Significance
The Staphylococcus aureus surface protein clumping factor A (ClfA) binds to the blood plasma protein fibrinogen (Fg) via molecular interactions that are poorly understood. Here, we unravel the forces guiding the interaction between ClfA and immobilized Fg, showing that it is dramatically enhanced by tensile loading. Our findings favor a model whereby ClfA interacts with Fg via two distinct binding sites, the adhesive function of which is tightly regulated by mechanical force. Reminiscent of a catch bond mechanism, this force-enhanced adhesion explains the ability of ClfA to promote S. aureus colonization of host tissues and biomedical devices under physical stress.
Keywords: Staphylococcus aureus, ClfA, fibrinogen, shear stress, atomic force microscopy
Abstract
Clumping factor A (ClfA), a cell-wall–anchored protein from Staphylococcus aureus, is a virulence factor in various infections and facilitates the colonization of protein-coated biomaterials. ClfA promotes bacterial adhesion to the blood plasma protein fibrinogen (Fg) via molecular forces that have not been studied so far. A unique, yet poorly understood, feature of ClfA is its ability to favor adhesion to Fg at high shear stress. Unraveling the strength and dynamics of the ClfA–Fg interaction would help us better understand how S. aureus colonizes implanted devices and withstands physiological shear stress. By means of single-molecule experiments, we show that ClfA behaves as a force-sensitive molecular switch that potentiates staphylococcal adhesion under mechanical stress. The bond between ClfA and immobilized Fg is weak (∼0.1 nN) at low tensile force, but is dramatically enhanced (∼1.5 nN) by mechanical tension, as observed with catch bonds. Strong bonds, but not weak ones, are inhibited by a peptide mimicking the C-terminal segment of the Fg γ-chain. These results point to a model whereby ClfA interacts with Fg via two distinct binding sites, the adhesive function of which is regulated by mechanical tension. This force-activated mechanism is of biological significance because it explains at the molecular level the ability of ClfA to promote bacterial attachment under high physiological shear stress.
The bacterial pathogen Staphylococcus aureus binds to host extracellular matrix proteins using a variety of cell-wall–anchored proteins (1). Among these, the fibrinogen (Fg)-binding microbial surface component recognizing the adhesive matrix molecule (MSCRAMM) protein clumping factor A (ClfA) is an important virulence factor of S. aureus involved in various infections (2–4). In addition, ClfA promotes bacterial attachment to plasma protein-coated biomaterials allowing the bacteria to colonize and form a biofilm (5). Vaccination with recombinant ClfA is protective against infection, and ClfA has been included as an antigen in a number of multivalent S. aureus vaccines currently in clinical trials (2, 6, 7). ClfA has an N-terminal A region composed of three separately folded subdomains: N1, N2, and N3. N2 and N3 form the minimum ligand-binding region and bind the carboxy-terminus of the γ-chain of Fg (Fig. 1A and refs. 8–10) through a variation of the multistep “dock, lock, and latch” (DLL) mechanism (11) first described for the binding of the Staphylococcus epidermidis protein SdrG to Fg (12). The carboxy-terminus of the γ-chain of Fg docks in a ligand-binding trench located between subdomains N2 and N3. The DLL mechanism involves dynamic conformational changes of the adhesin that result in a greatly stabilized adhesin-ligand complex. The overall affinity of the interaction of ClfA with Fg is increased through interactions at a recently described second site that lies at the top of subdomain N3 outside of the DLL ligand-binding trench (13).
Fig. 1.
Studying the ClfA–Fg interaction. (A) The N-terminal signal sequence (S) is followed by subdomains N1, N2, and N3 comprising the ligand-binding A region. At the junction between N2 and N3 is found a binding trench in which the ligand (red) inserts and is locked in place. A flexible serine-aspartate repeat region links region A to the C-terminal Wall (W) spanning region and the sorting sequence. The LPXTG motif allows anchoring of the protein to cell-wall peptidoglycan by sortase A. (B) Optical microscopy images of S. aureus bacteria expressing or not full-length ClfA [ClfA(+) and ClfA(−) cells] following incubation with Fg-conditioned substrates. (C) Force nanoscopy of the ClfA–Fg interaction: (Left) SCFS; (Right) SMFS. For clarity, the N1 domain of ClfA is not shown.
Antibiotic treatments have proven to be less and less effective over the years due to the emergence of multidrug-resistant strains (14, 15). Therefore, there is great interest in developing alternative strategies to fight bacterial infections (15, 16). An appealing approach is the use of antiadhesion compounds to block cell adhesion and biofilm development (17). A well-known example is the use of cranberry juice to treat urinary tract infections by uropathogenic Escherichia coli bacteria (18). The development of novel antiadhesion therapeutics targeting ClfA requires a detailed understanding of the ligand-binding mechanisms of this adhesin.
An interesting trait of ClfA is its ability to favor adhesion to blood proteins at high shear stress (19–21). Despite the biological relevance of such stress-induced adhesion, the underlying molecular mechanism has not yet been elucidated. We hypothesized that the interaction of ClfA with Fg might strengthen through a force-sensitive mechanism. To test this idea, we measured the strength of the bond between ClfA and immobilized Fg at various tensile loads, using atomic force microscopy (AFM) (Fig. 1C) (22–25). We found that, while the ClfA–Fg bond is weak at low applied force, the bond strength is dramatically increased at high force. These results provide compelling evidence that ClfA functions as a force-sensitive molecular switch that regulates the strength of adhesion of S. aureus to protein-conditioned biomaterials, thus emphasizing the role that physical forces play in activating the function of bacterial adhesins. This study holds promise for the design of antibacterial agents with the capacity to inhibit S. aureus adhesion at high shear rate.
Results
S. aureus Strongly Binds to Immobilized Fibrinogen.
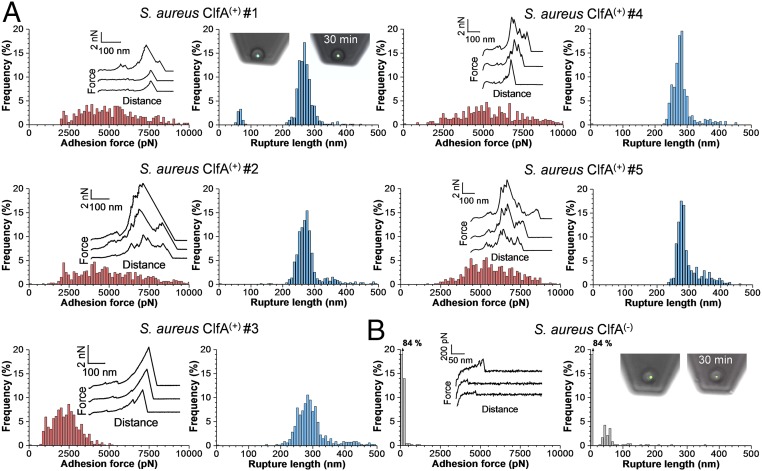
To investigate ClfA–Fg interactions in living bacteria, we used S. aureus SH1000 clfA clfB fnbA fnbB [hereafter called S. aureus ClfA(−) cells; ref. 26] and the same strain transformed with a plasmid expressing the entire clfA gene [ClfA(+) cells]. Fg was immobilized on solid substrates using N-hydroxysuccinimide (NHS) surface chemistry. Optical microscopy images confirmed that ClfA(+) cells adhered in large amounts to Fg-coated substrates, while no adhesion was seen with ClfA(−) cells (Fig. 1B). This indicates that ClfA is well-expressed and represents the only Fg-binding protein found at the cell surface of this strain. By means of single-cell force spectroscopy (SCFS; Fig. 1C, Left) (22, 27–29), we analyzed the ClfA–Fg-binding forces at the whole-cell level. Single bacteria were attached onto colloidal cantilevers, and the forces between the cell probes and Fg substrates were measured. Fig. 2A shows the maximum adhesion forces and rupture lengths obtained for five representative S. aureus ClfA(+) cells (for more cells, see SI Appendix, Fig. S1A). Most force curves feature adhesion force peaks ranging from ∼1,000 to ∼10,000 pN with rupture lengths of ∼250–300 nm. The characteristics of the curves did not substantially change when recording consecutive force curves on different spots of the substrate, meaning that force measurements did not alter the cell-surface properties. Adhesion forces were spread across a wide range, suggesting that a variable number of molecular bonds were probed. Most adhesive forces were larger than the forces measured for other staphylococcal adhesins (30, 31), suggesting that bacterial adhesion to Fg is very strong. As these large forces were abolished in ClfA(−) cells (Fig. 2B and SI Appendix, Fig. S1B), we conclude that they mostly reflect specific ClfA–Fg interactions. Most bonds ruptured at ∼250–300 nm, which is consistent with the length of fully unfolded adhesins. Assuming that the processed mature ClfA protein comprises 860 residues, that each amino acid contributes 0.36 nm to the contour length of the polypeptide chain, and that the ClfA folded length is ∼25 nm, we expect that the fully extended ClfA protein should be ∼285 nm long.
Fig. 2.
Forces guiding the adhesion of single bacteria to Fg substrates. (A) Maximum adhesion force (Left) and rupture length (Right) histograms with representative retraction force profiles (Insets) obtained by recording force-distance curves in PBS between five S. aureus ClfA(+) cells from different cultures and Fg substrates. (B) Force data obtained in the same conditions for a S. aureus ClfA(−) cell. Fluorescence images of the bacterial probes stained with the BacLight viability kit (Insets) confirmed that the cell membrane is intact even after 30 min of measurements.
How Strong Is the ClfA–Fibrinogen Interaction?
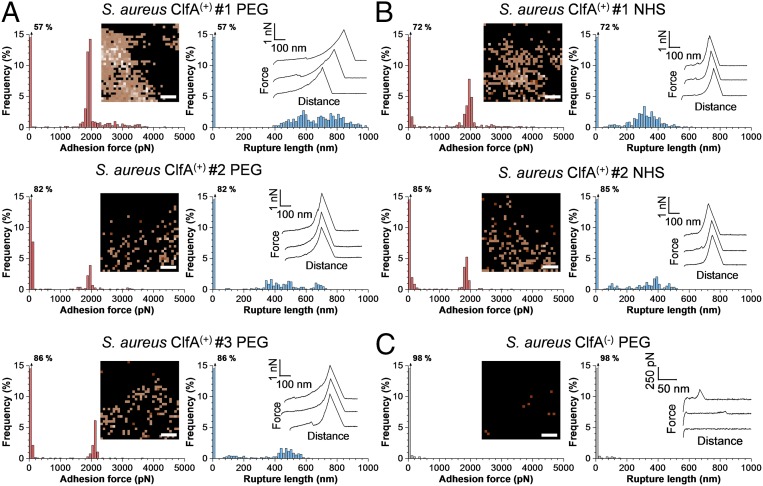
To quantify the strength of single ClfA-Fg bonds we used single-molecule force spectroscopy (SMFS; Fig. 1C, Right) (29, 32) with Fg-modified tips. Fg molecules were attached to the tips at low density using a PEG-benzaldehyde linker. In Fig. 3A we present the adhesion force maps, maximum adhesion forces, and rupture lengths obtained between three S. aureus ClfA(+) cells and AFM tips functionalized with PEG chemistry (for more cells, see SI Appendix, Fig. S2A). Strong adhesion peaks were detected with mean forces of 1,999 ± 267 pN (mean and SD of n = 401 adhesive curves), 1,909 ± 155 pN (n = 88), and 2,093 ± 68 pN (n = 107) for cell #1, cell #2, and cell #3, respectively. Similar adhesion forces were observed when the tips were modified via the NHS chemistry (Fig. 3B), indicating that the detected forces were not dependent on the grafting protocol. Adhesion was mediated by ClfA as it was abrogated in ClfA(−) cells (Fig. 3C and SI Appendix, Fig. S2B). Force maps revealed that ClfA was expressed at rather high density and heterogeneously distributed on the cell surface.
Fig. 3.
Binding strength of single ClfA–Fg bonds in living S. aureus bacteria. (A, Left) Maximum adhesion force histograms with force maps (Insets). (Scale bars: 100 nm.) (Right) Rupture length histograms with representative retraction force profiles (Insets) obtained by recording force-distance curves in PBS between three different S. aureus ClfA(+) cells and AFM tips functionalized with Fg using PEG chemistry. (B) Data obtained in the same conditions between two different S. aureus ClfA(+) cells and AFM tips functionalized with Fg using NHS chemistry. (C) Force data between a S. aureus ClfA(−) cell and a AFM tip functionalized with Fg using the PEG chemistry.
We believe that the ∼2-nN forces are associated with single DLL-like interactions for the following reasons. First, for all cells investigated, adhesion forces featured distributions that were narrow and centered near ∼2 nN, which strongly supports the idea that single bonds were probed. When multiple bonds break simultaneously, a wide force range reflecting multiples of the weakest unit force should be observed, which was not the case. Second, forces with very similar sharp distributions were obtained using two different tip chemistries (PEG vs. NHS) that are known to favor single-molecule detection. Third, strong forces are in the range of the strength measured for the DLL interaction between the structurally related SdrG protein and Fg, both on living bacteria (29) and on purified adhesins (33). Fourth, we generated an S. aureus strain (called ClfAPY) expressing ClfA carrying amino acid substitutions within the N2N3-binding trench (P336S and Y338A) that prevents ligand binding by the DLL mechanism. We found that these substitutions almost completely abolished Fg binding to ClfA (SI Appendix, Fig. S3), thus demonstrating that strong ClfA binding forces are due to a DLL-like interaction.
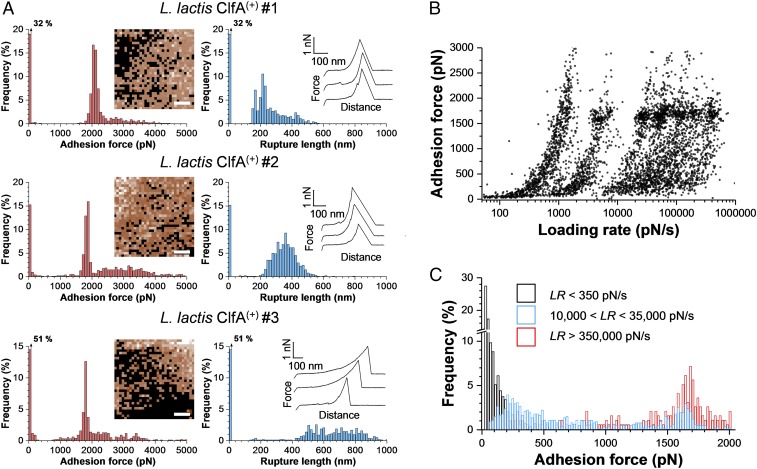
To study ClfA in the absence of other staphylococcal cell-wall components, we also examined cells from a Lactococcus lactis strain expressing ClfA [L. lactis ClfA(+) cells] (34). As illustrated in Fig. 4A (for more cells, see SI Appendix, Fig. S4), this strain featured the same behavior as the S. aureus ClfA(+) strain, with strong forces of 2,070 ± 93 pN (mean and SD on n = 508 adhesive curves), 1,848 ± 119 pN (n = 425), and 1,891 ± 225 pN (n = 333) for cell #1, cell #2, and cell #3, respectively. L. lactis ClfA(+) cells showed a higher adhesion frequency and sometimes large force values >2 nN. Whole-cell dot immunoblots (SI Appendix, Fig. S5) suggest that this may be due to a higher expression of the adhesin at the cell surface of L. lactis ClfA(+) compared with S. aureus ClfA(+).
Fig. 4.
ClfA behaves as a force-sensitive molecular switch. (A) SMFS data obtained by recording force-distance curves in PBS between Fg tips (PEG chemistry) and three different cells from a model L. lactis strain expressing ClfA [L. lactis ClfA(+) cells]. (Scale bars: 100 nm.) (B) Strength of the ClfA–Fg interaction measured at increasing loading rates (LRs) on L. lactis ClfA(+) cells (4,452 adhesive events from 10 cells). All adhesion peaks were analyzed to take into account all possible interactions. (C) The probability of forming strong bonds increases with the LR. From the data shown in B, discrete ranges of LRs were binned, and the force distributions were plotted as histograms (SI Appendix, Fig. S6). Shown here are the forces observed for low, medium, and high LRs.
One may argue that our experiments may not mimic in vivo conditions as Fg was attached to the tip. So we cannot extend our findings on immobilized Fg to conditions where the bacteria may interact with the soluble form of Fg. However, immobilized Fg is biologically relevant as it is associated with blood clots and, importantly, deposited on biomedical device surfaces. Given that the concentration of free Ca2+ in blood is equivalent to the IC50 for Ca2+ to inhibit Fg binding (35), 50% of ClfA molecules will not have soluble Fg bound and will be free to engage with immobilized Fg in a clot or on a surface.
The ClfA–Fibrinogen Interaction Is Dramatically Enhanced by Tensile Loading.
During colonization of host tissues and biomedical devices, S. aureus is subjected to physical forces, such as fluid flow and cell-surface contacts (36). An interesting but poorly understood feature of ClfA is its ability to promote S. aureus adhesion under shear stresses (19, 37). We postulated that the ClfA binding strength might be enhanced by mechanical force. To test this, the strength of the ClfA–Fg interaction (F) was measured while varying the rate at which force increases (loading rate, LR; the effective LR was estimated from the force vs. time curves) (38). Fig. 4B shows that the dynamic force spectroscopy data obtained on L. lactis ClfA(+) cells (data pooled from 4,452 adhesive curves on 10 cells) featured a fuzzy distribution with several clouds arising from the different pulling speeds used. Discrete ranges of LRs were binned, and the force distributions were plotted as histograms (SI Appendix, Fig. S6, from which Fig. 4C was obtained). Strikingly, we observed a major switch in force distribution with the LR (Fig. 4C): when the load was applied slowly, weak forces of 88 ± 69 pN (n = 277 events) were detected, whereas at fast LR only strong forces of 1,687 ± 180 pN (n = 155) were probed. Both weak and strong forces were seen when intermediate LRs were used. That strong bonds are favored when the force is increased quickly demonstrates that the strength of the ClfA–Fg interaction is dramatically enhanced with tensile loading.
How does the physical stress applied in AFM compare with that experienced in the body? Undoubtedly, S. aureus is exposed to many different levels of shear depending on its location in vivo—from low-shear conditions on implanted biomaterials to high-shear conditions in the blood. The loading rates that bacterial cells experience in biological flowing fluids can exceed 100,000 pN/s (39). So our results showing that a LR of 10,000 pN/s is sufficient to favor strong binding suggest that force-induced ClfA adhesion will occur in vivo in a number of situations.
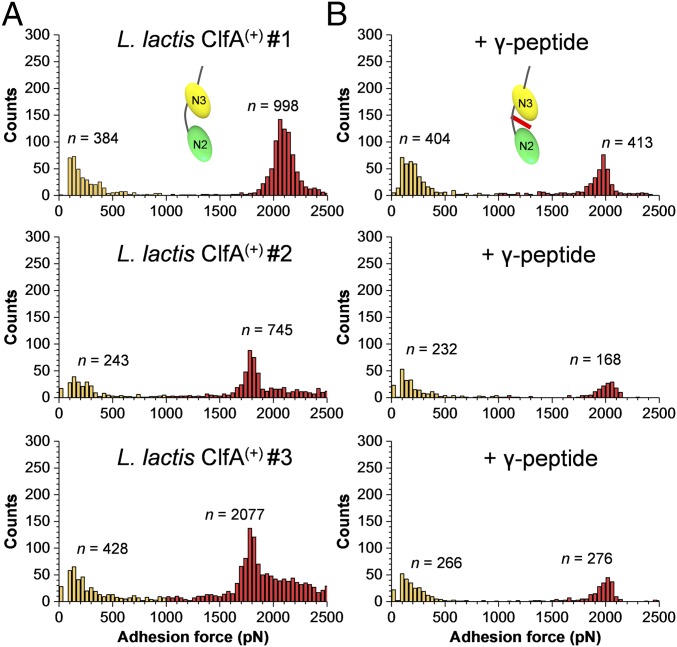
Finally, we asked whether weak and strong forces could be associated with two distinct binding sites. Previous studies have shown that Fg binding involves a DLL interaction between the trench of ClfA N2N3 subdomains and the extreme C terminus of the γ-chain of Fg (11, 13). We therefore tested the ability of a short γ-chain peptide to block the adhesive forces of L. lactis ClfA(+) cells. Fig. 5 and SI Appendix, Fig. S7 show that incubation of L. lactis ClfA(+) cells with the γ-chain peptide led to a dramatic inhibition of high forces while leaving low forces essentially unchanged. This means that strong forces, but not weak ones, are due to DLL binding between the ClfA trench and the Fg γ-chain. This finding supports a two-site mechanism and provides a direct confirmation of earlier observations that, in addition to the γ-peptide–binding trench, there is a second binding site located at the top of the N3 domain that is critical for an overall high-affinity Fg–ClfA interaction (13, 40).
Fig. 5.
Adhesion to Fg involves two ClfA-binding sites. (A and B) Adhesion forces obtained by recording force-distance curves in PBS between three different L. lactis ClfA(+) cells and Fg tips in the absence (A) or presence (B) of the C-terminal segment of the Fg γ-chain (0.2 mg⋅mL−1). All adhesion peaks were analyzed to take into account all possible interactions.
Discussion
ClfA is an important virulence factor of S. aureus that binds to Fg-coated biomaterials. This adhesin favors bacterial adhesion under high physical stress, but how this is achieved at the molecular level is not known. We have demonstrated that ClfA is a force-sensitive molecular switch that activates adhesion of S. aureus to Fg-conditioned surfaces under high mechanical forces. Our results emphasize the role of mechanobiology in regulating S. aureus adhesion and may contribute to the development of antiadhesion strategies using soluble inhibitors.
To understand the molecular origin of the force-induced strengthening of the ClfA–Fg bond, we recall that theory predicts—and experiments confirm—that unbinding force between receptors and ligands increases with the rate at which force is applied. The Bell–Evans theory (41) describes a log linear relationship between the LR and the rupture force. More recently, Friddle et al. (42) developed a model that adequately describes nonlinear trends in rupture forces, considering that nonlinearity arises either through the reforming of a single bond at slow loading rates or as a consequence of asynchronous fluctuations of several independent interactions. So current models and data all show continuous increases in rupture force with the LR. By contrast, the ClfA–Fg bond features an unusual switch in force distribution; that is, weak bonds (∼100 pN) dominate at low LR while strong bonds (∼1,500 pN) are favored at high LR. Because the strengths of weak and strong bonds differ by an order of magnitude and intermediate forces were rarely observed, we believe that strong bonds do not result from the simultaneous rupture of multiple weak bonds.
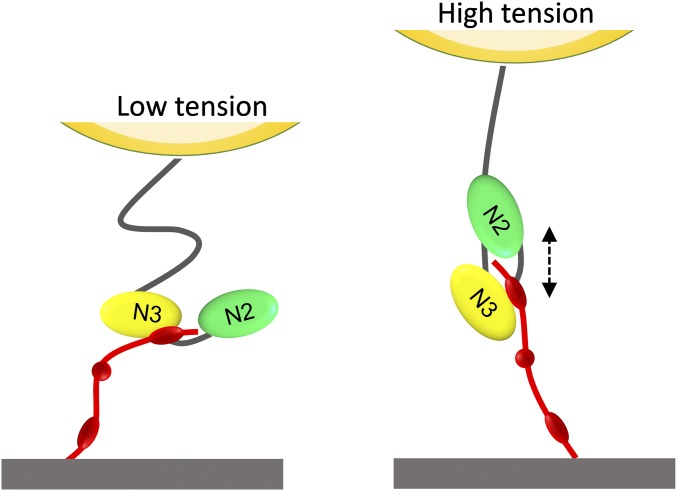
We propose that the unusual force-dependent strengthening of the ClfA–Fg bond involves a force-induced conformational change in the adhesin, from a weak- to a strong-binding state. That the γ-chain peptide of Fg inhibits high forces but not low forces favors a two binding site model, where the activity of the two binding sites is tightly regulated by tensile force (Fig. 6). Under low tensile force, Fg binds to the top of the ClfA N3 domain via weak bonds. Under high mechanical tension, extension and conformational changes in the ClfA molecule trigger the ultrastrong DLL interaction by the N2N3 subdomains. Such a mechanism may help us to identify soluble ligands capable of inhibiting bacterial adhesion under high flow conditions.
Fig. 6.
Proposed model for the force-activated adhesion of ClfA. (Left) Under low mechanical force, Fg weakly binds to the top of the ClfA N3 domain. (Right) Under high force, extension and conformational changes of the ClfA N2N3 subdomains enable the γ-chain of Fg to dock in the ligand-binding trench and form strong DLL-like interactions.
The force-dependent ClfA–Fg interaction is reminiscent of a catch-bond behavior, that is, a specific bond that is reinforced by mechanical stress (43). A well-documented example is the E. coli FimH adhesin that binds mannose residues on epithelial cells (39). The FimH–mannose bond is weak and relatively short lived at low flow, whereas this bond is strengthened at high flow. This is explained by an allosteric model in which tensile mechanical force induces an allosteric switch from a low- to a high-affinity conformation of the adhesin (43, 44). Perhaps the ClfA–Fg interaction involves such an allosterically controlled mechanism, whereby stretching of the subdomains would suppress allosteric interplay and trigger strong DLL binding.
The strength of the ClfA bond at high tensile load is in the range of that of covalent bonds, despite a moderate affinity value (8–10). This discrepancy suggests that the unbinding pathway of the adhesin may change when mechanical force is applied (45). So, when studying the mechanisms of bacterial adhesion under physiological shear, force measurements performed at nonequilibrium might be more relevant than equilibrium assays. That the ClfA–Fg complex resists very high forces is counterintuitive as rupture of the polypeptide backbones is expected to occur first. A possible explanation is that the complex may direct force along pathways nonparallel to the pulling direction, as shown for the mechanically stable multidomain cellulosome protein complex (45).
The high binding strength provides a molecular framework to explain how ClfA promotes S. aureus adhesion on blood protein-coated surfaces under high shear stress conditions (19, 37). Fg binding to ClfA expressed on the surface of S. aureus or L. lactis facilitates platelet capture and thrombus formation under high shear conditions but not when low shear rates are applied (19, 37). ClfA binding to Fg under shear conditions creates a bridge between the bacterium and integrin receptors expressed by endothelial cells (46). In addition, ClfA is involved in the shear-dependent adhesion of S. aureus to von Willebrand factor, thereby allowing the bacteria to resist shear forces of flowing blood (20, 21). It is possible that S. aureus has evolved force-dependent adhesion mechanisms such as the one unraveled here to help the bacteria resist physical stress during host colonization, whereas weak adhesion forces at low shear stress would favor cell detachment and thus the colonization of new sites.
Methods
S. aureus ClfA(−) is a S. aureus SH1000 clfA clfB fnbA fnbB strain defective in both clumping factors A and B and fibronectin-binding proteins A and B (26) whereas S. aureus ClfA(+) is SH1000 clfA clfB fnbA fnbB transformed with the plasmid pALC2073::clfA (47). To study the effect of amino acid substitutions within the N2N3-binding trench, we used S. aureus ClfAWT, which is SH1000 clfA clfB fnbA fnbB carrying the plasmid pCF77 expressing ClfA from its own promoter (48), and S. aureus ClfAPY, which is the same strain with P336S and Y338A substitutions in the N2 subdomain of ClfA (49). Growth conditions and AFM methods are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank David Alsteens and Timothy Foster for fruitful discussion. Work at the Université Catholique de Louvain was supported by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (Grant 693630); the Walloon Excellence in Life Sciences and Biotechnology (Grant WELBIO-CR-2015A-05); the National Fund for Scientific Research (FNRS); and the Research Department of the Communauté Française de Belgique (Concerted Research Action). Y.F.D. is Research Director at the FNRS.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718104115/-/DCSupplemental.
References
- 1.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Josefsson E, Hartford O, O’Brien L, Patti JM, Foster T. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J Infect Dis. 2001;184:1572–1580. doi: 10.1086/324430. [DOI] [PubMed] [Google Scholar]
- 3.McAdow M, et al. Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog. 2011;7:e1002307. doi: 10.1371/journal.ppat.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreillon P, et al. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaudaux PE, et al. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect Immun. 1995;63:585–590. doi: 10.1128/iai.63.2.585-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaffer AC, Lee JC. Staphylococcal vaccines and immunotherapies. Infect Dis Clin North Am. 2009;23:153–171. doi: 10.1016/j.idc.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Proctor RA. Recent developments for Staphylococcus aureus vaccines: Clinical and basic science challenges. Eur Cell Mater. 2015;30:315–326. doi: 10.22203/ecm.v030a22. [DOI] [PubMed] [Google Scholar]
- 8.McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 9.McDevitt D, Francois P, Vaudaux P, Foster TJ. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 10.McDevitt D, et al. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem. 1997;247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 11.Ganesh VK, et al. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008;4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponnuraj K, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 13.Ganesh VK, et al. Lessons from the crystal structure of the S. aureus surface protein clumping factor A in complex with Tefibazumab, an inhibiting monoclonal antibody. EBioMedicine. 2016;13:328–338. doi: 10.1016/j.ebiom.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 15.Bassetti M, et al. Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: A visionary approach. Intensive Care Med. 2017;43:1464–1475. doi: 10.1007/s00134-017-4878-x. [DOI] [PubMed] [Google Scholar]
- 16.Czaplewski L, et al. Alternatives to antibiotics: A pipeline portfolio review. Lancet Infect Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 17.Geoghegan JA, Foster TJ, Speziale P, Dufrêne YF. Live-cell nanoscopy in antiadhesion therapy. Trends Microbiol. 2017;25:512–514. doi: 10.1016/j.tim.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerrigan SW, et al. Molecular basis for Staphylococcus aureus-mediated platelet aggregate formation under arterial shear in vitro. Arterioscler Thromb Vasc Biol. 2008;28:335–340. doi: 10.1161/ATVBAHA.107.152058. [DOI] [PubMed] [Google Scholar]
- 20.Claes J, et al. Adhesion of Staphylococcus aureus to the vessel wall under flow is mediated by von Willebrand factor-binding protein. Blood. 2014;124:1669–1676. doi: 10.1182/blood-2014-02-558890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claes J, et al. Clumping factor A, von Willebrand factor-binding protein and von Willebrand factor anchor Staphylococcus aureus to the vessel wall. J Thromb Haemost. 2017;15:1009–1019. doi: 10.1111/jth.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helenius J, Heisenberg CP, Gaub HE, Müller DJ. Single-cell force spectroscopy. J Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 23.Xiao J, Dufrêne YF. Optical and force nanoscopy in microbiology. Nat Microbiol. 2016;1:16186. doi: 10.1038/nmicrobiol.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufrêne YF, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12:295–307. doi: 10.1038/nnano.2017.45. [DOI] [PubMed] [Google Scholar]
- 25.Dufrêne YF. Microbial nanoscopy: Breakthroughs, challenges, and opportunities. ACS Nano. 2017;11:19–22. doi: 10.1021/acsnano.6b08459. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill E, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaussart A, et al. Single-cell force spectroscopy of probiotic bacteria. Biophys J. 2013;104:1886–1892. doi: 10.1016/j.bpj.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beaussart A, et al. Quantifying the forces guiding microbial cell adhesion using single-cell force spectroscopy. Nat Protoc. 2014;9:1049–1055. doi: 10.1038/nprot.2014.066. [DOI] [PubMed] [Google Scholar]
- 29.Herman P, et al. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol Microbiol. 2014;93:356–368. doi: 10.1111/mmi.12663. [DOI] [PubMed] [Google Scholar]
- 30.Herman-Bausier P, et al. Mechanical strength and inhibition of the Staphylococcus aureus collagen-binding protein Cna. MBio. 2016;7:e01529-16. doi: 10.1128/mBio.01529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feuillie C, et al. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc Natl Acad Sci USA. 2017;114:3738–3743. doi: 10.1073/pnas.1616805114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinterdorfer P, Dufrêne YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 33.Milles LF, Schulten K, Gaub HE, Bernardi RC. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science. 2018;359:1527–1533. doi: 10.1126/science.aar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien L, et al. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: Roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 35.O’Connell DP, et al. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J Biol Chem. 1998;273:6821–6829. doi: 10.1074/jbc.273.12.6821. [DOI] [PubMed] [Google Scholar]
- 36.Otto M. Physical stress and bacterial colonization. FEMS Microbiol Rev. 2014;38:1250–1270. doi: 10.1111/1574-6976.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerrigan SW, Loughmann A, Meade G, Foster TJ, Cox D. Staphylococcus aureus clumping factor mediates rapid thrombus formation under high shear. Blood. 2006;108:1816. [Google Scholar]
- 38.Alsteens D, et al. Imaging G protein-coupled receptors while quantifying their ligand-binding free-energy landscape. Nat Methods. 2015;12:845–851. doi: 10.1038/nmeth.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakovenko O, et al. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geoghegan JA, et al. Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J Biol Chem. 2010;285:6208–6216. doi: 10.1074/jbc.M109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 42.Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci USA. 2012;109:13573–13578. doi: 10.1073/pnas.1202946109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokurenko EV, Vogel V, Thomas WE. Catch-bond mechanism of force-enhanced adhesion: Counterintuitive, elusive, but ... widespread? Cell Host Microbe. 2008;4:314–323. doi: 10.1016/j.chom.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer MM, et al. Catch-bond mechanism of the bacterial adhesin FimH. Nat Commun. 2016;7:10738. doi: 10.1038/ncomms10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoeler C, et al. Mapping mechanical force propagation through biomolecular complexes. Nano Lett. 2015;15:7370–7376. doi: 10.1021/acs.nanolett.5b02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonnell CJ, et al. Inhibition of major integrin αV β3 reduces Staphylococcus aureus attachment to sheared human endothelial cells. J Thromb Haemost. 2016;14:2536–2547. doi: 10.1111/jth.13501. [DOI] [PubMed] [Google Scholar]
- 47.McCormack N, Foster TJ, Geoghegan JA. A short sequence within subdomain N1 of region A of the Staphylococcus aureus MSCRAMM clumping factor A is required for export and surface display. Microbiology. 2014;160:659–670. doi: 10.1099/mic.0.074724-0. [DOI] [PubMed] [Google Scholar]
- 48.Hartford OM, Wann ER, Höök M, Foster TJ. Identification of residues in the Staphylococcus aureus fibrinogen-binding MSCRAMM clumping factor A (ClfA) that are important for ligand binding. J Biol Chem. 2001;276:2466–2473. doi: 10.1074/jbc.M007979200. [DOI] [PubMed] [Google Scholar]
- 49.Loughman A, et al. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol Microbiol. 2005;57:804–818. doi: 10.1111/j.1365-2958.2005.04731.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.