Significance
The most abundant modification in mRNA is the N6-methylation of internal adenosines (m6A), but m6A’s physiological function is unknown for most mRNAs. Here we show that Casein Kinase 1 Delta mRNA (Ck1δ), coding for a critical kinase in the control of circadian rhythms, is regulated by m6A. When m6A is inhibited, the expression of two CK1δ isoforms, uncharacterized until now, increases due to enhanced translation. This increase in CK1δs leads to a slower clock because of increased phosphorylation of the clock protein PER2 at a key residue, leading to the stabilization of PER2 protein.
Keywords: casein kinase, circadian, methylation, splicing, m6A
Abstract
The N6-methylation of internal adenosines (m6A) in mRNA has been quantified and localized throughout the transcriptome. However, the physiological significance of m6A in most highly methylated mRNAs is unknown. It was demonstrated previously that the circadian clock, based on transcription-translation negative feedback loops, is sensitive to the general inhibition of m6A. Here, we show that the Casein Kinase 1 Delta mRNA (Ck1δ), coding for a critical kinase in the control of circadian rhythms, cellular growth, and survival, is negatively regulated by m6A. Inhibition of Ck1δ mRNA methylation leads to increased translation of two alternatively spliced CK1δ isoforms, CK1δ1 and CK1δ2, uncharacterized until now. The expression ratio between these isoforms is tissue-specific, CK1δ1 and CK1δ2 have different kinase activities, and they cooperate in the phosphorylation of the circadian clock protein PER2. While CK1δ1 accelerates the circadian clock by promoting the decay of PER2 proteins, CK1δ2 slows it down by stabilizing PER2 via increased phosphorylation at a key residue on PER2 protein. These observations challenge the previously established model of PER2 phosphorylation and, given the multiple functions and targets of CK1δ, the existence of two isoforms calls for a re-evaluation of past research when CK1δ1 and CK1δ2 were simply CK1δ.
Since 2012, the N6-methylation of internal adenosines (m6A) has regained tremendous interest due to the use of affinity immunoprecipitation of methylated mRNA coupled with next-generation RNA-Seq (1, 2), confirming earlier reports (3, 4). m6A accounts for 50% of all methylated nucleotides in polyadenylated mRNA (5) and for 80% of all base methylations (3). It is the most prevalent internal mRNA modification in higher eukaryotes and is widely conserved from yeast to mammals (6, 7). m6A has been involved in the regulation of mRNA splicing, processing, and translation (8, 9) and is required for survival and development in multiple species (10–12).
The circadian clock is composed of clock genes interlocked in transcription-translation negative feedback loops (TTFL) regulating their own expression (13). This molecular oscillator, robust in normal conditions yet sensitive to perturbations affecting gene expression and RNA processing, allowed us to demonstrate the importance of m6A for its function (14), but which clock genes are directly regulated by m6A remains unknown.
While the core TTFL has been the focus of early circadian research, protein phosphorylation was quickly shown to be essential for molecular timekeeping (15, 16). Phosphorylation of clock proteins is mediated mainly by the Casein Kinases 1 Delta and Epsilon. The first human inherited circadian syndrome to be described, the Familial Advanced Sleep Phase Syndrome (FASPS), originates from a mutated CK1δ (17) or a mutated CK1δ-target serine in PER2 protein (18), causing early sleep onset and offset. The first circadian mutation to be discovered in a mammalian model is a mutation in CK1ε (16, 19).
Here, we show that the 3′-UTR of Ck1δ is heavily m6A-methylated and negatively controls the translation of two CK1δ isoforms, uncharacterized until now. These two CK1δ isoforms are expressed in a tissue-specific manner and antagonistically regulate the circadian clock.
Results
The 3′-UTR of Ck1δ Is Methylated.
To identify clock-related transcripts regulated by m6A, we quantified m6A levels in mouse liver mRNA at two time points, CT4 and CT16 (CT is circadian time, and CT0 and CT12 correspond to the beginning of the rest and the active phase, respectively). Significant m6A peaks were found at CT4 (8,414) and at CT16 (8,821). Most peaks (72%) were detected at both time points (Dataset S1). Several transcripts involved in circadian regulation showed significant peaks at CT4 and CT16 (SI Appendix, Fig. S1A). Since Clock (20), Dbp (21), Ck1δ (17), and Fbxl3 (22, 23) are potent regulators of the circadian period, m6A methylation of these transcripts is likely to be of importance for clock function. Of these four transcripts, the most significant peak, according to the q value of the peak-calling analysis (SI Appendix, Fig. S1A), was the single peak in the 3′-UTR of Ck1δ (SI Appendix, Fig. S1B), but no significant peaks were detected in the mRNA of its close homolog, Ck1ε (SI Appendix, Fig. S1B). Given these observations, together with the relevance of Ck1δ for human FASPS and its role as a hub kinase (24), we selected Ck1δ for further investigations.
m6A Regulates the Expression of Two CK1δ Isoforms.
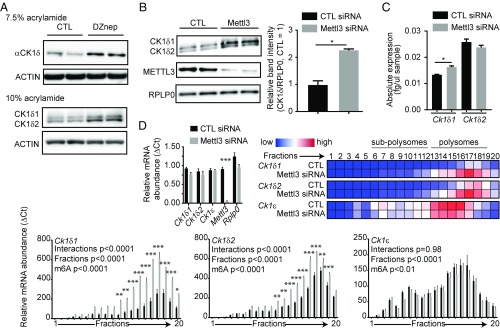
To investigate the role of m6A in the control of Ck1δ expression, we treated mouse embryonic fibroblasts (MEFs) with Deazaneplanocin A (DZnep), an inhibitor of methylation known to inhibit m6A and elongate the circadian period length in these cells (14, 25), and observed an increase in CK1δ proteins (Fig. 1A). This was opposite to our expectations, since available evidence published so far in mammals has associated higher CK1δ expression with a faster clock (26). While replicating these results, we noticed that, under certain conditions during migration of proteins in the acrylamide gel, two bands, not one, were seen on the membrane, both bands increasing under DZnep treatment (Fig. 1A). By comparing these two bands with the bands obtained from cells transfected with CK1δ expression vectors, we later identified the two bands as two isoforms of CK1δ (Fig. 3), CK1δ1 (415 aa) and CK1δ2 (409 aa), reported in databases but of functions unknown, sharing 99.75% identity with human CK1δs, and conserved in virtually all vertebrates (SI Appendix, Fig. S1 C and D). The only difference between these isoforms is a small 63-bp exon in mouse, retained in Ck1δ2 immediately upstream of the 3′-UTR and containing a STOP codon (SI Appendix, Fig. S1B), thus coding for the smaller kinase.
Fig. 1.
Two Ck1δ alternative transcripts are under m6A control. (A) A 50-μM DZnep treatment for 2 h increased CK1δ protein levels, and two CK1δ isoforms were observed when run on a 10% acrylamide gel. ACTIN was used as a loading control. (B) Mettl3 silencing increases CK1δ protein levels. Knock-down of METTL3 was confirmed, and RPLP0 was used as a loading control. (Right) Quantification of the total intensity of CK1δ bands relative to RPLP0, analyzed by Student t test, shown as mean ± SD, *P < 0.05. (C) Mettl3 silencing causes little changes in Ck1δ mRNA steady-state levels. Only Ck1δ1 showed a mild but significant increase. Data for each transcript were analyzed by Student t test and are shown as mean ± SEM, n = 3, *P < 0.05. (D) Mettl3 silencing increases the translated fraction of Ck1δ transcripts. (Top Right) Heatmap visualization of mean relative mRNA abundance for each transcript in each fraction. (Top Left) Quantification of transcripts in input cytoplasmic lysates, showing significant knock-down of Mettl3 but no significant differences in Ck1δ1, Ck1δ2, Ck1ε, and Rplp0 mRNA levels; ΔCT data, analyzed by multiple t tests, are shown as mean ± SEM, n = 3, ***P < 0.001. (Bottom) Bar charts show mean ΔCT ± SEM of indicated transcripts in each fraction, with significance levels in two-way ANOVA followed by Bonferroni post hoc test, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001.
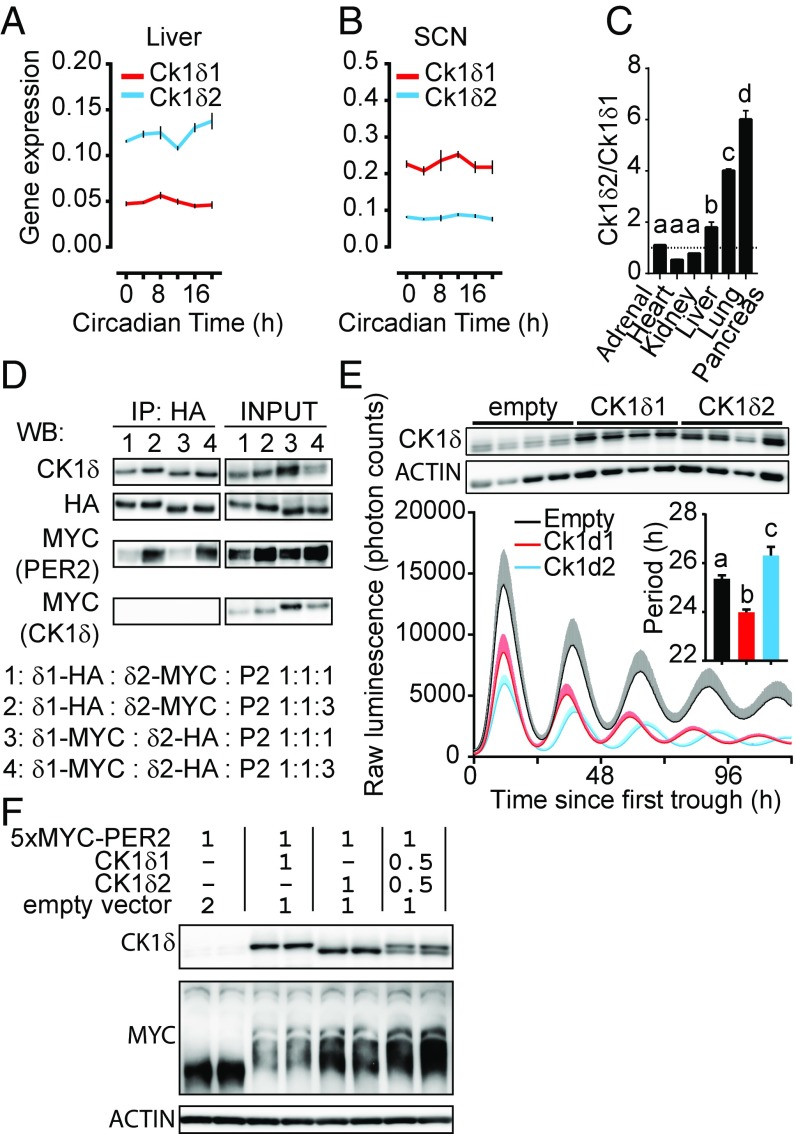
Fig. 3.
CK1δ1 and CK1δ2 are antagonistic. Ck1δ1 and Ck1δ2 mRNA levels in the liver (A) and the SCN (B) across a circadian day, with opposite Ck1δ2/Ck1δ1 ratios mainly due to different levels of Ck1δ1. Mean ± SEM analyzed by two-way ANOVA, n = 3, significant for Ck1δ1 vs. Ck1δ2 (P < 0.0001) and for time, only in the liver for Ck1δ2 (P < 0.05). (C) Tissue-specific Ck1δ2/Ck1δ1 ratios measured in six different tissues. Analyzed by one-way ANOVA, P < 0.0001, followed by Bonferroni post hoc, a vs. b vs. c vs. d, indicating at least P < 0.05 significance. Results shown are mean ± SD of n = 2. (D) Coimmunoprecipitation shows that both CK1δ1-HA and CK1δ2-HA interact with 5xMYC-PER2. Two CK1δ1-tag:CK1δ2-tag:5xMYC-PER2 ratios were used at 1:1:1 and 1:1:3. (E) Stable overexpression of CK1δ1 and CK1δ2 in PER2::LUC MEFs leads to period shortening and lengthening, respectively. (Inset) Period analyzed by one-way ANOVA, P < 0.001, with a vs. b vs. c in Bonferroni post hoc at least P < 0.05; results shown are mean ± SEM of n = 4 monoclonal cell lines for each group. (Top) CK1δ and ACTIN immunoblots for each cell line. (F) Cotransfection of 5xMYC-PER2 with untagged CK1δ1 and/or CK1δ2 shows higher 5xMYC-PER2 phosphorylation (and/or higher stability) when CK1δ2 is present.
To confirm that the results above were dependent upon mRNA m6A inhibition, we silenced in MEFs the catalytic subunit of the mRNA m6A methyltransferase, Mettl3 (27, 28), and measured CK1δ protein levels by immunoblotting. Again, two specific bands were detected. Silencing of Mettl3 increased the intensity of the two bands, demonstrating m6A negatively regulates CK1δ expression (Fig. 1B). At the mRNA level, Mettl3 silencing mildly increased Ck1δ1 and had little effect on Ck1δ2 (Fig. 1C) and significantly delayed the initial steps in Ck1δ1 and Ck1δ2 degradation (SI Appendix, Fig. S2A). Surprisingly, the half-life of Ck1δ2 was significantly longer than that of Ck1δ1, which we confirmed in a longer time course (SI Appendix, Fig. S2B).
Since the increase in CK1δs under m6A inhibition cannot be readily explained by changes in steady-state transcript levels or mRNA stability, we measured the impact of Mettl3 silencing on translation of Ck1δ transcripts by polysome fractionation (Fig. 1D). Consistent with a recent report showing that m6A attenuates translation efficiency (29), Mettl3 silencing significantly increased the amount of Ck1δ transcripts only in fractions containing the heavier polysomes, while Ck1ε was not significantly affected.
The 3′-UTR of Ck1δ transcripts is over 2,000 nucleotides long, but its function in the regulation of CK1δ expression is unknown. We designed reporter mRNAs in which the luciferase coding sequence was flanked by the 5′- and 3′-UTRs of Ck1δ. To closely mimic Ck1δ1 and Ck1δ2, the end of the coding sequence of each Ck1δ was also included downstream of the luciferase STOP codon. We also designed Ck1δluc transcripts lacking most of the 3′-UTR. These reporter mRNAs were transcribed in vitro and then added to reticulocyte lysates to assess the efficiency of their translation by luminometry (SI Appendix, Fig. S2C). The deletion of the 3′-UTR caused a significant increase in Ck1δ1luc (approximately sixfold) and Ck1δ2luc (approximately threefold) luminescence, demonstrating the role of the 3′-UTR as a negative regulator of CK1δ translation.
Ablation of Ck1δ m6A Methylation by CRISPR-Cas9 in Vivo.
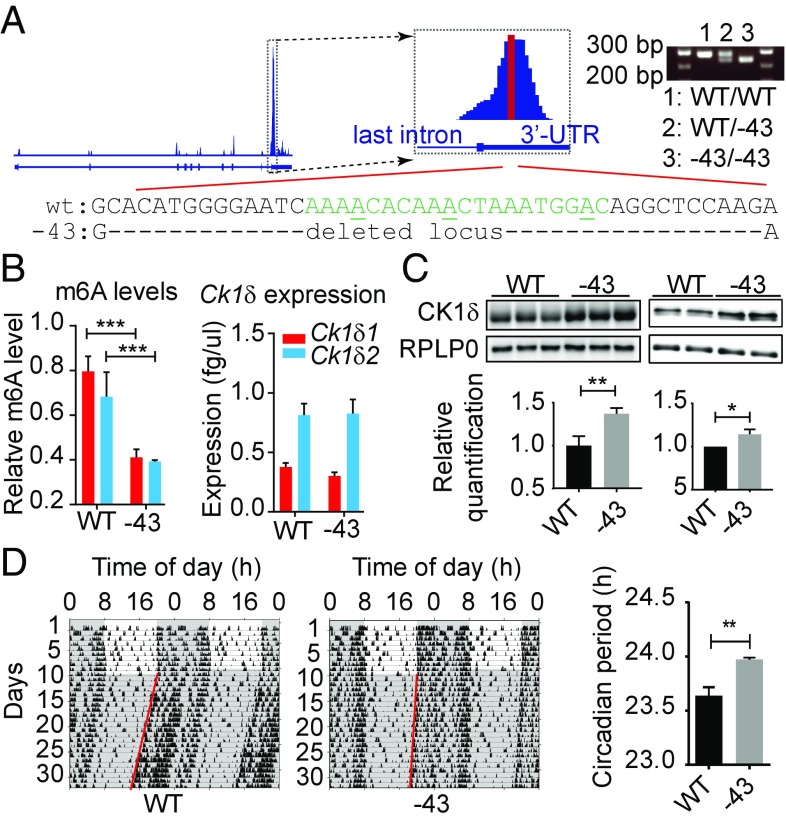
Next, to investigate the physiological role of the methylated locus in the 3′-UTR of Ck1δ, we generated a mutant mouse strain with a 43-bp deletion in the genomic region corresponding to the methylated peak (Fig. 2A). This deletion does not affect the coding sequence but removes the N6-methylated adenosine at GGm6ACA, located previously by screening of m6A sites in brain and liver transcriptomes (30). The consequences of this mutation on Ck1δ m6A levels, mRNA, and protein expression were investigated in the liver, since it is known to have high mRNA m6A levels (2). As expected, Ck1δ transcripts had lower m6A levels in −43/−43 animals compared with wild type (Fig. 2B), but the level of Ck1δ mRNAs was not significantly affected (Fig. 2B). In contrast, CK1δ protein levels increased significantly: homozygous −43 mutant mice had higher CK1δ levels in the liver and the brain (Fig. 2C), despite an inversed CK1δ1/CK1δ2 ratio (Fig. 3), and had a longer period (23.97 h ± 0.02) of locomotor activity behavior rhythms compared with wild-type mice (23.64 h ± 0.08, Fig. 2D). Although noncoding, this mutation, weakening m6A-dependent posttranscriptional processes limiting CK1δ expression, was sufficient to cause an observable increase in CK1δ proteins, accompanied by a change in circadian locomotor activity behavior. How an increase in the expression of endogenous CK1δs is able to elongate period length is investigated in the next part of this work.
Fig. 2.
Deletion of m6A locus from Csnk1d 3′-UTR is sufficient to elongate the circadian period in vivo. (A) Location of the mutation and genotyping of mice bearing a −43 deletion in the Csnk1d gene. The region of the zoomed m6A peak highlighted in red corresponds to the peak summit identified by MACS2 peak calling analysis. (B) Quantification of Ck1δ mRNA m6A levels (Left) and of Ck1δ1 and Ck1δ2 mRNA expression in the liver by qPCR (Right). Analyzed by two-way ANOVA (wild type vs. −43, P < 0.0001, n = 3) followed by Bonferroni post hoc, ***P < 0.001. (C, Left) Representative immunoblotting for CK1δ and loading control RPLP0 with liver samples. Mean ± SEM quantification of CK1δ band intensity analyzed by Student t test is shown below; n = 3, **P < 0.01. (C, Right) Representative immunoblotting for CK1δ and loading control RPLP0 with n = 2 brain samples. (Bottom) Mean ± SEM quantification of data aggregated from three independent experiments for a total of n = 6 animals, analyzed by Student t test; *P < 0.05. (D) Circadian locomotor activity recording from one representative animal of each genotype is shown. (Right) Period was analyzed by χ2 periodogram; mean period ± SEM was analyzed by Student t test of n = 6 animals.
CK1δ1 and CK1δ2 Are Two Different Kinases.
Data presented above go against expectations based on period elongation observed in liver explants from Ck1δ knock-out mice (31) and on CK1δ overexpression causing a shorter period (26). The overexpression studies, however, did not specify which isoform was used. The existence of two isoforms has been suggested before in a circadian context (32), but has been ignored until now. Surprisingly, since CK1δ1 is considered canonical and has been the de facto focus of past works, CK1δ2 is the major isoform in the liver, where both transcripts show relatively constant levels across 24 h (Fig. 3A). In the suprachiasmatic nucleus of the hypothalamus (SCN), this relationship is reversed (Fig. 3B; see also Fig. 2C), and the molar ratio of Ck1δ2/Ck1δ1 is tissue-specific (Fig. 3C).
Both CK1δs can interact with PER2 (Fig. 3D), but no evidence of CK1δ1/CK1δ2/PER2 complexes was observed. To test the role of CK1δs in the circadian clock, immortalized MEFs prepared from PER2::LUCIFERASE knock-in reporter mice (33) were stably transfected with CK1δ1 or CK1δ2 expression vectors. Overexpression of CK1δ1 led to a shorter period (Fig. 3E) compared with colonies stably transfected with the empty vector. In contrast, CK1δ2 led to an ∼1-h period elongation. While both isoforms could phosphorylate PER2 (Fig. 3F), the intensity of the phosphorylated bands was higher with CK1δ2 and CK1δ1 + CK1δ2, suggesting higher kinase activity for CK1δ2 and/or faster PER2 degradation with CK1δ1. A picture emerges in which CK1δ isoforms may not simply be two redundant kinases but may phosphorylate PER2 differently and in cooperation.
Work on CK1δ and -ε have shown that PER2 phosphorylation can either accelerate or decelerate the clock depending on which residue is targeted (15, 34, 35). pS478 in mice leads to faster degradation of PER2 by the ubiquitin ligase β-Trcp (36–38), while pS659, named FASPS because its mutation in humans (S662G) causes FASPS, stabilizes PER2 and elongates period (16, 18, 34, 38–40). S659 and three to four nearby downstream residues are phosphorylated in vivo, but work in vitro using CK1ε or CK1δ has shown that CK1 needs a priming phosphorylation at S659 by an unknown “priming” kinase before phosphorylating downstream residues in the cluster, which is necessary for PER2 stabilization. In vitro experiments concluding that CK1δ could not phosphorylate S659, however, used a truncated CK1δ containing only the N-terminal kinase domain and thus gave no insights into the respective functions of CK1δ isoforms (41).
CK1δ1 and CK1δ2 Lead to Different PER2 Phosphorylation Patterns.
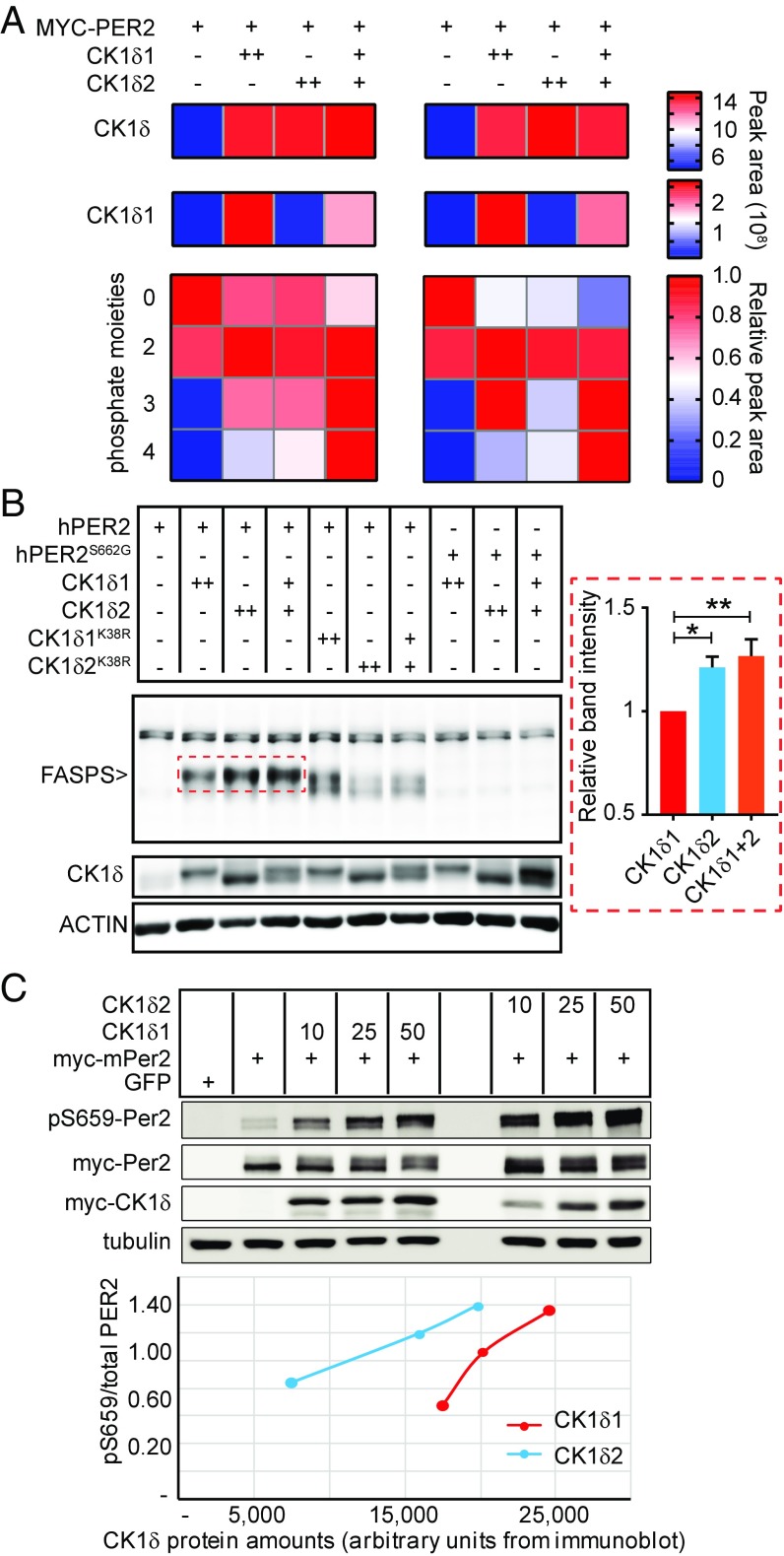
To gain insights into the respective functions of CK1δs, we refined a previously published mathematical model of the regulation of PER2 by branched phosphorylation fates (38) to allow the rate of each phosphorylation step to be independently tuned (SI Appendix, Fig. S3A). As explained in SI Appendix, Supplemental Results, our model predicted that CK1δ2 could be the priming kinase. To test this possibility, we transfected PER2::LUC MEFs with the vector mixes used in Fig. 3E, immunoprecipitated MYC-PER2, and quantified PER2 phosphopeptides after trypsin digestion. Tetra-, tri, di-, and unphosphorylated FASPS peptides were detected (Fig. 4A, SI Appendix, Fig. S4, and Dataset S2). The amount of unphosphorylated FASPS peptide was highest without CK1δs, and lowest with CK1δs together. The diphosphorylated peptide, always detected without pS659, had similar levels with or without CK1δs. The level of triphosphorylated peptides showed some variations between replicates but was always highest when both CK1δ1 and CK1δ2 were present. Remarkably, the tetraphosphorylated FASPS peptide, with confirmed pS659 (SI Appendix, Fig. S4B), showed the most pronounced isoform-dependent differences in abundance among all PER2 phosphopeptides detected (SI Appendix, Fig. S4A and Dataset S2). It was undetected without CK1δ and highest when CK1δs were transfected together, indicating that both isoforms are required for full FASPS phosphorylation.
Fig. 4.
CK1δ1 and CK1δ2 cooperate in phosphorylating PER2. (A) CK1δ isoform-dependent PER2 FASPS phosphopeptides analysis. Data from two technical replicates are provided, side by side. CK1δ peptides were also detected: the first row shows total CK1δ, and the second row corresponds to the CK1δ1-specific VASSGLQSVVHR peptide. The 4 × 4 heatmaps show quantification of the FASPS peptide, sorted by number of phosphate moieties detected. The bar on the Right is the color legend for each corresponding heatmap. (B) Immunoblotting of hPER2 FASPS phosphorylation by CK1δ isoforms. CK1δ and actin immunoblotting are shown as reference and loading control, respectively. Graph on the Right shows mean FASPS band intensity ± SEM of n = 4 independent replicate wild-type hPER2 transfections with CK1δ1, CK1δ2, and 1 + 2. *P < 0.05; **P < 0.01. (C) mPER2 was expressed in HEK293 cells in the presence of increasing amounts of vectors (10, 25, 50 ng) encoding CK1δ splice variants as indicated. Forty-eight hours after transfection, lysates were probed with anti-pS659 mPER2, anti-Myc, and anti-tubulin antibodies. (Bottom) The ratio of pS659/total PER2 as a function of CK1δ amounts.
Next, we confirmed the results above with immunoblotting using an anti-triphospho FASPS antibody (39). Since this antibody was raised against the human FASPS sequence, we used an untagged human PER2 (hPER2) expression vector. The phosphorylated FASPS cluster was not detected when hPER2 was transfected alone (Fig. 4B), indicating that endogenous kinases are not sufficient to noticeably phosphorylate exogenous PER2 (same with Figs. 3F and 4A). When transfected with CK1δ1, a new band appeared corresponding to FASPS phosphorylation. This band increased in intensity with CK1δ2 and with CK1δ1 + CK1δ2. Validating these results, CK1δs mutated to K38R, which has been reported to blunt kinase activity (42), were unable to normally phosphorylate the FASPS cluster (Fig. 4B). In addition, wild-type CK1δs could not phosphorylate a S662G-mutated hPER2. Together, these data demonstrate that CK1δ1 and CK1δ2 cooperate in the phosphorylation of PER2, with CK1δ2 promoting FASPS phosphorylation.
Next, the activity of CK1δs on the phosphorylation of S659 in mouse PER2 was investigated. To validate our results independently, we transfected HEK293 cells with different vectors encoding myc-tagged CK1δ1 or CK1δ2, together with myc-tagged mPER2, and probed cell lysates with a novel antibody specifically raised against pS659 (43). Despite lower exogenous expression of CK1δ2, these data revealed clearly that the activity of CK1δ2 on S659 is remarkably higher than that of CK1δ1 (Fig. 4C). Together, these data demonstrate that the activity of CK1δ2 as a priming kinase is higher than that of CK1δ1, but that both isoforms are required for the full phosphorylation of the FASPS cluster.
Lack of CK1δ2 Shortens the Period.
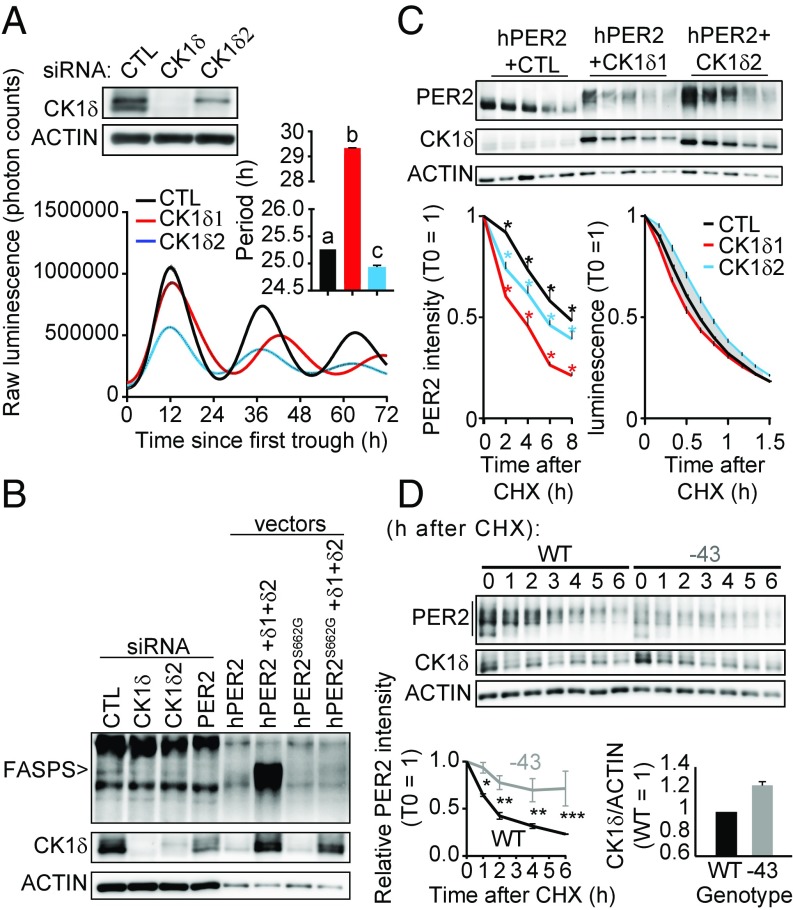
We next sought to silence endogenous Ck1δ2 by siRNA in PER2::LUC MEFs. While Ck1δs knock-down elongated the circadian period in a manner consistent with previous observations (31), silencing only Ck1δ2 significantly shortened the period (Fig. 5A).
Fig. 5.
CK1δ2 stabilizes PER2 and slows the clock. (A) Silencing of Ck1δ2 leads to short period, while silencing of both Ck1δs leads to long period. Knock-down efficiency of Ck1δ and Ck1δ2 was first confirmed by immunoblotting (Top). Main graph shows mean ± SEM of real-time luminometry traces for each group; n = 2. (Right Inset) Mean period ± SEM analyzed by one-way ANOVA, P < 0.0001. a vs. b vs. c, at least P < 0.01 in Bonferroni post hoc. (B) Silencing of endogenous Ck1δ2 in human U-2 OS cells is sufficient to decrease FASPS phosphorylation, as identified by immunoblotting. The identity of the FASPS band was confirmed by transfection of mutated or wild-type expression vectors. (C) The stability of hPER2 phosphorylated by CK1δ2 is higher than that of CK1δ1-phosphorylated hPER2, measured in transiently transfected cells. Representative PER2, CK1δ, and ACTIN immunoblots are shown. (Bottom Left) Mean relative band intensity ± SEM of total PER2, analyzed by two-way ANOVA, all sources of variations at least P < 0.01, n = 3; *at least P < 0.05 in Bonferroni’s post hoc. (Bottom Right) The effect of CK1δs on PER2::LUC stability in the stably transfected cell lines used in Fig. 3E, showing mean relative luminescence ± SEM, analyzed by two-way ANOVA, with all sources of variations P < 0.0001, n = 6. The grayed area shows where significance reached P < 0.0001 between cell lines in Bonferroni’s post hoc test. (D) PER2 is more stable in −43/−43 primary adult skin fibroblasts than in wild type, consistent with higher CK1δ expression, represented mainly by CK1δ2 in these cells. The lower ACTIN signal from −43/−43 cells was due to lower cell counts at the time of sampling, i.e., mean ± SD, 3.68 ± 0.15 × 105 for −43/−43 cells versus 4.42 ± 0.27 × 105 for wild type. (Bottom Left) Quantification of the entire PER2 signal at 0, 1, 2, 4, and 6 h after CHX from n = 3 experiments and analyzed by two-way ANOVA followed by Bonferroni post hoc; *P < 0.05; **P < 0.01; ***P < 0.001. (Bottom Right) CK1δ/ACTIN signal ratio averaged across the entire time course shown above, with the relative intensity in wild type for each time point equal to 1.
The effect of Ck1δ2 knock-down on endogenous FASPS phosphorylation and PER2 levels was also measured. Knocking down Ck1δ2 in human U-2 OS cells caused an observable decrease in FASPS, despite CK1δ1 being still present. To ascertain that the band that we detected is truly FASPS-phosphorylated endogenous PER2, as positive controls Ck1δs and Per2 were also silenced, leading, as expected, to a decrease in the FASPS band intensity. In addition, diluted lysate of cells transfected with wild-type or S662G mutant hPER2 expression vector, with or without CK1δ expression vectors, confirmed the identity of the endogenous FASPS band (Fig. 5B).
According to our results so far, the consequences of PER2 phosphorylation by CK1δ1 or CK1δ2 on PER2 stability should be different: more stable when phosphorylated by CK1δ2 than by CK1δ1 due to increased FASPS phosphorylation. We tested this by transfecting hPER2 together with CK1δ1 or CK1δ2 and then treating the cells with the translation inhibitor cycloheximide (CHX). PER2 stability was highest without cotransfected CK1δ (Fig. 5C). When comparing CK1δ1 vs. CK1δ2, not only the intensity of phosphorylated PER2 was higher with CK1δ2, as observed in Fig. 3F, but the stability of total PER2 was higher in the presence of CK1δ2 (Fig. 5C), which explains the former observation. Surprisingly, CK1δ2 did not stabilize transfected hPER2 as expected. Since the large amounts of transfected vectors may affect normal regulation or PER2 proteins, we sought to investigate the stability of PER2::LUC proteins in stably transfected cells described in Fig. 3E. These control, CK1δ1-, and CK1δ2-overexpressing PER2::LUC cell lines were synchronized by a dexamethasone shock, and after ∼30 h (time of the first PER2 peak), were treated with CHX to assess PER2::LUC stability. In agreement with our hypothesis, while CK1δ1 destabilized PER2::LUC, CK1δ2 had a remarkable effect on its stabilization (Fig. 5C).
Finally, we sought to confirm that the long-period phenotype of the −43/−43 mice originates from a more stable PER2 protein due to an increase in CK1δ (2) expression. Primary skin fibroblasts were prepared from adult −43/−43 and wild-type mice, synchronized, and then treated after 30 h with CHX and sampled every hour to measure PER2 stability. Consistent with previous observations, −43/−43 cells had increased CK1δ expression compared with wild type, both wild-type and −43 cells mostly expressing CK1δ2 (Fig. 5D). In agreement with our hypothesis, PER2 was more stable in −43/−43 cells (Fig. 5D).
Discussion
We have shown that two alternative Ck1δ transcripts are regulated by m6A. When the methylation of Ck1δ 3′-UTR is inhibited or prevented by mutation, the expression of CK1δ isoforms increases, affecting the circadian clock. It is likely, given that Dbp, Fbxl3, and Clock transcripts are also methylated, that global inhibition of mRNA m6A methylation would still affect the clock in −43/−43 animals, as the general disruption of mRNA processing caused by the lack of m6A would still occur.
While the lack of CK1δ is embryonic lethal (31), higher expression or increased activity has been linked to cancer progression, migraine, and circadian disruption (24). As we demonstrated here, the expression of CK1δ isoforms is kept to a low level by its methylated 3′-UTR. The significance of these observations goes beyond the circadian field, given the manifold functions of CK1δ. Further investigations should reveal what the respective physiological functions of CK1δ isoforms are, and additional discussion on the role and regulation of CK1Ds can be found in SI Appendix, Supplemental Discussion.
Materials and Methods
Reagents were from Nacalai Tesque unless stated otherwise.
Animals.
All experiments were approved by the animal experimentation committee of Kyoto University. Twelve-week-old C57BL/6 males were maintained at 23 °C ± 1 °C with 50% ± 10% relative humidity, on a 12-h light/12 h dark cycle (lights on 8:00, lights off 20:00), food and water ad libitum. For Fig. 3 A and B, lights were permanently switched off from 8 PM on day 0, and animals were sampled from 8 AM (corresponding to circadian time 0) on day 2.
Mice with a −43-bp deletion in the 3′-UTR of Csnk1d by CRISPR-Cas9 were generated with the same method used for similar genetic editing (44) as briefly explained in SI Appendix, Supplemental Methods. Locomotor activity recording and analysis were obtained from animals housed individually in light-tight, ventilated cages with ad libitum access to food and water. The animals were entrained on a 12-h light (∼200 lx fluorescent light)/12-h dark cycle for at least 1 wk to synchronize the circadian clock to the ambient light–dark cycle, before turning lights off for the remainder of the experiment. Locomotor activity was recorded in 5-min bins with passive infrared sensor (FA-05 F5B; Omron) and analyzed with Clocklab software (Actimetrics) on MatLab (Mathworks). Free-running period was quantified by χ2 periodogram over the entire recording in constant darkness.
In Vitro Experiments.
All in vitro experiments are described in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank the nonprofit organization Biotechnology Research and Development for technical assistance. This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency Grant CREST/JPMJCR14W3 (to H.O.); by the Kato Memorial Bioscience Foundation (J.-M.F.); by the Senri Life Science Foundation (J.-M.F.); by the Mochida Memorial Foundation for Medical and Pharmaceutical Research (J.-M.F.); by the Kobayashi International Scholarship Foundation (H.O.); by a Grant-in-Aid for Scientific Research on Innovative Areas (26116713) (to J.-M.F.); by a Grant-in-Aid for Young Scientists (26870283) (to J.-M.F.); by a Grant-in-Aid from the Japan Society for the Promotion of Science Research Fellows (15F15081) (to H.-Y.C.); and by a Grant-in-Aid for Scientific Research A (15H01843) (to H.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721371115/-/DCSupplemental.
References
- 1.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 2.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 5.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 6.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci USA. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci USA. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 16.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 18.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 19.Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 20.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 23.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knippschild U, et al. The CK1 family: Contribution to cellular stress response and its role in carcinogenesis. Front Oncol. 2014;4:96. doi: 10.3389/fonc.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vizirianakis IS, Tsiftsoglou AS. Induction of murine erythroleukemia cell differentiation is associated with methylation and differential stability of poly(A)+ RNA transcripts. Biochim Biophys Acta. 1996;1312:8–20. doi: 10.1016/0167-4889(96)00012-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 28.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slobodin B, et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169:326–337.e12. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Etchegaray JP, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishida Y, et al. Constitutive expression and delayed light response of casein kinase Iepsilon and Idelta mRNAs in the mouse suprachiasmatic nucleus. J Neurosci Res. 2001;64:612–616. doi: 10.1002/jnr.1114. [DOI] [PubMed] [Google Scholar]
- 33.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloss B, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 36.Isojima Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, Kim JK, Eng GW, Forger DB, Virshup DM. A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol Cell. 2015;60:77–88. doi: 10.1016/j.molcel.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Shanware NP, et al. Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J Biol Chem. 2011;286:12766–12774. doi: 10.1074/jbc.M111.224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanselow K, et al. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlosser A, Vanselow JT, Kramer A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal Chem. 2005;77:5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- 42.Tan ST, Xue HW. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014;9:1692–1702. doi: 10.1016/j.celrep.2014.10.047. [DOI] [PubMed] [Google Scholar]
- 43.Narasimamurthy R, et al. CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci USA. 2018;115:5986–5991. doi: 10.1073/pnas.1721076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oji A, et al. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci Rep. 2016;6:31666. doi: 10.1038/srep31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.