GENETICS Correction for “CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles,” by Brenda R. Lemos, Adam C. Kaplan, Ji Eun Bae, Alexander E. Ferrazzoli, James Kuo, Ranjith P. Anand, David P. Waterman, and James E. Haber, which was first published February 13, 2018; 10.1073/pnas.1716855115 (Proc Natl Acad Sci USA 115:E2040–E2047).
The authors note that Fig. 1 and Fig. 2 appeared incorrectly. The corrected figures and their legends appear below.
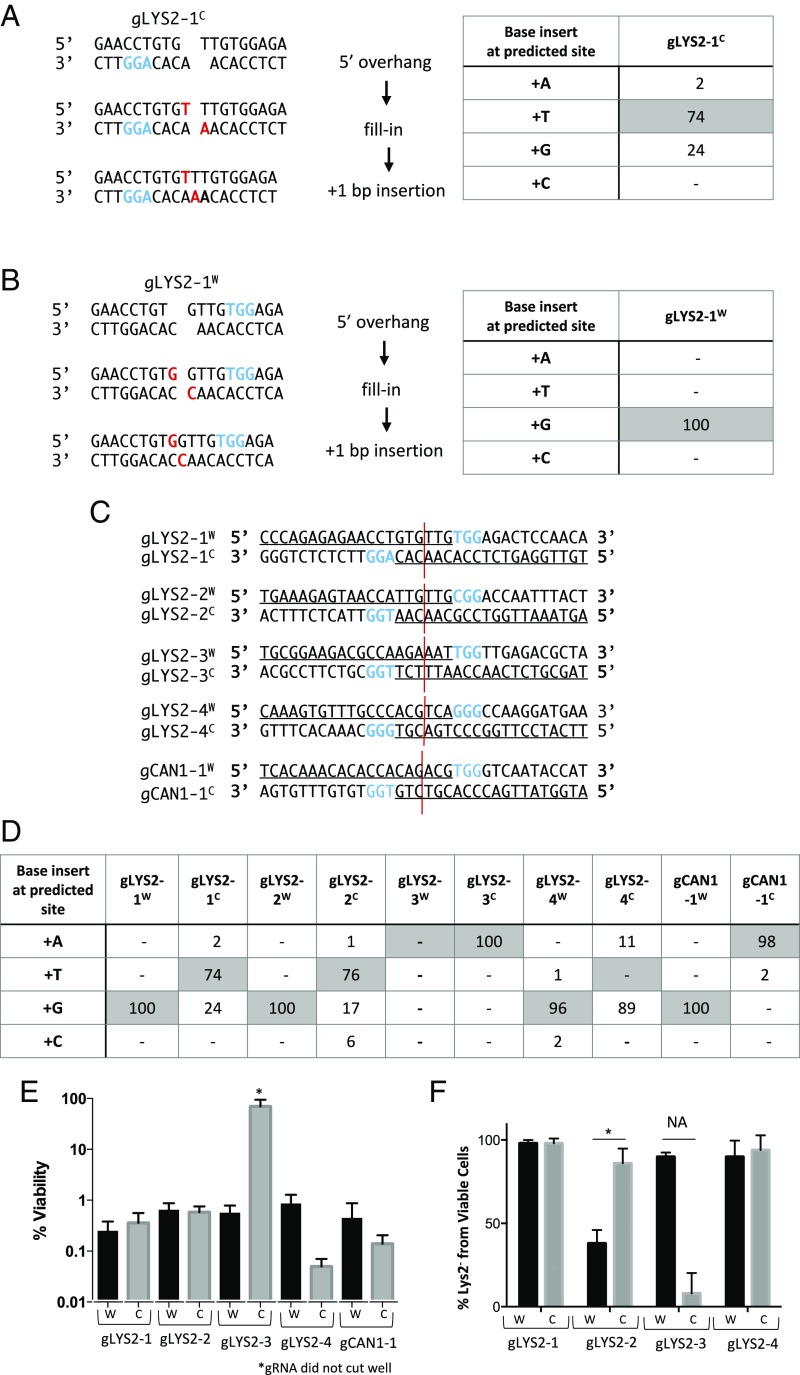
Fig. 1.
Templated insertions and nonrandom deletions at CRISPR-Cas9 DSBs. (A) Templated 1-nt insertions following Cas9-induced DSB, resulting in a 5′ 1-nt overhang-mediated gLYS2-1C. The table shows the percentage of +1 insertions, with shaded values representing expected templated insertions. (B) Templated 1-nt insertions following Cas9 induced DSB resulting in a 5′ 1-nt overhang mediated gLYS2-1W. The table shows the percentage of +1 insertions, with shaded values representing expected templated insertion. (C) Five pairs of gRNAs targeting either the LYS2 or CAN1 locus were designed to cleave five different genomic sites. Paired gRNAs were designed to create a DSB in the same DNA location. (D) Percentage of +1 insertions following cleavage from 10 different gRNAs targeting different iPAMs. Shaded values represent the predicted templated insertions for each guide. (E) Percent viability from gRNAs targeting the LYS2 or CAN1 locus. W and C reflect PAM location on Watson and Crick strands as shown. (F) Percentage of Lys2− for each gRNA targeting LYS2 from viable cells. A statistically significant difference in the percentage of Lys2− cells was found for gRNA pair 2 (P = 0.002). NA denotes statistics not applicable because gRNA did not cut well.
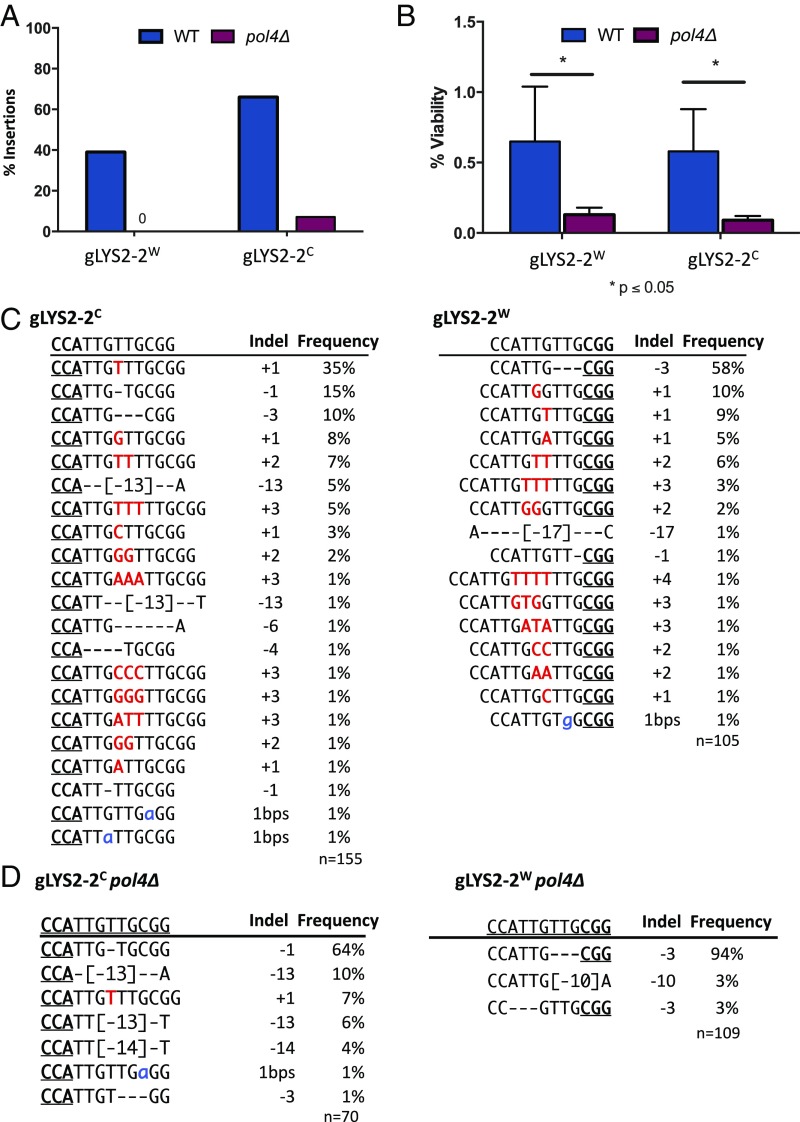
Fig. 2.
Pol4 is required for templated and nontemplated insertions. (A) Insertions during repair of gLYS2-2W and gLYS2-2C DSBs are mostly lost in the absence of POL4. (B) Effect on NHEJ survival in the absence of POL4. (C) Indel profile from colonies recovered following DSB from gLYS2-2W and gLYS2-2C. (D) Indel profile in the absence of pol4Δ for gLYS2-2W and gLYS2-2C.
The authors also note that, in SI Appendix, Supplemental Table 2, data for the iPAM sequences in the CAN1 gene were omitted. The SI has been corrected online. These errors do not affect the conclusions of the article.