Significance
Climate change is challenging plant agriculture and our ability to manage food security. Crop growth and yield are controlled by several phytohormones and their overlapping signal networks. We report here an unexpected aspect of the abscisic acid (ABA) signal network that directly impacts rice productivity. Simultaneously mutating the genes encoding the ABA receptors pyrabactin resistance 1-like 1 (PYL1), PYL4, and PYL6 causes improved growth and increased grain yield in rice. Our work thus reveals an important role of these ABA receptors in growth control and a genetic strategy to improve rice yield.
Keywords: hormone, signaling, stress, crop, yield
Abstract
Abscisic acid (ABA) is a key phytohormone that controls plant growth and stress responses. It is sensed by the pyrabactin resistance 1 (PYR1)/PYR1-like (PYL)/regulatory components of the ABA receptor (RCAR) family of proteins. Here, we utilized CRISPR/Cas9 technology to edit group I (PYL1–PYL6 and PYL12) and group II (PYL7–PYL11 and PYL13) PYL genes in rice. Characterization of the combinatorial mutants suggested that genes in group I have more important roles in stomatal movement, seed dormancy, and growth regulation than those in group II. Among all of the single pyl mutants, only pyl1 and pyl12 exhibited significant defects in seed dormancy. Interestingly, high-order group I mutants, but not any group II mutants, displayed enhanced growth. Among group I mutants, pyl1/4/6 exhibited the best growth and improved grain productivity in natural paddy field conditions, while maintaining nearly normal seed dormancy. Our results suggest that a subfamily of rice PYLs has evolved to have particularly important roles in regulating plant growth and reveal a genetic strategy to improve rice productivity.
Increasing the productivity of food and fiber has been a continual effort of human cultures since the advent of agriculture nearly 10,000 y ago. Abscisic acid (ABA) is a crucial stress hormone that enhances plant adaptation to abiotic and biotic stresses, partly by controlling aspects of plant growth and development, including productivity (1–4). Abiotic stresses, especially water deficit, induce ABA accumulation, which triggers rapid biochemical and physiological responses that enhance stress adaptation. Although ABA may promote root growth in response to drought stress, it generally has inhibitory roles in affecting overall plant growth (1–9). ABA is perceived by the soluble pyrabactin resistance 1 (PYR1)/PYR1-like (PYL)/regulatory components of the ABA receptor (RCAR) family of proteins (9–12). The binding of ABA to PYLs triggers conformational changes that enable interactions between the PYLs and clade A type 2C protein phosphatases (PP2Cs). This leads to the inhibition of the PP2Cs, thus releasing the sucrose nonfermenting 1-related protein kinase 2s (SnRK2s) from inhibition by the PP2Cs (9–19). Activated SnRK2s then phosphorylate, among other proteins, ABA-responsive element-binding factors and components of the machinery regulating stomatal aperture (20–24). These components include the slow anion channel SLAC1 and the NADPH oxidase AtrbohF (21–24). The phosphorylation ultimately results in activation of many responses that contribute to stress adaptation.
In Arabidopsis, the PYL family consists of 14 genes (11). High-order mutants, from several types of triple mutants to the pyr1pyl1pyl2pyl4pyl5pyl8 sextuple mutant, have been generated (11, 17, 25). The high-order mutants exhibit reduced vegetative growth and strong ABA-insensitive phenotypes (25). Although extensive gene redundancy exists in the PYL family, specific functions and characteristics have been attributed to several Arabidopsis PYLs. For example, the Arabidopsis PYL8 plays particularly important roles in regulating root growth (26–28). Thirteen PYLs have been predicted in the rice genome (29, 30). These PYLs have different inhibitory activities on PP2Cs in in vitro assays (29, 30). Overexpression of rice PYL3, PYL5, PYL9, or PYL11 improved drought stress resistance and led to hypersensitivity to ABA for inhibition of seed germination and seedling growth (29, 31, 32). Among the rice PYLs, PYL12 is unique because it is unable to bind ABA but shows potent constitutive inhibition on the activities of PP2Cs in vitro (30).
To date, no high-order pyl mutants have been reported in rice, and the function of rice PYLs remains to be identified. Here, we utilized CRISPR/Cas9 technology to systematically mutate the PYL genes. Through analyses of the pyl mutants, we found that combinatorial mutations of group I genes (PYL1–PYL6 and PYL12) promote rice growth, whereas mutations of group II genes (PYL7–PYL11 and PYL13) have no significant effects on growth. Among group I mutants, pyl1/4/6 exhibited the most robust growth and improved grain productivity, while maintaining near-normal seed dormancy and other agronomic traits. These results show functional differentiation of rice PYLs and provide a genetic strategy to improve rice productivity.
Results
Multigene Knockouts of Rice PYLs.
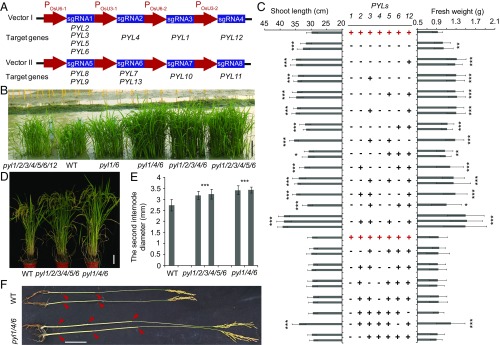
Our real-time RT-PCR analyses showed a differential expression pattern for each of the 13 rice PYLs in various tissues (SI Appendix, Fig. S1), suggesting functional differences between rice PYLs. We constructed two CRISPR/Cas9 vectors to edit the 13 genes (Fig. 1A and specific target sites on each PYL gene are shown in SI Appendix, Table S1). Each vector targets a group of neighboring genes on the phylogenetic tree (Fig. 1A and SI Appendix, Fig. S2). One vector (vector I) targets group I genes, including PYL1, PYL2, PYL3, PYL4, PYL5, PYL6, and PYL12, and another (vector II) targets group II genes, including PYL7, PYL8, PYL9, PYL10, PYL11, and PYL13 (Fig. 1A). We also constructed individual vectors to edit each PYL independently (target sequences are shown in SI Appendix, Table S2).
Fig. 1.
Group I pyl mutations promote rice growth. (A) Strategy of vector construction for editing rice PYL genes. (B) Comparison of wild-type, pyl1/6, pyl1/4/6, pyl1/2/3/4/6, pyl1/2/3/4/5/6, and pyl1/2/3/4/5/6/12 seedlings. (C) Shoot lengths and fresh weights of 21-d-old group I pyl seedlings. Every column represents an independent line. The “+” and “−” represent the wild-type and mutated genes, respectively. (D) Morphological features of wild-type, pyl1/2/3/4/5/6, and pyl1/4/6 plants at the mature stage. (E) Diameters of wild-type, pyl1/2/3/4/5/6, and pyl1/4/6 second internodes. (F) Comparison of the panicles and internodes of the wild type and pyl1/4/6. The arrowheads indicate the stem nodes. (Scale bars, 10 cm.) WT, wild type. Data are presented as means ± SD. P values (versus the wild type) were calculated by the Student’s t test: ***P < 0.001; **P < 0.01; *P < 0.05.
Through Agrobacterium-mediated transformation of the rice variety Nipponbare, seedlings of the first (T0) generation with specific mutations in PYLs were obtained. In the T2 generation, 153 lines (A1–A153) of homozygous mutants for group I genes and 84 lines (B6–B89) of homozygous mutants for group II genes were identified (Dataset S1; A1–A108 and B6–B51 were identified from the multiplex gene-edited lines, and A109–A153 and B52–B89 were obtained from the single gene-edited lines). Since pyl7/8/9/10/11/13 mutants were not found among the multiplex gene-edited lines, we crossed pyl7/8/9/10/13 lines with pyl11 and identified five homozygous pyl7/8/9/10/11/13 lines from the segregating second (F2) population (Dataset S1; B1–B5). In subsequent analyses, we focused on plants with frameshift mutations since they are most likely knockout mutants. To ascertain the genotype and heritability of every line, we sequenced the target genes to confirm the genotype in the following generations and selected CRISPR/Cas9-free plants for seed harvesting.
High-Order Group I, but Not Group II, Mutants Exhibit Improved Growth Compared with the Wild Type.
Phenotypic analyses of the mutants were made in the paddy fields of Shanghai and Lingshui County of Hainan Island under natural conditions. We did not observe significant morphological differences between the wild type and any single mutant of the 13 PYLs. During the entire rice life cycle, high-order group II mutants also appeared to have similar morphological phenotypes to the wild type (SI Appendix, Fig. S3A). Interestingly, we found that many group I mutants had more robust growth than the wild type (Fig. 1 B–D and SI Appendix, Fig. S3 B–D). During the seedling stage (one- to five-leaf stage) in both Shanghai and Hainan, among pyl1/6 to pyl1/2/3/4/5/6/12, most mutants had a larger stature than the wild type (Fig. 1 B and C and SI Appendix, Fig. S3B). Among the mutants, pyl1/4/6 had the most robust growth, indicating important roles for PYL1, PYL4, and PYL6 in inhibiting rice growth (Fig. 1 B and C). All lines of pyl1/4/6 seedlings had larger fresh weight and longer shoot length than the wild type and appeared larger than the other mutants overall (Fig. 1C). Among the double (pyl1/6, pyl1/4, pyl1/2, and pyl1/12) and triple (pyl1/4/6, pyl1/2/4, pyl1/4/5, and pyl1/3/4) mutants, only pyl1/6 and pyl1/4/6 seedlings were larger than the wild type (Fig. 1C). The other high-order group I mutants were all larger than the wild type. From the triple to sextuple group I mutants, all of the seedlings that contained a mutation in PYL6 had longer shoot length and larger fresh weight than those without the pyl6 mutation (Fig. 1C), indicating that PYL6 plays a crucial role in controlling rice plant growth.
In Hainan, after the seedling stage, high-order group I mutants continued to exhibit more robust growth than the wild type. However, in Shanghai in the year 2016, phenotypic differences from those in Hainan appeared. In Shanghai in 2016, from mid-July, a high-temperature climate (about 41 °C at midday) appeared and lasted for more than 2 wk. Passing through this high-temperature period, higher order group I mutant plants (∼30–60 d postgermination) suffered more severely retarded growth (SI Appendix, Fig. S4A), suggesting hypersensitivity of the higher order mutants to the high-temperature climate. During and after this high-temperature period, the pyl1/4/6 lines were still larger than the wild type (SI Appendix, Fig. S4A).
After the heading stage, most high-order group I mutant plants in both Shanghai and Hainan were larger than the wild type (Fig. 1D and SI Appendix, Fig. S3 C and D). We measured the plant height at this stage on Hainan Island, and the data revealed a generally similar growth pattern to that during the seedling stage (SI Appendix, Fig. S3D). From pyl1/6 to pyl1/2/3/4/5/6/12, most mutant lines were taller than the wild type (Fig. 1D and SI Appendix, Fig. S3D). Among the high-order group I mutant lines, the pyl1/4/6 lines were often the tallest (Fig. 1D and SI Appendix, Fig. S3D). In Shanghai in 2016, despite the high-temperature episode, the group I mutants regained their faster growth phenotype after the heading stage (SI Appendix, Fig. S4B). Except for pyl1/2/3/4/5/6/12 mutants, all of the plants in Shanghai showed a similar phenotype to those in Hainan at the mature stage (SI Appendix, Fig. S4B). The pyl1/2/3/4/5/6/12 plants in Shanghai showed similar plant height to the wild type at the mature stage (SI Appendix, Fig. S4B), whereas they were slightly taller than the wild type in Hainan (SI Appendix, Fig. S3D). At the mature stage, we also measured the internode diameters of wild-type, pyl1/2/3/4/5/6, and pyl1/4/6 plants. The pyl1/2/3/4/5/6 and pyl1/4/6 mutants had larger internode diameters than the wild type (Fig. 1 E and F). Compared with the wild type, the length of every internode in pyl1/4/6 was also increased, especially the lower internodes (Fig. 1F and SI Appendix, Fig. S3E).
To further characterize the increased growth in group I mutants, we compared the cell size of pyl1/4/6 and the wild type at the seedling stage. The results showed that the epidermal cells of pyl1/4/6 leaf blades and sheaths were longer than those of the wild type (SI Appendix, Fig. S5 A–D). However, no significant differences (P > 0.1) in the cell width between pyl1/4/6 and the wild type were observed (SI Appendix, Fig. S5 A–D). At the seed-filling stage, the pyl1/4/6 flag leaf blades were longer and wider than the wild type (SI Appendix, Fig. S5 H and I). Consistent with this phenotype, the epidermal cells of the flag leaf blades in pyl1/4/6 were longer than the wild type, whereas the epidermal cell widths of the flag leaf blades showed no significant differences between pyl1/4/6 and the wild type (SI Appendix, Fig. S5J). The epidermal cells of flag leaf sheaths in pyl1/4/6 were longer but narrower than those of the wild type (SI Appendix, Fig. S5K). Histological analyses of cross-sections of leaf blades and internodes revealed no obvious differences in the sizes of the cells between pyl1/4/6 and the wild type (SI Appendix, Fig. S5E). Using flow cytometry, we examined DAPI-stained nuclei isolated from the bud tips of germinated seeds (germinated for 2 d). The results showed that pyl1/4/6 had a higher percentage of cells in G2/M phase than the wild type (SI Appendix, Fig. S5 F and G), indicating a faster cell cycle in pyl1/4/6. Our data suggest that mutations in group I PYLs promote rice plant growth through accelerating cell division and increasing cell elongation.
Group I, but Not Group II, Mutations Delay Heading Dates.
Heading date is an important trait for adapting crops to different cultivating regions and cropping seasons. In many of the group I mutants, a substantial delay in heading date was observed (SI Appendix, Fig. S3F). Compared with the wild type, the heading date of pyl1/2/3/4/5/6 and pyl1/2/3/4/5/6/12 was delayed by about 9 d, pyl1/2/3/4/6 by about 7 d, pyl1/2/3/4 by about 5 d, and pyl1/4/6 by about 1 d. No differences in heading dates were observed between the wild type and other group I triple, double, and single mutants. The group II mutants had heading dates similar to the wild type.
Group I Mutations Result in Seed Dormancy Defects and Preharvest Sprouting.
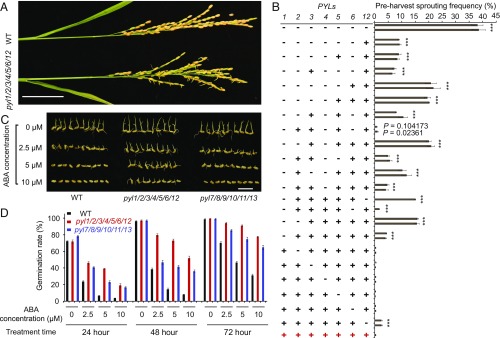
Seeds sometimes fail to enter a period of strong dormancy and may germinate while still attached to the parent plants. This condition is known as preharvest sprouting (PHS). PHS is usually associated with reduced levels of ABA or ABA signaling in the maturing seeds. Among the high-order group I, but not group II, mutants, significant PHS was often observed, especially in Shanghai (Fig. 2A). In Shanghai, during the harvest time of Nipponbare (late September and early October), there was often very abundant rainfall that can promote PHS. Therefore, we investigated the PHS of the pyl mutants in Shanghai in the year 2016. The highest frequency of PHS was observed in pyl1/2/3/4/5/6/12 (Fig. 2B). Among the single mutants, only pyl1 and pyl12 had notably higher frequencies of PHS than the wild type (Fig. 2B), indicating that PYL1 and PYL12 play critical roles in establishing seed dormancy. Consistent with the critical role of PYL12 in seed dormancy, the frequency of PHS in pyl1/2/3/4/5/6/12 was much higher than that in pyl1/2/3/4/5/6 (Fig. 2B). The important role of PYL12 in seed dormancy is interesting, since PYL12 cannot bind to ABA in in vitro assays (30). Interestingly, we found that the frequency of PHS was lower in pyl1/6 than in pyl1, and lower in pyl1/4/6 than in pyl1 and pyl1/4 (Fig. 2B). Thus, there appears to be an antagonistic function between PYL6 and the other group I PYL genes in establishing seed dormancy. Among the triple mutants (pyl1/4/6, pyl1/2/4, pyl1/4/5, and pyl1/3/4), pyl1/4/6 had the lowest frequency of PHS (Fig. 2B). In fact, among the high-order mutants of group I, pyl1/4/6 had the lowest PHS frequency, which was nearly comparable to the wild type (P values of 0.104173 and 0.02361 for two independent lines) (Fig. 2B). In other planting seasons, we did not observe obvious PHS in pyl1/4/6. The most robust growth and lowest PHS frequency observed in pyl1/4/6 indicated that this combination of mutations may be used to generally improve rice productivity. No obvious PHS was observed in group II mutants at the normal harvest time. However, we did find higher frequencies of seed germination on panicles in pyl7/8/9/10/13 and pyl7/8/9/10/11/13 than in the wild type when the seed harvesting was delayed by about 25 d in the year 2016 in Shanghai (SI Appendix, Fig. S6). Thus, there appears to be some defects in seed dormancy in these group II mutants.
Fig. 2.
PHS and seed dormancy of the pyl mutants. (A) PHS of pyl1/2/3/4/5/6/12 mutant. (Scale bar, 5 cm.) (B) PHS frequencies in group I mutants. Every column represents an independent line. In every line, all of the seeds of three plants were counted. The “+” and “−” represent the wild-type and mutated genes, respectively. P values (versus the wild type) were calculated by the Student’s t test: ***P < 0.001. (C) Germinating seeds of the wild type, pyl1/2/3/4/5/6/12, and pyl7/8/9/10/11/13 treated with ABA for 4 d. (Scale bar, 2 cm.) (D) Effect of ABA on the seed germination rates of the wild type, pyl1/2/3/4/5/6/12, and pyl7/8/9/10/11/13. In every treatment, 100 seeds were used. The treatment was repeated three times. WT, wild type. Data are presented as means ± SD.
The observed PHS implies that differences in the sensitivity of seed germination to ABA may exist between the pyl mutants and the wild type. Therefore, we treated the germinating seeds of the wild type, pyl1/2/3/4/5/6/12, and pyl7/8/9/10/11/13 with 2.5, 5, and 10 μM ABA for 4 d, and examined the germination status each day. These assays revealed the highest germination rate in pyl1/2/3/4/5/6/12 under the ABA treatment conditions (Fig. 2 C and D). Notably higher germination rates in pyl7/8/9/10/11/13 compared with the wild type were also observed during the treatments with each ABA concentration (Fig. 2D). These results indicate that both group I and group II PYLs control seed dormancy but that group I genes have more important functions in seed dormancy than group II genes.
The Growth of Group I, but Not Group II, Mutants Is Less Sensitive to ABA than the Wild Type.
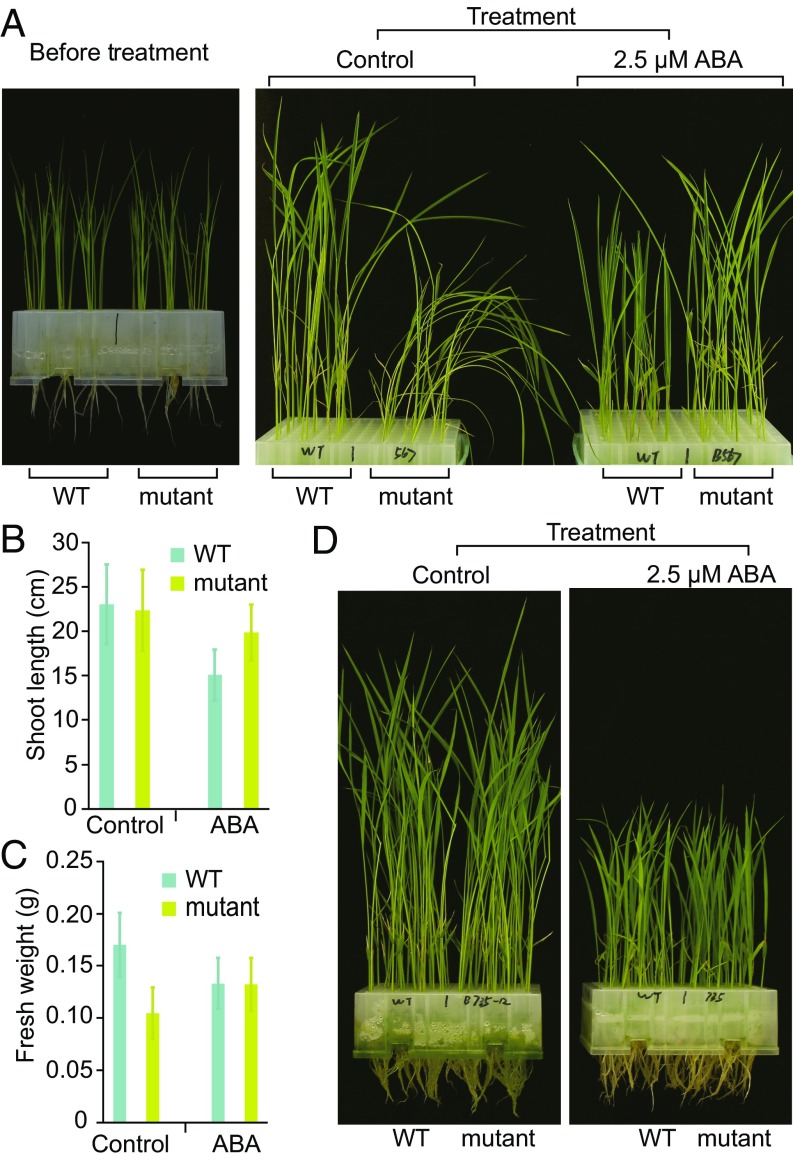
To investigate the sensitivity to ABA at the postgermination stage, 8- to 9-d-old seedlings with similar sizes were treated with 2.5 μM ABA. Five days after the treatment, compared with the untreated control, the shoot length of the wild type was reduced by about 35%, whereas the ABA treatment reduced the shoot length of pyl1/2/3/4/5/6/12 by only 11%, indicating less sensitivity of pyl1/2/3/4/5/6/12 seedlings to ABA (Fig. 3 A and B). The ABA treatment reduced the fresh weight of the wild type by 22%, whereas the fresh weight of pyl1/2/3/4/5/6/12 increased by 27% after the treatment (Fig. 3 A and C). This suggests that a basal level of ABA signaling promotes plant growth. In fact, without ABA treatment, pyl1/2/3/4/5/6/12 seedlings were not robust when grown in the greenhouse (26 °C, 80% humidity, and 12 h light/12 h dark), and it was even difficult for them to stand erect (Fig. 3A). ABA treatment made pyl1/2/3/4/5/6/12 stronger physically and more robust (Fig. 3 A and C). No obvious differences in ABA response were observed between wild-type and pyl7/8/9/10/11/13 seedlings (Fig. 3D).
Fig. 3.
Sensitivity of wild-type and pyl seedlings to ABA. (A) Wild-type and pyl1/2/3/4/5/6/12 (mutant) seedlings grown for 5 d in 2/3 Murashige and Skoog (MS) liquid medium without (control) or with 2.5 μM ABA. (B) Shoot lengths of the wild-type and pyl1/2/3/4/5/6/12 (mutant) seedlings treated with 0 (control) and 2.5 μM ABA for 5 d. Twenty-four seedlings of each material in the control and ABA treatments were measured. (C) Fresh weights of the wild-type and pyl1/2/3/4/5/6/12 (mutant) seedlings treated with 0 (control) and 2.5 μM ABA for 5 d. Twenty-four seedlings of each material in the control and ABA treatments were measured. (D) Wild-type and pyl7/8/9/10/11/13 (mutant) seedlings grown for 5 d in 2/3 MS liquid medium without (control) or with 2.5 μM ABA. WT, wild type. Data are presented as means ± SD.
Group I PYL Knockouts Lead to Severe Defects in Stomatal Movement and Water Loss Control.
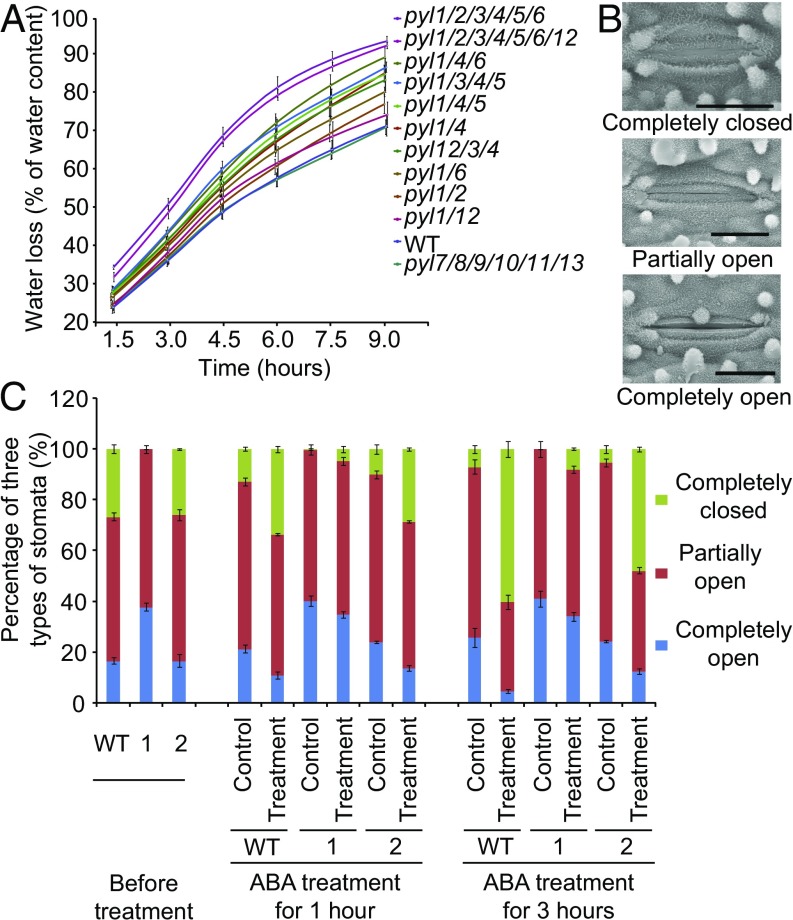
Given that ABA controls transpirational water loss by inducing stomatal closure, we investigated the water loss rate of detached flag leaves. Gravimetric measurement of water loss from detached leaves indicates that high-order group I, but not group II, mutants lose water more quickly than the wild type (Fig. 4A and SI Appendix, Fig. S7A). Among all of the pyl mutants tested, pyl1/2/3/4/5/6 and pyl1/2/3/4/5/6/12 lost water most quickly (Fig. 4A). Infrared imaging revealed a lower leaf surface temperature in pyl1/4/6 than in the wild type in the greenhouse (26 °C, 80% humidity, and 12 h light/12 h dark) (SI Appendix, Fig. S7B), consistent with more rapid transpiration in group I mutants. Water withholding assays showed that pyl1/4/6 mutant plants were more sensitive to drought than the wild type (SI Appendix, Fig. S7C). Next, we examined whether ABA induction of stomatal closure was affected in the pyl mutants. Before treatment with 30 μM ABA, ∼27% and ∼26% of the stomata in the wild type and pyl7/8/9/10/11/13 were closed, respectively, whereas no stomata of pyl1/2/3/4/5/6/12 leaves were closed (Fig. 4 B and C and SI Appendix, Table S4). After treatment in 30 μM ABA for 1 h, stomata of the wild type and pyl7/8/9/10/11/13 began to close, reaching around 60% and 48% completely closed after 3 h, respectively (Fig. 4C and SI Appendix, Table S4). Only 8% of the stomata of pyl1/2/3/4/5/6/12 were closed completely after the ABA treatment for 3 h (Fig. 4C and SI Appendix, Table S4). These results clearly show that ABA treatment induces much less stomatal closure in pyl1/2/3/4/5/6/12 than in the wild type and pyl7/8/9/10/11/13, suggesting that the group I PYLs have a more important function in stomatal movement than group II PYLs.
Fig. 4.
Stomatal movement in the wild type and pyl mutants. (A) Cumulative transpirational water loss from the detached flag leaves of the wild type and pyl mutants. Five flag leaf blades of each mutant were used for this assay. (B) Scanning electron microscopy images of three levels of stomatal opening. (Scale bars, 10 μm.) Stomata with a similar or wider opening than the stoma as pictured in the Bottom panel were considered completely open. (C) Effect of pyl mutations on ABA-induced closure of stomata. 1, pyl1/2/3/4/5/6/12; 2, pyl7/8/9/10/11/13; Control, treatment in stomata opening buffer without ABA; WT, wild type. Data are presented as means ± SD.
Improving Productivity with pyl1/4/6.
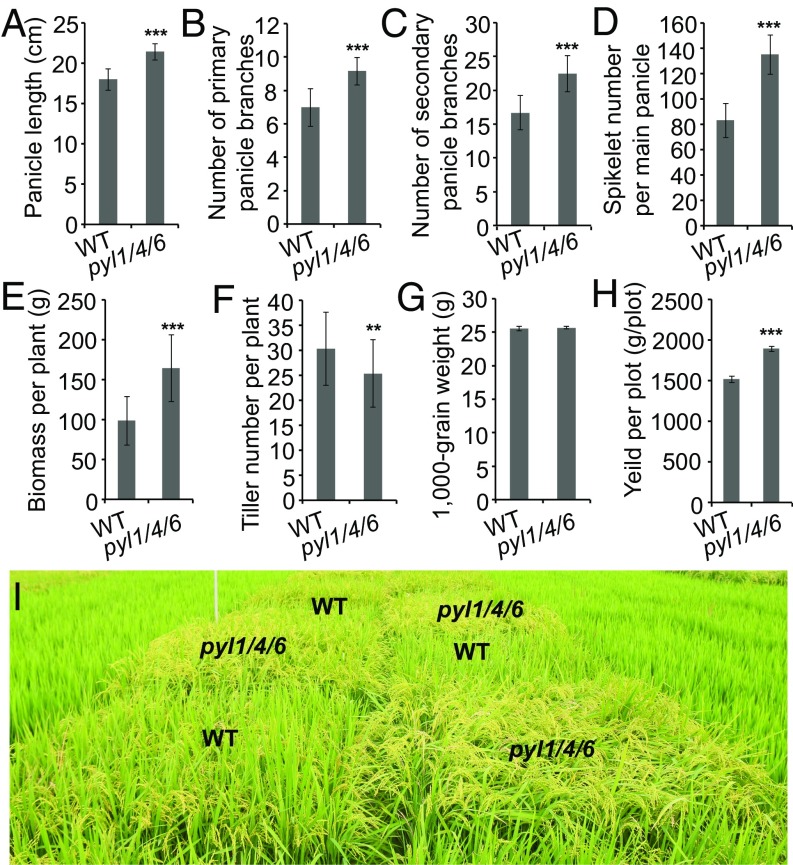
The above results revealed that pyl1/4/6 lines exhibited the most robust growth among the pyl mutants, while maintaining nearly normal seed dormancy and heading date, suggesting that this mutant has important potential for use in agriculture. So, we compared several agronomic characteristics of pyl1/4/6 with the wild type. The seed density of the panicles in pyl1/4/6 was not obviously different from that in the wild type, but the panicles of pyl1/4/6 were longer than the wild type (Figs. 1F and 5A). There were more primary and secondary branches in the panicles of pyl1/4/6 (Fig. 5 B and C), and the spikelet number per main panicle was also increased in pyl1/4/6 (Fig. 5D). Compared with the wild type, the biomass of pyl1/4/6 at the mature stage was significantly increased (Fig. 5E), even though the tiller number was decreased (Fig. 5F). The 1,000-grain weight of pyl1/4/6 plants showed no significant differences from the wild type (Fig. 5G; P > 0.2).
Fig. 5.
Agronomic characteristics and yield test of the wild type and pyl1/4/6. (A) Main panicle lengths of the wild type and pyl1/4/6. Numbers of primary (B) and secondary (C) branches per main panicle are shown. (D) Spikelet numbers per main panicle of the wild type and pyl1/4/6. (E) Biomasses of the wild type and pyl1/4/6 at the mature stage. (F) Tiller numbers of the wild type and pyl1/4/6. (G) One thousand-grain weights of the wild type and pyl1/4/6. (H) Grain yields per plot of the wild type and pyl1/4/6 in Shanghai in the year 2016. The planting density was 15 × 15 cm, with one plant one hill. One hundred forty-four (12 × 12) plants were cultivated in every plot. (I) Plot yield test in Shanghai in the year 2016. WT, wild type. Data are presented as means ± SD. P values (versus the wild type) were calculated with the Student’s t test: ***P < 0.001; **P < 0.01.
The productivity of pyl1/4/6 in paddy field conditions was examined using plot yield tests. Our test in Shanghai in 2016 showed that compared with the wild type, pyl1/4/6 had ∼25% higher grain yield (Fig. 5 H and I). We repeated the test in Hainan Island. Here, we observed an increase of about 31% in grain production (a yield per plot of 1,727.1 ± 34.9 g for pyl1/4/6 versus 1,317.5 ± 25.7 g of yield per plot for the wild type, 15 × 15-cm plant density, 144 plants per plot). These results indicate that pyl1/4/6 has significantly improved yields under natural paddy field conditions.
Discussion
The growth-repressing function of ABA has been thought to be a trade-off for improving stress adaptation (3, 33, 34). Since the plant growth and stress adaptation processes are contrary to each other in many ways, it is important to establish a balance between them that is appropriate for the environment. Therefore, ABA levels and signaling should have significant effects on both growth and adaptation. Genetic adjustment of the balance by manipulating ABA levels and/or signaling may generate useful crop varieties to improve productivity in specific environments.
The PYLs are currently the largest plant hormone receptor family known (35). In Arabidopsis, 14 PYL members have been identified, and redundant as well as differential functions of the genes have been documented (11, 26–28, 35, 36). In rice, 13 PYL members were predicted (29, 30). Our results indicate differential and redundant functions for different members as well. The functional differentiation and redundancy provide the possibility to adjust the balance between growth and stress resistance to improve crop productivity through editing certain PYLs. In rice, we found that among the pyl mutants, pyl1/4/6 showed the best growth, while maintaining nearly normal seed dormancy. During the heat wave of the 2016 summer in the paddy field in Shanghai, although the growth of quintuple to septuple pyl mutants was increasingly retarded, the pyl1/4/6 mutants still grew better than the wild type. This suggests that the pyl1/4/6 lines may be more tolerant to the hot weather than the quintuple, sextuple, and septuple group I mutants. These results indicate that the balance between growth and stress adaptation is altered in high-order group I pyl mutants, and that the new balance in pyl1/4/6 plants favors more growth, while stress adaptation is less compromised than in higher order group I pyl mutants.
Previous studies showed that Arabidopsis high-order pyl mutants exhibited retarded growth, and that this growth defect can be ameliorated by increasing the humidity of the growth environment (25, 37). Still, improved growth (compared with the wild type) was not observed in Arabidopsis pyl mutants (25). Although, overall, rice PYLs have similar functions to Arabidopsis PYLs in promoting seed dormancy and stomatal closure, they seem to differ in their impacts on plant growth. It is likely that under the paddy field growth conditions, some of the rice PYLs have been selected to have a particularly important role in restraining plant growth. In the future, it will be of interest to determine how PYL1, PYL4, and PYL6 (particularly PYL6) are linked to growth regulation in rice. In the paddy field, where water is not limiting, the higher transpiration of pyl1/4/6 may contribute to faster growth and higher yield by facilitating rapid CO2 absorption, while avoiding the negative effects of excessive water loss. Plant breeders have long suggested that the improvement of crop performance in certain climates may be achieved by reducing responses to ABA or endogenous ABA level (34). The pyl1/4/6 rice mutants may represent an effective approach to achieve this historically coveted result.
Materials and Methods
Details are provided in SI Appendix, SI Materials and Methods, including vector construction and plant cultivation, shoot length and fresh weight measurements, heading date comparison, histological analyses and epidermal cell observations, cell cycle comparison, seed dormancy analyses, stomatal movement and water loss assays, ABA sensitivity assays of seedling growth, real-time RT-PCR, plot field test, and accession numbers.
Supplementary Material
Acknowledgments
This work was supported by the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804774115/-/DCSupplemental.
References
- 1.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Vishwakarma K, et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front Plant Sci. 2017;8:161. doi: 10.3389/fpls.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P, et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell. 2018;69:100–112.e6. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christmann A, et al. Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 2006;8:314–325. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, et al. Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front Plant Sci. 2017;8:1121. doi: 10.3389/fpls.2017.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, et al. Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 2013;197:139–150. doi: 10.1111/nph.12004. [DOI] [PubMed] [Google Scholar]
- 7.Rowe JH, Topping JF, Liu J, Lindsey K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016;211:225–239. doi: 10.1111/nph.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C-W, Yang Y-W, Lur H-S, Tsai Y-G, Chang M-C. A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol. 2006;47:1–13. doi: 10.1093/pcp/pci216. [DOI] [PubMed] [Google Scholar]
- 9.Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21:R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J-K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S-Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 13.Hao Q, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell. 2011;42:662–672. doi: 10.1016/j.molcel.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 16.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. The HAB1 PP2C is inhibited by ABA-dependent PYL10 interaction. Sci Rep. 2015;5:10890. doi: 10.1038/srep10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim N, et al. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front Plant Sci. 2015;6:614. doi: 10.3389/fpls.2015.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiger D, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirichandra C, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geiger D, et al. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal. 2011;4:ra32. doi: 10.1126/scisignal.2001346. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Guzman M, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing L, Zhao Y, Gao J, Xiang C, Zhu J-K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep. 2016;6:27177. doi: 10.1038/srep27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, et al. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal. 2014;7:ra53. doi: 10.1126/scisignal.2005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoni R, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161:931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian X, et al. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice (N Y) 2015;8:28. doi: 10.1186/s12284-015-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, et al. Identification and characterization of ABA receptors in Oryza sativa. PLoS One. 2014;9:e95246. doi: 10.1371/journal.pone.0095246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, et al. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot. 2012;63:1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, et al. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J Exp Bot. 2014;65:453–464. doi: 10.1093/jxb/ert397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claeys H, Inzé D. The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013;162:1768–1779. doi: 10.1104/pp.113.220921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum A. Towards a conceptual ABA ideotype in plant breeding for water limited environments. Funct Plant Biol. 2015;42:502–513. doi: 10.1071/FP14334. [DOI] [PubMed] [Google Scholar]
- 35.Dupeux F, et al. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 2011;30:4171–4184. doi: 10.1038/emboj.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui F, et al. Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol Plant. 2016;9:926–938. doi: 10.1016/j.molp.2016.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.