Significance
Heterosis is a fundamental biological phenomenon and important to crop and animal production. However, molecular mechanisms for heterosis remain elusive. Ethylene promotes fruit ripening but inhibits vegetative growth. Here we report that diurnal down-regulation of ethylene biosynthesis genes in Arabidopsis thaliana F1 hybrids corresponds to the decrease of ethylene production and biomass heterosis. Expression of ACS genes is diurnally and indirectly regulated by CCA1 during the day and directly regulated by PIF5 at night. Disruption of ethylene biosynthesis or signaling transduction leads to higher biomass, while application of exogenous ethylene reduced growth vigor, further validating the negative roles of ethylene in biomass heterosis. Thus, ethylene can integrate circadian rhythms and light signaling to regulate growth vigor in plant hybrids.
Keywords: hybrid vigor, ACS, circadian clock, PIF, epigenetics
Abstract
Heterosis is widely applied in agriculture; however, the underlying molecular mechanisms for superior performance are not well understood. Ethylene biosynthesis and signaling genes are shown to be down-regulated in Arabidopsis interspecific hybrids. Ethylene is a plant hormone that promotes fruit ripening and maturation but inhibits hypocotyl elongation. Here we report that application of exogenous ethylene could eliminate biomass vigor in Arabidopsis thaliana F1 hybrids, suggesting a negative role of ethylene in heterosis. Ethylene biosynthesis is mediated by the rate-limiting enzyme, 1-aminocyclopropane-1-carboxylate synthase (ACS). Down-regulation of ACS genes led to the decrease of ethylene production, which was associated with the high-vigor F1 hybrids, but not with the low-vigor ones. At the mechanistic level, expression of ACS genes was down-regulated diurnally and indirectly by Circadian Clock Associated 1 (CCA1) during the day and directly by Phyotochrome-Interacting Factor 5 (PIF5) at night. Consistent with the negative role of ethylene in plant growth, biomass vigor was higher in the acs mutants than in wild-type plants, while increasing endogenous ethylene production in the hybridizing parents reduced growth vigor in the hybrids. Thus, integrating circadian rhythms and light signaling into ethylene production is another regulatory module of complex biological networks, leading to biomass heterosis in plants.
Heterosis or hybrid vigor describes a phenomenon of superior growth and fitness in the F1 hybrid offspring relative to the parents (1). Although several genetic models, including dominance and overdominance, are used to explain heterosis, the molecular basis remains poorly understood (1–3). In tomato, a single-locus overdominance is shown to affect fruit yield, probably through changes in floral architecture (4). In Arabidopsis interspecific and intraspecific hybrids, altered circadian rhythms increased photosynthesis and starch metabolism (5, 6) and decreased stress responses (5, 7–9), leading to biomass heterosis (2, 10). Mechanisms for these changes could be related to epigenetic perturbations of key regulatory genes in the F1 hybrids (2, 11–14). In addition, basal expression levels of stress-responsive genes can be used as genetic distance indicators between the parents to predict heterosis levels in the F1 hybrids (5). These data suggest that either increasing carbon metabolism and/or decreasing energy cost for defense can promote heterosis (15). Consistent with the notion, many genes involved in the ethylene biosynthesis and signaling pathway including those encoding 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ethylene response factor (ERF) are down-regulated in the Arabidopsis allotetraploids that are formed between Arabidopsis thaliana and Arabidopsis arenosa (16). However, the mechanism for ethylene in biomass heterosis is largely unknown.
Ethylene is a plant hormone that regulates growth and developmental processes; disruption of ethylene biosynthesis or signal transduction affects growth and development of roots, hypocotyls, and seeds (17). Ethylene biosynthesis begins with the conversion of methionine to S-AdoMet (SAM) by S-AdoMet synthetase. SAM is then converted to ACC by ACS, which is a rate-limiting step (17). Finally, ACC is oxidized to ethylene by ACC oxidase (ACO). The Arabidopsis genome contains 12 annotated ACS genes, 8 of which (ACS2, ACS4–9, and ACS11) show functional activities (17). Ethylene production is circadian regulated by CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and TIME OF CAB EXPRESSION 1 (TOC1) in A. thaliana (18), but the biological significance of circadian-mediated ethylene production remains unknown.
Here we show that applying excessive exogenous ethylene could eliminate biomass heterosis, suggesting negative effects of ethylene on heterosis. Consistently, ACS genes were transcriptionally down-regulated in Arabidopsis hybrids, and consequently ethylene production was reduced in the hybrids. In contrast to previous findings (19), all ACS genes tested were subject to diurnal but not circadian expression. ACS genes were indirectly regulated by CCA1 during the day and directly by PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) at night, both of which were also down-regulated in the hybrids. Consistent with the down-regulation of ethylene production and increased growth vigor in the hybrids, disruption of ethylene biosynthesis or signaling transduction resulted in higher growth traits, including rosette areas and biomass. Increasing endogenous ethylene production could reduce biomass heterosis in the hybrids, further validating a critical role of ethylene in heterosis.
Results
Ethylene Production Is Down-Regulated in A. thaliana Hybrids.
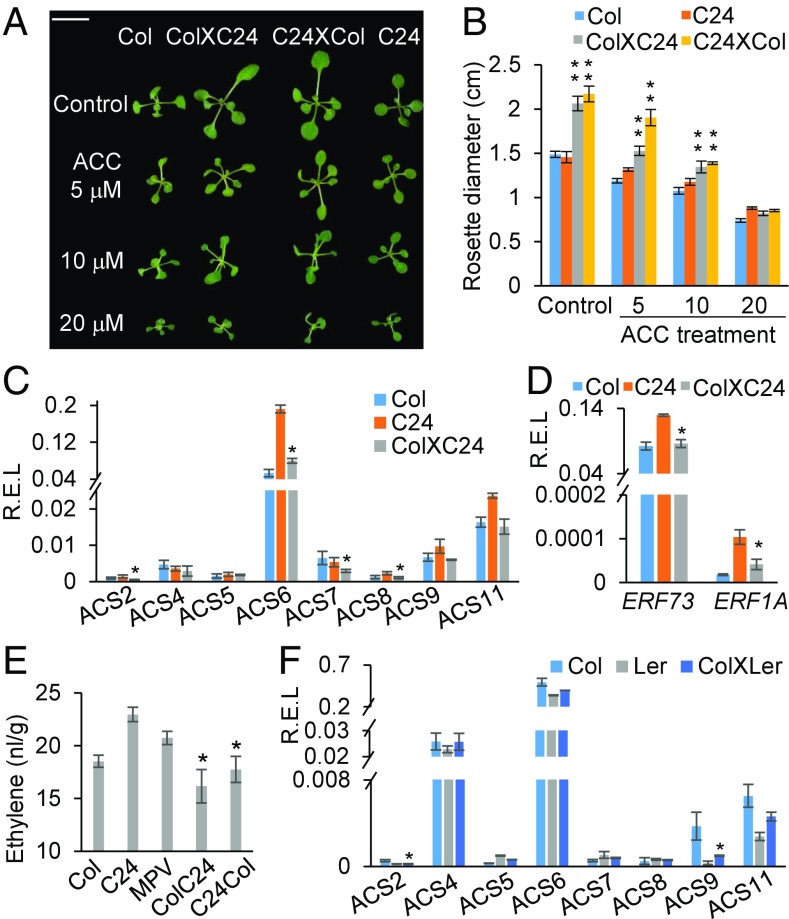
A previous study has found an overall down-regulation of ethylene biosynthetic and signaling pathway genes in Arabidopsis interspecific hybrids or allotetraploids (16), suggesting a potential role for ethylene in hybrid vigor. To determine the relationship between ethylene and heterosis, we applied exogenous ACC, the precursor of ethylene, to the growth media to test the effect of excessive ethylene on growth vigor in F1 hybrids and their parents. In the control, the better parent heterosis (BPH) value was 39–45% in the hybrids, which was reduced in a manner depending on ACC concentrations (Fig. 1 A and B). As the ACC concentration increased from 5 μM to 10 μM, BPH values gradually decreased from 16–44% to 14–15%. When the ACC concentration increased to 20 μM, the heterosis was nearly eliminated (Fig. 1 A and B). These data indicate that ethylene has a negative effect on heterosis.
Fig. 1.
Ethylene biosynthesis is down-regulated in Arabidopsis hybrids. (A) Representative images of seedlings of Col-0, C24, reciprocal F1 hybrids (ColXC24 and C24XCol) with or without ACC treatments. (Scale bar, 1 cm.) (B) Quantitative analysis of rosette diameter in A. Double asterisks indicate statistical significance levels at P < 0.01 compared with the midparent value (MPV). (C) Relative expression levels (R.E.L.) of ACS genes at ZT0 in Col-0, C24, and F1 hybrid (ColXC24). Asterisks indicate down-regulation in the hybrids at the statistically significant level (P < 0.05) relative to MPV. (D) R.E.L. of ERF1A and ERF73 as the materials used in C. (E) Ethylene production in F1 and F1r hybrids and their parents. (F) R.E.L. of ACS genes in hybrid (ColXLer).
This negative effect could be reversed by repressing ACS genes in A. thaliana intraspecific hybrids, as in the Arabidopsis interspecific hybrids (16). Indeed, among eight ACS genes, ACS2 and ACS6–8 were significantly down-regulated below the midparent value (MPV) in the F1 intraspecific hybrid (ColXC24) (Fig. 1C). To further validate down-regulation of ACS genes in heterosis, we examined ACS expression changes in several other F1 hybrids produced from different A. thaliana ecotypes, including Col-0 (United States), C24 (Portugal), Ws (Russia), Ler (Germany), and Est-1 (Russia), which displayed high biomass heterosis (5). All eight ACS genes tested (ACS2, ACS4–9, and ACS11) were down-regulated below the MPV in all or most hybrid combinations (SI Appendix, Fig. S1). Moreover, some examined ethylene-responsive genes, including ERF1A and ERF73, were also down-regulated below the MPV in the high-vigor hybrids (Fig. 1D and SI Appendix, Fig. S2). As a result, ethylene production was significantly reduced in the high-vigor F1 hybrids (Fig. 1E). These data indicate a role for repressing ACS genes in heterosis. Consistent with this notion, in the F1 hybrid (ColXLer) with low-level or no biomass heterosis, most ACS genes, except for ACS2 and ACS9, were expressed at similar levels to the MPV (Fig. 1F).
Since ethylene is a negative regulator for plant vegetative growth (17), down-regulation of ACS expression and ethylene production in the hybrids could increase growth vigor. Indeed, all single acs mutants and the double mutants with erf, and the pentuple mutant acs24569 showed higher shoot length than the wild type (Col-0) (Fig. 2A). The double mutants acs6 erf1a and acs6 erf73 grew larger than the single acs mutants (Fig. 2A). Biomass traits, including rosette size and dry weight, were significantly increased in the double mutants acs6 erf1a and acs6 erf73 and in the pentuple mutant acs24569 (Fig. 2 B and C). These data provide independent genetic evidence that suppression of individual ACS and ERF genes in the hybrids could increase biomass heterosis as in the diploid mutants.
Fig. 2.
Growth vigor is reduced in ACS gene mutants. (A–C) Comparison of Col-0, C24, F1 (ColXC24), and reciprocal F1 (C24XCol) hybrids, acs and erf single mutants, acs pentuple mutant (acs24569), and acs erf double mutants (acs6 erf1a and acs6 erf73), showing shoot length in mature plants (A), rosette size (B), and dry weight (C) in plants at 18 d after sowing. Asterisks indicate statistically significant levels (hybrid vs. MPV or mutant vs. wild type). Vertical dashed lines indicate phenotypic values of the wild type (Col-0). *P < 0.05, **P < 0.01. (Scale bar, 1 cm.)
Expression of ACS Genes Is Subject to Diurnal but Not Circadian Regulation.
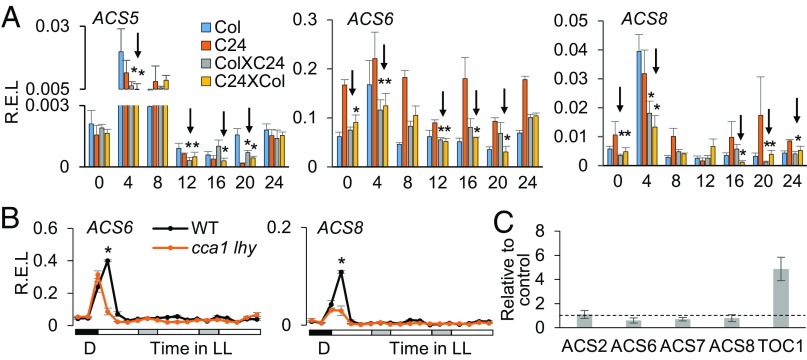
As ethylene production is diurnally regulated in both short-day (12/12 h of light/dark cycle) and long-day (16/8 h of light/dark cycle) conditions (18), we further examined expression of ACS genes in a diurnal cycle. All ACS genes, except for ACS4, were down-regulated in the A. thaliana F1 (ColXC24) and F1 reciprocal (F1r, C24XCol) hybrids in both day and night, especially at zeitgeber time 0 (ZT0 = 6 AM, dawn, or beginning of the light cycle), ZT4, and ZT16 (Fig. 3A and SI Appendix, Fig. S3).
Fig. 3.
ACS genes are regulated diurnally and indirectly by CCA1 during the day. (A) Relative expression levels (R.E.L.) of ACS genes every 4 h in a 24-h period (ZT0 = dawn) in the Col-0, C24, and reciprocal F1 hybrids (ColXC24 and C24XCol). Asterisks indicate down-regulation (arrows) in the hybrids at the statistically significant level (P < 0.05) relative to MPV. ACT7 was used as an internal control. (B) R.E.L. of ACS6 and ACS8 in the wild type (Ws) and cca1 lhy double mutant under a diurnal cycle (24 h) followed by constant light (LL) (48 h) conditions. Black, white, and gray boxes indicate dark, light, and subjective night, respectively. The asterisk indicates the statistically significant level (P < 0.05). (C) ChIP-qPCR showed the fold enrichment (y axis) of CCA1-binding fractions in the promoters of ACS2 and ACS6–8. The TOC1 promoter containing evening elements was used as the positive control. UBQ10 was used as a nonbinding control for normalization. *P < 0.05.
Given the role of circadian rhythms in ethylene production (18), we proposed that the circadian clock regulates expression of ACS genes, in addition to photosynthetic, metabolic, and stress-responsive genes in the hybrids and allotetraploids (5, 6). To test this, we examined ACS gene expression subjected to diurnal (24 h, 16 h light/8 h dark) and circadian conditions (48 h, constant light) in the wild-type (Ws) and cca1 lhy mutant (Ws background), respectively. Under the diurnal condition, all ACS genes showed expression peaks at ZT4 in the wild type, and they were significantly down-regulated in the cca1 lhy mutant compared with the wild type during the day, particularly during the peak expression time at ZT4 (Fig. 3B and SI Appendix, Fig. S4A). However, under the constant light condition, all ACS genes were expressed at lower levels and did not show rhythmic peaks in the wild type, while ACS expression levels were similar between the cca1 lhy mutant and the wild type (Fig. 3B and SI Appendix, Fig. S4A). The data suggest that ACS expression is subject to diurnal but not circadian regulation. This notion was supported by the analysis of chromatin immunoprecipitation (ChIP) quantitative PCR (qPCR). Using antibodies against CCA1, ChIP-qPCR results showed that CCA1 was significantly enriched in the TOC1 promoter (as a positive control, which was normalized to UBQ10), but not in the promoters of four ACS genes (ACS2 and ACS6–8) (Fig. 3C and SI Appendix, Fig. S4C), although each has at least one CCA1-binding site (CBS) in its promoter sequence (SI Appendix, Fig. S4B). These data suggest an indirect role of CCA1 in altering ACS expression.
ACS Genes Are Directly Regulated by PIF5 at Night.
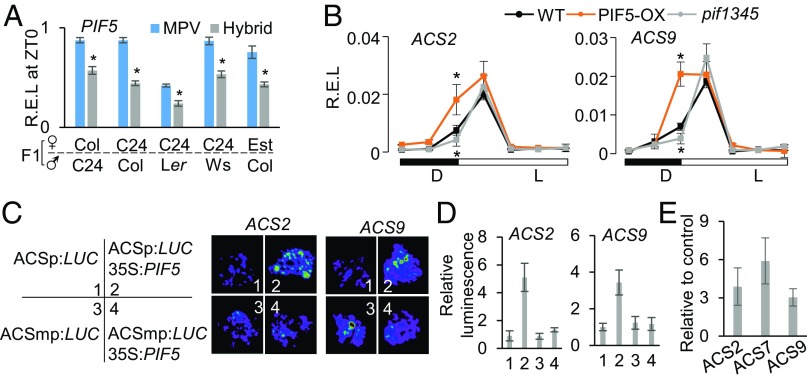
CCA1 was repressed in the hybrids during the day (SI Appendix, Fig. S5), and ACS genes were also down-regulated in the cca1 lhy mutant during the day but not at night (Fig. 3 B and C and SI Appendix, Fig. S4A), indicating that down-regulation of CCA1 in the hybrids contributes to the down-regulation of ACS genes during the day. However, ACS genes were down-regulated both during the day and at night in the hybrids (Fig. 3A and SI Appendix, Fig. S3). This suggests that other genes are responsible for ACS down-regulation at night in the hybrids. The other factors could be PIF4 and PIF5 because they affect ethylene levels, ACS expression, and hypocotyl elongation in darkness (19–21). PIF4 and PIF5 normally activate ACS expression at night and inhibit hypocotyl elongation. Consistent with this notion, PIF4 and PIF5 were down-regulated at night (ZT0 and ZT18, 16 h light/8 h dark) but not during the day (ZT12) in the F1 hybrids derived from different combinations of A. thaliana ecotypes (Fig. 4A and SI Appendix, Fig. S6), which could reduce ACS expression in the hybrids. Indeed, in PIF5 overexpression (PIF5-OE) plants, all ACS genes were significantly up-regulated, whereas in the pif1 pif3 pif4 pif5 (pif1345) mutant, several ACS genes (ACS2, ACS5, and ACS9) were significantly down-regulated at night (ZT0), but not during the day (Fig. 4B and SI Appendix, Fig. S7). These data suggest a role for PIFs in ACS and ethylene regulation at night in the hybrids as in the diploids (19).
Fig. 4.
ACS genes are regulated indirectly by PIF5 at night. (A) R.E.L. of PIF5 at ZT0 in F1 hybrids between different combinations of ecotypes. Asterisks indicate down-regulation in the hybrids at the statistically significant level (P < 0.05) relative to MPV. (B) R.E.L. of ACS2 and ACS9 in Col-0 (WT), PIF5-OX, and pif1345 mutant lines in diurnal conditions. Asterisks Above and Below the graphs, respectively, indicate up-regulation in PIF5-OX and down-regulation in pif1345 at the statistically significant level (P < 0.05) relative to WT. (C) Transient luciferase expression assays indicate that PIF5 directly activated expression of ACS2 and ACS9. The infiltrated constructs are ACS promoter fused with luciferase (ACSp:LUC, 1), ACSp:LUC cotransfected with 35S:PIF5 (2), ACS mutated promoter:luciferase (ACSmp:LUC, 3), and ACSmp:LUC with 35S:PIF5 (4). Luciferase activity images of N. benthamiana leaves are shown for ACS2 and ACS9. (D) Relative luminescence intensity (y axis) for each comparison in C. (E) ChIP-qPCR showed fold enrichment (y axis) of PIF5 in the promoters of ACS2, ACS7, and ACS9. Coding region of PP2A was used as a nonbinding control for normalization.
The nightly expression change of ACS genes is dependent on PIFs. Three ACS genes (ACS2, ACS7, and ACS9) contain at least one PIF-binding motif G-box within 2-kb promoter sequences (SI Appendix, Fig. S4B). In the Nicotiana benthamiana transient expression experiment, overexpression of PIF5 could elevate expressions of ACS2, ACS7, and ACS9 under the control of native promoters; this activation was abolished when the G-box in the promoters was mutated (Fig. 4 C and D and SI Appendix, Fig. S8). ChIP-qPCR analysis also showed that PIF5 bound to G-box in vivo in the promoters of ACS2, ACS7, and ACS9 (Fig. 4E). A previous study also observed an enrichment of PIF4 in promoters of ACS5 and ACS7–9 (22). Together, these results suggest that reduced expression of PIF4 and PIF5 in hybrids led to down-regulation of ACS genes at night to promote hypocotyl elongation and growth vigor.
Excessive Endogenous Ethylene Could Eliminate Heterosis in F1 Hybrids.
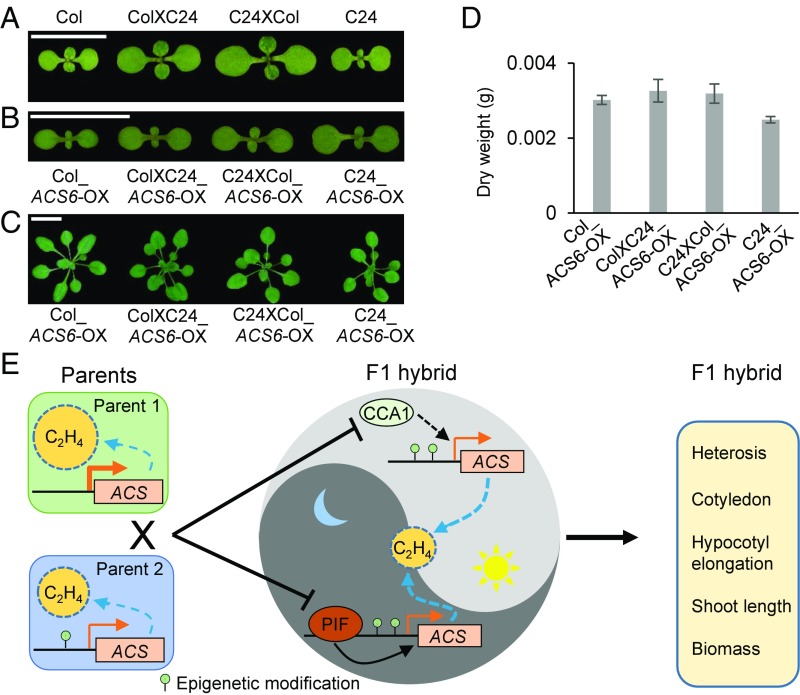
Higher growth traits resulting from disrupting ethylene biosynthesis or signaling transduction prompted us to test whether increasing endogenous ethylene production could inhibit heterosis. As constitutively expressing ACS6DDD, a gain of function for ACS6, results in constitutive ethylene production (23), we tested the effect of ACS6DDD overexpression on heterosis in the hybrids. Transgenic plants overexpressing ACS6DDD in Col (Col_ACS6-OX) and in C24 (C24_ACS6-OX) were used to generate reciprocal F1 hybrids (ColXC24_ ACS6-OX and C24XCol_ ACS6-OX), which all showed more ethylene production (11- to 15-fold) than the wild type (Fig. 1E and SI Appendix, Fig. S9A). Biomass traits, including cotyledon area, rosette size, and dry weight in the hybrids ColXC24_ACS6-OX and C24XCol_ACS6-OX were significantly lower than the hybrids between the wild-type parents (Figs. 2 and 5 A–D). The BPH value of cotyledon area was 47–85% in the hybrids of wild-type parents (Fig. 5A). However, in the F1 hybrids of ACS6DDD-overexpression parents, no heterosis was observed in the cotyledon area (Fig. 5B) or in the rosette size and dry weight (Fig. 5 C and D). These results indicate a negative effect of the constitutive ethylene production on heterosis during plant growth and development.
Fig. 5.
Production of excessive endogenous ethylene inhibits heterosis. (A and B) Representative cotyledon images of F1 reciprocal hybrids of the wild type (A) and ACS6DDD-overexpression (B) parents. (Scale bars, 1 cm.) (C and D) Representative rosette-size images (C) and quantified dry weight (D) of Col_ACS6-OX, C24_ACS6-OX, and their F1 (ColXC24_ACS6-OX) and reciprocal F1 (C24XCol_ACS6-OX) hybrids at 18 d after sowing. (Scale bar, 1 cm.) (E) Model showing the role of ethylene in heterosis. Different ethylene levels between the parents result in the reduced level of ethylene production in the hybrids. The down-regulation of ethylene production is associated with down-regulation of ACS genes, which is mediated indirectly by CCA1 during the day and directly by PIF at night. Decrease in ethylene biosynthesis (yellow circle) promotes superior performance of growth and biomass traits (cotyledon area, hypocotyl elongation, and shoot length) in the hybrids, leading to heterosis.
Discussion
Our data collectively support a model for CCA1-mediated ethylene emission during the day and PIF-mediated ACS expression at night to promote growth vigor in hybrids (Fig. 5E). In A. thaliana F1 hybrids and Arabidopsis allotetraploids, circadian clock genes such as CCA1 and LHY are repressed by DNA methylation and histone modifications during the day (6, 14). As a result, repressing CCA1 during the day decreases ethylene production by indirectly inhibiting ACS expression in the hybrids. At night, PIF genes, which regulate skotomorphogenesis (etiolation) (19), were repressed in hybrids, and repressing PIFs could decrease ethylene production through direct regulation of ACS genes. As PIF proteins are degraded under light (24), and CCA1 cannot directly bind promoters of ACS genes, other circadian clock regulators such as ELF3 could also regulate ACS expression during the day and night (25). Taken together, our data indicate that decrease of ethylene production leads to superior growth traits in the hybrids.
Current studies have revealed an emerging complexity of biomass heterosis under the control of regulatory networks involving circadian rhythms, which mediate photomorphogenesis, PIF signaling, skotomorphogenesis, ethylene production, and stress responses. Altered circadian rhythms in the hybrids could not only gate the timing of photosynthesis and starch biosynthesis and degradation, which stimulates carbon metabolism and photomorphogenesis and growth phenotypes (2, 6), but also gate the timing of stress responses, which balances the tradeoff between defense and growth (5). Ethylene emission is also regulated by diurnal rhythms (18). In the hybrids, ethylene biosynthesis and signaling pathways are repressed through integration with circadian rhythms and PIFs to promote growth vigor in F1 hybrids, which reveals another regulatory module of complex networks that lead to heterosis.
In Arabidopsis, more than 7,000 genes display diurnal expression patterns (26). As a result, circadian-regulated genes could exhibit opposite trends of expression changes during day and night in the hybrids or allotetraploids (5, 6). A recent study reported up-regulation of PIF4 in the F1 hybrid (C24XLer) during the day (ZT7) (27). In our study, PIF4 and PIF5 were repressed at night (ZT18) in the F1 hybrids produced from several different A. thaliana ecotypes, and their expression changes during the day were not obvious in the hybrids relative to the parents (Fig. 4A and SI Appendix, Fig. S6). This discrepancy could result from different hybrids and time points tested in two studies. It is notable that PIF proteins are rapidly degraded by light-activated phytochromes, and they are more potent at night to regulate hypocotyl growth (24). Consistent with this notion, expression of ACS genes is significantly up-regulated in the PIF5-OX plants and down-regulated in pif1345 mutant at night but not during the day (Fig. 4B and SI Appendix, Fig. S7). We also noticed an expression increase of several ACS genes (ACS4–7 and ACS9) during the day in the pif1345 mutant and PIF5-OX lines as in the wild-type plants (Fig. 4B and SI Appendix, Fig. S7), suggesting that ACS genes are required for ethylene production and related biological activities independent of PIFs during the day. Indeed, constantly overexpressing or repressing PIFs affected overall growth as observed in the PIF5_OX lines and pif1345 mutant, which showed severe and moderate reductions of vegetative growth, respectively (SI Appendix, Fig. S9 B and C). In the pif1345 mutant, the positive effect of reducing PIFs at night on hypocotyl growth could be counterbalanced and uncoupled by the increased ethylene activities during the day. We predict that altered expression of PIFs in the hybrids at night is likely more biologically relevant to growth vigor, while their roles during the day remain to be investigated.
Ethylene is also known as a stress-related hormone (28), and many ecotypes used for the study have different basal expression levels of stress-responsive genes (5). C24 is more stress tolerant, especially to biotic stresses than Col-0 (29), and consequently more ethylene is produced in C24 than in Col-0, which is less sensitive to the ethylene treatment (Fig. 1 A, B, and E). Natural variation of ethylene production and sensitivity suggests that plants can adapt to local environments by adjusting ethylene biosynthetic and signaling pathways. This adaptation could be related to altered expression of ACS genes through epigenetic modifications, as observed for some stress-responsive genes (5). Notably, many stress-responsive genes are also repressed in the F1 hybrids to promote growth vigor (5, 9). Thus, optimizing diurnal and circadian expression of ethylene and stress-responsive genes under stress and nonstress conditions provides a better resource allocation for promoting growth vigor as well as environmental adaption in the hybrids as in the parents.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana ecotypes including Col-0, C24, Ws, Ler, and Est-1 (CS22629) were used to generate F1 hybrids by hand pollination. The cca1 lhy (CS9380), pif1345 (CS66049), acs2 (CS16564), acs4 (CS16566), acs5 (CS16568), acs6 (CS16569), acs9 (CS16571), acs24569 (CS16644), erf1a (SALK_036267), and erf73 (SALK_012913) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). The PIF5-OX line (35S:PIF5-myc) was obtained from a previous report (30). Transgenic plants that overexpressed ACS6DDD in Col (Col_ACS6-OX) were generously provided by Shuqun Zhang, University of Missouri, Columbia, MO. Transgenic plants that overexpressed ACS6DDD in C24 (C24_ACS6-OX) were generated by Agrobacterium tumefaciens-mediated transformation. For diurnal conditions, plants were grown under the light/dark (L/D) cycle of 16 h/L at 22 °C and 8 h/D at 20 °C. For circadian conditions, plants were grown under the constant light of 16 h/L at 22 °C and 8 h/L at 20 °C. Plant transformation was performed in 4- to 5-wk-old flowering plants following the floral dipping method (31).
Plasmid Constructs.
The N. benthamiana transient expression assay employed luciferase reporter constructs, each containing 2-kb sequence upstream from the ATG codon of ACS2 and ACS6–9 that was cloned into pFAMIR-LUC vector (5). For 35S-driven overexpression constructs, each contained coding sequence of PIF5 or CCA1 that was cloned into pF35SE vector (5). For generating ACS6DDD-overexpression transgenic lines in C24 backgrounds, coding sequences of ACS6DDD were amplified from Col_ACS6DDD-OX and cloned into pF35SE vector. Each construct was cloned into Agrobacterium strain GV3101 for plant transformation (31) or N. benthamiana transient expression assay (32). Primer sequences for individual inserts are listed in SI Appendix, Table S1.
RNA Extraction and qRT-PCR.
Total RNA was isolated from aerial rosette tissues of ∼18-d-old seedlings using Plant RNA Reagent (Invitrogen). After digestion by RNase-Free DNase (Promega), total RNA (1 μg) was used to produce first-strand cDNA with the Omniscript RT Kit (Qiagen). The cDNA was used as the template for qRT-PCR using FastStart Universal SYBR Green Master (Roche). The reaction was run on the LightCycler 96 System (Roche). The relative expression level was quantified using internal control ACT7 (At5g09810) (6). Three biological replicates were performed for each sample and three technical replicates were used for each biological replicate in qRT-PCR experiments. The MPV was calculated as the mean of parental expression value for each replicate. One-way ANOVA was used to determine the significance in each comparison. The primer sequences are listed in SI Appendix, Table S1.
Ethylene Measurement by Gas Chromatography.
For each genotype, 12 seeds were germinated in 40 mL in each gas chromatography (GC) vial containing 30 mL of Murashige and Skoog agar. These vials were sealed with permeable tape. After 10 d, the vials were capped and the plants were incubated for another 36 h in capped GC vials in a growth chamber. The ethylene level was measured using GC-2014 (Shimadzu). Ethylene production was normalized to fresh weight of seedlings with five biological replicates.
N. benthamiana Transient Expression Assay.
The transient expression assays were performed in leaves of N. benthamiana as previously described (32). The A. tumefaciens strain GV3101 containing the luciferase reporter construct or 35S-driven overexpression construct was incubated in Luria–Bertani medium and finally resuspended in infiltration buffer (10 mM Mes, 0.2 mM acetosyringone, 10 mM MgCl2) to an ultimate concentration of OD600 = 1.0. Equal amounts of different combined bacterial suspensions were infiltrated into the young leaves of the 5-wk-old plants using a needleless syringe. After infitration, the plants were grown first in the dark for 12 h and then with a cycle of 16 h/8 h (light/dark) for 48 h at 24 °C before imaging. The leaves were sprayed with 100 µM luciferin (Promega) and placed in the dark for 5 min. Luciferase activity was observed using NightOWL LB 983 in Vivo Imaging System (Berthold). Each experiment was performed using three independent biological replicates.
ChIP-qPCR.
ChIP was performed as previously described (33). Briefly, 2 g of rosette leaves from 2-wk-old Col-0 or 35S:PIF5-myc seedlings were used as starting materials for ChIP experiments. Antibodies against CCA1 (Abiocode) were used to immunoprecipitate the protein–DNA complex. For PIF5, antibodies against MYC (Cell Signaling) were used for immunoprecipitation. The isolated chromatin without antibodies was used as input control. UBQ10 and PP2A were used as internal controls for normalization. Primers used for ChIP-qPCR are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Xiaole Chen (Center for Electrochemistry, The University of Texas at Austin) for help on GC analysis and Dr. Shuqun Zhang (University of Missouri) for Col_ACS6-OX seeds. This work was supported by Grants 91631302 and 31621001 from the National Natural Science Foundation of China, the Peking-Tsinghua Center for Life Sciences, and the State Key Laboratory of Protein and Plant Gene Research, and Grants GM109076 (to Z.J.C.), GM114297 (to E.H.), and GM115879 (to H.Q.) from the National Institutes of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722068115/-/DCSupplemental.
References
- 1.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22:2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen ZJ. Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet. 2013;14:471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- 3.Schnable PS, Springer NM. Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol. 2013;64:71–88. doi: 10.1146/annurev-arplant-042110-103827. [DOI] [PubMed] [Google Scholar]
- 4.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 5.Miller M, Song Q, Shi X, Juenger TE, Chen ZJ. Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat Commun. 2015;6:7453. doi: 10.1038/ncomms8453. [DOI] [PubMed] [Google Scholar]
- 6.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA. 2012;109:7109–7114. doi: 10.1073/pnas.1204464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Lu H, Erickson JW. Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- 9.Groszmann M, et al. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci USA. 2015;112:E6397–E6406. doi: 10.1073/pnas.1519926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dapp M, et al. Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants. 2015;1:15092. doi: 10.1038/nplants.2015.92. [DOI] [PubMed] [Google Scholar]
- 12.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24:875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greaves IK, et al. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA. 2012;109:3570–3575. doi: 10.1073/pnas.1201043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng DW, et al. A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell. 2014;26:2430–2440. doi: 10.1105/tpc.113.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JS, Venable DL. Polyploidy and the evolution of gender dimorphism in plants. Science. 2000;289:2335–2338. doi: 10.1126/science.289.5488.2335. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H, Ecker JR. The ethylene signaling pathway: New insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Thain SC, et al. Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 2004;136:3751–3761. doi: 10.1104/pp.104.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna R, et al. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh E, et al. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bours R, Kohlen W, Bouwmeester HJ, van der Krol A. Thermoperiodic control of hypocotyl elongation depends on auxin-induced ethylene signaling that controls downstream PHYTOCHROME INTERACTING FACTOR3 activity. Plant Physiol. 2015;167:517–530. doi: 10.1104/pp.114.254425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2017;114:E3555–E3562. doi: 10.1073/pnas.1703179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bechtold U, et al. Constitutive salicylic acid defences do not compromise seed yield, drought tolerance and water productivity in the Arabidopsis accession C24. Plant Cell Environ. 2010;33:1959–1973. doi: 10.1111/j.1365-3040.2010.02198.x. [DOI] [PubMed] [Google Scholar]
- 30.Sakuraba Y, et al. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Song QX, et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J Exp Bot. 2013;64:4329–4341. doi: 10.1093/jxb/ert238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi N, et al. PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book. 2014;12:e0170. doi: 10.1199/tab.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.