Abstract
Experimental data offers empowering constraints for structure prediction. These constraints can be used to filter equivalently scored models or more powerfully within optimization functions toward prediction. In CASP12, Small Angle X-ray Scattering (SAXS) and Cross-Linking Mass Spectrometry (CLMS) data, measured on an exemplary set of novel fold targets, were provided to the CASP community of protein structure predictors. As HT, solution-based techniques, SAXS and CLMS can efficiently measure states of the full-length sequence in its native solution conformation and assembly. However, this experimental data did not substantially improve prediction accuracy judged by fits to crystallographic models. One issue, beyond intrinsic limitations of the algorithms, was a disconnect between crystal structures and solution-based measurements. Our analyses show that many targets had substantial percentages of disordered regions (up to 40%) or were multimeric or both. Thus, solution measurements of flexibility and assembly support variations that may confound prediction algorithms trained on crystallographic data and expecting globular fully-folded monomeric proteins. Here, we consider the CLMS and SAXS data collected, the information in these solution measurements, and the challenges in incorporating them into computational prediction. As improvement opportunities were only partly realized in CASP12, we provide guidance on how data from the full-length biological unit and the solution state can better aid prediction of the folded monomer or subunit. We furthermore describe strategic integrations of solution measurements with computational prediction programs with the aim of substantially improving foundational knowledge and the accuracy of computational algorithms for biologically-relevant structure predictions for proteins in solution.
Keywords: flexibility, unstructured regions, disorder, unfolded regions, assembly, crystallography, SAXS, SAS, experimental restraints, modeling, protein folding, solution scattering, prediction accuracy, combined methods, solution structure
1 INTRODUCTION
Many advances have been made with protein structure prediction algorithms, which are now capable of predicting certain atomic structures to angstrom accuracy.1,2 While these algorithms work effectively for target proteins that have close homologs, predicting structures without a template or fold knowledge remains particularly challenging. These challenges are an especially great problem for flexible regions and for multi-domain proteins and complexes, essentially limiting the effective protein size that can be predicted. The state-of-the-art for protein structure prediction algorithms is assessed every two years by the Critical Assessment of Protein Structure Prediction (CASP) experiments [REFERENCE to the introductory article, Moult et al., this issue]. Predictors are given an amino acid (AA) sequence of an unreleased structure and are given two weeks to predict its structure. Structure predictions are compared to the unreleased structure. One of the main goals of CASP, when assessing the capabilities of structure prediction algorithms, is to drive future innovation and accuracy. More recently CASP began collaborating with CAPRI [REFERENCE to the CAPRI article, Lensink et al., this issue], to address difficulties in predicting interfaces, multimeric structure, and formation.
Barriers to accurate atomic structure prediction include inaccurate quality scoring functions of atomic models, limited sampling, incomplete knowledge of the energy folding landscape, protein flexibility, and inability to unambiguously identify domains or subunits that fold as independent units within an oligomeric complex. Logically, these barriers can be reduced by incorporating experimental data to either validate a predicted model or provide a driving energetic factor in the algorithm. Towards this end, the two experimental high-throughput (HT) techniques that provided solution data to predictors in CASP12 were cross-linking/mass spectrometry (CLMS or XL-MS) and Small Angle X-ray Scattering (SAXS).
CLMS and SAXS are ideal experimental techniques to complement structure prediction algorithms. Both methods provide HT structural information, short time to obtain data, low costs, and no need for crystallization, large quantities of protein, or special labeling.
CLMS has excelled as a low-resolution structural biology technology that can be readily combined with other structural biology techniques,3,4 including X-ray crystallography, cryo-electron microscopy and nuclear magnetic resonance spectroscopy, as well as providing valuable information by itself.5–7 CLMS relies on the capture of proximity via newly introduced covalent bonds, both within and between proteins. The application of photoactivatable cross-linkers, with reduced reaction specificity, leads to high-density data.8,9 This high-density CLMS data can then be combined with computational biology, to produce accurate protein structure models (up to 2.5 Å, RMSD to X-ray crystal structure).9 CLMS has the important advantage of small sample requirements (nano to micromolar).
In CASP11, the Rappsilber group provided the first experimental data in CASP history, in the form of high-density CLMS data.10–12 CLMS data was released to prediction groups worldwide under the assisted structure prediction category, for four CASP targets (Tx781, Tx808, Tx767 and Tx812). The CLMS data released consisted of lists of identified linked residue pairs, representing distance constraints, with an associated upper boundary and identification confidence (based on false discovery rate (FDR)13) estimation. Crucially, CLMS data were acquired without any knowledge of protein structure; they thus functioned as a true blind test of the potential for CLMS data to be used as a hybrid method in CASP. Yet, the improvements to the model quality were slight. The CASP11 experience revealed key experimental challenges to overcome in combining CLMS with structure prediction, namely full coverage along the protein sequence, and coverage within beta-sheet regions. 10,11
Whereas CLMS provides a limited set of assigned distances, SAXS uniquely provides a histogram of all the electron pair distances in the solution ensemble. As such, SAXS has great potential as an experimental restraint for protein modeling by providing metrics based upon all electron pairs of a protein that can furthermore be directly calculated from and compared with an atomic model. SAXS is particularly powerful when combined with crystallography structures or predicted atomic based models.14,15 Thus, although an atomic model cannot currently be uniquely determined from SAXS data alone, SAXS results can identify and rank atomic models that generate similar scattering curves.16–18
CASP12 included both CLMS and SAXS data. The Rappsilber group committed to provide CLMS data for CASP12,10 which culminated in the release of CLMS data on three CASP12 targets (Tx892, Tx894 and Tx895). SAXS data was collected for the first time for CASP12. In total, SAXS measurements were provided on 10 CASP targets. Two of the structures have been published19,20 and crystal structures are available for all ten from the CASP Protein Structure Prediction web site (http://predictioncenter.org). Analysis of the prediction models based on CLMS and SAXS are described in the accompanying paper by Tamò and colleagues [REFERENCE to the Assessment of data assisted modeling paper, Tamò et al., this issue]. Here we report details of the data provided for CASP12. For the SAXS, the SAXS profiles were consistent with the crystallographic results, albeit when the crystal structure is considered as part of the full biological unit. However, there were challenges in effectively incorporating SAXS into prediction models. SAXS is measured in solution, and many targets multimerized or contained substantial unstructured regions, or both: findings that are frequently ambiguous in crystallography. Based upon the CLMS and SAXS data integrated with CASP12 results, we discuss proposed changes to data collection and processing of SAXS data for future CASPS along with suggested interpretation tools and strategies aimed at enabling more accurate computational approaches and prediction.
2 METHODS
2.1 CLMS Data Collection
Proteins (UDP-glucose-glycoprotein N-term (UGGT, Tx892), CDI204-E1 (Tx894) and CDI204-E2 (Tx895)) were received from the respective crystallographers (Table 1). UGGT was received at 8.18 mg/mL concentration in 20 mM HEPES, 150 mM NaCl, 100 µM EDTA, pH 7.4, and was diluted to 1 mg/mL concentration prior to cross-linking using 20 mM HEPES, 20 mM NaCl, 5 mM MgCl2, pH 7.8. CDI204-E1 and CDI204-E2 were supplied as one sample, at 24 mg/mL concentration in 20 mM Tris, 150 mM NaCl, 2 mM DTT, pH 8.0. Buffer was exchanged to 20 mM HEPES, 20 mM NaCl, 5 mM MgCl2 and proteins diluted to 1 mg/mL.
TABLE 1.
Information on CLMS and SAXS Protein Targets.
| Labels | Target Name | Conc. (mg/mL) |
PDB | Buffer Condition |
|---|---|---|---|---|
| YfgC | Ts0942 | 5.8 | - | 50 mM MES pH 6.0, 200 mM NaCl |
| Bd0412 | Ts0899 | 6.2 | - | 50 mM MES pH 7.5, 200 mM NaCl, 1 mM MgCl2 |
| Bd1483 | Ts0905 | 4.8 | - | 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM MgCl2 |
| Bd3099 | Ts0901 | 5.0 | - | 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM MgCl2 |
| Bd0553 | Ts0947 | 6.4 | - | 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM MgCl2 |
| Bd0886 | Ts0896 | 3.1 | - | 50 mM tri-sodium citrate, pH 6.0 |
| Bd3702 | Ts0941 | 6.1 | 50 mM tri-sodium citrate, pH 6.0 | |
| SnLH3 | Ts0909 | 6.1 | 5g5n | 10 mM Tris-HCl pH 8.5, 50 mM NaCl |
| CDI204 | Ts0894 /Ts0895 | 25 | 5hkq | 20mM Tris pH8.0, 150 mM NaCl 2mM DTT |
| MlaD | Ts0866 | 5 | 5uw2 | 20 mM Tris pH 8.0, 150 mM NaCl |
| FliD | Ts0886 | 9.6 | 5fhy | 20mM CAPS, 80mM NaCl, pH 11 |
Ts0886 was provided by Sandra Postel, Ts0909 by Mark J. van Raaij, Ts0894/895 by Karolina Michalska, Ts0866 by Damian C. Ekiert, and Ts0899, Ts0905, Ts0901, Ts0947, Ts0896, and Ts0941 by Andrew Lovering.
Proteins were cross-linked separately (Tx894 and Tx895 were cross-linked as one sample) using eight different cross-linker to protein ratios (0.13:1, 0.19:1, 0.25:1, 0.38:1, 0.5:1, 0.75:1, 1:1 and 1.5:1 (w/w)). Cross-linking was carried out in two-stages: firstly sulfo-SDA (Thermo Scientific Pierce, Rockford, IL), dissolved in cross-linking buffer (25 µL, 20 mM HEPES-OH, 20 mM NaCl, 5 mM MgCl2, pH 7.8), was added to target protein (25 µg, 1 µg/µL) and left to react in the dark for 50 minutes at room temperature. This was followed by photoactivation of the diazirine group using UV irradiation, at 365 nm, from a UVP CL-1000 UV Crosslinker (UVP Inc.). Samples were spread onto the inside of Eppendorf tube lids by pipetting (covering the entire surface of the inner lid), placed on ice at a distance of 5 cm from the tubes and irradiated for 20 minutes. Following cross-linking, 10 µg equivalent of each reaction condition (for either Tx892 or Tx894/Tx895) was combined and mixed (80 µg), and loaded (10 µg per lane) onto NuPAGE 4–12% Bis-Tris gels for electrophoresis. A second gel was loaded with individual reaction conditions to be run separately (10 µg per lane). Proteins were separated using constant voltage at 190 V, using an XCell SureLock™ Mini-Cell Electrophoresis system (Thermo Fisher Scientific), with MES SDS running buffer. Proteins were stained using Imperial Protein Stain (Coomassie blue stain) (Thermo Scientific). and the band corresponding to the monomer was digested using trypsin via standard protocols.21 Resulting peptides were desalted using StageTips.22,23
Samples were analyzed using an HPLC (UltiMate 3500RS Nano LC system, Thermo Fisher Scientific, San Jose, CA) coupled to a tribrid mass spectrometer (Orbitrap Fusion Lumos Tribrid Mass Spectrometer, fitted with an EASY-Spray Source, Thermo Fisher Scientific, San Jose, CA). Peptides were loaded onto a 500 mm C-18 EASY-Spray LC column (Thermo Fisher Scientific, San Jose, CA), operating at 50 °C. Mobile phase A consisted of water and 0.1% formic acid, mobile phase B of 80% acetonitrile, 0.1% formic acid and 19.9% water. Peptides were loaded at a flow-rate of 0.3 µL/min and eluted at 0.2 µL/min, using a linear gradient starting at 2% mobile phase B and increasing over 109 min to 40%, followed by a linear increase over 11 min, from 40% to 95% mobile phase B.
MS data were acquired in the Orbitrap at resolution 120,000, using the top-speed, data-dependent mode. Precursor automatic gain control (pAGC) target value was set to 4 × 105, maximum injection time at 50 ms, precursor priority was set to highest charge state then most intense, charge range was from 3–8, scan range between 300–1700 m/z, dynamic exclusion was set at 60 s duration and mass tolerance was set at 10 ppm. Precursor ion isolation was carried out with the quadrupole and an m/z window of 1.6 Th. Selected precursor ions were fragmented using higher-energy collisional dissociation (HCD), using a normalized collision energy of 30%. Fragmentation spectra were then recorded in the Orbitrap at resolution 15,000, AGC target set to 5 × 104 and maximum injection time of 60 ms.
2.2 CLMS Data analysis
Raw files were processed with MaxQuant (v. 1.5.2.8)24 to generate peak files (APL format), with “Top MS/MS peaks per 100 Da” set to 100. Peak files were searched against FASTA sequence files using Xi25 (https://github.com/Rappsilber-Laboratory/XiSearch), with the following settings: MS accuracy, 6 ppm; MS/MS accuracy, 20 ppm; enzyme, trypsin; maximum missed cleavages, 4; maximum number of modifications, 3; fixed modifications, none; variable modifications, carbamidomethylation (Cys), oxidation (Met) and loop-links (“SDA-loop”, mass modification: 82.041865); Sulfo-SDA cross-linking reactions were assumed to conjoin the side chains of lysine, serine, threonine, tyrosine or the protein N-terminus at one end, with any amino acid at the other end. False discovery rates (FDR) were estimated following a modified target-decoy search strategy.13,21,26
2.3 SAXS Sample Preparation and Data Collection
Samples for collection generally arrived frozen with the following concentrations and conditions (Table 1). Just prior to data collection, samples were prepared in 96-well plates, where 20 µL of the consecutive protein concentrations were bracketed with two 20 µL protein-free buffer samples. The protein concentrations used for data collection consisted of the original protein concentration, a 1:2 dilution, and a 1:4 dilution.
SAXS data were collected at the SIBYLS beamline (12.3.1) at the Advanced Light Source, part of the Lawrence Berkeley National Laboratory.27 Samples are transferred from a 96-well plate at 10 °C to the sample cuvette, where they are exposed to an X-ray beam for a total of 10 seconds28. By collecting data on three protein concentrations, we were able to correct for concentration-dependent behavior. Scattering images are collected by a PILATUS 2M detector every 0.3 seconds, for a total of 33 sample images. The sample-to-detector distance is 1.5 m. The wavelength of the beam was 1 Å, and the flux was 1013 photons per second. For each sample collected, two protein-free buffer samples were also collected to reduce error in subtraction. Each collected image was circularly integrated and normalized for beam intensity to generate a one-dimensional scattering profile by beamline specific software.
2.4 SAXS Data Analysis
The one-dimensional scattering profile of each protein sample were buffer-subtracted by each of the two corresponding buffers, producing two sets of buffer subtracted sample profiles. Profiles were examined for radiation damage. Scattering profiles over the ten-second exposure were sequentially averaged together until radiation damage affects were seen to begin changing the scattering curve. Averaging was performed with web-based software (sibyls.als.lbl.gov/ran).
Parameters such as radius of gyration (RG), the Radius of the cross-section (RXC), and the volume of correlation (Vc) were calculated using Scatter.29–31 The P(r), Rg2, and DMax were calculated using GNOM.32 Molecular envelope calculations were performed using GASBOR.33
2.5 Native Gels and re-collection of SAXS data
Added efforts were taken when samples did not meet basic quality control conditions. Samples suspected of poor buffer subtraction were re-dialyzed and re-collected. Samples with RG values of greater than 70Å were spun through a 1 MDa centrifugation filter and also re-collected. All samples were run on a native gel though interpretation was challenging as seven remained in the loading well or ran as long streaks. Ts0899 ran as a single band corresponding to near monomeric size. Ts0909 ran as a single band with trimeric molecular weight. Ts0901 ran as a single band with molecular weight 4 times larger than a monomer.
2.6 Predictor SAXS Data Packages
From data collection to analysis, data for ten SAXS samples were passed to CASP in under a 3-week period. All data are available at the CASP 12 web address (predictioncenter.org) for download in the “Targets” tab under “Assisted structure prediction”. Eight of the targets, have a discrepancy between the SAXS sample and the sequence provided to predictors. Ts0899, Ts0896, Ts0901, Ts0941, Ts0942, Ts0947 were shorter by 20, 29, 20 105, 27, and 36 residues at the N-terminus, respectively. Ts0866 is the same as the deposited crystal structure and was longer by 18 residues. Ts0909 was similar in length and termini to the predictor sequence, but there was an error in an internal sequence which is now fixed in the deposited crystal structure. All models shown are given for the predictor-provided sequence.
3 RESULTS
3.1 Cross-linking and Mass Spectrometry
Data was provided for 3 targets in CASP12: Tx892 (UGGT, UDP-glucose-glycoprotein N-term), Tx894 (CDI204-E1) and Tx895 (CDI204-E2) (Figure 1). Importantly, Tx892 is only a 193-residue section (residues 26-220) of the protein that was analyzed by CLMS, UGGT (1509 residues). Similarly, Tx894 and Tx895 were analyzed as a protein complex, yet used as individual targets in CASP12.
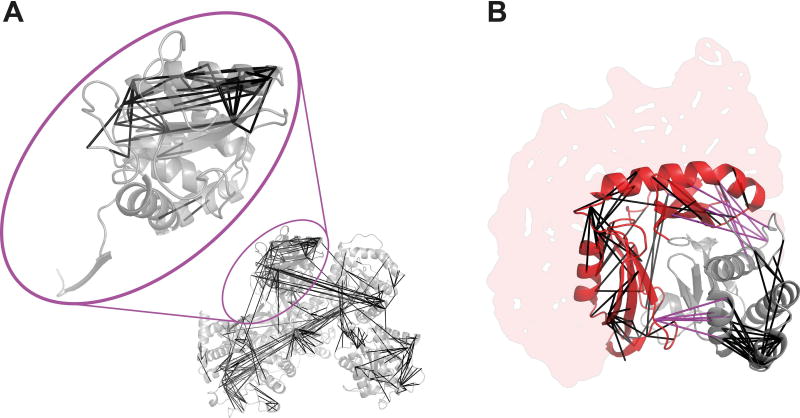
Figure 1.
Unique cross-linked residue pairs identified at 5% FDR. (A) Cross-links plotted in UGGT. Tx892 (within highlighted oval) is a 193-residue section of UGGT. (B) Cross-links plotted in PDB|5HKQ, covering Tx894 (red) and Tx895 (grey). The structure covers residues 181-323 only of Tx894, and missing structure is represented by the red shaded area. Links between Tx894 and Tx895 are shown in pink.
Data was acquired from 26 × 160 min LC-MS runs (2.9 days) in the case of UGGT (Tx892), and 17 × 160 min LC-MS runs (1.9 days) in the case of the Tx894/Tx895 complex. Over the whole structure of UGGT, 433/541/982 unique residue pairs were identified at respectively 5%/10%/20% False Discovery Rate (FDR) (0.29–0.65 links per residue). Of these, 56/68/100 unique residue pairs fell into the region of Tx892 at respectively 5%/10%/20% FDR (0.29–0.52 links per residue). Links that fell between Tx892 and the rest of UGGT were not used in CASP12, despite likely constraining the structure modeling. Nor was the fact that Tx892 was actually not surrounded by water, but in parts by the rest of UGGT (Figure 1A). For the complex of Tx894 and Tx895, 232/424/621 unique residue pairs were identified at respectively 5%/10%/20% FDR. Within this list, respectively 195/370/556 unique residue pairs were within Tx894 (respectively 50/90/138 fell within the available structure) and respectively 29/42/52 unique residue pairs were within Tx895.
The CLMS experimental data did fit well to the crystal structures for all FDR values, within the target regions (94% – 100%, see Table 2). The high agreement reduces to more expectable readings when considering the whole of UGGT (70–90%). Filtering to a spatially confined region effectively removes the false identifications, since these were more likely to occur across the entire protein, and therefore extend past the upper distance boundary of 25 A. Note that only 25% of cross-links of the protein containing Tx894 can be fitted to the crystal structure of the Tx894/Tx895 complex, as only a fragment of the whole protein is resolved in the available structure (Figure 1B) as also shown in the SAXS data.
TABLE 2.
Percentages of unique residue pairs identified by CLMS that fit the structures available within 25 Å (unique residue pair numbers shown in brackets).
| Target protein | 5% FDR | 10% FDR | 20% FDR |
|---|---|---|---|
| Tx892 (193 AA) | 98% (55/56) | 97% (66/68) | 95% (95/100) |
| Tx894 (143 AA) | 96% (48/50) | 96% (86/90) | 94% (129/138) |
| Tx895 (120 AA) | 100% (29/29) | 100% (42/42) | 100% (52/52) |
Ts894/895 were provided by by Karolina Michalska and Tx892 by Pietro Roversi.
3.2 CASP SAXS targets
Demonstrating the throughput necessary for CASP and protein predication in general, SAXS data were collected at the SIBYLS beamline 12.3.1 in the Advanced Light Source Synchrotron and analyzed on all targets sent.27,29,34 CASP target providers (Postel, Ekiert, Lovering, van Rajj, and Michalska) sent 11 samples in total. Out of the 11 targets sent, all but one were analyzed and reported to CASP predictors with a set of scalar values, the primary SAXS curves (Intensity vs momentum transfer (q)), the pair distribution function P(r), and three dimensional shapes.
For CASP12, data collection and analysis were conducted over 2 weeks between sample arrival and CASP timelines for prediction. Despite the high-throughput of SAXS measurements per se, the predefined short time frame presented logistical challenges as it compromised the ability to completely perform and test all controls and then track down any sources of inconsistencies. Moreover, communication between sample providers and the beamline was minimized so as to avoid compromising the CASP experiment by preventing information known from crystal structures to be passed down through the SAXS analysis.
Samples were of high quality as only one of the 11 were significantly aggregated after thawing the frozen samples. However, the samples sent were prepared predominantly for crystallization with only a few prepared for the purpose of SAXS. Most of the samples went through one or more potentially damaging freeze thaw cycles. For optimal SAXS data collection for protein structure predictions, buffer must be properly subtracted and the protein should be multimerically and conformationally homogeneous. Scattering curves were examined for buffer blank mismatches and when apparent, dialysis was performed to exchange buffer. To assess homogeneity, native gels were conducted with mixed results. Several proteins had multiple bands while some ran counter to the electric field confounding native gel analysis with the available apparatus. As each protein had unique buffer conditions, sample quantity was limited and the CASP time requirements were short, on-site re-purification with size exclusion chromatography (SEC) was not attempted. (Suggested changes in sample data collection for a future CASP are discussed later.) Several scattering curves required trimming of the low q data to remove contributions from a small population of aggregates contaminating the sample. Nevertheless the ten data sets impressively passed this first quality control and had linear Guinier regions with 20 or more measured points up to and mostly smaller than q = 0.02Å−1, indicating the robustness of the HT SAXS method and that the samples were of sufficient quality for further processing. With these challenges, data was provided to predictors with our best efforts on data quality. The reported SAXS scalars are defined in Table 3 and tabulated in Table 4 for each construct.
TABLE 3.
SAXS Experimentally Extracted Scalar Data types
| Experimentally Extracted Scalar Data types | Symbol | Units |
|---|---|---|
| Radius of Gyration from Guinier + Error | Rg | Angstroms (Å) |
| Degree of Flexibility | PD | Range (2 – 4) (Unfolded – Globular) |
| Experimental Mass | MSAXS | Daltons (Da) |
| Maximum Dimension + Error | DMax | Angstroms (Å) |
| Radius of Cross-Section | RXC | Angstroms (Å) |
| Experimental Volume | VSAXS | Cubic Angstroms (Å3) |
| Radius of Gyration From P(r) Function | Rg2 | Angstroms (Å) |
TABLE 4.
List of SAXS scalars and % of residues ordered in crystals for each SAXS target
| Target | Rg (Å) |
PD | Mass SAXS (kDa) |
Mass seq (kDa) |
Dmax (Å) |
Rxc (Å) |
Vol. (103 Å3) |
Rg2 (Å) |
Multimeric | %crystal /SAXS |
|---|---|---|---|---|---|---|---|---|---|---|
| Ts0866 | 42 | 4 | 142 | 13 | 125 | 37.6 | 390 | 43 | Large Heteromer | 73% |
| Ts0886 | 61 | 3.1 | 54 | 39 | 212 | 25 | 170 | 58 | Dimer | 66% |
| Ts0894/Ts0895 | 31 | 4 | 30 | 51 | 118 | 15 | 71 | 32 | ||
| Ts0896 | 27 | 2.9 | 24 | 53.5 | 114 | 19 | 89 | 53 | Monomer | 92% |
| Ts0899 | 38 | 4 | 57 | 47 | 170 | 18.6 | 116 | 42 | Monomer | 82% |
| Ts0901 | 47 | 4 | 216 | 36 | 216 | 18 | 150 | 53 | Filament | 89% |
| Ts0905 | Aggregated | |||||||||
| Ts0909 | 29 | 4 | 85 | 37 | 80 | 28 | 134 | 28 | Trimer | 96% |
| Ts0941 | 42 | 4 | 45 | 51 | 200 | 18 | 110 | 49 | Monomer | 73% |
| Ts0942 | 33 | 4 | 65 | 54 | 101 | 23 | 155 | 33 | Monomer | 79% |
| Ts0947 | 24 | 3.7 | 18 | 25 | 78 | 16.3 | 57 | 27 | Monomer | 80% |
The %crystal/SAXS is the number of ordered residues in the crystal over the number of residues provided to predictors in the SAXS category.
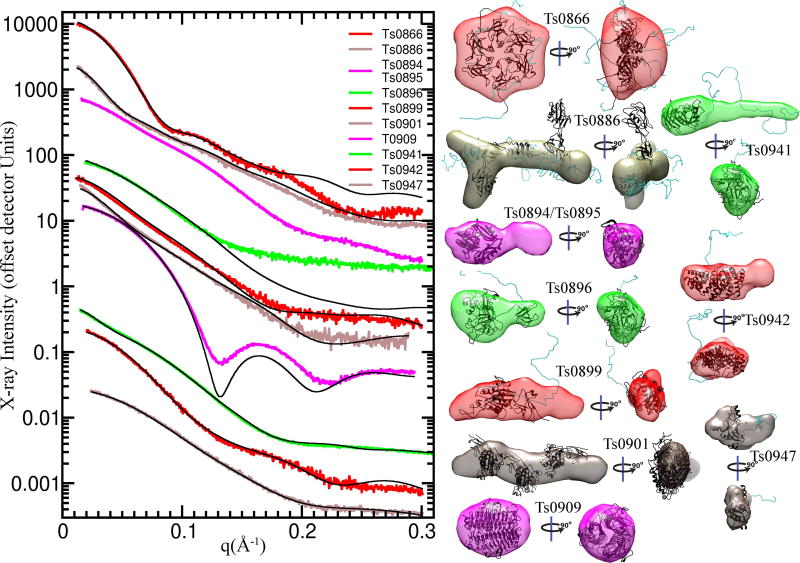
Scattering curves or profiles (I vs q), shape predictions based on SAXS data, and the P(r) plots for the ten targets show the diversity of target sizes and shapes and the quality of the measured profiles (Figures 2 and 3). The curves for Ts0866 and Ts0909 show well featured curves predicting a spherical globular structure, while on the other side of the spectrum, those for Ts0941 and Ts0901 have few features, indicating an elongated and possibly flexible structure. In the P(r) plots, Ts0886, Ts0899, Ts0901, and Ts0941 have “tails” at long distances (r), consistent with elongated and/or flexible structures. The shoulders in the P(r) plots for Ts0894, Ts0896, Ts0899, and Ts0901 suggested multiple domains.
Figure 2. Scattering curves and shape envelopes of CASP SAXS targets.
(Left Panel) Experimental scattering curves (colored) are shown for ten CASP SAXS targets and overlaid with the predicted scattering (black) from an ensemble of atomic models, found to best match the experimental data. SAXS curves can be scaled without losing information content, so the SAXS curves have been offset for visual clarity. The atomic model(s) are full-length models, based on the crystal structure or when appropriate, multimeric models based on the crystallographic lattice. (Right panel) Ab initio shape reconstructions based on the SAXS data and overlaid with a single representative atomic model.
Figure 3. Panels of the real space P(r) distributions of the CASP SAXS targets.
The P(r) distribution calculated from the experimental SAXS data (colored) are shown, overlaid with those from the ensemble of atomic models, described in Figure 1.
Extraction of global parameters (scalars) from X-ray scattering provide insights into structure and assembly. The Radius of Gyration (RG) characterization of the first moment of inertia for the samples ranged from 24 to 61Å, indicating that all assemblies were medium to large. The RG was estimated two ways. First through use of the Guinier region in reciprocal space, and second (Rg2) through analysis of the real space P(r) function. All samples had comparable values from both methods, passing this added data quality control.
The Porod-Debye (PD or PE) value provides objective insights into flexibility.31 PD is determined from the rate of decay as a function of q in the mid q range (0.05 < q < 0.2 Å−1) and depends on the volume of the protein. A q−2 dependence indicates and unfolded structures while a q−4 indicates a globular one. The PD is represented as the negative of the exponent, and most proteins were near 4, indicating a high proportion of folded regions. Yet, for Ts0886, Ts0896, and Ts0947 the PD indicated significant flexibility in the entire protein system. The mass of the folded region can be estimated from SAXS (MassSAXS) by defining the PD range and calculating the Volume of correlation (Vc).30 When the MassSAXS is greater than the theoretical mass of the protein sequence, multimerization is indicated. When smaller, proteolysis or partial disorder is suspected.
The radius of cross-section (Rxc) characterizes the second moment of inertia of the protein. When Rxc values are comparable to RG, the protein is globular. When Rxc is significantly smaller, the protein is elongated. Ts0886, Ts0894/Ts0895, Ts0899, Ts0901, and Ts0941 had small Rxc values relative to RG indicating a high proportion of elongated structures in this set, consistent with the tails at high q in the P(r) plots. SAXS data were also used to estimate the volume of the protein through integration of the curve. This volume was used to later set the contour level and display the shapes determined from SAXS. These scalar values could be advantageous for predictors to constrain models.
3.3 Multimerization in SAXS targets
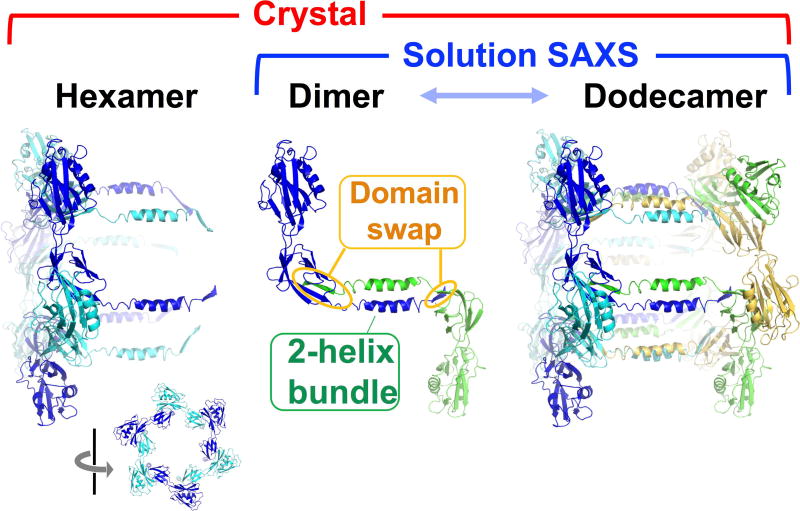
Multimerization posed a challenge to both modeling and the use of SAXS data in CASP12. Fifty percent of the targets collected (Ts0866, Ts0886, Ts0894, Ts0901 and Ts0909) were multimeric. Thus, modeling needed to take into account a buried surface area, and multimer or mixed multimer models were required to fit SAXS results. Additionally, as SAXS intensity is related to the square of the overall mass of the particle in solution, larger multimers will contribute more to the scattering than the monomer. For example, a dimer will contribute 4-fold more to the scattering than a monomer. Unfortunately, based on a post-CASP12 survey, not all participants realized the need to model multimers. The extent of multimerization was not surprising given similar findings on proteins from a SAXS assessment of structural genomics target proteins.29 As all targets are known to form crystals and therefore to undergo “ordered aggregation”, multimerization is not unexpected. Importantly, SAXS can reliably define biological assembly state; these multimers are also found in the crystal lattice, but are often obscured by larger crystal contacts. For example, the larger crystallographic interface in the abscisic acid binding receptor initially identified by the crystallographers was not the functional interface identified by SAXS, and the change in dimer assignment altered the biological interpretation of the structure.35 Also, CASP-CAPRI reported ambiguous and even inaccurate assignments of oligomerization interfered with their docking assessments.36 Here, three of the SAXS-determined multimerizations appear biologically relevant, where contacts between multimers are important for maintaining folds in those regions. Indeed, Ts0886 provides a potential example where the multimerization was ambiguous in the crystal lattice (Figure 4). The crystallographers assigned Ts0886 as a hexamer20; in the SAXS experiment, Ts0886 was dimeric in solution. A dimer in the crystal lattice that was not within the assigned hexamer best matched the scattering data and contained a β-sheet domain swap and a two-helix bundle interface. Two multimers (Ts0894 and Ts0901) appear to be non-specifically forming filaments and are best fit as mixtures of monomers, dimer, trimers and higher multimers. The application of SEC-coupled SAXS to separate out multimers, SEC-SAXS, would provide greater clarity, as seen in a recent study of multiprotein complexes37. Algorithms that use SAXS data from multimers by fitting folds of monomers alone will be led astray as global parameters of monomers can only approach those of multimers by assuming expanded and likely unrealistic folds. Using the SAXS data to predict the fold of monomeric units (subunits within oligomers) would require the added challenge of predicting the fold in the context of a multimer.
Figure 4. Determination of Multimerization in Solution by SAXS.
Based on the crystallographic lattice and electron microscopy images, the crystallographers assigned FliD (Ts0886) as a hexamer. However, the molecular weight calculated from the SAXS data of the same sequence suggested that FliD was a dimer. Comparison of all possible dimers in the crystallographic lattice identified one as the most likely dimer. The involvement in domain swapping and in a two-helix bundle supported this dimer as biologically relevant. Another construct of FliD showed strong concentration dependence of assembly (dimer to dodecamer) and thus was not included as a target in CASP. We postulate that the dodecamer, observed also in the crystallographic lattice, is created from assembly of the dimeric form.
3.4 Flexibility in SAXS targets
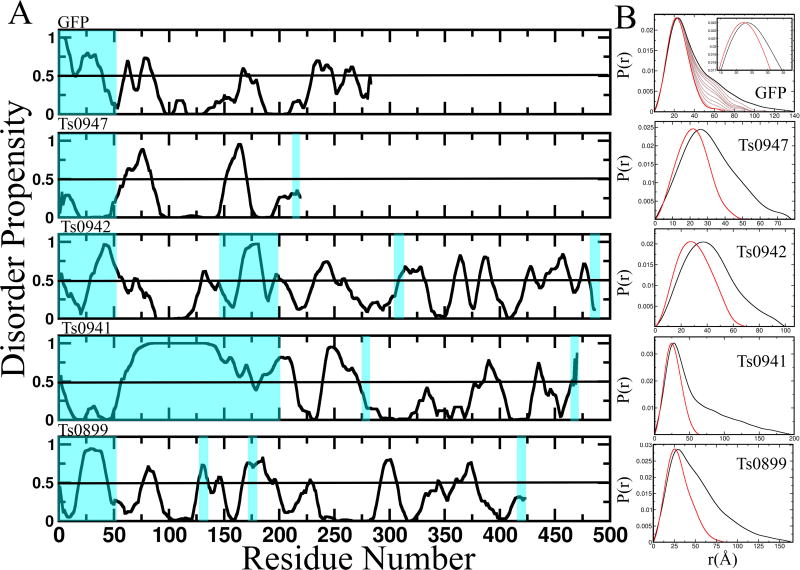
More surprising than multimerization was the extent of missing residues in the available crystal structures (Table 4). On average crystal structures were missing 20% of their sequence (44% in an extreme case), suggesting prediction algorithms include the possibility of unstructured regions. Missing residues are not resolvable in crystal structures often because of flexibility that also exists in solution, but is measured in the SAXS data. E.g. ten amino acids can add ~30Å to the maximum dimension of a structure if relatively extended. Such extensions significantly affect the global parameters that characterize a structure and are therefore reflected in the SAXS data. Four target proteins (Ts0899, Ts0941, Ts0942 and Ts0947) were monomeric and their crystal structures, without adding residues, poorly fit the SAXS data (Figure 5). This poses a significant challenge in using SAXS data to predict structure that may be overcome through a variety of approaches.
Figure 5. Disorder in the CASP SAXS targets.
(Left) PONDR analysis of four of the targets and a GFP “disorder” construct shows that these proteins are likely to have disordered regions. Blue highlight shows the regions missing in the globular crystal models and generally correlate to regions predicted to be disordered. (Right) Comparison of the P(r) predicted from the ordered crystallographic region overlaid with that calculated from the experimental data of the corresponding full-length protein illustrates how flexibility contributes to the scattering. In the GFP example, we measured experimental data for GFP alone (thick black) and for a GFP “disorder” construct with an added 50 residue C-terminus (red). We identified an ensemble of atomic models of GFP with a 50 residue C-terminus whose predicted scatter matches the experimental data. The predicted P(r) when we then removed 5, 10, 15, 20, 25, 30, 35, 40, and 45 residues from these atomic models, shown as light black lines, illustrate the additive effect of disordered residues, suggesting that DMax and perhaps the P(r) without flexibility could be estimated.
Analysis of disorder plots predicted from sequence can identify flexible regions. The program PONDR, among others, predicts the amino-acid disorder. In Figure 5, we map missing residues in the crystal structure onto PONDR generated plots showing the degree of agreement between predicted disorder and crystallographically missing residues (shaded blue). In three of the four cases, there is a direct correlation.
3.5 Generation of models for comparison to the SAXS experimental data
Importantly, the SAXS profile can be accurately estimated from crystal structures serving as a basis for comparison.16–18 After the CASP12 experiment concluded, the crystallographically determined structures were made available. As SAXS detects the presence of the mass in solution, including disordered regions not modeled, simple comparisons to the crystal structure are often not fully useful due to these missing regions. Adding any missing flexible regions to the crystal structures significantly improves their fits to the SAXS data (Figures 2–4). We generated full-length models based on the crystal structures and included regions that were missing from the crystal model but that were present in the SAXS-analyzed sample (Table 4). We used BILBOMD to create a population of conformers with the disordered regions allowed to move while the crystal structure regions were maintained as a rigid body. A minimal ensemble search (MES) implemented in FOXS identified three conformers that added together fit the data.18,38–40 While the resolved ensembles are not unique, they do provide a means to estimate the contribution of flexibility: a real factor in protein structure-function relationships that therefore needs to accounted for computationally. For multimeric assemblies, we created a stoichiometrically mixed population and again identified a minimal ensemble of monomers/multimers that fit the experimental data. We also combined flexibility and multimerization, when appropriate. Overlay of full-length and/or multimeric models based on the respective crystal structures into the envelopes illustrate how well the SAXS shapes predict the crystal structure models. One exception was the model for Ts0901, which showed small albeit significant deviation in the low q region of the scattering curve and large deviation in the P(r) plot, indicating that our model does not adequately predict the experimental scatter. The other exception was Ts0896, which highlights a potential deficiency in the experiment. Of the ten structures for which data was provided, Ts0896 was noted in the accompanying paper by Tamò and colleagues that the quaternary structure of the crystal model did not fit in the envelope [REFERENCE to the Assessment of data assisted modeling paper, Tamò et al., this issue]. Trapping a conformation in the crystal lattice occasionally occurs, and we allowed one domain to move as a separate rigid body and obtained a decent fit in the low q region. However, Ts0896 appears to have had a poor buffer subtraction, based upon our experience at the beamline with many datasets and the observation that no manipulation of the crystal structure yielded a profile matching the high q behavior. Nevertheless, poor buffer subtraction will primarily affect the high q region, so the low q region, which provides most of the information on the CASP-provided scalars, remains valid. The remaining 9 could be fit with structures based on those found in crystals (Figures 2 and 3), suggesting that modeling of multimers and/or disordered regions by predictors may lead to better fits of models to the experimental data.
4 DISCUSSION
CASP12 was the first CASP to include SAXS data as an experimental restraint and the inclusion of SAXS turned out to be an experiment unto itself. Participants were judged on similarity of the model structure within the region that crystallized. Although SAXS has the potential to provide restraints for computational algorithms41–43, the SAXS data collected was complicated by a high level of flexibility in the targets and/or multimerization. Modelers needed to include this information, in order to generate models that matched the SAXS data. Yet, in a survey of CASP participants who used the SAXS data, only half were aware that many of the targets were flexible and/or multimers, suggesting that the other half were trying to model a globular monomer without substantial flexible regions. Ideally for CASP13 if SAXS or other solution data reflecting the entire sequence is included, it would be optimal to have some targets that are monomeric without extended flexibility in solution as benchmarks.
However, the argument for inclusion of complex solution data in CASP is that flexibility and multimerization are innate, functionally relevant properties of proteins in solution and biology. Flexible regions are often required for stability and solubility, as seen for NEIL1.44,45 Ignoring multimerization is potentially disregarding contacts that may be just as important for folding as those made within the peptide itself and could thus mislead protein algorithms to identify surfaces that are exposed in subunits but buried in the solution state assembly as exposed surfaces. Similarly, forcing tertiary structure positioned by the crystal lattice, as in Ts0901, could mislead prediction algorithms into scoring these weak interfaces as stable. Thus, it is possible that algorithms have been handicapped in part by targeting “crystallographic monomers” without substantial disordered regions and assembly interfaces for training and optimization.
In support of the feasibility for computational programs to handle the challenges of flexibility and multimerization and to leverage the information in SAXS data, we show in Figures 2 and 3 how simplistic models that incorporate missing regions and/or are multimeric can generally recapitulate the experimental SAXS curves. SAXS data with mixed multimers should be avoided for CASP where possible, as the mixed stoichiometry adds a complexity likely to hinder prediction of novel folds. In the case of proteins with biologically relevant multimerization, predictors may be able to model mixed populations, if the association constants are determined in advance and if there is little change in conformation upon association.
Given the value for including some monomeric globular targets and stoichiometrically monodisperse samples, the following strategies for SAXS data collection and analysis are suggested to improve target data for predictors. First, priority should be given to identify proteins with low percentages of residues missing from the crystal structure and those whose biological unit is predicted to be monomeric. Second, samples should be filtered to remove non-specific aggregation, improving monodispersity of the sample and data quality. Third, targets that initially multimerize should be screened for buffer conditions (pH, salt) that reduce multimerization. Fourth, stable multimers should be purified and analyzed by SEC-SAXS, so that data is not from mixed stoichiometric populations but a single multimeric population. Fifth, for targets with large unfolded regions, novel approaches need to be developed that reduce the contribution of disordered regions to the SAXS data such as limited proteolysis prior to data collection, “cloaking” the unfolded regions using high density buffer for subtraction, and or computational subtraction.
Towards this last goal of “subtracting disorder” from the SAXS curve, we are working on developing test systems with engineered flexibility. In Figure 5, we engineered green fluorescent protein (GFP) with a 50 residue C-terminal tail predicted to be disordered (GFP50). Comparison of the P(r) of experimental data for GFP (black) and GFP50 (red) reveals that the disordered C-terminal tail adds to an increase of the histogram at longer distances and shifts the peak. Using BILBOMD, we identified three GFP models with C-terminal that can recapitulate the SAXS data of GFP50. Using these conformers, we simulated what the effect of different length tails by removing 5 residues at a time (brown curves). We observe a steady decrease in DMax, suggesting that the contribution of the flexible region can theoretically be subtracted from the SAXS data, to provide CASP predictors with either modified DMax or P(r) curves for the globular portion.
On the computational side, there is a need to develop novel approaches that take advantage of the rich information encoded in SAXS data. A survey of the predictors who participated in the SAXS data-assisted category showed that most of the groups used SAXS as an energetic factor to drive their prediction algorithms, with only one using it as an end-filter. Such direct incorporation of a SAXS data discrepancy function as a pseudo-potential energy term into refinement has notable advantages over using SAXS to filter structural solutions, as SAXS can provide a driving force toward the correct structure that may not be present in even the largest set of conformers for novel folds.14 Interestingly, investigators were almost evenly divided by what information they used: the scattering curve, the P(r) plot, and envelope. More tests of these different types of SAXS restraint or their combination seem merited as it is unclear what may prove most powerful for fold prediction. For those using the scattering curve, all used the most common model comparison metric for SAXS, χ2 to quantitate the difference between the experimental data and the model.
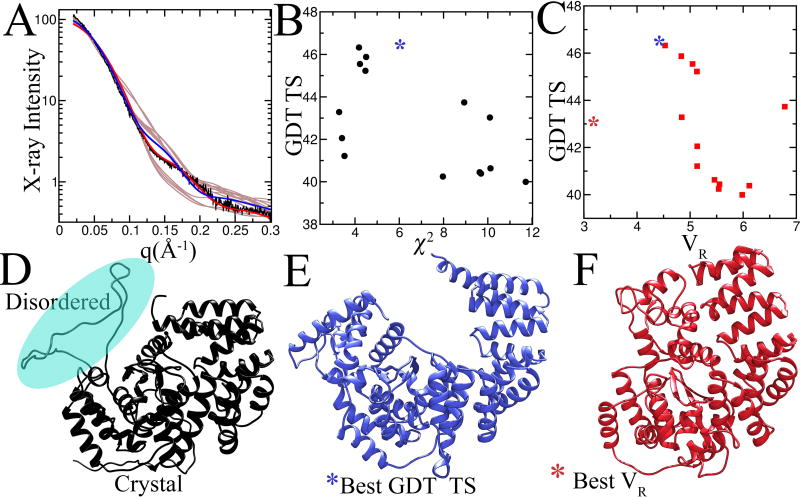
We caution a reliance on subtraction methods such as χ2. We have found that χ2 is biased by global parameters that typically weight the lowest angle data over data in the higher q range and fail to take full advantage of key information in the higher q range.46 Notably, the scattering curve can decrease 100–1000 fold in intensity from low q to high q, leading to this bias. Thus, ratio methods that take into account the intensity range of the scattering curve, such as VR, capture more information and better correlate with RMSD and other structural similarity indices.46 Indeed, we tested the GDT_TS correlation between VR and χ2 for one test CASP target (Figure 6). While χ2 did not show a strong correlation among prediction models, low VR was more predictive of high GDT_TS. The one outlier with the best VR was insightful, as outliers can be: that model predicted localization of certain residues that was not predicted in the highest scoring GDT_TS model. Notably, this plot demonstrates that VR, in contrast to GDT_TS, is responsive to the global shape first and fold second.
Figure 6. SAXS as a measure of the quality of prediction using χ2 and VR.
The calculated SAXS profile from the top 19 GDT_TS scored unassisted predictions for target Ts0942 were compared against the measured SAXS profile (A). The agreement between the experimental data and data predicted from the models was scored using χ2 (A) and VR (B) and plotted against GDT_TS. (D) The full-length model based on the crystal structure that best matches the experimental SAXS. The modeled region is highlighted by cyan sphere. (E) The highest GDT_TS scored model has a cavity not found in the full-length model. (F) The highest VR-scored model captures the overall shape of the full-length model.
Methods are needed that incorporate the hydration layer in calculation of the P(r) from atomic models, as we have seen its importance in calculation of the scattering curve.47 Flexibility is another parameter that can inform protein structure algorithms and can depend upon small changes in sequence or even ligand binding status.14 SAXS provides multiple measures of flexibility that merit consideration for inclusion in computational prediction: lack of convergence in Kratky plot, invalidation Porod-Debye law, Flory’s inequality, low particle density, and inability to model data with a single model. Combining protein volume derived from SAXS data with measured mass allows calculation of the SAXS-based density, which can vary significantly for flexible proteins48: flexible proteins are likely to have densities of 0.9–1.0 g·cm−3, which is far below the canonical value of 1.37 g·cm−3 for folded compact proteins.49 In fact, there is a huge opportunity for computational prediction methods that employ SAXS data and incorporate the bound hydration layer and local flexibility, as these two features result in all crystal structures having substantially more error in the refined crystallographic models than in the measured diffraction data50. Thus, computationally modeling suitably combined with SAXS to incorporate bound water and local disorder may offer a means to improve all known crystal structures: this opportunity for computational prediction certainly merits investigation.
Looking forward, computational algorithms will be key to timely and impactful predictions for biologically relevant structures and importantly, mutant structures leading to disease. Without experimental feedback, it will be difficult to get such predictions right. Consider that single amino acid mutations, small ligands, and pH can drastically alter protein conformation. SAXS can accurately show the conformation and assembly state that can vary with ligand binding, as seen for proteins such as abscisic acid receptor and apoptosis inducing factor 35,51. SAXS can similarly distinguish differences in aggregation or assembly resulting from single residue changes, as found for superoxide dismutase mutations that correlate to Amyotrophic Lateral Sclerosis prognosis,52 for macromolecular interactions controlling pathogenesis,53 for nanomachines orchestrating genetic integrity,54 and for design of mega-protein assemblies.55 In fact, SAXS and CLMS are poised to provide enabling high-throughput experimental data to improve folding accuracy and algorithms. Inversely, computational modeling efforts are needed for improved interpretation of SAXS and CLMS data. The impact of the DNA double helix model consistent with low-resolution X-ray scattering data56 underscores how integration with sequence-based modeling algorithms is a critical goal for further development of SAXS and CLMS analysis and refinement. Computational prediction combined with experimental data thus holds great promise for achieving the goal of robust sequence-level interpretations of SAXS and CLMS data for proteins as well as for more accurate computational models.
Supplementary Material
Acknowledgments
For financial support, we thank NIH (PO1CA92584 to J.A.T., R01GM110387 to S.T., and R01GM100482 to KF); and KAUST for CRG3 funding to J.A.T. J.A.T. acknowledges support by a Robert A. Welch Chemistry Chair, the Cancer Prevention and Research Institute of Texas, and the University of Texas System Science and Technology Acquisition and Retention. This research used resources of the Advanced Light Source, which are DOE Office of Science User Facilities under contract no. DE-AC02-05CH11231. The SIBYLS beamline 12.3.1 is supported by the IDAT program from the DOE BER and KAUST.
References
- 1.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Arakaki AK, Skolnick J. TASSER: an automated method for the prediction of protein tertiary structures in CASP6. Proteins. 2005;(61 Suppl 7):91–98. doi: 10.1002/prot.20724. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. The EMBO journal. 2010;29(4):717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci. 2016;41(1):20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339(6122):913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lossl P, van de Waterbeemd M, Heck AJ. The diverse and expanding role of mass spectrometry in structural and molecular biology. The EMBO journal. 2016;35(24):2634–2657. doi: 10.15252/embj.201694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belsom A, Mudd G, Giese S, Auer M, Rappsilber J. Complementary Benzophenone Cross-Linking/Mass Spectrometry Photochemistry. Anal Chem. 2017;89(10):5319–5324. doi: 10.1021/acs.analchem.6b04938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsom A, Schneider M, Fischer L, Brock O, Rappsilber J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol Cell Proteomics. 2016;15(3):1105–1116. doi: 10.1074/mcp.M115.048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belsom A, Schneider M, Brock O, Rappsilber J. Blind Evaluation of Hybrid Protein Structure Analysis Methods based on Cross-Linking. Trends Biochem Sci. 2016;41(7):564–567. doi: 10.1016/j.tibs.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belsom A, Schneider M, Fischer L, Mabrouk M, Stahl K, Brock O, Rappsilber J. Blind testing cross-linking/mass spectrometry under the auspices of the 11th critical assessment of methods of protein structure prediction (CASP11) Wellcome Open Res. 2016;1:24. doi: 10.12688/wellcomeopenres.10046.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider M, Belsom A, Rappsilber J, Brock O. Blind testing of cross-linking/mass spectrometry hybrid methods in CASP11. Proteins. 2016;(84 Suppl 1):152–163. doi: 10.1002/prot.25028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer L, Rappsilber J. Quirks of Error Estimation in Cross-Linking/Mass Spectrometry. Anal Chem. 2017;89(7):3829–3833. doi: 10.1021/acs.analchem.6b03745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual review of biophysics. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 15.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly reviews of biophysics. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 16.Svergun D, Barberato C, Koch MHJ. CRYSOL- a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. Journal of applied crystallography. 1995;28(6):768–773. [Google Scholar]
- 17.Forster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. Journal of molecular biology. 2008;382(4):1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research. 2010;38:W540–544. doi: 10.1093/nar/gkq461. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. Architectures of Lipid Transport Systems for the Bacterial Outer Membrane. Cell. 2017;169(2):273–285. doi: 10.1016/j.cell.2017.03.019. e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postel S, Deredge D, Bonsor DA, Yu X, Diederichs K, Helmsing S, Vromen A, Friedler A, Hust M, Egelman EH, Beckett D, Wintrode PL, Sundberg EJ. Bacterial flagellar capping proteins adopt diverse oligomeric states. Elife. 2016;5 doi: 10.7554/eLife.18857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6(12):2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Giese SH, Fischer L, Rappsilber J. A Study into the Collision-induced Dissociation (CID) Behavior of Cross-Linked Peptides. Mol Cell Proteomics. 2016;15(3):1094–1104. doi: 10.1074/mcp.M115.049296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nature methods. 2012;9(9):901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 27.Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, Tainer JA. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. Journal of applied crystallography. 2013;46(Pt 1):1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyer KN, Hammel M, Rambo RP, Tsutakawa SE, Rodic I, Classen S, Tainer JA, Hura GL. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods in molecular biology. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nature methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes FE, Schwartz CR, Tainer JA, Rambo RP. Methods for using new conceptual tools and parameters to assess RNA structure by small-angle X-ray scattering. Methods in enzymology. 2014;549:235–263. doi: 10.1016/B978-0-12-801122-5.00011-8. [DOI] [PubMed] [Google Scholar]
- 32.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of applied crystallography. 1992;25:495–503. [Google Scholar]
- 33.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophysical journal. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Classen S, Rodic I, Holton J, Hura GL, Hammel M, Tainer JA. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. Journal of synchrotron radiation. 2010;17(6):774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326(5958):1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Rie Lee G, Seok C, Qin S, Zhou HX, Ritchie DW, Maigret B, Devignes MD, Ghoorah A, Torchala M, Chaleil RA, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JP, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond AS, Visscher K, Kastritis PL, Bonvin AM, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim HR, Roy A, Han X, Esquivel-Rodriguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jimenez-Garcia B, Moal IH, Fernandez-Recio J, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Roy Burman SS, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchigrechko A, Wodak SJ. Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment. Proteins. 2016;(84 Suppl 1):323–348. doi: 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, Kuzdovich M, Remesh SG, Fang S, Tomkinson AE, Lees-Miller SP, Tainer JA. An Intrinsically Disordered APLF Links Ku, DNA-PKcs, and XRCC4-DNA Ligase IV in an Extended Flexible Non-homologous End Joining Complex. The Journal of biological chemistry. 2016;291(53):26987–27006. doi: 10.1074/jbc.M116.751867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. General physiology and biophysics. 2009;28(2):174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic acids research. 2016;44(W1):W424–429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneidman-Duhovny D, Kim SJ, Sali A. Integrative structural modeling with small angle X-ray scattering profiles. BMC structural biology. 2012;12:17. doi: 10.1186/1472-6807-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annual review of biophysics. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W, Doniach S. Protein structure prediction constrained by solution X-ray scattering data and structural homology identification. Journal of molecular biology. 2002;316(1):173–187. doi: 10.1006/jmbi.2001.5324. [DOI] [PubMed] [Google Scholar]
- 43.Xu X, Yan C, Wohlhueter R, Ivanov I. Integrative Modeling of Macromolecular Assemblies from Low to Near-Atomic Resolution. Comput Struct Biotechnol J. 2015;13:492–503. doi: 10.1016/j.csbj.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandaru V, Cooper W, Wallace SS, Doublie S. Overproduction, crystallization and preliminary crystallographic analysis of a novel human DNA-repair enzyme that damage recognizes oxidative DNA damage. Acta Crystallogr D. 2004;60:1142–1144. doi: 10.1107/S0907444904007929. [DOI] [PubMed] [Google Scholar]
- 45.Hegde PM, Dutta A, Sengupta S, Mitra J, Adhikari S, Tomkinson AE, Li GM, Boldogh I, Hazra TK, Mitra S, Hegde ML. The C-terminal Domain (CTD) of Human DNA Glycosylase NEIL1 Is Required for Forming BERosome Repair Complex with DNA Replication Proteins at the Replicating Genome DOMINANT NEGATIVE FUNCTION OF THE CTD. Journal of Biological Chemistry. 2015;290(34):20919–20933. doi: 10.1074/jbc.M115.642918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, Tainer JA. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nature methods. 2013;10(6):453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophysical journal. 2013;105(4):962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95(8):559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voss NR, Gerstein M. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. Journal of molecular biology. 2005;346(2):477–492. doi: 10.1016/j.jmb.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 50.Holton JM, Classen S, Frankel KA, Tainer JA. The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures. The FEBS journal. 2014;281(18):4046–4060. doi: 10.1111/febs.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brosey CA, Ho C, Long WZ, Singh S, Burnett K, Hura GL, Nix JC, Bowman GR, Ellenberger T, Tainer JA. Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor. Structure. 2016;24(12):2067–2079. doi: 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt AJ, Shin DS, Merz GE, Rambo RP, Lancaster WA, Dyer KN, Borbat PP, Poole FL, 2nd, Adams MW, Freed JH, Crane BR, Tainer JA, Getzoff ED. Aggregation propensities of superoxide dismutase G93 hotspot mutants mirror ALS clinical phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(43):E4568–4576. doi: 10.1073/pnas.1308531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammel M, Amlanjyoti D, Reyes FE, Chen JH, Parpana R, Tang HY, Larabell CA, Tainer JA, Adhya S. HU multimerization shift controls nucleoid compaction. Sci Adv. 2016;2(7):e1600650. doi: 10.1126/sciadv.1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brosey CA, Ahmed Z, Lees-Miller SP, Tainer JA. What Combined Measurements From Structures and Imaging Tell Us About DNA Damage Responses. Methods in enzymology. 2017;592:417–455. doi: 10.1016/bs.mie.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai YT, Hura GL, Dyer KN, Tang HY, Tainer JA, Yeates TO. Designing and defining dynamic protein cage nanoassemblies in solution. Sci Adv. 2016;2(12):e1501855. doi: 10.1126/sciadv.1501855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franklin RE, Gosling RG. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953;172(4369):156–157. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.