activity. This view is lent further support by previous studies suggesting that MeCP2 globally alters the chromatin state in neurons to a more repressive state to dampen transcriptional noise genomewide (6). Recent studies suggest that the MeCP2/NCoR interaction plays a critical role in the repressor function of MeCP2, and that the RTT-causing mutation R306C located at the NCoR interaction domain (NID) abolishes the interaction of the NCoR repressor complex with MeCP2 (30–32). Furthermore, in WT mice, neuronal activity induces phosphorylation of T308, located in the NID, allowing the dissociation of MeCP2/NCoR interaction and the efficient up-regulation of activity-dependent genes (30). However, insertion of T308A mutation, which blocks phosphorylation, leads to a decreased induction of activity-dependent genes, such as Npas4 and Bdnf, likely due to the persistent interaction of NCoR with MeCP2 during neuronal activity (30). The dynamic interaction of MeCP2/NCoR via phosphorylation/dephosphorylation of MeCP2 during neuronal stimulation most likely modulates the repressive state of the chromatin to allow efficient induction of activity-dependent genes while fine-tuning the level of their expression. Based on this mechanism, the loss of the MeCP2/NCoR interaction and its repressor activity, as in the Mecp2-null mouse model in our study, could explain the higher levels of expression of activity-dependent genes we observed. It will be intriguing to explore whether activity-dependent genes are up-regulated to higher levels during neuronal activity also in mice harboring RTT-causing mutations, which abolish the interaction of NCoR with MeCP2, such as R306C mutation and others.
We also showed that the mutant RTT mice are significantly more susceptible to seizures, followed by lethality, in response to neuronal excitation induced by KA, consistent with the network hyperexcitability observed in the cortex and hippocampus of RTT mice (21–23). It is possible that the altered levels of activity-dependent genes we observed in the RTT mice mediate, at least in part, the hyperexcitability. Interestingly, our transcriptome analysis clearly indicates a higher activity-dependent stress response in RTT mice compared with WT mice. For example, some of the stress-response genes, such as genes encoding HSPs and others, were activated only in RTT mice and not in WT mice. Such stress response could be cell-autonomous and activated in the neurons but could also be non-cell-autonomous and activated by glia in response to neuronal dysfunction or neuronal stress during their excitation. It is likely that the activity-dependent stress response exacerbates the symptoms initiated by neurons in RTT.
Alternative splicing plays important roles in the structure/function of neurons, such as synapse formation (neurexins) (33), fast excitatory neurotransmission (AMPAR subunits Gria1–4) (34), and neurotransmitter release and presynaptic plasticity (Cav2.1) (35). For example, the two major spliced isoforms of the voltage-gated Ca2+ channel, Cav2.1[EFa] and Cav2.1[EFb], regulate neurotransmitter release and short-term synaptic plasticity in opposite directions (35). Furthermore, neuronal activity regulates the abundance of these two isoforms and thus the function associated with each isoform (35). Therefore, a tight regulation of the ratios of different alternatively spliced isoforms is highly important for proper functioning of the neuronal networks. Indeed, aberrant ratios between the flip and flop isoforms of the Gria1–4 subunits of AMPAR have been linked recently to abnormal synaptic transmission in RTT (12). Our study showed widespread aberrations in alternative splicing, specifically in response to neuronal activity in RTT mice. Interestingly, although the different types of alternative splicing events, such as ES, IR, A3SS, and A5SS as well as AFE and ALE, are regulated by different mechanisms (36), the dysregulation of each one of them was apparent during neuronal activity in the hippocampi of mutant RTT mice. Importantly, we noted a significant bias in the inclusion/exclusion ratios of IR events (increased retained introns in mutant) and ES events (increased skipped exons in mutant), many of which underwent splicing in opposite directions in mutant mice. Furthermore, the number of ES events with significant change, upon neuronal activity, was more than threefold higher in mutant mice than in the WT mice. These findings are supported by previous studies demonstrating that in human cell lines the loss of MeCP2 results in increased retained introns (37) and increased skipped exons (38).
Notably, the mRNAs of several important splicing factors/regulators (e.g., Ptbb1, Ptbp3, Srrm4, Srsf7, Clk1, and Clk4) not only were differentially expressed in response to neuronal activity in mutant mice, but some of these splicing regulators and others also underwent aberrant ES or IR events themselves, including splicing in opposite directions [e.g., Clk1 (IR), Clk4 (IR, ES), Srsf3 (ES), Dusp11 (ES), and Rbfox1 (ES)]. The abnormal splicing and/or expression levels of these splicing factors could potentially further induce a second wave of aberrations in downstream splicing events and explain the increase in splicing aberrations we observed in mutant mice, upon neuronal stimulation. For example, the splicing factor Rbfox1 has been shown to control neuronal excitability, and reduction in its expression leads to hyperexcitability (36). Moreover, changes in the expression of Rbfox1 have been linked to aberrations in alternative splicing in the brains of individuals with autism spectrum disorder (39, 40). Thus, aberrations not only in the activity-dependent gene transcription program but also in the activity-dependent alternative splicing program could potentially lead to the hyperexcitability observed in RTT mice. MeCP2 is known to interact not only with transcriptional regulators but also with splicing factors/regulators, such as YB-1, Prpf3, LEDGF, DHX9, Tra2b, and others (11, 12, 37, 41, 42). The loss of these interactions due to the loss of MeCP2 in mutant mice could also explain the aberrant splicing we observed in response to neuronal activity. In support of this view, it was suggested that the aberrant ratios of the flip/flop alternative splicing of AMPAR subunits in Mecp2-null mice are likely due to the loss of LEDGF/MeCP2 interaction (12). Further, it has been shown that reduced MeCP2 binding near splice junctions facilitates retention of intron (IR) via reduced recruitment of splicing factors, such as Tra2b (37).
Because activity-induced modifications of the transcriptome play a crucial role in regulating neuronal properties and synaptic plasticity, significant aberrations in the activity-induced transcriptome when MeCP2 function is lost could negatively affect the structure/function of the neuronal networks. Indeed, our GO analyses suggest that, upon neuronal activity, several pathways involved in neuronal properties and synaptic plasticity are affected by the aberrant expression of activity-dependent genes and alternative splicing events occurred in the MeCP2-mutant mice. In addition, accumulating evidence indicates that many of the genes, which are mutated in autism spectrum disorder, represent crucial components of the activity-dependent signaling networks that regulate synaptic plasticity (28, 39, 43, 44). Thus, aberrations in the activity-dependent molecular program likely play a fundamental role in the etiology of autism spectrum disorders in general and RTT in particular.
Materials and Methods
All animal studies were approved by the Institutional Animal Care and Use Committees at Stony Brook University and were in line with the guidelines established by the National Institutes of Health. Neuronal activity was induced by either KCl (in vitro) or KA (in vivo) using WT and Mecp2-null mice. RNA-Seq was performed on an Illumina HiSeq 2500 sequencer, using 2 × 125-bp cycles, and the data were analyzed for expression and alternative splicing (45, 46). Validations of the RNA sequencing analyses were performed by quantitative real-time RT-PCR and semiquantitative RT-PCR. Additional details are provided in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank the New York Genome Center for assistance with the RNA-Seq analysis, Dr. Hammell (Cold Spring Harbor Laboratory) for valuable advice about RNA-Seq analysis, and Drs. Colognato and Tsirka (Stony Brook University) for the glial-type-specific antibodies. This work was supported by the National Institutes of Health Grant MH102711 (to N.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no.GSE113477).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722546115/-/DCSupplemental.
References
- 1.Chahrour M, Zoghbi HY. The story of Rett syndrome: From clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol Cell Biol. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugino K, et al. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J Neurosci. 2014;34:12877–12883. doi: 10.1523/JNEUROSCI.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson BS, et al. Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat Med. 2017;23:1203–1214. doi: 10.1038/nm.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young JI, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, et al. Misregulation of alternative splicing in a mouse model of Rett syndrome. PLoS Genet. 2016;12:e1006129. doi: 10.1371/journal.pgen.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng TL, et al. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Mellios N, et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry. 2017;23:1051–1065. doi: 10.1038/mp.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, et al. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2010;107:18161–18166. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen MV, et al. MeCP2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. J Neurosci. 2012;32:10021–10034. doi: 10.1523/JNEUROSCI.1316-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGraw CM, Samaco RC, Zoghbi HY. Adult neural function requires MeCP2. Science. 2011;333:186. doi: 10.1126/science.1206593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du F, et al. Acute and crucial requirement for MeCP2 function upon transition from early to late adult stages of brain maturation. Hum Mol Genet. 2016;25:1690–1702. doi: 10.1093/hmg/ddw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheval H, et al. Postnatal inactivation reveals enhanced requirement for MeCP2 at distinct age windows. Hum Mol Genet. 2012;21:3806–3814. doi: 10.1093/hmg/dds208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calfa G, Hablitz JJ, Pozzo-Miller L. Network hyperexcitability in hippocampal slices from Mecp2 mutant mice revealed by voltage-sensitive dye imaging. J Neurophysiol. 2011;105:1768–1784. doi: 10.1152/jn.00800.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calfa G, Li W, Rutherford JM, Pozzo-Miller L. Excitation/inhibition imbalance and impaired synaptic inhibition in hippocampal area CA3 of Mecp2 knockout mice. Hippocampus. 2015;25:159–168. doi: 10.1002/hipo.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Peterson M, Beyer B, Frankel WN, Zhang ZW. Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J Neurosci. 2014;34:2754–2763. doi: 10.1523/JNEUROSCI.4900-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kron M, et al. Brain activity mapping in Mecp2 mutant mice reveals functional deficits in forebrain circuits, including key nodes in the default mode network, that are reversed with ketamine treatment. J Neurosci. 2012;32:13860–13872. doi: 10.1523/JNEUROSCI.2159-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kline DD, Ogier M, Kunze DL, Katz DM. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J Neurosci. 2010;30:5303–5310. doi: 10.1523/JNEUROSCI.5503-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, et al. Enhanced susceptibility to kainate-induced seizures, neuronal apoptosis, and death in mice lacking gangliotetraose gangliosides: Protection with LIGA 20, a membrane-permeant analog of GM1. J Neurosci. 2005;25:11014–11022. doi: 10.1523/JNEUROSCI.3635-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLeod F, et al. Reduced seizure threshold and altered network oscillatory properties in a mouse model of Rett syndrome. Neuroscience. 2013;231:195–205. doi: 10.1016/j.neuroscience.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 28.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauger O, Lemoine F, Scheiffele P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016;92:1266–1278. doi: 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 30.Ebert DH, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyst MJ, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillotson R, et al. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature. 2017;550:398–401. doi: 10.1038/nature24058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommer B, et al. Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 35.Thalhammer A, et al. Alternative splicing of P/Q-Type Ca2+ channels shapes presynaptic plasticity. Cell Rep. 2017;20:333–343. doi: 10.1016/j.celrep.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 36.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong JJ, et al. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat Commun. 2017;8:15134. doi: 10.1038/ncomms15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damianov A, et al. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell. 2016;165:606–619. doi: 10.1016/j.cell.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Z, Ambigapathy G, Keifer J. MeCP2 regulates Tet1-catalyzed demethylation, CTCF binding, and learning-dependent alternative splicing of the BDNF gene in Turtle. eLife. 2017;6:e25384. doi: 10.7554/eLife.25384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long SW, Ooi JY, Yau PM, Jones PL. A brain-derived MeCP2 complex supports a role for MeCP2 in RNA processing. Biosci Rep. 2011;31:333–343. doi: 10.1042/BSR20100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irimia M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quesnel-Vallières M, et al. Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol Cell. 2016;64:1023–1034. doi: 10.1016/j.molcel.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 45.Yu P, Shaw CA. An efficient algorithm for accurate computation of the Dirichlet-multinomial log-likelihood function. Bioinformatics. 2014;30:1547–1554. doi: 10.1093/bioinformatics/btu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Yu P. Genome-wide transcriptome analysis identifies alternative splicing regulatory network and key splicing factors in mouse and human psoriasis. Sci Rep. 2018;8:4124. doi: 10.1038/s41598-018-22284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.