Summary
The absence of cancer-restricted surface markers is a major impediment to antigen-specific immunotherapy using chimeric antigen receptor (CAR) T cells. For example, targeting the canonical myeloid marker CD33 in acute myeloid leukemia (AML) results in toxicity from destruction of normal myeloid cells. We hypothesized that a leukemia-specific antigen could be created by deleting CD33 from normal hematopoietic stem and progenitor cells (HSPC), thereby generating a hematopoietic system resistant to CD33-targeted therapy and enabling specific targeting of AML with CAR T cells. We generated CD33-deficient human HSPC and demonstrated normal engraftment and differentiation in immunodeficient mice. Autologous CD33 KO HSPC transplantation in rhesus macaques demonstrated long-term multilineage engraftment of gene-edited cells, with normal myeloid function. CD33-deficient cells were impervious to CD33-targeting CAR T cells, allowing for efficient elimination of leukemia without myelotoxicity. These studies illuminate a novel approach to antigen-specific immunotherapy by genetically engineering the host to avoid on-target, off-tumor toxicity.
eTOC
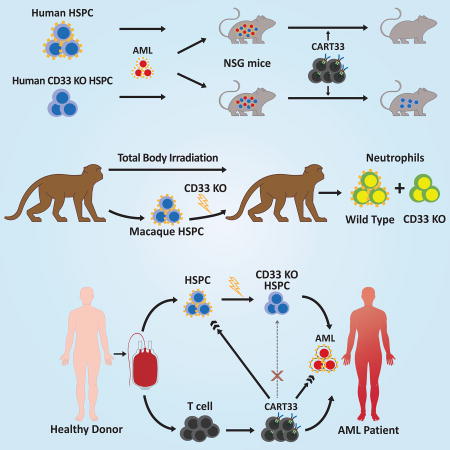
Reconstitution of the immune system with CD33 negative human hematopoietic stem cells enables anti-CD33 CAR-T cell killing of acute myeloid leukemia while sparing myeloid development and function.
Introduction
Recent advances in cancer immunotherapy with chimeric antigen receptor (CAR) T cells have enabled eradication of cells expressing a specific surface antigen. While this approach has been successful in targeting CD19 in B-cell neoplasms (Davila et al., 2014; Kochenderfer et al., 2015; Lee et al., 2015; Maude et al., 2014; Turtle et al., 2016), it does not discriminate between normal and malignant B cells, and thus the feasibility of this therapy rests on the tolerability of prolonged B cell aplasia. However, most malignancies do not have an expendable normal tissue counterpart, and whether the success of CAR T cells can be extrapolated beyond B-cell neoplasms will depend on the ability to develop strategies to mitigate toxicity to normal cells.
The majority of acute myeloid leukemia (AML) patients relapse despite intensive therapy. AML cell surface antigens are shared with normal myeloid progenitors (Levine et al., 2015; Taussig et al., 2005), and hence targeting AML also generates toxicity to the myeloid system (Gill et al., 2014; Leong et al., 2017; Mardiros et al., 2013; Pizzitola et al., 2014; Tashiro et al., 2017). While strategies to produce transient CAR T cells are being explored in clinical trials to avoid longterm myeloablation while targeting AML (e.g. #NCT03126864, #NCT02159495), this negates a fundamental strength of this therapy, namely its long-term anti-tumor immune surveillance. Therefore, in the absence of a truly AML-specific antigen, novel approaches are required to definitively target AML while sparing normal hematopoiesis.
Here we propose a new paradigm for antigen-specific immunotherapy against myeloid neoplasms, in which potent antigen-directed therapy can be administered without causing prolonged myeloid aplasia. We use gene-edited HSPC to regenerate an antigen-negative myeloid system that is resistant to antigen-targeting therapy.
CD33 is a member of the sialic acid binding immunoglobulin-like lectin (siglec) family. CD33 is expressed on all normal myeloid cells downstream of the common myeloid progenitor, and is used as both a diagnostic marker and a therapeutic target for AML (Laszlo et al., 2014). We show here that CD33 knockout (KO) HSPC can generate functional hematopoiesis that is resistant to CD33-targeted AML therapy in two relevant preclinical models and provide evidence for the clinical feasibility and efficacy of combining allogeneic transplantation of CD33 KO HSPC with CD33 CAR T therapy to treat patients with otherwise incurable AML.
Results
Human CD33 KO HSPC remain functional
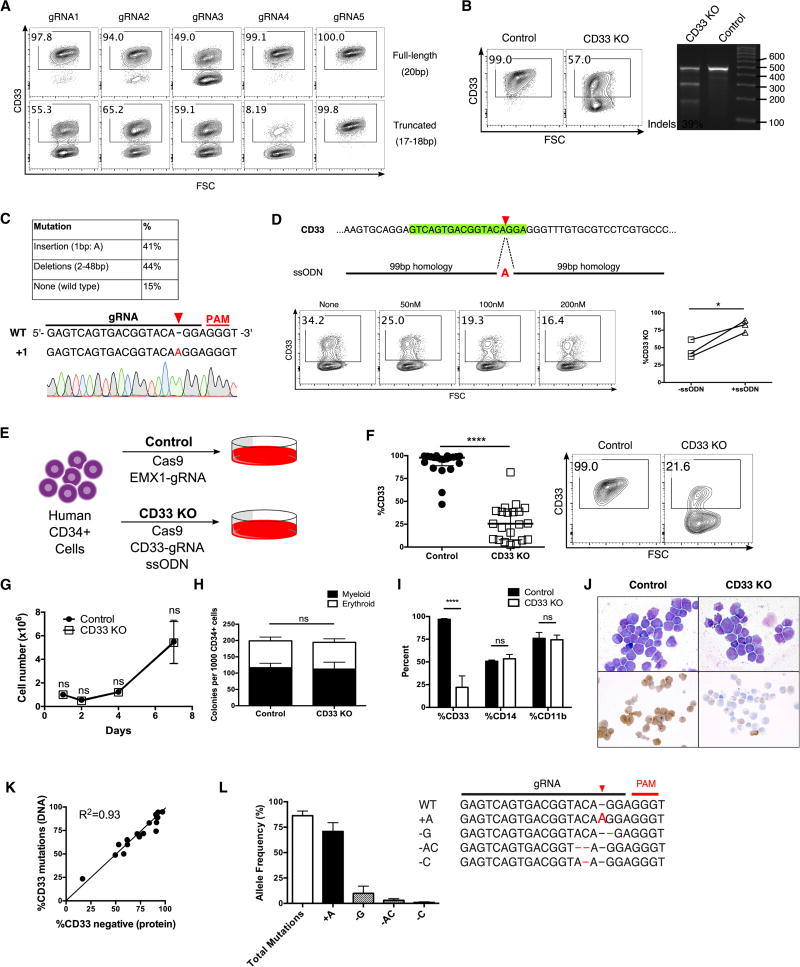
We disrupted the CD33 gene in primary human CD34+ cells using truncated guide RNAs (gRNAs) (Fu et al., 2014), which have been reported to increase specificity of gene editing. We found that truncated gRNAs increased the efficacy of CD33 KO compared to their full-length counterparts (Fig 1A). Initially, we achieved around 40% KO in primary CD34+ cells (Fig 1B), of which a large proportion of mutations consisted of a single adenine (A) nucleotide insertion (Fig 1C). We further enhanced editing by adding a single-stranded oligodeoxynucleotide (ssODN) template containing the A insertion, leading to dose-dependent increase in frequency of KO (Fig 1D). Subsequent experiments were performed via transfection of Cas9 protein complexed with a truncated gRNA targeting CD33 and the ssODN template to generate CD33 KO HSPC, while control HSPC were treated with Cas9 and a gRNA targeting the irrelevant gene EMX1 (Fig 1E). In vitro differentiated CD33 KO HSPC had markedly reduced levels of surface CD33 protein expression (27±4%) compared to controls (92±3%) (Fig 1F), without impairment of growth or differentiation (Fig 1G–J). High frequency of mutations in CD33 was confirmed by tracking of indels by decomposition (TIDE) and next-generation sequencing (Fig 1K–L).
Fig. 1. Generation of CD33 KO human HSPC in vitro.
(A) Full-length (20bp) and truncated (17–18bp guide RNAs (gRNAs) targeting CD33 were tested in for editing efficiency in a human AML cell line, Molm14, after electroporation with Cas9. (B) Human CD34+ cells were electroporated with Cas9 protein and truncated CD33-targeting gRNA4 (panel A), and surface CD33 protein expression was assessed by flow cytometry (left) and DNA mutations were quantified by SURVEYOR assay (right) following 7 days of in vitro myeloid differentiation culture. (C) Sanger sequencing of individual alleles reveals a high proportion of mutations containing a single A insertion (top); representative chromatogram is shown (bottom). (D) A single-stranded oligodeoxynucleotide (ssODN) was designed to contain 99bp homology arms around the gRNA cut site and the A insertion mutation (top). CD34+ cells were electroporated with Cas9 protein/gRNA and different concentrations of the ssODN, after which CD33 expression was assessed by flow cytometry (bottom left) following 7 days of culture. Addition of the ssODN increases the frequency of CD33 mutations in three different donors in three independent experiments (bottom right). (E) Experimental schema: Human CD34+ cells were electroporated with either Cas9 complexed with EMX1-gRNA (Control), or Cas9/CD33-gRNA4 and the ssODN (CD33 KO) and cultured for 7 days. (F) Cultured CD33 KO CD34+ cells have decreased levels of surface CD33 expression by flow cytometry compared to controls (n=21, 18 different donors, 14 independent experiments). Representative flow cytometry plot is shown on right. (G) Control and CD33 KO CD34+ cells maintained in StemSpan SFEM with SCF 100ng/ml, Flt3L 100ng/ml, TPO 50ng/ml, IL-6 50ng/ul show similar growth kinetics. (n=3, 3 donors, 2 independent experiments) (H) CD34+ cells were plated in Methocult H4435 one day after electroporation and scored for colony formation after 14 days. (I) Methocult colonies were resuspended in liquid media and flow cytometry was performed for the myeloid markers CD33, CD14 and CD11b. (n=3, 3 donors, 2 independent experiments) (J) In vitro differentiated control and CD33 KO HSPC show normal myeloid morphology by Wright-Giemsa staining with decreased CD33 expression by immunocytochemistry in the CD33 KO cells only. (K) Comparison of levels of CD33 protein loss by flow cytometry and CD33 gene mutations by TIDE analysis show a high level of correlation (n=18). %CD33 negative (protein) was calculated as follows: {1 − (%CD33+ in KO)/(%CD33+ in control)}*100. (L) Targeted amplicon sequencing of the gRNA site from CD33 KO cell DNA confirms high levels of total mutations, with the majority consisting of A insertions.
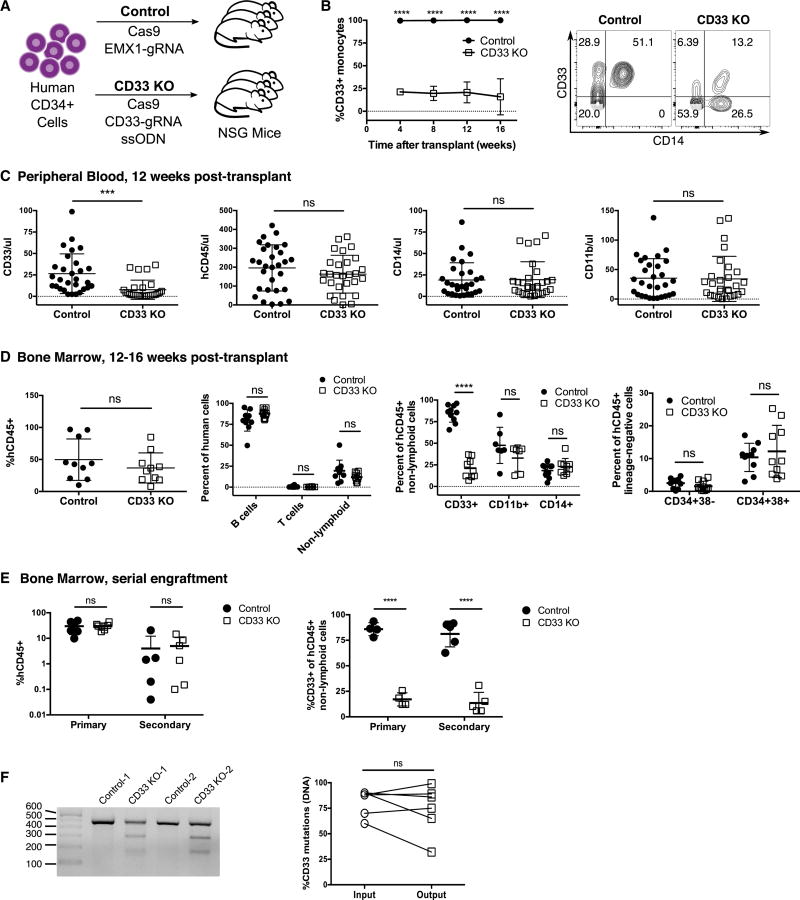
We injected NOD/SCID/IL2rγnull (NSG) mice with control (Cas9/EMX1-gRNA) or CD33 KO (Cas9/CD33-gRNA/ssODN) HSPC and followed human cell engraftment in the peripheral blood over time (Fig 2A). We found that human monocytes in the peripheral blood of mice engrafted with CD33 KO HSPC had the expected diminished expression of CD33 that was sustained over time (Fig 2B). Total human CD33+ cells were significantly decreased in the CD33 KO HSPC-engrafted mice compared to controls, while numbers of total human CD45+ cells as well as myeloid subsets (CD11b+ and CD14+ cells) were identical between the two groups (Fig 2C). Bone marrow analysis confirmed similar levels of human engraftment and differentiation between control and CD33 KO HSPC-engrafted mice, while decreased expression of CD33 was observed engrafted myeloid cells. Levels of the most primitive (CD34+38−) and more mature (CD34+38+) human HSPC in the marrow were not significantly different between the two groups (Fig 2D).
Fig. 2. CD33 KO human HSPC show sustained loss of CD33 in vivo without impairment of growth and differentiation.
(A) NSG mice were injected with 1–5×105 control or CD33 KO HSPC and human CD45+ hematopoiesis was analyzed over time. (B) Longitudinal monitoring of human CD14+ monocytes show a decreased fraction of CD33+ monocytes in CD33 KO HSPC-engrafted mice compared to controls. Representative flow plot is shown on right. ns: not significant (p>0.05); ****p<0.0001 (unpaired t-test) (C) CD33 KO HSPC-engrafted mice have significantly decreased numbers of CD33+ cells in the peripheral blood at 12 weeks post-transplant, while total numbers of cells expressing CD45, CD14 and CD11b are similar to controls (n=57 mice, 4 independent experiments, 5 donors). ns: not significant (p>0.05); ***p<0.001 (unpaired t-test) (D) Bone marrow harvested from mice after 12–16 weeks of HSPC engraftment show equivalent levels of human CD45+ cell engraftment, with differentiation into both lymphoid (B cells: CD19+, T cells: CD3+) and myeloid lineages, while myeloid cells have markedly decreased levels of CD33. Human hematopoietic stem (CD34+38−) and progenitor (CD34+38+) cell levels are comparable between control and CD33 KO HSPC engrafted mice. ****p<0.001; ns: not significant (p>0.05) (unpaired t-test) (n=20 mice, 3 independent experiments, 3 donors). (E) Marrow was harvested from mice engrafted with control or CD33 KO HSPC after 16 weeks (primary), and human CD45+ cells were purified and injected into secondary recipients (secondary) (each donor transplanted into a single recipient) and monitored for an additional 12 weeks. Both groups of mice show sustained human hematopoiesis after long-term engraftment (left), and the CD33 KO HSPC-engrafted group has persistent loss of CD33 expression (right). (n=12 mice, 3 independent experiments, 4 donors). ****p<0.001; ns: not significant (p>0.05) (unpaired t-test) (F) PCR performed on marrow from secondary recipients confirms continued presence of mutations in the CD33 gene by SURVEYOR assay (left). Mutation levels as quantified by TIDE are similar between the initial infusion product (input) and the bone marrow 28 weeks after transplantation (output) (right). ns: not significant (p>0.05) (paired t-test). All data are represented as means ± SD.
To document that CD33 KO HSPC retain long-term repopulating function, we harvested marrow cells from mice engrafted with control or CD33 KO HSPC after 16 weeks and transferred these cells into secondary recipients. Bone marrow analysis of the secondary recipients after 12 additional weeks showed sustained human engraftment, while CD33 expression remained diminished in CD33 KO HSPC-recipients (Fig 2E), and PCR confirmed the presence of mutations in CD33 at similar levels to the initial infusion product (Fig 2F). These results indicate that CD33 KO occurred in cells capable of long-term and serial engraftment.
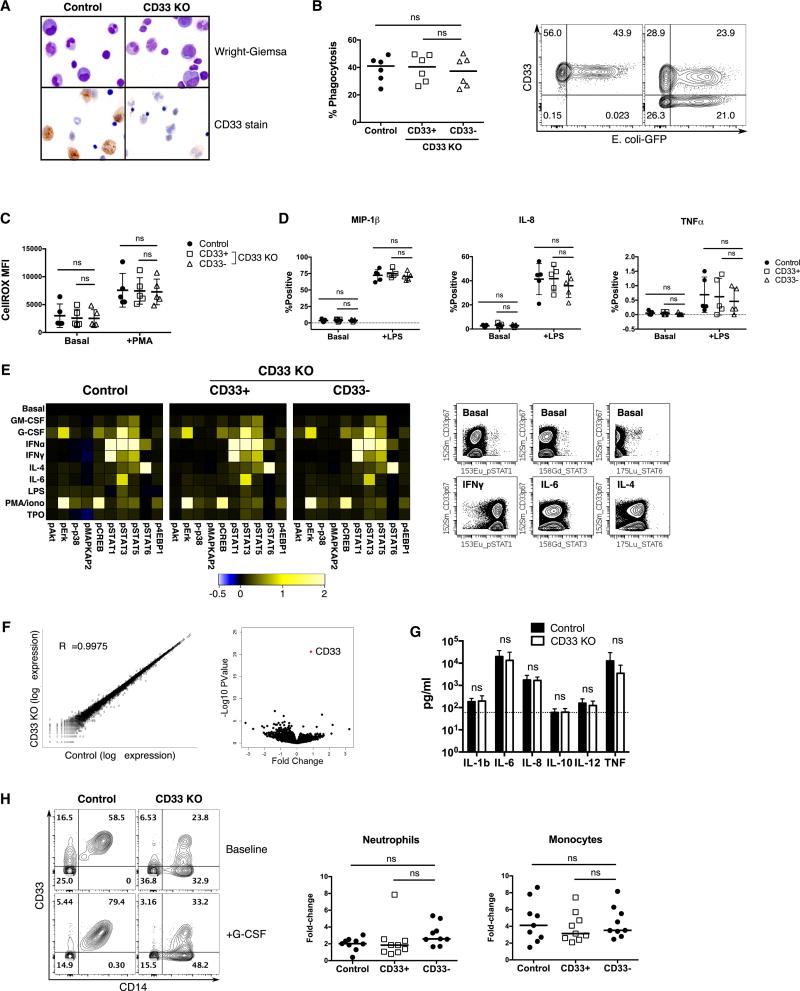
We next performed several studies to probe the function of CD33 KO myeloid cells. Human cells in the marrow of mice engrafted with CD33 KO HSPC were morphologically normal, and absence of CD33 was confirmed by immunocytochemistry (Fig 3A). We interrogated the ability of in vitro differentiated CD33 KO myeloid cells to perform phagocytosis, generate reactive oxygen species, and secrete inflammatory cytokines/chemokines (Fig 3B–D). We used HSPC that had approximately 50% CD33 KO and gated on the CD33+ and CD33− cells separately, using the CD33+ cells as an internal control, while also comparing to control HSPC. We also performed these studies on cells sorted for CD33+/− expression to exclude any non-cell autonomous function of CD33 (Fig S1). We found no significant difference in any of our studies when comparing the function of CD33− myeloid cells to control cells that were uniformly CD33+.
Fig. 3. CD33 is not essential for human myeloid cell function.
(A) Bone marrow cells from NSG mice engrafted with control or CD33 KO human CD34+ cells exhibit normal human myeloid cell morphology, with decreased CD33 expression confirmed by immunocytochemistry. (B–F) Control and CD33 KO CD34+ cells were differentiated in vitro with SCF, TPO, Flt3L, IL-3, IL-6 and GM-CSF for 7–14 days prior to functional assays. CD33+/− cells within the CD33 KO group were gated separately by flow cytometry for analysis, as compared to ungated control cells which are all CD33+. (B) In vitro differentiated CD33 KO myeloid cells retain phagocytosis ability as measured by internalization of pHrodo green E. coli bioparticles (n=6/group, one-way ANOVA). (C) Levels of reactive oxygen species (ROS) production after phorbol myristate acetate (PMA) stimulation are similar among the three groups of cells. ROS production was measured by fluorescence of CellROX Green reagent (n=5/group, one-way ANOVA). (D) Cytokine production as measured by intracellular staining is not significantly different in CD33− cells as compared to internal CD33+ or unedited controls, whether under basal conditions or after lipopolysaccharide (LPS) stimulation (n=5/group, one-way ANOVA). (E) Cells were treated with the indicated stimuli (GM-CSF, G-CSF, IFNα, IFNγ, IL-4, IL-6, LPS, PMA/ionomycin, or TPO) for 15 min followed by mass cytometry analysis using antibodies to 20 surface markers and 10 phosphoproteins for a comprehensive analysis of signaling pathways (n=3/group). Control (ungated) and CD33+/− gated cells within the CD33 KO cell group display identical signaling profiles within the myeloid progenitor population as defined by SPADE analysis (see fig. S2 for SPADE diagram). Representative plots of CD33 KO cells show that CD33− and residual CD33+ cell populations respond to the indicated stimuli to the same degree (gated on live CD64+HLA-DR+ events). (F) Gene expression profile of in vitro differentiated control and CD33 KO CD34+ cells (with 70–85% CD33 KO) were analyzed by RNA-seq (n=5/group). Log-scale scatter plot of mean gene expression values of control and CD33 KO samples show high correlation between the two groups (left), while volcano plot shows CD33 as the most significant differentially expressed gene (right). (G) LPS injection induces similar levels of human inflammatory cytokine secretion in the serum of mice engrafted with control or CD33 KO HSPC (control: n=10, CD33 KO: n=12). ns: not significant (p>0.05) (unpaired t-test) (H) NSG mice engrafted with control or CD33 KO HSPC were injected with G-CSF and numbers of peripheral blood human myeloid cells were evaluated before and after treatment (n=9/group, 2 donors, 2 independent experiments). Representative flow cytometry plot shows increase in both %CD14+CD33+ and %CD14+CD33− cells in the CD33 KO group after G-CSF injection (left), gating on single live human CD45+ events. Increased numbers of neutrophils (CD66b+) and monocytes (CD14+) were detected in the peripheral blood of both cohorts after G-CSF, and within the CD33 KO HSPC-engrafted mice no difference in CD33+ or CD33− cell populations was detected (right) (one-way ANOVA). ns: not significant (p>0.05).
We also performed high dimensional functional profiling of control and CD33 KO HSPC using mass cytometry (Behbehani et al., 2015; Levine et al., 2015). Other than the expected differences in CD33 expression, we found no differences in the differentiation profile between control and CD33 KO cells by SPADE analysis (Fig S2A). Cells were exposed to relevant ex vivo perturbations (GM-CSF, G-CSF, IFNα, IFNγ, IL-4, IL-6, LPS, PMA/ionomycin, TPO) and activation of intracellular signaling pathways in relevant hematopoietic subpopulations was quantified. Remarkably, control and CD33 KO cells responded identically to external stimuli, and within the CD33 KO cell population both the CD33+ and CD33− cells responded to the same degree (Fig 3E, S2B–C).
To exclude a major impact of CD33 loss on downstream gene expression, we performed RNA sequencing of CD33 KO myeloid cells and controls, which showed a highly concordant gene expression profile. Paired analysis of matched control and CD33 KO samples showed that CD33 was the most significantly differentially expressed gene (Fig 3F).
Finally, we performed studies in mice engrafted with CD33 KO or control human hematopoiesis to evaluate their functional properties in vivo. To model gram-negative sepsis, we injected mice with lipopolysaccharide, which induced high levels of human inflammatory cytokines in the serum, with no difference detected between control and CD33 KO (Fig 3G). We also challenged the mice with G-CSF, which induced an equivalent increase of human neutrophils (CD66b+) and monocytes (CD14+) in the peripheral circulation, while in the CD33 KO HSPC-engrafted mice, both CD33+ and CD33− myeloid cells increased to the same degree (Fig 3H). These observations confirmed our hypothesis that human myeloid cells would tolerate loss of CD33.
For additional evidence that CD33 may be redundant in human health, we examined several databases of naturally occurring homozygous loss-of-function mutations in large population cohorts (Lek et al., 2016; Narasimhan et al., 2016; Saleheen et al., 2017; Sulem et al., 2015). We found 51 individuals reported to have homozygous loss-of-function mutations in the CD33 gene (Table S1), of whom the majority (86%) consisted of a deletion of 4 base pairs in exon 3 (rs201074739), resulting in a frameshift mutation in all protein-coding transcripts of CD33. In the Iceland cohort (Sulem et al., 2015), 4 of these individuals were reported to have reached 64 to 93 years of age.
To evaluate whether our CRISPR/Cas9-based CD33 KO system would cause any off-target mutagenesis, we performed cytogenetic analysis of multiple control/CD33 KO primary samples for gross chromosomal rearrangements, which were not detected (Fig S3A). We further analyzed 6 donor samples by PCR and found that all showed evidence of deletion of the 14kb fragment between CD33 and SIGLEC22P, a pseudogene that has 100% homology with CD33 at the gRNA binding site (Fig S3B). These two genes are directly adjacent to each other with no known genetic element in the intervening sequence. We then performed targeted deep sequencing of the top 10 predicted off-target sites in primary CD34+ cells after CD33 KO in comparison with control cells from the same donor (Fig S3C). In all samples the on-target site, CD33, had high levels of mutations (86±4%), and SIGLEC22P harbored a similar extent of mutations as the target gene (85±9%). Only one additional site, SIGLEC6, was found to have low frequency mutations (0.41–1.72%) in 3 of the 5 donors evaluated.
As a more comprehensive screen for all potential off-target mutation sites in a genome-wide unbiased manner, we performed Circularization for In vitro Reporting of Cleavage Effects by sequencing (CIRCLE-seq) (Tsai et al., 2017) on genomic DNA from human primary cells, and identified 95 sites cleaved by the Cas9/CD33-gRNA complex in vitro (Fig S3D,E). We then performed targeted deep sequencing in primary CD34+ cells in 16 sites that had high read count on CIRCLE-seq and/or had 3 or less nucleotide mismatch to the on-target sequence. Most of the sites had no detectable mutations in primary cells.
Transplantation of CD33 KO HSPC in Rhesus macaques
We next turned to a non-human primate (NHP) model to rigorously evaluate the long-term effects and functional consequences of CRISPR/Cas9-mediated CD33 KO on HSPC. CD33 was expressed uniformly on over 95% of neutrophils from both rhesus macaques (RM) and humans, but expression on monocytes was more heterogeneous in RM (Fig S4A); thus we focused on the neutrophil population for further studies. Of note, following in vitro expansion in the presence of myeloid cytokines for 10 days, virtually all human CD34+ cells expressed CD33, in contrast to only 20–30% of cultured RM CD34+ cells (Fig S4B), highlighting some species-specific differences in CD33 expression.
We identified gRNA sequences targeting sites in exons 2 or 3 of the RM CD33 gene that showed activity in vitro in primary RM CD34+ cells. As levels of CD33 KO in RM CD34+ cells were not as robust as in human cells, we used two methods to increase the frequency of mutations: first, we used two gRNAs in combination, and second, we added a ssODN template analogous to that used in the human studies (Fig S4C, D). Both approaches were utilized in in vivo studies.
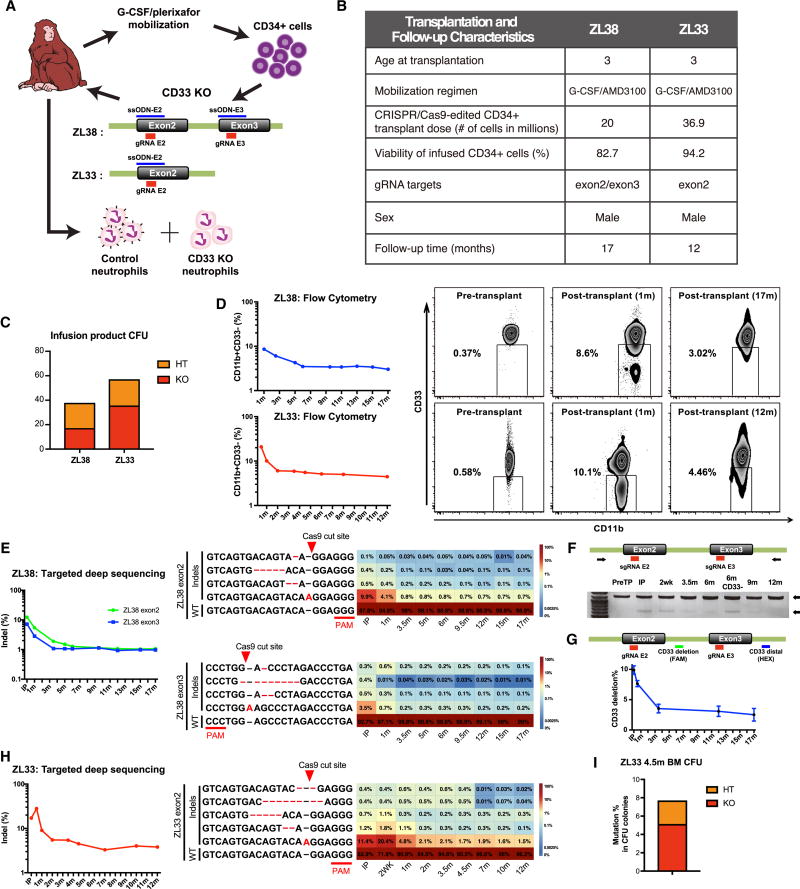
We performed apheresis collection and purification of CD34+ HSPC in two monkeys, ZL38 and ZL33. Autologous CD34+ HPSC were electroporated with the CD33-targeting gRNAs/Cas9 protein complexes and ssODN homologous to the targeted exons, and reinfused into the donor following myeloablative total body irradiation (Fig 4A). Fig 4B summarizes transplantation parameters for ZL38 and ZL33. Following autologous transplantation of CD33 KO HSPC, both RM engrafted promptly without transplantation-related complications. The pattern and rate of recovery of blood counts, including neutrophils and monocytes, was similar in these two animals compared to three historical controls (Wu et al., 2017; Wu et al., 2014) (Fig S5A–D). Furthermore, we found similar morphology and myeloperoxidase (MPO) expression in the marrow between ZL38/ZL33 and a control RM that underwent autologous transplantation with lentivirally transduced CD34+ cells (Fig S5E).
Fig. 4. CD33 KO HSPC show long-term multi-lineage engraftment in rhesus macaques.
(A) Schema of the rhesus macaque (RM) autologous CD33 KO transplantation model. RM were mobilized with G-CSF and plerixafor followed by leukapheresis, and CD34+ cells were selected and electroporated with Cas9 protein, CD33-targeting gRNA(s), and ssODN homologous to the targeted exons. For the first RM, ZL38, two gRNAs targeting the CD33 exon 2 and 3 coding sequence were used, with ssODNs designed with a single A insertion for each gRNA (ssODN-E2, ssODN-E3). For the second RM, ZL33, a single gRNA targeting the CD33 exon 2 with ssODN-E2 was used. Grey boxes indicate exons; Green lines indicate introns. Following gene editing, autologous CD34+ cells were re-infused back into the RM and presence of control (CD33+CD11b+) and CD33 KO (CD33−CD11b+) neutrophils was serially monitored in the peripheral blood by flow cytometry. (B) Transplantation characteristics of ZL38 and ZL33. (C) Cells from the infusion product were plated in MethoCult and individual colonies were harvested after 14 days. Each CFU colony was analyzed by PCR and sequencing of the CD33 gRNA targeted region. Percentage of CFU with heterozygous (HT) and homozygous (KO) mutations were scored. (D) Loss of CD33 expression on CD11b+ cells was longitudinally monitored on neutrophils from ZL38 and ZL33 by flow cytometry. Representative flow cytometry plot of pre-transplant, early (1 month) and late time point (ZL38; 17 months, ZL33; 12 months) post-transplantation are shown. (E) Left: Targeted deep sequencing of CD33 exon2 and 3 was performed in the infusion product and neutrophils of ZL38. Indel frequency of each region was plotted over time starting from the infusion product (IP), demonstrating the percentage of reads containing indels within 18bp gRNA window. Right: Individual indel types were identified by targeted deep sequencing analysis in ZL38 for exon2 and exon3 targets. The most abundant indel types in at least one of the depicted samples are plotted over time in all samples. PAM sequence and expected Cas9 cut site are shown. Insertions or deletions are shown in red font. Heatmap showing the read count contribution of indels over time during hematopoietic reconstitution in ZL38. Each row in the heatmap corresponds to an individual indel type and each column to a sample. Color gradient depicts the fractional contribution of individual indel to each sample. IP: infusion product. WT: wild type. (F) PCR was performed in pre-transplant and post-transplant neutrophils of ZL38 at indicated time points. Intact locus 1175 bp PCR product and a shorter 614 bp PCR product resulting from a deletion between the CD33 exon2 and exon3 target sites. (G) Deletion rate between the two gRNA cut sites was quantified by digital droplet PCR (ddPCR) with probes targeting the CD33 deletion and distal regions. Relative expression of each probe was calculated and plotted over time as CD33 deletion %. Error bars represent the standard deviation of the replicates. (H) Left: Targeted deep sequencing of CD33 exon2 was performed on the infusion product and neutrophils of ZL33. Indel frequency of CD33 exon2 was plotted over time. Right: Individual indel types identified by targeted deep sequencing analysis is depicted as in (E). (I) Bone marrow was collected 4.5 months post-transplantation from ZL33. CD34+ cells were selected and plated in MethoCult. After 14 days, individual CFU were analyzed by PCR and sequencing of gRNA targeting region (CD33 exon 2). Percentage of heterozygous (HT) and homozygous (KO) mutations were scored.
ZL38 received cells exposed to gRNAs targeting both exon 2 and exon 3, while ZL33 received cells exposed to the exon 2 targeting gRNA only. An aliquot of the HSPC infusion product showed 17% and 36% homozygous KO colony-forming units (CFU) in ZL38 and ZL33 respectively (Fig 4C). We were able to detect clear populations of CD33-negative neutrophils in the blood of both animals by flow cytometry following engraftment (Fig 4D). The levels were highest at 2–4 weeks following transplantation, and then decreased, plateauing by 6 months post-transplantation at levels of 3–6% in both animals, with no further significant decrease by 12 and 17 months post-transplantation in ZL38 and ZL33, respectively. This pattern suggests more efficient editing of short-term progenitors than long-term HSC.
To confirm editing at the CD33 target sequences, we tracked the edited allelic fractions over time using targeted deep sequencing to identify the specific indels introduced by CRISPR/Cas9. In ZL38, two gRNAs were used, and the target sites of each were sequenced individually. The frequencies of indels in the infusion product were 12% and 7% for exons 2 and 3 respectively, and highest levels were observed at 1 month in vivo, then plateaued at a level of about 1% for each exon over time (Fig 4E). The most frequent indel at both exon2 and exon3 targets was an A insertion at the Cas9 cut site presumably resulting from recombination with the ssODN template. Large deletions of the area between exon2 and exon3 were detected in approximately 2% of the alleles and Sanger sequencing confirmed this fragment represented fusions between sequences upstream of the exon 2 cut site and downstream of the exon 3 cut site (Fig 4F). The frequency of the deletion between the two gRNA cut sites of the CD33 gene was quantified by digital droplet PCR (ddPCR). Similar to flow cytometry or deep sequencing data, the CD33 deletion frequency dropped early during the post-transplantation period, and a deletion rate of 2–4% persisted long-term (Fig 4G). ZL33 was treated with a single gRNA and exhibited the same pattern of higher initial engraftment with edited cells, followed by stabilization at a level of approximately 4%, again with the A insertion most prominent (Fig 4H). We analyzed indel frequency in CFU grown from the marrow of ZL33 at 4.5 months post-transplant and observed 5% homozygous and 3% heterozygous mutations, within the range observed in circulating cells at the same time point (Fig 4I).
We sorted neutrophils based on CD33 expression. As expected, the frequency of indels in purified CD33-negative neutrophils was significantly higher than in the bulk neutrophil population, and CD33-positive neutrophils had the fewest indels (Fig S6A, B). We performed studies to assess maturation and differentiation of CD33 KO HSPC in ZL33. First, we examined the distribution of CD33 mutations across purified cells from various hematopoietic lineages in the blood. All showed similar CD33 indel frequencies, suggesting that CD33 KO HSPC maintain their ability to differentiate to all lineages (Fig S6C). We analyzed CD33 mutations in CD34+ cells, CD34− cells and granulocytes from marrow aspirates, and compared to CD33 mutations in circulating neutrophils, and found indel frequencies to be similar between progenitors and mature cells in the marrow or in the circulation (Fig 4H), suggesting normal myeloid maturation and release (Fig S6D). In summary, CD33 KO HSPC successfully engrafted and maintained long-term stable repopulation capacity, without evidence of perturbation with respect to hematopoietic lineage differentiation capacity.
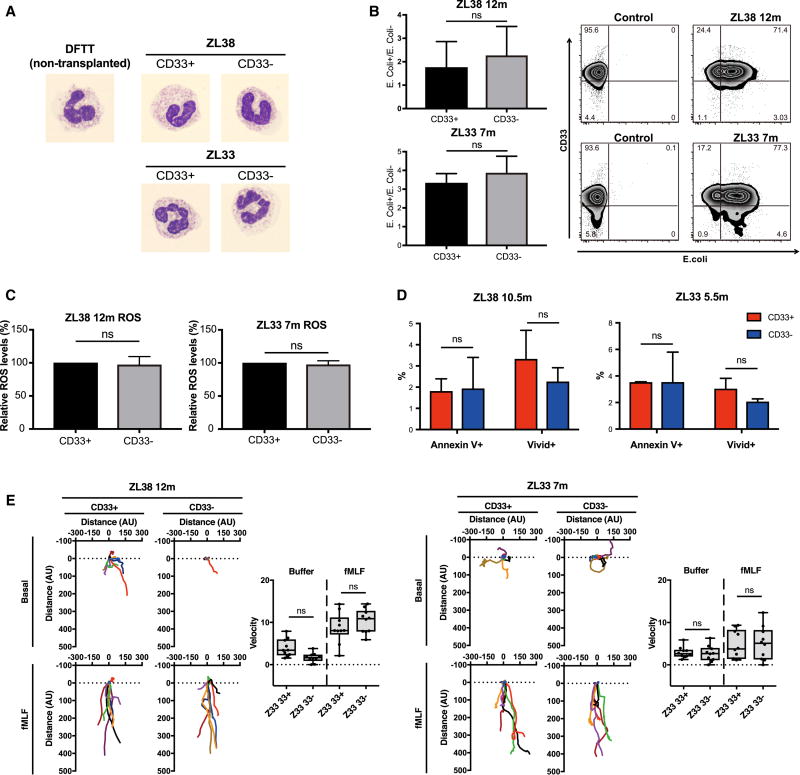
We next assessed the physiologic functions of CD11b+CD33− and CD11b+CD33+ neutrophils purified from the blood of ZL38 and ZL33. There were no differences in morphology between CD33+ and CD33− bands and neutrophils from the gene-edited animals, and all samples were similar to neutrophils from control RM (Fig 5A). We also studied phagocytosis and ROS production capabilities of CD33+ and CD33− neutrophils and found no differences (Fig 5B, C). Apoptosis and necrosis were also similar between CD33+ and CD33− neutrophils (Fig 5D). Finally, we tested neutrophil chemotaxis, and found no defect in chemotaxis of CD33− neutrophils at baseline or towards formyl-methionyl-leucyl-phenylalanine (fMLF), a prototypic chemotactic factor (Fig 5E). Taken together, our data suggest that CD33-deficient myeloid progenitors and mature neutrophils traffic and function normally, and that CD33 KO HSPC should be able to safely maintain myelopoiesis.
Fig. 5. Loss of CD33 does not perturb rhesus macaque neutrophil functions.
(A) Representative images of neutrophils from a non-transplanted control (DFTT) and sorted CD11b+CD33+ and CD11b+CD33− neutrophils from ZL38 and ZL33 stained with Giemsa and visualized by light microscopy at 1000× magnification. (B) Bacterial phagocytosis of internalized pHrodo Green E. coli bioparticles by CD11b+CD33+ or CD11b+CD33− cells from ZL38 and ZL33 at 7 or 12 months post-transplantation respectively. Bar graph (left) shows the mean ± SD, n=3. ns: not significant and the panel (right) shows representative flow cytometric plots. (C) Cellular reactive oxygen species (ROS) levels were detected by flow cytometry (CellROX™ Green Flow Cytometry Assay Kit) within CD11b+CD33+ or CD11b+CD33− gated cells at 7 or 12 months from ZL33 or ZL38, respectively. The relative ROS level was defined as the measured ROS level within the CD11b+CD33− cells relative to the level within the CD11b+CD33+ cells, which was set to 100%. (D) Percentage of annexin V+ or ViVid+ cells within the CD11b+CD33+ or CD11b+CD33− gates in blood cells collected at 10.5 months for ZL38 and 5.5 months for ZL33. Mean ± SD, n=3. ns: not significant (p>0.05). (E) Neutrophil chemotaxis of sorted CD11b+CD33+ or CD11b+CD33− cells measured using an EZ-TAXIScan on cells collected at 7 or 12 months post-transplantation from ZL33 or ZL38, respectively. Basal and fMLF conditions represent random migration and directed migration of neutrophils respectively. Ten randomly chosen cells were electronically traced using the acquired images and the paths of the cells plotted. Individual tracks are represented by different colors and were recorded over 1 hour. Scattergrams summarize the average velocities of the individual cells. ns: not significant. All data are represented as means ± SD.
CD33 KO HSPC are resistant to CD33-targeted therapy
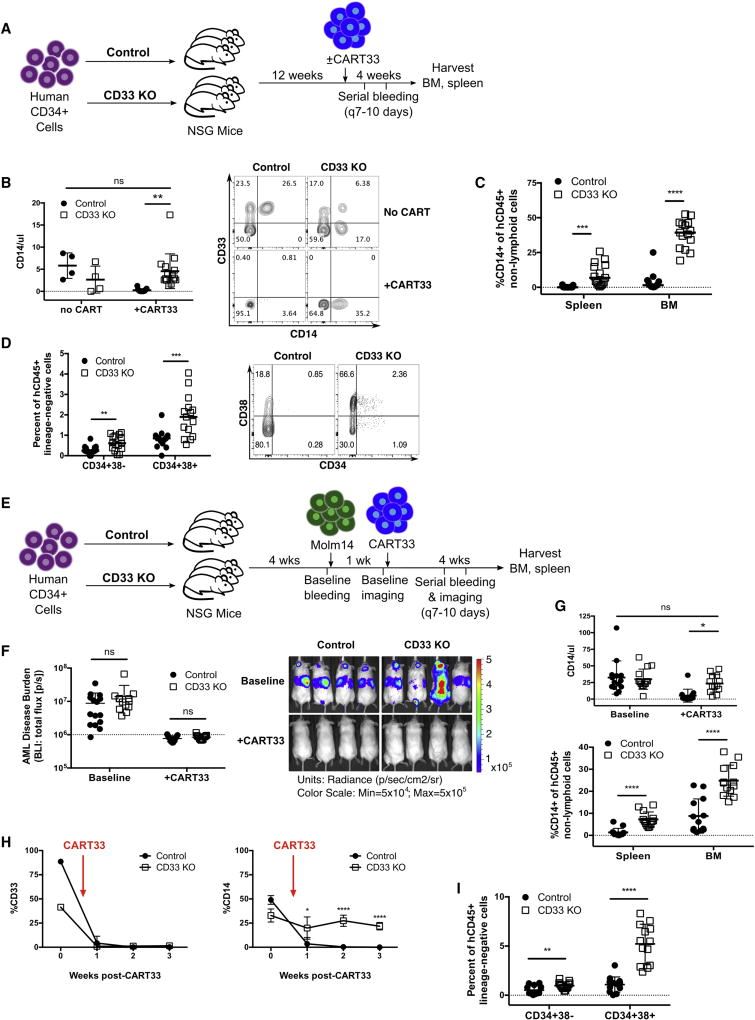
To demonstrate resistance of CD33 KO HSPC to CD33-targeted therapy, we turned to our human xenograft models. CAR T cells targeting CD33 (CART33) have been shown to eradicate AML, while also eliminating normal CD33+ myeloid cells (Dutour et al., 2012; Kenderian et al., 2015; Pizzitola et al., 2014; Wang et al., 2015). We treated mice engrafted with control or CD33 KO HSPC with autologous T cells transduced with lentivirus encoding an anti-CD33-41BB-CD3z CAR as previously reported (Kenderian et al., 2015) (Fig 6A). As expected, all CD33+ cells were eliminated in both groups, leading to complete disappearance of CD14+ human monocytes in the control mice (Fig 6B). Crucially, in the mice engrafted with CD33 KO HSPC, CD14+ cells were retained in the peripheral blood, spleen and marrow (Fig 6B, C). We also found sparing of human hematopoietic stem cells (CD34+38−) and progenitors (CD34+38+) in the marrow of mice engrafted with CD33 KO HSPC compared to controls, showing that the CD33 KO HSPC and their myeloid progeny are indeed resistant to CD33-targeted therapy (Fig 6D).
Fig. 6. Human myeloid cells lacking expression of CD33 are resistant to CD33-targeted therapy.
(A) Experimental schema. NSG mice engrafted with control or CD33 KO HSPC were treated with autologous anti-CD33 CAR T cells (CART33), followed by serial retro-orbital bleeding. Mice were euthanized and bone marrow (BM) and spleen were analyzed 4 weeks after CART33 infusion. A subset of mice in each group did not receive CART33. (Control-no CART33: n=4, CD33 KO-no CART33: n=4, Control+CART33: n=15, CD33 KO+CART33: n=15, 2 independent experiments, 2 donors). (B) Numbers of CD14+ monocytes in the peripheral blood of CD33 KO HSPC-engrafted mice after CART33 treatment are similar to control HSPC mice without CART33 treatment (one-way ANOVA) (left). Representative flow cytometry plot (right) shows complete eradication of CD33+ cells in both groups after CART33 treatment, which leads to loss of CD14+ monocytes in the control HSPC-engrafted mice, while CD14+CD33− cells are still detected in CD33 KO HSPC-engrafted mice. Gated on live singlet human CD45+CD19−CD3− cells. (C) CD14+ cells are present in significantly higher numbers in the spleen and bone marrow of CD33 KO HSPC-engrafted mice after CART33 treatment compared to controls (unpaired t-test). (D) Bone marrow human stem cells (CD34+38−) and progenitors (CD34+38+) are significantly higher after CART33 treatment in CD33 KO HSPC mice compared to controls (unpaired t-test). Representative flow cytometry plot showed on right, gated on live singlet human CD45+ lineage-negative cells. (E) Experimental schema. Control or CD33 KO HSPC-engrafted mice were injected with Molm14-GFP/luciferase prior to treatment with CART33 (n=26 mice, 2 independent experiments, 3 donors). (F) Mice show the expected reduction in AML burden as measured by bioluminescent imaging (BLI) after CART33 treatment (unpaired t-test). (G) Co-engraftment of AML does not impair the survival of CD33− negative myeloid cells in vivo after CART33 treatment in the peripheral blood (top; one-way ANOVA), spleen and bone marrow (bottom; unpaired t-test). (H) Peripheral blood CD33 levels decline after CART33 treatment in both cohorts of mice, resulting in undetectable levels of CD14+ cells in the control mice, while CD14+ cells persist in CD33 KO HSPC-engrafted mice (shown is one representative cohort of mice, single donor, n=4/group). (I) Bone marrow human stem and progenitor cells continue to survive CART33-mediated attack even in the setting of coexisting AML (unpaired t-test). All data are represented as means ± SD. ns: not significant (p>0.05); **p<0.01; ***p<0.001; ****p<0.0001.
The presence of CD33+ AML has the potential to stimulate CART33 proliferation and activation and thus cause bystander myelotoxicity. Therefore, we further interrogated the resistance of CD33 KO HSPC by injecting mice engrafted with control or CD33 KO HSPC with Molm14, a CD33+ AML cell line. We confirmed engraftment of Molm14 by bioluminescent imaging and then treated the mice with CART33 (Fig 6E). Leukemia responded to CART33 treatment as expected in all mice (Fig 6F). Importantly, we again found rapid clearance of CD33+ cells leading to myeloablation of non-leukemic human cells in the control HSPC mice, while mice engrafted with CD33 KO HSPC continued to sustain differentiated human myeloid cells and HSPC despite clearance of leukemic cells (Fig 6G–I). These studies show that CD33 KO HSPC can be used to circumvent the myeloid toxicity of potent CD33-targeted therapy while still permitting persistent anti-tumor efficacy.
Discussion
Our results provide the first proof-of-principle that a leukemia-specific antigen can be synthesized by removal of a shared antigen from normal tissue. Specifically, we have shown that the combination of CD33 KO HSPC with CART33 can generate a functional hematopoietic system that allows specific targeting of AML. The recent regulatory approvals of anti-CD19 CART for B-cell malignancies has accelerated the development of CART cell therapies for other malignancies, but tumor specificity remains a major challenge.
The approach we have proposed here can be viewed as a next-generation transplant, delivering both anti-leukemic T cells as well as HSPC resistant to CART33, followed by definitive therapy with CART33 to ensure disease eradication and potent targeted immunosurveillance. CD33 has been pursued as a target for AML for 30 years, culminating in the recent re-approval of gemtuzumab, a toxin-conjugated anti-CD33 monoclonal antibody, (Appelbaum and Bernstein, 2017). We postulate that the use of CART cell therapy against CD33 will lead to higher response rates, analogous to the situation with anti-CD19 CART cells. We anticipate that our strategy could be rapidly translated to the clinic. Since patients in morphologic remission of AML may harbor otherwise-undetectable leukemic cells, we would obviate the possibility of inadvertently gene-editing residual AML cells by generating the gene-edited HSPC and CAR T cells from allogeneic healthy donors. Patients would undergo conditioning and infusion of allogeneic donor-derived CD33 KO HSPC, followed by administration of CART33 from the same donor.
We could not demonstrate a specific role for CD33 in myeloid differentiation or function in vitro or in vivo in human-murine xenografts or in macaques, with the caveat that we achieved relatively low-level gene editing in the non-human primates. Our work contrasts with previous studies where CD33 was demonstrated to have a constitutive inhibitory effect on myeloid cells through its ITIM signaling domain (Ishida et al., 2014; Paul et al., 2000; Taylor et al., 1999; Ulyanova et al., 1999). One explanation for this discrepancy could be that while CD33 does have a suppressive function, other inhibitory cell surface receptors can compensate for loss of CD33. For example, multiple receptors within the CD33-related siglec family are also expressed on myeloid cells and have overlapping functions (Cao and Crocker, 2011; Nguyen et al., 2006), suggesting functional degeneracy within this group of proteins. In addition to our findings, the absence of any discernable abnormalities in CD33-deficient mice (Brinkman-Van der Linden et al., 2003) and the existence of humans with homozygous loss of CD33 also indicate that loss of CD33 may be well tolerated. Nonetheless, we cannot definitively exclude the possibility of occult functional defects in CD33 KO myeloid cells that may manifest when this therapy is extended to human subjects and followed long-term, or if patients encounter a specific inflammatory stimulus. Therefore, translation of this approach to humans will have to be undertaken carefully.
Gene-edited HSPC are being investigated for a variety of genetic diseases, utilizing a number of nuclease platforms including zinc finger nucleases, TALENS, and most recently CRISPR/Cas9. We developed a CRISPR/Cas9 gene-editing protocol resulting in consistent and sustained NHEJ-mediated disruption of the CD33 gene, resulting in efficient homozygous knockout and complete loss of CD33 cell surface expression on human HSPC and myeloid progeny. Longitudinal tracking of surface CD33 protein expression and mutations in CD33 showed that the levels of deletion were stable over time, including in long-term repopulating HSC. In RM HSPC, we found that CD33 KO was more difficult to achieve compared with human HSPC, but we were able to demonstrate low-level CD33− myeloid cells and bone marrow HSPC for up to 17 months. Of note, in a previous NHP model using zinc finger nucleases to disrupt CCR5, levels of gene disruption decreased from 40% early post-transplant to 3–5% at 6 months (Peterson et al., 2016), concordant with prior reports that long-term HSC are less susceptible to nuclease-mediated editing than short-term progenitors (Genovese et al., 2014). The unremarkable recovery course in the RM after autologous CD33 KO HSPC transplant was comparable with that seen after autologous transplantation of lentivirally-transduced HSPC, an approach that has already accumulated several decades of clinical experience. The CD33 KO RM model demonstrates for the first time that CRISPR/Cas9 technology can be utilized for genome engineering in a clinically-relevant large animal model with significantly enhanced biallelic gene-editing efficiency compared with previously published NHP data.
The mitigation of hematologic toxicity from CART33 requires sufficient CD33 KO HSPC to sustain myelopoiesis. The average level of long-term engrafted human CD33− myeloid cells produced within the human hematopoietic compartment in NSG mice was approximately 75%. These levels sustained normal numbers of myeloid cells in engrafted NSG mice following CART33 administration, and would be predicted to be sufficient in humans, based on the ability of even single HSPC to sustain hematopoiesis in preclinical studies (Notta et al., 2011) and the observation that only a few thousand clones are required for stable multilineage hematopoiesis in patients undergoing HSPC gene therapy (Biasco et al., 2016). However, in a clinical trial we would endeavor to infuse at least 2×106/kg CD33-deficient HSPC, as this should provide a safe margin for engraftment and hematopoiesis (based on the lower limit of acceptable cells for bone marrow transplantation and assuming that none of the CD33-sufficient HSPC remain after CART-33).
Finally, extensive evaluation of off-target activity failed to reveal sites of potentially critical genotoxicity occurring in CD33 KO HSPC. However, this does not exclude the possibility of novel off-target activity occurring due to human genetic variation (Scott and Zhang, 2017; Tsai et al., 2017; Yang et al., 2014), or important consequences arising from mutations in genomic sites hitherto classified as benign. Therefore, individual assessment of genotoxicity risks and close monitoring post-therapy will be essential during clinical translation.
Antigen-negative escape has been seen after anti-CD19 CAR treatment of ALL (Sotillo et al., 2015), and could potentially occur after successful treatment of AML with the approach proposed here. If this occurs in clinical trials, we could develop a multiplex editing approach to allow simultaneous targeting of multiple antigens. More broadly, this work represents a significant advance in the field of antigen-specific immunotherapy, by providing a pathway to avoid on-target off-tumor toxicity when a tumor-specific target is lacking. While our investigations have focused on CD33, this strategy could be extended to other cell-surface antigens, or to other forms of antigen-specific immunotherapy such as monoclonal antibodies, antibody-drug conjugates or bi-specific T cell engagers, thus widening the therapeutic index of a variety of treatment modalities in AML.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Saar Gill (saargill@pennmedicine.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
The Molm14 cell line was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ); the cell line was established from a 20 year old male. Cells were cultured in RPMI media with 10% fetal calf serum, penicillin and streptomycin. Molm14 cells were lentivirally transduced with luciferase/GFP under the control of the EF1α promoter and sorted to 100% GFP+ for mouse experiments.
The LCL8664 rhesus macaque cell line was obtained from the American Type Culture Collection (ATCC); it was derived from a 5 year old male. Cells were cultured in RPMI media with 10% fetal calf serum, penicillin and streptomycin.
Primary human CD34+ cells
G-CSF mobilized peripheral blood samples were obtained from left-over clinical specimens according to a University of Pennsylvania IRB-approved consent form for clinical hematopoietic stem/progenitor cell donation. Human subjects were de-identified and thus age and sex information are not available. CD34+ selection was performed using the CD34 Microbead Kit (Miltenyi Biotec, 130-046-702), and purity was confirmed by flow cytometry to be >95%. Cells were cultured in StemSpan SFEM (Stem Cell Technologies, 09650) supplemented with human cytokines (SCF 100ng/ul, Flt3 ligand 100ng/ul, TPO 50ng/ul, IL-6 50ng/ul, all purchased from Peprotech).
Primary human T cells
Autologous peripheral blood mononuclear cells remaining from the G-CSF mobilized peripheral blood donors after CD34+ cell selection were expanded in vitro using anti-CD3/CD28 Dynabeads (Life Technologies) in X-Vivo 15 media with 5% human serum, penicillin, streptomycin and glutamax. T cells were transduced with lentivirus containing the CAR33 construct as previously described (Kenderian et al., 2015). The human CD33 scFv was derived from the gemtuzumab ozogamicin antibody (clone P67.6), and the light and heavy chains were cloned into the murine CART19 plasmid vector (Milone et al., 2009), a third generation lentiviral vector in which the CAR is expressed under control of the EF-1α promoter. The CAR construct uses a light-to-heavy orientation of the scFv, followed by the human CD8 hinge and transmembrane, 4-1BB costimulatory domain and CD3zeta intracellular signaling domain. Lentivirus was generated by transient transfection of 293T cells using Lipofectamine 2000 (ThermoFisher Scientific, Cat#11668500). T cells were activated with CD3/CD28 Dynabeads (ThermoFisher Scientific, Cat#11132D) at a 1:3 ratio, followed by transduction with lentivirus 1 day after stimulation at a multiplicity of infection of 3. T cells were grown for 10–14 days prior to cryopreservation. Prior to all experiments T cells were thawed and rested at 37°C for 4–16 hours.
Xenogeneic mouse transplantation model
Male and female 8–12 week old NOD-SCID-IL2rγ−/− (NSG) mice were purchased from Jackson laboratories or bred in-house. All experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania. Mice were maintained in dedicated BSL-2 animal barrier spaces. Age and sex-matched animals were randomly assigned to experimental groups; no gender-specific influences were detected in the experimental results.
1–5×105 control or gene-edited human CD34+ cells were infused by tail vein injection into the mice after prior conditioning with busulfan 30mg/kg intraperitoneally. Mice were evaluated by serial retro-orbital bleeding. Mice were also injected with Molm14 (1×106 cells) and/or autologous CAR T cells (5×106 cells) after CD34+ cell engraftment was confirmed (range: 4–12 weeks post-CD34+ cell injection). After Molm14 injection, mice underwent weekly bioluminescent imaging using a Xenogen IVIS-200 Spectrum camera. Images were acquired and analyzed using Living Image version 4.4 (Caliper LifeSciences, Inc., PerkinElmer).
Rhesus macaque animal care and oversight
All rhesus macaques were housed, handled and underwent procedures conforming to all regulatory standards, and as specified in a protocol approved by the National Heart, Lung and Blood Institute Animal Care and Use Committee. Both ZL38 and ZL33 were male, three years of age (older juveniles just before sexual maturity) at the time of transplantation (Figure 4B). They were heathy and specific pathogen free (SPF) at the time of initiation of transplantation procedures. Neither had been previously enrolled in any other experimental study, undergone prior procedures, or been treated with any medications other than for routine clinical care. Animals were visually monitored daily and underwent quarterly full physical exams. Monitoring of complete blood counts and serum chemistries were monitored at least three times a week following transplantation until complete recovery, and then at least quarterly.
Rhesus macaque mobilization, HSPC collection and autologous transplantation procedures
At least one week prior to HSPC collection, an indwelling venous catheter was placed surgically into the internal jugular vein and tunneled under the skin to an exit site between the shoulder blades. This catheter remains in place until the animal recovers normal blood counts and activity post-transplantation, and is used for administration of medications and blood transfusions, blood drawing, and for apheresis.
Rhesus macaque Mobilization, HSPC collection, and immunoselection were performed as previously described (Donahue et al., 2005; Wu et al., 2014). Briefly, HSPC mobilization was performed via administration of G-CSF (Amgen) 15 ug/kg/day subcutaneously (SC) for 6 days, and AMD3100 (Sigma) 1 mg/kg SC on day 5 and day 6, the days of apheresis, 2–4 hours before apheresis was initiated. Apheresis was performed using a Fenwal CS-3000 cell separator (no longer manufactured), using a saphenous vein temporary catheter as an inlet port and the central line catheter as an outlet port. Apheresis was performed for 2 hours on days 5 and 6 to generate a mononuclear cell products of 35–45 ml each. CD34+ immunoselection of the concentrated mononuclear cell products was performed using the anti-mouse IgM MicroBeads (Miltenyi Biotec, 130-047-301) and the anti-CD34 hybridoma clone 12.8 (a murine IgM anti-human CD34 cross-reactive with rhesus CD34). The antibody was derived at the Fred Hutchinson Cancer Center, Seattle, WA and is not commercially-available, but access can be arranged through Dr. Irwin Bernstein at the Fred Hutchinson Cancer Center.
After completion of HSPC collection, fractionated total body irradiation (TBI) at a dose of 5Gy rads daily for two days for a total dose of 10Gy was given at a dose rate of 0.5Gy/minute, using a cobalt source located at the Armed Forces Radiobiology Research Institute (Bethesda, MD). Following the 2nd dose of TBI, edited CD34+ cells (see below) were washed, resuspended in PBS/2% FBS/Heparin (10U/ml) and infused via the central line.
METHOD DETAILS
DNA/RNA Methods
Guide RNA design and production
Human codon optimized Cas9 expressed under the T7 promoter was kindly provided by Dr. Yangbing Zhao, and Cas9 mRNA was in vitro transcribed using the mMessage mMachine T7 Ultra kit (Ambion, AM1345). Cas9 protein was purchased from PNA Bio (CP02).
The human CD33-targeting gRNA was obtained by screening the top 5 gRNAs previously reported to have high KO efficiency for CD33 (Doench et al., 2014). Full length and truncated gRNA sequences are as follows:
| Full length | Truncated | |
|---|---|---|
| gRNA1 | tggggtgattatgagcaccg | ggtgattatgagcaccg |
| gRNA2 | tgagcatcgtagacgccagg | gcatcgtagacgccagg |
| gRNA3 | atccctggcactctagaacc | gcctggcactctagaacc |
| gRNA4 | gagtcagtgacggtacagga | gtcagtgacggtacagga |
| gRNA5 | tgtcacatgcacagagagct | gcacatgcacagagagct |
The control EMX1-targeting gRNA was previously reported (Cong et al., 2013); sequence is as follows: 5’-GAGTCCGAGCAGAAGAAGAA-3’. All gRNAs were generated using the pUC57-sgRNA expression vector (Addgene plasmid #51132) using overlapping DNA oligonucleotides to generate the gRNA insert; cloning was performed using standard molecular biology techniques. The gRNAs were in vitro transcribed using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, #K0441). RNA was purified using the RNeasy Mini Kit (Qiagen, 74104). Truncated gRNA4 was found to be most efficacious and was used for all subsequent experiments.
The rhesus macaque CD33-targeting gRNA E2 was selected due to its homology to the gRNA used for human, as it differs by only one nucleotide. The E3 gRNA was designed using Benchling (https://benchling.com); gRNAs with high on-target scores were screened in a rhesus macaque cell line, LCL8664, and E3 was identified as having high efficacy at generating mutations in the CD33 gene as determined by DNA PCR and TIDE analysis.
Single-stranded oligodeoxynucleotide (ssODN) design
We added a ssODN to the electroporation to increase the frequency of knockout mutations. The ssODN was designed to have 99 base pair homology arms around the Cas9 cut site of the CD33 gene, and an A nucleotide insertion that generates a frameshift mutation and premature truncation of the CD33 protein. The ssODN was ordered from IDT (Integrated DNA Technologies) as an 4nmole Ultramer DNA Oligo. Lyophilized oligo was resuspended in 40ul TE buffer and 1ul was used for each electroporation of 2×106 cells in 500ul OptiMEM.
Human CD33 locus DNA analysis
Genomic DNA was extracted from the CD34+ cells using the High Pure PCR Template Preparation Kit (Roche, 11796828001). PCR primer sequences are listed in Table S5. Surveyor Mutation Detection kit (Transgenomics) was used to detect the presence of mutations and band intensities were analyzed using ImageJ software. PCR amplicons were analyzed by Sanger sequencing and allele modification frequency was calculated using TIDE (Tracking of Indels by Decomposition) software available at http://tide.nki.nl.
Rhesus macaque CD33 locus DNA analysis
Genomic DNA from hematopoietic cells was extracted using the DNeasy Blood & Tissue kit (Qiagen, Germantown, MD). Deletions between gRNAs E2 and E3 were detected by PCR amplification using F1 and R2, then quantified with a digital droplet PCR (ddPCR) (Bio-Rad) following the manufacturer’s instructions. Briefly, genomic DNA from neutrophils was digested by HindIII (NEB) and diluted to 5ng/ul. Each reaction included 10 µl of 2× ddPCR Supermix for probes (no dUTP), 1 µl of CD33 deletion primer/probe mix (FAM), 1 µl of CD33 distal primer/probe mix (HEX), and 30–100 ng of genomic DNA. The samples were processed using the QX200™ Droplet Digital™ PCR system, and the CD33 deletion rate was calculated with the QuantaSoft software (Bio-Rad). All primer sequences are listed in Table S5.
Human Cell Methods
Cell line electroporation
An AML cell line, Molm1, was used for human gRNA screening. Cells were washed once and resuspended in Opti-MEM. Cas9 mRNA 10ug was added to cell suspension and electroporated with the BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus) using a single pulse of 400V and 5msec. Cells were incubated at 32°C overnight and re-electroporated with gRNA 5ug the next day using the same machine and settings. Cells were kept at 32°C until the following day, after which they were cultured at 37°C until analysis.
Human CD34+ cell electroporation
Cas9 protein and gRNA were mixed and incubated at room temperature for 10 minutes prior to electroporation. CD34+ cells were washed and resuspended in Opti-MEM and electroporated with the Cas9 ribonucleoprotein complex using the BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus) with a single pulse of 400V and 5msec. 10ug of Cas9 and 5ug of gRNA was used for 2×106 CD34+ cells in 750ul total volume. After electroporation, cells were incubated at 32°C until the following day, after which they were cultured at 37°C until analysis.
In vitro human CD34+ cell differentiation
CD34+ cells were cultured in IMDM+10% FBS and the following human cytokines: SCF 100ng/ul, Flt3 ligand 100ng/ul, TPO 50ng/ul, IL-6 50ng/ul, GM-CSF 100ng/ul, IL-3 10ng/ul (all purchased from Peprotech). Cells were maintained at 0.5–1×106 cells/ml for 5–7 days prior to analysis.
Morphological analysis
Cytospin specimens of in vitro differentiated control or CD33 KO human CD34+ cells were prepared and immunocytochemistry for CD33 was performed using an anti-CD33 antibody (Novocastra NCL-L-CD33). Staining was done on a Leica Bond-IIITM instrument using the Bond Polymer Refine Detection System (Leica Microsystems DS9800). Heat-induced epitope retrieval was done for 20 minutes with ER1 solution (Leica Microsystems AR9961). Incubation with the anti-CD33 antibody was performed at 1:150 dilution for 15 min. followed by 8 min post-primary step and 8 min incubation with polymer HRP. Endogenous peroxidase was blocked for 5 min, followed by 10 min incubation with DAB (3,3’-diaminobenzidine). All procedures were performed at room temperature. Slides are washed three times between each step with bond wash buffer or water.
Human colony forming cell (CFC) assay
One day after electroporation, 1000 CD34+ cells were plated in 1.1ml of methylcellulose (MethoCult H4435 Enriched, Stem Cell Technologies) on 6 well plates in duplicate and cultured for two weeks, after which colonies were counted and scored. Individual colonies were picked and heat lysed in 40ul of lysis buffer containing 50mM NaOH and 0.2mM EDTA. Samples were heated to 95°C for 20 minutes then cooled down, after which 1ul of 1M TrisCl was added. 2ul of reaction was used for PCR with AccuPrime Pfx SuperMix (Invitrogen, 12344-040) as per manufacturer’s instructions. Alternatively, MethoCult wells were solubilized with RPMI media overnight and flow cytometry was performed on single-cell suspensions.
Flow cytometry
In vitro differentiated CD34+ cells were analyzed by flow cytometry 5–7 days after electroporation using the following antibodies: CD11b-FITC (BioLegend, 301329), CD14-APC (BD, 340436), CD33-PE (Thermo Fisher Scientific, 12-0339-41), CD45-BV421 (BioLegend, 304032), and Live/Dead Fixable Aqua (Life Technologies, L34957).
For in vivo studies assessing human hematopoietic engraftment, the following panel was used: mouse CD45-APC/Cy7 (BioLegend, 103116), human CD45-BV421 (BioLegend, 304032), Live/Dead Fixable Aqua (Life Technologies, L34957), CD19-PE/Cy7 (BioLegend, 115520), CD3-BV605 (BioLegend, 317322), CD11b-FITC (BioLegend, 301329), CD14-APC (BD, 340436), CD33-PE (Thermo Fisher Scientific, 12-0339-41). For the in vivo studies using Molm14-GFP/luciferase, CD11b-FITC was removed from the panel. For the in vivo studies looking at the effects of G-CSF stimulation, CD66b-PerCP/Cy5.5 (BioLegend, 305107) and CD56-BV711 (BioLegend, 318335) were added. Countbright beads (Invitrogen) were used to quantify cell numbers.
Analysis of bone marrow human HSPC subsets was performed using the following panel: mouse CD45-APC/Cy7 (BioLegend, 103116), human CD45-BV421 (BioLegend, 304032), Live/Dead Fixable Aqua (Life Technologies, L34957), lineage-FITC (Thermo Fisher Scientific, 22-7778-72), CD34-APC (BioLegend, 343608), CD38-BV711 (BioLegend, 303528), CD33-PE (Thermo Fisher Scientific, 12-0339-41). All flow cytometry was performed on a BD Fortessa, and analysis was performed using FlowJo X10.
Intracellular cytokine assay of human myeloid cells
Cells were incubated with either monensin (BioLegend, 420701) or monensin and LPS for 4 hours at 37°C. Subsequently, cells were harvested and surface staining with CD33-PE was performed, followed by fixation with Invitrogen Fixation Medium A (GAS001S5) for 15 minutes. Cells were then incubated for 20 minutes with Permeabilization Medium B (GAS002S5) containing the following antibodies: MIP1b-PE/Cy7 (BD Pharmingen, 560687), IL-8-AF488 (BioLegend, 511412), and TNFa-AF700 (BioLegend, 502928).
Phagocytosis assay
In vitro differentiated control or CD33 KO human myeloid cells were incubated with pHrodo green E. coli bioparticles (Thermo Fisher Scientific, P343666) for 1 hour at 37°C. Cells were then washed once with Flow Buffer (PBS with 1% FBS and 0.1% sodium azide) and stained with CD33-PE (Thermo Fisher Scientific, 12-0339-41) and Live/Dead Fixable Aqua (Life Technologies, L34957) for 15 minutes at room temperature. Cells were then washed again with Flow Buffer and acquired by flow cytometry to quantify phagocytosis.
Cellular reactive oxygen species (ROS) analysis
In vitro differentiated control or CD33 KO human myeloid cells were stained with CellROX™ Green Flow Cytometry Assay Kit and evaluated for ROS by flow cytometry according to the manufacturer’s protocol (Thermo Fisher Scientific, C10492). In brief, cells were incubated with PMA (5ng/ul) for 15 minutes at 37°C, after which the CellROX reagent was added at a final concentration of 500 nM. Cells were further incubated for 30 minutes at 37°C, then washed once with Flow Buffer and stained with CD33-PE (Thermo Fisher Scientific, 12-0339-41) and Live/Dead Fixable Aqua (Life Technologies, L34957) for 15 minutes at room temperature. Cells were then washed again with Flow Buffer, and flow cytometry was performed to quantify ROS.
In vivo functional studies of xenografted human HSPC
NSG mice engrafted with human CD34+ cells were injected with G-CSF 300ug/kg intraperitoneally twice daily for 2 days at 8 weeks post-transplantation. Frequency of human CD66b+ cells (neutrophils) and CD14+ cells (monocytes) were analyzed before and after G-CSF treatment. At 12 weeks post-transplantation, mice were given 10ug of LPS intraperitoneally, and serum was collected at 0 and 3 hours after injection. Levels of human inflammatory cytokines were measured by BD cytometric bead array, Human Inflammatory Cytokine Kit (BD Biosciences, 551811).
Cytogenetics of human hematopoietic cells
Primary human CD34+ cells electroporated with Cas9 and either control or CD33-targeted gRNA and cultured for 7 days in IMDM+10% FBS with the following human cytokines: SCF 100ng/ul, Flt3 ligand 100ng/ul, TPO 50ng/ul, IL-6 50ng/ul, GM-CSF 100ng/ul, IL-3 10ng/ul. Samples were then treated with colcemid (Life Technologies, 15210-040) and hypotonic solution (homemade solution of EGTA 0.4g (Sigma #4378), Hepes buffer powder 9.6g (Life Technologies, 11344-025), KCl 6.0g in 2L of distilled water; pH adjusted to 7.4 with 10mM NaOH) for 55 minutes, and fixed with a 3:1 methanol/acetic acid solution. Slides were prepared and G-banding was performed using trypsin-Wright staining (Trypsin-EDTA: Life Technologies, 15400-054; Wright stain stock solution: add 3g wright stain powder (Sigma, W0625-25g) to 1L methanol; Wright stain working solution: add 1ml stock solution to 3ml pHydrion 6.8 buffer working solution). Metaphases were analyzed using Cytovision chromosome analysis software (Leica Biosystems) and the results were described according to the International System for Human Cytogenetic Nomenclature (ISCN 2016). A clone is defined by conventional cytogenetics as the presence of at least two cells with the same chromosome gain or structural rearrangement, or at least three cells with the same chromosome loss.
Mass cytometry analysis of human hematopoietic cells
In vitro differentiated control or CD33 KO human HSPC were generated from three normal donors and cultured in cytokine-deplete media (1:32 dilution of differentiation media with cytokine-free IMDM) overnight prior to the assay. Cells were incubated with cisplatin at 50µM concentration for 1 minute at room temperature for viability staining, then washed and resuspended in cytokine-deplete media. Cells were divided into 10 groups and following cytokines were added to each: none (basal), TPO (50ng/ml), G-CSF (20ng/ml), GM-CSF (20ng/ml), IFNα (5000U/ml), IFNγ (20ng/ml), IL-4 (20ng/ml), IL-6 (50ng/ml), PMA/ionomycin (50nM and 1ug/ml each), LPS (100ng/ml). Cells were incubated at 37°C for 15 minutes after which they were immediately fixed by addition of 1/10th volume of 16% paraformaldehyde (PFA) for 10 minutes at room temperature. Cells were then snap frozen in PBS with 0.5% FBS, 0.02% sodium azide and 10% DMSO and stored at −80°C.
Cells were thawed at 4°C and then washed once with cell staining media (CSM; 1× PBS with 0.5% BSA and 0.02% sodium azide). Cells were then barcoded in groups of 20 cell samples from each donor (2 million cells from each of the 10 stimulation conditions for the control and CD33 KO cells) using a previously described protocol (Zunder et al., 2015). Briefly, cells were washed twice in PBS at 4°C followed by a 15 minute incubation with palladium barcoding reagent (Fluidigm); the barcoding incubation (and all subsequent steps unless otherwise noted) were performed at room temperature. This barcoding protocol permanently labels each of the 20 samples with a unique combination of 3 of the 6 stable Pd isotopes which allows the cells from each sample to be identified. All 20 samples from each donor were then washed three times in CSM and combined into a single tube for antibody staining and mass cytometry analysis. Mass cytometry staining of the 40 million combined cells was performed in a staining volume of 2mL for 50 minutes at room temperature with gentle agitation using separate staining steps for the surface antigens and intracellular antigens. The antibodies utilized for staining are listed in Supplemental Table 2. Antibodies were either purchased from Fluidigm with metal tags attached, or were metal tagged using the Fluidigm MaxPar X8 polymer kit according to the manufacturer’s protocol. After each staining step, cells were washed twice in CSM. After surface staining, cells were fixed with 1.5% PFA and then permeabilized with 100% methanol for 15 minutes at −20°C prior to the intracellular stain. After completion of intracellular staining cells were washed twice in CSM and then incubated in Irridium intercalator solution (PBS with a 1:5,000 dilution of the iridium intercalator pentamethylcyclopentadienyl-Ir(III)-dipyridophenazine [Fluidigm] and 1.5% PFA) at 4°C for 16–72 hours prior to analysis.
Prior to analysis excess intercalator solution was removed with one CSM wash and two washes in pure water. Cells were then resuspended in pure water at a concentration of approximately 1 million per mL and mixed with mass cytometry measurement standard beads (Fluidigm). Cell events were acquired on a Helios mass cytometer (Fluidigm) at an event rate of 100 to 300 events per second with instrument-calibrated dual-count detection (Ornatsky et al., 2008). Instrument setting were as follows: noise reduction was on, event duration of 8 to 150, lower convolution threshold of 600, sigma = 3, dual count start = 1. After data acquisition, the mass bead signal was used to correct short-term signal fluctuation during the course of each experiment, and bead events were removed (Finck et al., 2013). An average of approximately 800,000 cell events were collected for each sample (range 568,274 – 1,029,292 events).
Mass cytometry analysis was performed using Cytobank (www.cytobank.org; (Kotecha et al., 2010)). SPADE analysis (Qiu et al., 2011) was performed using Cytobank on a sample of approximately 40% of total events (to facilitate computational analysis). Clustering markers are indicated in table S2, down-sampling was performed to a target 8% of total events, clustering was performed to a target of 100 nodes, and fold change was calculated relative to the average of the basal condition of the control cells from the three donors. Approximate immunophenotypic annotations are indicated on each SPADE tree and assignment was performed manually based on a best approximation of the cell immunophenotype and signaling responses; given the artificial nature of the in vitro differentiation, the annotations represent a best estimation and may not be perfectly accurate. Mass cytometry data are plotted on an arcsinh transformed scale.
Rhesus Macaque Cell Methods
Rhesus macaque HSPC electroporation and autologous transplantation
Guide RNAs and Cas9 protein were mixed and incubated for 10 minutes before electroporation. Rhesus macaque CD34+ cells were washed with phosphate buffered saline (PBS) and resuspended in Opti-MEM. Subsequently, the cell suspension was mixed with the Cas9 RNP, then electroporated using the BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus) with a single pulse of 400V for 5msec. 30–60ug of Cas9 protein and 15–30ug of gRNA were used for electroporation of aliquots of 3–5×106 CD34+ cells in a total volume of 750ul. After electroporation, cells were pooled and incubated in X-VIVO™ 10 (Lonza, BE04-743Q) supplemented with 1% HSA (Baxter, 2G0012) and cytokines (SCF 100ng/mL, FLT3L 100ng/mL and TPO 100ng/mL; all purchased from PeproTech) at 32°C until the following day, when they were reinfused into the autologous macaque following total body irradiation (500 rad/day for 2 days).
Rhesus macaque peripheral blood flow cytometric analysis
Rhesus macaque PB was processed via centrifugation over Lymphocyte Separation Medium (MP Biomedicals, 0850494X). PBMNC (peripheral blood mononuclear cells) and neutrophils were separated and treated with ACK lysing buffer (Quality Biological, 118-156-101). Cells were stained with the following antibodies: CD45-BV605 (BD Biosciences, 564098), CD45-APC (BD Biosciences, 561290), CD11b-FITC (BioLegend, 301329), CD14-pacific blue (Thermo Fisher Scientific, MHCD1428), CD3-BV786 (BD Biosciences, 563918), CD20-APC-cy7 (BD Biosciences, 335794), CD56-PE (BD Biosciences, 555516), CD16-APC (BioLegend, 302012), and CD33-PE (Miltenyi Biotec, 130-091-732). Flow cytometric analysis and/or sorting were performed on a FACSARIA-II flow sorter (BD), and data were analyzed using FlowJo software (Tree-star Inc.).
Rhesus macaque colony forming unit (CFU) assay
Single CD34+ cell-derived CFUs were grown for 14 days in MethoCult™ H4435 Enriched (STEMCELL Technologies). For infusion product CFUs, CD34+ cells electroporated with Cas9 RNP were incubated for 24 hours at 32°C, then plated with 1.1ml of MethoCult H4435 in 35-mm CFU dishes at a density of 2000 cells/dish. For ZL33’s 4.5m BM CFUs, 20 ml of BM aspirate was collected, and CD34+ cells were immunoselected as previously described (Donahue et al., 2005; Wu et al., 2014). BM-derived CD34+ cells were plated with 1.1ml of MethoCult H4435 in 35-mm CFU dishes at a density of 2000 cells/dish. The remaining cells were used for targeted deep sequencing. Single CFU colonies were picked, and DNA was extracted with the Maxwell® 16 Cell LEV DNA Purification Kit (Promega, AS1140) on day 14 of plating.
Rhesus bone marrow immunohistochemistry
Rhesus macaque bone marrow trephine biopsies were fixed in B-Plus fixative, decalcified, embedded in paraffin, and processed for morphologic evaluation using standard procedures. Bone marrow sections were stained with hematoxylin and eosin (H&E), and immunohistochemistry for MPO (Ventana Medical Systems, 760-2659) was performed using the Ventana Benchmark Ultra platform (Ventana Medical Systems). Images were using an Olympus BX-41 microscope (Olympus America) equipped with a DPlan 10/50 numeric aperture objectives and captured using an Olympus DP70 digital camera system.
Rhesus macaque neutrophil apoptosis and necrosis analysis
RM neutrophils were stained with Annexin V-PE and evaluated for apoptosis by flow cytometry according to the manufacturer’s protocol (BD, 559763). Briefly, cells were washed with PBS and stained with 5 µl of Annexin V-PE in 1 × binding buffer for 15 minutes at room temperature in the dark. For the ViViD assay, 1 × 106 cells were washed with PBS and resuspended in 1ml of PBS. 1 µl of reconstituted fluorescent reactive dye (Thermo Fisher Scientific, L34964) was added to the cell suspension, and the cells were incubated on ice for 30 minutes in the dark. Cells were then washed with PBS, and apoptotic/necrotic cells were determined using a FACSARIA-II flow sorter (BD).
Rhesus macaque neutrophil chemotaxis assay
Neutrophil chemotaxis assay was performed on freshly isolated CD33+ and CD33− cells using the EZ-TAXIScan (Effector Cell Institute, Tokyo, Japan). In brief, 5 × 103 neutrophils were added to the “Cell” well of the EZ-TAXIScan and 1 µl of either buffer or fMLF (5×10−8 M) was added to the opposing “Chemoattractant” well. Images of cellular migration were captured every 30 sec for 60 min at 37°C and ten randomly selected cells were tracked digitally using the ImageJ plug-in, MTrackJ. The paths of the migrating cells were plotted with the position of each cell at t = 0 anchored at the origin. Using the coordinates of the individual cells in each image, the distance that each cell migrated and the average velocity were calculated using the distance formula.
Rhesus macaque phagocytosis assay
CD33 KO HSPC-transplanted rhesus macaques peripheral blood cells were incubated with pHrodo green E. coli bioparticles (Thermo Fisher Scientific, P343666) for 1 hour at 37°C. Cells were washed once and stained with CD33-PE for 15 minutes at room temperature. Cells were then washed again and acquired by flow cytometry to quantify phagocytosis.
Rhesus macaque reactive oxygen species (ROS) analysis
Rhesus macaque peripheral blood cells were stained with CellROX™ Green Flow Cytometry Assay Kit and evaluated for ROS by flow cytometry according to the manufacturer’s protocol (Thermo Fisher Scientific, C10492) (see human ROS analysis for details).
Next-Generation Sequencing Methods
RNA-sequencing of human hematopoietic cells
RNA was isolated from 5 matched samples of in vitro differentiated control/CD33 KO human HSPC using Ambion RiboPure RNA purification kit (Thermo Fisher Scientific, AM1924). RNA-seq libraries were generated using TruSeq RNA library prep kit (Illumina, RS-122-2001/2) per the manufacturer’s instructions. The libraries were sequenced as 100bp single end using a HiSeq 2500 sequencer (Illumina). Sequencing reads were aligned to the human genome (hg19) using RUM, and differential expression was analyzed using edgeR.
Off-target evaluation: In silico sites
Off-targets sites were selected based on two web tools: crispr.mit.edu and CCTop (crispr.cos.uniheidelberg.de). We selected the top 5 coding sites and the top 3 non-coding sites from each web tool that were predicted to have off-target activity based on sequence homology to the CD33− targeting gRNA. We PCR amplified these off-target loci from genomic DNA extracted from CD34+ cells treated with CRISPR/Cas9, using 5 matched control and CD33 KO donors. PCR products were designed to be 385±24bp around the gRNA cut site. PCR products from each donor were pooled and libraries were generated using NEBnext. Sequencing was performed on the MiSeq 500 (Illumina) with 250bp paired end reads. Read counts obtained per sample and per target are shown in Table S3. Point mutations and INDELs were identified through the in-house pipeline. Raw reads (fastq) were mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA). Low complexity regions (LCRs) were removed from the mapped reads using bedtools and then realigned for the INDEL region by GATK IndelRealigner. GATK BaseRecalibrator was then use to recalibrate the filtered reads based on quality. Point mutations and INDELs were called by the variant caller, VarScan2 limited to the mutations that had an allele frequency > 0.1%, a minimum read depth > 8 and existence only in CD33 KO samples. The variants were then annotated by the ANNOVAR. Sequences with indels of ≥ 1 bp located within 5 bp of the predicted Cas9 cut site on either side were considered CRISPR/Cas9 induced genome modifications.
CIRCLE-seq
Genomic DNA was isolated using Gentra Puregene Kit (Qiagen) according to manufacturer’s instructions. CIRCLE-seq was performed as previously described (Tsai et al., 2017). Briefly, purified genomic DNA was sheared with a Covaris E200 instrument to an average length of 300 bp. The fragmented DNA was end repaired, A tailed and ligated to a uracil-containing stem-loop adaptor, using KAPA HTP Library Preparation Kit PCR Free (KAPA Biosystems). Adaptor ligated DNA was treated with Lambda Exonuclease (NEB) and E. coli Exonuclease I (NEB) and then with USER enzyme (NEB) and T4 polynucleotide kinase (NEB). Intramolecular circularization of the DNA was performed with T4 DNA ligase (NEB) and residual linear DNA was degraded by Plasmid-Safe ATP-dependent DNase (Epicentre). In vitro cleavage reactions were performed with 125 ng of Plasmid-Safe-treated circularized DNA, 90 nM of SpCas9 protein (NEB), Cas9 nuclease buffer (NEB) and 90 nM of in vitro transcribed gRNA, in a 50 µl volume. Cleaved products were A tailed, ligated with a hairpin adaptor (NEB), treated with USER enzyme (NEB) and amplified by PCR with barcoded universal primers NEBNext Multiplex Oligos for Illumina (NEB), using Kapa HiFi Polymerase (KAPA Biosystems). Libraries were quantified by droplet digital PCR (Bio-Rad) and sequenced with 150 bp paired-end reads on an Illumina MiSeq instrument. CIRCLE-seq data analyses were performed using open-source CIRCLE-seq analysis software (https://github.com/tsailabSJ/circleseq).
Off-target evaluation: CIRCLE-seq sites
We selected the top 7 sites with highest CIRCLE-seq read count (c10orf20, DGKG, EPS8, ALDH3B1, RN7SL601P, DAPK2, TSPAN2) and all sites with 3 or less mismatches to the on-target site to validate in primary CD33 KO cell samples. PCR amplification, sequencing and analysis was performed in a similar fashion to the in silico predicted off-target evaluation. Read counts obtained per sample and per target are shown in Table S4.
Targeted deep sequencing of rhesus macaque CD33 locus
Next generation sequencing was conducted using the Miseq sequencer (Illumina) with 300bp paired end reads for indel identification. PCR products were amplified using F1 and R1 for the gRNA E2 targeting region and F2 and R2 for the gRNA E3 targeting region as described above. Sequencing depth was generally between 50,000 – 300,000 reads/gRNA target site. To account for single nucleotide polymorphisms unique to individual monkeys, a consensus sequence for each targeted gene in each edited monkey was generated from the pre-transplantation sequencing data with a custom Python pipeline. CRISPResso (http://crispresso.rocks) was then used to align sequencing reads from various time points to the appropriate consensus and to quantify the number of reads for each unique sequence. Sequences that, at all other time points, do not change appreciably in frequency (< |0.1%|) compared to the precut sample and sequences that only appear once across all time points were excluded from further analysis with custom Python code. Custom Python code was also used to apply an 18 bp window positioned where the gRNA complimentary base pairs with the consensus sequence. Heatmaps were then generated using a custom R application.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 6. Unpaired Student’s t-test was performed for pairwise comparisons and one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparison post-test for three or more groups, as indicated.
DATA AND SOFTWARE AVAILABILITY
RNA-seq data has been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE113670 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113670).
Custom Python code for Rhesus macaque targeted deep sequencing analysis is available at https://github.com/shirleychen2/crispr_analysis.git.
Supplementary Material
Table S1, related to Figure 3. Individuals with CD33 mutations identified from human knockout investigations.
Supplementary Figure 5. Hematologic recovery following infusion of CRISPR/Cas9 CD33− edited autologous CD34+ cells, Related to Figure 4. (A–D) Absolute total number of neutrophils (A), platelets (B), lymphocytes (C), and monocytes (D) were determined in the peripheral blood of macaques at the indicated time points after autologous transplantation. Cell counts of ZL38 (red line) and ZL33 (blue line) are shown in comparison to three historical control macaques receiving similar CD34+ cell doses/kg of lentivirally-transduced cells (black lines). The y axis shows the number of cells per uL peripheral blood. (E) Immunohistochemical (IHC) staining of H&E and myeloperoxidase (MPO) are shown in CD33 KO macaques (ZL38, ZL33) and a control macaque (ZI67). Scale bar = 20um.
Supplementary Figure 6. Targeted sequencing to confirm presence of indels within the CD33 gene in different rhesus macaque blood and bone marrow cell populations, Related to Figure 4. (A) Targeted deep sequencing of CD33 exon2 and 3 was performed on three populations of neutrophils (total neutrophils; CD11b+CD33+ neutrophils; CD11b+CD33− neutrophils) at 6 months post-transplantation (6m) in ZL38. The percentage of indels within the 18bp gRNA window for each target site is shown. (B) Targeted deep sequencing of CD33 exon2 was performed on three populations of neutrophils (total neutrophils; CD11b+CD33+ neutrophils; CD11b+CD33− neutrophils) at 1 and 4.5 months post-transplantation (1m, 4.5m) in ZL33. The percentage of indels within the 18bp gRNA window in exon 2 is shown. (C) Targeted deep sequencing of CD33 exon 2 in ZL33 was performed in sorted blood cells from the following lineages; neutrophils, total mononuclear cells, CD3+ T cells, CD20+ B cells, CD14+ monocytes, CD56dim/-16+NK cells, and CD56+CD16− NK cells at 3.5 months post-transplantation. The percentage of indels within the 18bp gRNA window in exon 2 is shown. (D) Targeted deep sequencing of CD33 exon 2 was performed in neutrophils, CD34+ cells and CD34− cells at 4.5 months post-transplantation from bilateral BM aspirates in ZL33. The percentage of indels within the 18bp gRNA exon 2 window is shown.
Table S2, related to Figure STAR Methods. Antibodies used for mass cytometry experiments.
Table S3, related to Figure STAR Methods. Read counts obtained for in silico off-target sites.
Table S4, related to Figure STAR Methods. Read counts obtained for CIRCLE-seq off-target sites.
Table S5, related to Figure STAR Methods. Primers and Probes used in this study
Supplementary Figure 1. CD33-negative human cells show no functional defects compared to CD33-positive controls, Related to Figure 3. CD33 KO CD33+ cells were differentiated in vitro with SCF, TPO, Flt3L, IL-3, IL-6 and GM-CSF for 7 days after which cells were sorted based on CD33 expression, and functional assays were performed as in Fig. 3 (n=5/group, 2 independent experiments). (A) CD33− cells can perform phagocytosis of E. coli bioparticles to the same degree as CD33+ cells. (B) ROS production at basal and after PMA stimulation is almost identical between CD33− and CD33+ cells. (C) Intracellular cytokine production is similar between CD33+ and CD33− cells after LPS stimulation. (D) Cytokine/chemokine secretion was measured in the supernatant before and after LPS stimulation for 24 hours. CD33− negative cells show the same degree of cytokine/chemokine secretion after LPS stimulation. All statistical tests were performed with unpaired Student’s t-test. ns=not significant.
Supplementary Figure 2. Mass cytometry analysis of control and CD33 KO primary human CD34+ cells show no difference in surface markers or intracellular signaling profile, Related to Figure 3. (A) SPADE diagram of a representative donor showing CD33 expression of in vitro differentiated control and CD33 KO primary CD34+ cells. SPADE clustering was performed on all 6 samples (3 donors, each with control and CD33 KO) simultaneously to generate a single tree structure for all samples, and all events from each sample were mapped to the common tree structure. CD33 expression (as depicted by node color) is globally decreased across all groups in the CD33 KO cells, while differentiation profile (as depicted by node size) is similar to control cells. (B) Heatmaps showing signaling profile of myeloid cell clusters within control and CD33 KO cells in response to external stimuli. In vitro differentiated control and CD33 KO HSPC from three donors were treated with various stimuli (GM-CSF, G-CSF, IFNα, IFNγ, IL-4, IL-6, LPS, PMA/ionomycin, and TPO), and mass cytometry was performed with antibodies to 20 surface markers and 10 phosphoproteins for comprehensive analysis of signaling. CD33+ and CD33− cells were analyzed separately within the CD33 KO cell population and compared to ungated controls. Groups are defined by the SPADE tree shown in (A). Scaled to arcsinh ratio vs. basal of the same donor. One representative donor is shown. (C) Additional flow cytometry plots of CD33 KO cells showing the indicated phosphoprotein response (on X-axis) to stimuli depicted in box (gated on live CD64+HLA-DR+ events). CD33 is depicted on Y axis. Shown are plots from one representative donor.
Supplementary Figure 3. CD33 KO human HSPC do not display any significant off-target events, Related to Figure 3. (A) Cytogenetic analysis of contol and CD33 KO CD34+ cells cultured in vitro showed no gross chromosomal abnormalities. Shown is one representative karyogram of 6 different donors assessed. (B) Evaluation of chromosomal rearrangements between SIGLEC22P and CD33 from CRISPR/Cas9-mediated genetic modification. Left panel: Schematic of PCR primers (A, B, C, D) used to detect deletion or inversion of the DNA segment between the two genes. Guide RNA cut site is depicted by arrows within each gene. Right panel: PCR of DNA extracted from 6 control/CD33 KO HSPC human donors shows that deletion between the two genes (AD) occurs in all CD33 KO samples, while inversion (AC/BD) is not detected. (C) Targeted deep sequencing of on-target and in silico predicted off-target sites in genomic DNA extracted from 5 human HSPC donors (A–E) after CD33 KO. (D) Targeted deep sequencing of on-target and CIRCLE-seq predicted off-target sites in 3 human HSPC donors (E–G) after CD33 KO. 16 off-target sites were selected based on high CIRCLE-seq read count and/or low numbers of mismatch with the target site. (E) Visualization of all on-target and off-target sites detected by CIRCLE-seq in a primary human genomic DNA sample that is identical to donor G in (D). CIRCLE-seq was performed by in vitro digestion of genomic DNA with Cas9/CD33-gRNA; validation by targeted deep sequencing was performed on genomic DNA from cells that underwent the CD33 KO procedure then differentiated in vitro (donors E,G) as outlined in Figure 1E, or differentiated in mice (donor F) as outlined in Figure 2A.
Supplementary Figure 4. Evaluation of CD33 expression and in vitro editing in rhesus macaque cells, Related to Figure 4. (A) Peripheral blood from RM and humans was evaluated by flow cytometry for CD33 expression on neutrophils and monocytes (rhesus, n=5; human, n=3). Representative flow cytometry plot is shown on right. (B) CD33 expression was evaluated on in vitro differentiated rhesus and human CD34+ cells after culture with SCF, TPO, FLT3L, IL-3, IL-6, GM-CSF for 7–10 days. (C) CD34+ cells from a rhesus macaque donor (ZK29) were electroporated with SpCas9 protein and the indicated gRNAs (group A: gRNA Exon 2, group B: gRNA Exon 3, group C: gRNA Exon 2+gRNA Exon 3 combined), and CD33 expression was evaluated by flow cytometry after 6 days of in vitro culture. Mutations in the CD33 gene were assessed by PCR around the gRNA site followed by TIDE (tracking of indels by decomposition) analysis. As the distance between the two gRNA cut sites is ~500bp, PCR was performed separately for each gRNA with the primers indicated. For group C, in which CD34+ cells were electroporated using both gRNAs, an additional PCR was performed using primers flanking both gRNAs, to detect deletions between the two gRNA cut sites. (D) CD34+ cells from a different rhesus macaque donor (ZK39) were electroporated as in (A) with either gRNA E2 alone or with the addition of a single-stranded oligodeoxynucleotide (ssODN) template containing 99 base pair homology arms around the Cas9 cut site and a single nucleotide (adenine) insertion, followed by flow cytometry evaluation of CD33 expression after 10 days of in vitro culture.
Highlights.
CD33 is not required for human myeloid development and function.
CD33-deficient non-human primate myeloid cells are fully functional.
Anti-CD33 CAR T cells can eradicate AML while sparing CD33-deficient hematopoiesis.
This is a synthetic biology approach to generating a leukemia-specific antigen.
Acknowledgments
We thank Peter Klein, Robert Vonderheide, and Carl June for constructive comments; Yangbing Zhao, Jiangtao Ren and Jian Huang for technical assistance; Sam Sadigh for photomicroscopy; Amy Ziober and Li Ping for immunocytochemistry; Vania Aikawa for cytogenetic analysis; Taehyong Kim, Zhiping (Paul) Wang and Jonathan Schug for assistance with analysis of NGS; Mary Sell and Sam Mignogno for providing human apheresis specimens; NHLBI DNA Sequencing and Genomics and the NHLBI Flow Cytometry Cores; Naoya Uchida and John Tisdale for providing control bone marrow; Mark Raffeld, Liqiang Xi, and Chingiz Underbayev for ddPCR consultation and technical assistance; Keyvan Keyvanfar, Stephanie Sellers, Lemlem Alemu, and Jun Zhu for technical assistance; Ilker Tunc for off-target prediction; Aylin Bonifacino for RM CD34+ immunoselections; Allen Krouse, Mark Metzger, and Sandra Price and the veterinary staff for care of the macaques. This work was supported by the Leukemia & Lymphoma Society (SCOR grant; CHJ/SG), the National Institute of Health (1 K08 CA194256-01; SG), the Korean Visiting Scientist Training Award (K-RY) and the Intramural Research Program of the NHLBI.
Declaration of Interests: M.Y.K and S.G. have filed patent PCT/US2016/060273 relating to methods for gene editing in hematopoietic stem cells to enhance the therapeutic efficacy of antigen-specific immunotherapy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Conceptualization MYK, K-RY, CED, and SG; Methodology MYK, KRY; Investigation MYK, K-RY, SSK, MR, SC, T-HS, DS, MK, OS, MSK, KDC, XS, PS, CRL, KRC, DBK, RED, GKB, SQT; Resources AB, and JJDM; Formal Analysis AAA and MS; Writing–Original Draft MYK, K-RY; Writing–Review & Editing MYK, K-RY, CED and SG; Visualization MYK, K-RY, GKB, CRL, SQT; Funding Acquisition, CED and SG; Supervision CED and SG.
References
- Appelbaum FR, Bernstein ID. Gemtuzumab ozogamicin for acute myeloid leukemia. Blood. 2017;130:2373. doi: 10.1182/blood-2017-09-797712. [DOI] [PubMed] [Google Scholar]
- Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass cytometric functional profiling of acute myeloid leukemia defines cell cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discovery. 2015 doi: 10.1158/2159-8290.CD-15-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasco L, Pellin D, Scala S, Dionisio F, Basso-Ricci L, Leonardelli L, Scaramuzza S, Baricordi C, Ferrua F, Cicalese MP, et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell. 2016;19:107–119. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Angata T, Reynolds SA, Powell LD, Hedrick SM, Varki A. CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Molecular and cellular biology. 2003;23:4199–4206. doi: 10.1128/MCB.23.12.4199-4206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Crocker PR. Evolution of CD33-related siglecs: regulating host immune functions and escaping pathogen exploitation? Immunology. 2011;132:18–26. doi: 10.1111/j.1365-2567.2010.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:224ra225. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue RE, Kuramoto K, Dunbar CE. Large animal models for stem and progenitor cell analysis. Curr Protoc Immunol. 2005;Chapter 22(Unit 22A):21. doi: 10.1002/0471142735.im22a01s69. [DOI] [PubMed] [Google Scholar]
- Dutour A, Marin V, Pizzitola I, Valsesia-Wittmann S, Lee D, Yvon E, Finney H, Lawson A, Brenner M, Biondi A, et al. In Vitro and In Vivo Antitumor Effect of Anti-CD33 Chimeric Receptor-Expressing EBV-CTL against Acute Myeloid Leukemia. Advances in Hematology. 2012;2012:10. doi: 10.1155/2012/683065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe'er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Tasian SK, Ruella M, Shestova O, Li Y, Porter DL, Carroll M, Danet-Desnoyers G, Scholler J, Grupp SA, et al. Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood. 2014;123:2343–2354. doi: 10.1182/blood-2013-09-529537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida A, Akita K, Mori Y, Tanida S, Toda M, Inoue M, Nakada H. Negative Regulation of Toll-like Receptor-4 Signaling through the Binding of Glycosylphosphatidylinositol-anchored Glycoprotein, CD14, with the Sialic Acid-binding Lectin, CD33. J Biol Chem. 2014;289:25341–25350. doi: 10.1074/jbc.M113.523480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenderian SS, Ruella M, Shestova O, Klichinsky M, Aikawa V, Morrissette JJ, Scholler J, Song D, Porter DL, Carroll M, et al. CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia. 2015;29:1637–1647. doi: 10.1038/leu.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer JN, Dudley ME, Kassim SH, Somerville RPT, Carpenter RO, Stetler-Stevenson M, Yang JC, Phan GQ, Hughes MS, Sherry RM, et al. Chemotherapy-Refractory Diffuse Large B-Cell Lymphoma and Indolent B-Cell Malignancies Can Be Effectively Treated With Autologous T Cells Expressing an Anti-CD19 Chimeric Antigen Receptor. J Clin Oncol. 2015;33:540–U531. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Current protocols in cytometry. 2010;Chapter 10(Unit10):17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo GS, Estey EH, Walter RB. The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Reviews. 2014;28:143–153. doi: 10.1016/j.blre.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YZK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SR, Sukumaran S, Hristopoulos M, Totpal K, Stainton S, Lu E, Wong A, Tam L, Newman R, Vuillemenot BR, et al. An anti-CD3/anti-CLL-1 bispecific antibody for the treatment of acute myeloid leukemia. Blood. 2017;129:609–618. doi: 10.1182/blood-2016-08-735365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-aD, Tadmor M, Litvin O, Fienberg H, Jager A, Zunder E, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardiros A, Dos Santos C, McDonald T, Brown CE, Wang XL, Budde LE, Hoffman L, Aguilar B, Chang WC, Bretzlaff W, et al. T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood. 2013;122:3138–3148. doi: 10.1182/blood-2012-12-474056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng ZH, Lacey SF, et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. New Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone MC, Fish JD, Carpenito C, Carroll RG, Binder GK, Teachey D, Samanta M, Lakhal M, Gloss B, Danet-Desnoyers G, et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17:1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, Barnett AH, Bates C, Bellary S, Bockett NA, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Ball ED, Varki A. Myeloid precursors and acute myeloid leukemia cells express multiple CD33-related Siglecs. Experimental hematology. 2006;34:728–735. doi: 10.1016/j.exphem.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Ornatsky OI, Kinach R, Bandura DR, Lou X, Tanner SD, Baranov VI, Nitz M, Winnik MA. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. Journal of analytical atomic spectrometry. 2008;23:463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SP, Taylor LS, Stansbury EK, McVicar DW. Myeloid specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood. 2000;96:483–490. [PubMed] [Google Scholar]
- Peterson CW, Wang J, Norman KK, Norgaard ZK, Humbert O, Tse CK, Yan JJ, Trimble RG, Shivak DA, Rebar EJ, et al. Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman primates. Blood. 2016;127:2416–2426. doi: 10.1182/blood-2015-09-672337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, Lassailly F, Tettamanti S, Spinelli O, Biondi A, Biagi E, Bonnet D. Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia. 2014;28:1596–1605. doi: 10.1038/leu.2014.62. [DOI] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotech. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, Won H-H, Karczewski KJ, O’Donnell-Luria AH, Samocha KE, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Zhang F. Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med. 2017;23:1095–1101. doi: 10.1038/nm.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et al. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov. 2015;5:1282–1295. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, Hjartarson E, Sigurdsson GT, Jonasdottir A, Jonasdottir A, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- Tashiro H, Sauer T, Shum T, Parikh K, Mamonkin M, Omer B, Rouce RH, Lulla P, Rooney CM, Gottschalk S, et al. Treatment of Acute Myeloid Leukemia with T Cells Expressing Chimeric Antigen Receptors Directed to C-type Lectin-like Molecule 1. Molecular Therapy. 2017;25:2202–2213. doi: 10.1016/j.ymthe.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, Luongo JL, Danet-Desnoyers GA, Bonnet D. Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood. 2005;106:4086–4092. doi: 10.1182/blood-2005-03-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor VC, Buckley CD, Douglas M, Cody AJ, Simmons DL, Freeman SD. The myeloid-specific sialic acid-binding receptor, CD33, associates with the protein-tyrosine phosphatases, SHP-1 and SHP-2. J Biol Chem. 1999;274:11505–11512. doi: 10.1074/jbc.274.17.11505. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Meth. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, et al. CD19 CAR-T cells of defined CD4(+): CD8(+) composition in adult B cell ALL patients. J Clin Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulyanova T, Blasioli J, Woodford-Thomas TA, Thomas ML. The sialoadhesin CD33 is a myeloid-specific inhibitory receptor. Eur J Immunol. 1999;29:3440–3449. doi: 10.1002/(SICI)1521-4141(199911)29:11<3440::AID-IMMU3440>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang Q-s, Wang Y, Lv H-y, Han Q-w, Fan H, Guo B, Wang L-l, Han W-d. Treatment of CD33-directed Chimeric Antigen Receptor-modified T Cells in One Patient With Relapsed and Refractory Acute Myeloid Leukemia. Molecular Therapy. 2015;23:184–191. doi: 10.1038/mt.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Espinoza DA, Koelle SJ, Potter EL, Lu R, Li B, Yang D, Fan X, Donahue RE, Roederer M, et al. Geographic clonal tracking in macaques provides insights into HSPC migration and differentiation. J Exp Med. 2017 doi: 10.1084/jem.20171341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Li B, Lu R, Koelle SJ, Yang Y, Jares A, Krouse AE, Metzger M, Liang F, Lore K, et al. Clonal tracking of rhesus macaque hematopoiesis highlights a distinct lineage origin for natural killer cells. Cell Stem Cell. 2014;14:486–499. doi: 10.1016/j.stem.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang C-Z, Chari R, Homsy J, Cai X, Zhao Y, Fan J-B. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nature communications. 2014;5 doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 3. Individuals with CD33 mutations identified from human knockout investigations.
Supplementary Figure 5. Hematologic recovery following infusion of CRISPR/Cas9 CD33− edited autologous CD34+ cells, Related to Figure 4. (A–D) Absolute total number of neutrophils (A), platelets (B), lymphocytes (C), and monocytes (D) were determined in the peripheral blood of macaques at the indicated time points after autologous transplantation. Cell counts of ZL38 (red line) and ZL33 (blue line) are shown in comparison to three historical control macaques receiving similar CD34+ cell doses/kg of lentivirally-transduced cells (black lines). The y axis shows the number of cells per uL peripheral blood. (E) Immunohistochemical (IHC) staining of H&E and myeloperoxidase (MPO) are shown in CD33 KO macaques (ZL38, ZL33) and a control macaque (ZI67). Scale bar = 20um.
Supplementary Figure 6. Targeted sequencing to confirm presence of indels within the CD33 gene in different rhesus macaque blood and bone marrow cell populations, Related to Figure 4. (A) Targeted deep sequencing of CD33 exon2 and 3 was performed on three populations of neutrophils (total neutrophils; CD11b+CD33+ neutrophils; CD11b+CD33− neutrophils) at 6 months post-transplantation (6m) in ZL38. The percentage of indels within the 18bp gRNA window for each target site is shown. (B) Targeted deep sequencing of CD33 exon2 was performed on three populations of neutrophils (total neutrophils; CD11b+CD33+ neutrophils; CD11b+CD33− neutrophils) at 1 and 4.5 months post-transplantation (1m, 4.5m) in ZL33. The percentage of indels within the 18bp gRNA window in exon 2 is shown. (C) Targeted deep sequencing of CD33 exon 2 in ZL33 was performed in sorted blood cells from the following lineages; neutrophils, total mononuclear cells, CD3+ T cells, CD20+ B cells, CD14+ monocytes, CD56dim/-16+NK cells, and CD56+CD16− NK cells at 3.5 months post-transplantation. The percentage of indels within the 18bp gRNA window in exon 2 is shown. (D) Targeted deep sequencing of CD33 exon 2 was performed in neutrophils, CD34+ cells and CD34− cells at 4.5 months post-transplantation from bilateral BM aspirates in ZL33. The percentage of indels within the 18bp gRNA exon 2 window is shown.
Table S2, related to Figure STAR Methods. Antibodies used for mass cytometry experiments.
Table S3, related to Figure STAR Methods. Read counts obtained for in silico off-target sites.
Table S4, related to Figure STAR Methods. Read counts obtained for CIRCLE-seq off-target sites.
Table S5, related to Figure STAR Methods. Primers and Probes used in this study
Supplementary Figure 1. CD33-negative human cells show no functional defects compared to CD33-positive controls, Related to Figure 3. CD33 KO CD33+ cells were differentiated in vitro with SCF, TPO, Flt3L, IL-3, IL-6 and GM-CSF for 7 days after which cells were sorted based on CD33 expression, and functional assays were performed as in Fig. 3 (n=5/group, 2 independent experiments). (A) CD33− cells can perform phagocytosis of E. coli bioparticles to the same degree as CD33+ cells. (B) ROS production at basal and after PMA stimulation is almost identical between CD33− and CD33+ cells. (C) Intracellular cytokine production is similar between CD33+ and CD33− cells after LPS stimulation. (D) Cytokine/chemokine secretion was measured in the supernatant before and after LPS stimulation for 24 hours. CD33− negative cells show the same degree of cytokine/chemokine secretion after LPS stimulation. All statistical tests were performed with unpaired Student’s t-test. ns=not significant.
Supplementary Figure 2. Mass cytometry analysis of control and CD33 KO primary human CD34+ cells show no difference in surface markers or intracellular signaling profile, Related to Figure 3. (A) SPADE diagram of a representative donor showing CD33 expression of in vitro differentiated control and CD33 KO primary CD34+ cells. SPADE clustering was performed on all 6 samples (3 donors, each with control and CD33 KO) simultaneously to generate a single tree structure for all samples, and all events from each sample were mapped to the common tree structure. CD33 expression (as depicted by node color) is globally decreased across all groups in the CD33 KO cells, while differentiation profile (as depicted by node size) is similar to control cells. (B) Heatmaps showing signaling profile of myeloid cell clusters within control and CD33 KO cells in response to external stimuli. In vitro differentiated control and CD33 KO HSPC from three donors were treated with various stimuli (GM-CSF, G-CSF, IFNα, IFNγ, IL-4, IL-6, LPS, PMA/ionomycin, and TPO), and mass cytometry was performed with antibodies to 20 surface markers and 10 phosphoproteins for comprehensive analysis of signaling. CD33+ and CD33− cells were analyzed separately within the CD33 KO cell population and compared to ungated controls. Groups are defined by the SPADE tree shown in (A). Scaled to arcsinh ratio vs. basal of the same donor. One representative donor is shown. (C) Additional flow cytometry plots of CD33 KO cells showing the indicated phosphoprotein response (on X-axis) to stimuli depicted in box (gated on live CD64+HLA-DR+ events). CD33 is depicted on Y axis. Shown are plots from one representative donor.
Supplementary Figure 3. CD33 KO human HSPC do not display any significant off-target events, Related to Figure 3. (A) Cytogenetic analysis of contol and CD33 KO CD34+ cells cultured in vitro showed no gross chromosomal abnormalities. Shown is one representative karyogram of 6 different donors assessed. (B) Evaluation of chromosomal rearrangements between SIGLEC22P and CD33 from CRISPR/Cas9-mediated genetic modification. Left panel: Schematic of PCR primers (A, B, C, D) used to detect deletion or inversion of the DNA segment between the two genes. Guide RNA cut site is depicted by arrows within each gene. Right panel: PCR of DNA extracted from 6 control/CD33 KO HSPC human donors shows that deletion between the two genes (AD) occurs in all CD33 KO samples, while inversion (AC/BD) is not detected. (C) Targeted deep sequencing of on-target and in silico predicted off-target sites in genomic DNA extracted from 5 human HSPC donors (A–E) after CD33 KO. (D) Targeted deep sequencing of on-target and CIRCLE-seq predicted off-target sites in 3 human HSPC donors (E–G) after CD33 KO. 16 off-target sites were selected based on high CIRCLE-seq read count and/or low numbers of mismatch with the target site. (E) Visualization of all on-target and off-target sites detected by CIRCLE-seq in a primary human genomic DNA sample that is identical to donor G in (D). CIRCLE-seq was performed by in vitro digestion of genomic DNA with Cas9/CD33-gRNA; validation by targeted deep sequencing was performed on genomic DNA from cells that underwent the CD33 KO procedure then differentiated in vitro (donors E,G) as outlined in Figure 1E, or differentiated in mice (donor F) as outlined in Figure 2A.
Supplementary Figure 4. Evaluation of CD33 expression and in vitro editing in rhesus macaque cells, Related to Figure 4. (A) Peripheral blood from RM and humans was evaluated by flow cytometry for CD33 expression on neutrophils and monocytes (rhesus, n=5; human, n=3). Representative flow cytometry plot is shown on right. (B) CD33 expression was evaluated on in vitro differentiated rhesus and human CD34+ cells after culture with SCF, TPO, FLT3L, IL-3, IL-6, GM-CSF for 7–10 days. (C) CD34+ cells from a rhesus macaque donor (ZK29) were electroporated with SpCas9 protein and the indicated gRNAs (group A: gRNA Exon 2, group B: gRNA Exon 3, group C: gRNA Exon 2+gRNA Exon 3 combined), and CD33 expression was evaluated by flow cytometry after 6 days of in vitro culture. Mutations in the CD33 gene were assessed by PCR around the gRNA site followed by TIDE (tracking of indels by decomposition) analysis. As the distance between the two gRNA cut sites is ~500bp, PCR was performed separately for each gRNA with the primers indicated. For group C, in which CD34+ cells were electroporated using both gRNAs, an additional PCR was performed using primers flanking both gRNAs, to detect deletions between the two gRNA cut sites. (D) CD34+ cells from a different rhesus macaque donor (ZK39) were electroporated as in (A) with either gRNA E2 alone or with the addition of a single-stranded oligodeoxynucleotide (ssODN) template containing 99 base pair homology arms around the Cas9 cut site and a single nucleotide (adenine) insertion, followed by flow cytometry evaluation of CD33 expression after 10 days of in vitro culture.