Significance
Leaf senescence is regulated in a complex manner, involving time-dependent interactions with developmental and environmental signals. Genetic screens have identified key regulators of senescence, particularly late-stage senescence regulators. Recently, time-course gene-expression and network analyses, mostly analyses of static networks, have predicted many senescence regulators. However, senescence is defined by time-evolving networks, involving the temporal transition of interactions among senescence regulators. Here, we present time-evolving networks of NAM/ATAF/CUC (NAC) transcription factors, central regulators of leaf senescence in Arabidopsis, via time-course gene-expression analysis of NACs in their mutants. These time-evolving networks revealed a unique regulatory module of NACs that controls the timely induction of senescence-promoting processes at a presenescent stage of leaf aging.
Keywords: leaf senescence, time-evolving network, NAC, presenescent repressors, salicylic acid response
Abstract
Senescence is controlled by time-evolving networks that describe the temporal transition of interactions among senescence regulators. Here, we present time-evolving networks for NAM/ATAF/CUC (NAC) transcription factors in Arabidopsis during leaf aging. The most evident characteristic of these time-dependent networks was a shift from positive to negative regulation among NACs at a presenescent stage. ANAC017, ANAC082, and ANAC090, referred to as a “NAC troika,” govern the positive-to-negative regulatory shift. Knockout of the NAC troika accelerated senescence and the induction of other NACs, whereas overexpression of the NAC troika had the opposite effects. Transcriptome and molecular analyses revealed shared suppression of senescence-promoting processes by the NAC troika, including salicylic acid (SA) and reactive oxygen species (ROS) responses, but with predominant regulation of SA and ROS responses by ANAC090 and ANAC017, respectively. Our time-evolving networks provide a unique regulatory module of presenescent repressors that direct the timely induction of senescence-promoting processes at the presenescent stage of leaf aging.
In plants, aging involves age-dependent developmental changes during the lifespan. At the end of aging, plants undergo senescence followed by death of cells, tissues, organs, or the entire organism. Leaf senescence is crucial for the fitness and survival of plants (1). During growth, leaves harvest solar energy and accumulate chemical energy and biomass through carbon fixation. When leaves enter senescence after a prolonged productive photosynthetic period, leaf cells undergo drastic physiological, biochemical, and metabolic changes in an orderly manner. The most obvious phenotypic change during senescence is the yellowing of leaves caused by the breakdown of chlorophyll in chloroplasts. Other metabolic changes include increased oxidation and hydrolysis of macromolecules, such as proteins, lipids, and nucleic acids. These hydrolyzed molecules in senescing leaves are recycled as materials and energy to newly developing organs or offspring in annual plants. Thus, the timely onset of leaf senescence is critical for effective recycling of materials and energy.
Leaf senescence is regulated over a long period of time in a complex manner, involving time-dependent interactions with various developmental and environmental signals (1). Genetic screens have revealed mutants with accelerated or delayed leaf senescence and thus have uncovered key regulators of leaf senescence (2–4). However, these screens are limited to the identification of one regulator per mutant rather than a regulatory module of multiple genes. Moreover, these approaches identify individual senescence regulators based on only a few phenotypes in a narrow time window of senescence, mostly at senescent stages, thus limiting insights into the regulators acting at presenescent or early stages of leaf senescence. Alternative screening approaches are needed to discover these early regulators or regulatory modules of leaf senescence.
Time-course gene-expression profiling of Arabidopsis leaves during aging was used recently as an alternative approach to uncover senescence regulators (5, 6). Woo et al. (5) and Breeze et al. (6) provided thousands of senescence-associated genes with differential expression during leaf aging. Potential early regulators [e.g., receptors, kinases, phosphatases, and transcription factors (TFs)] can be predicted from genes with altered expression at early stages. Moreover, regulatory modules can also be proposed as subsets of the potential early regulators. However, it is difficult to prioritize potential regulators and regulatory modules for functional validation. To resolve this issue, network analysis has often been combined with time-course gene-expression analysis. For instance, Penfold et al. (7) built a regulatory network model of senescence-regulator candidates using time-course gene-expression data (6) and prioritized key senescence regulators based on their degrees of centrality in the network model. Also, Allu et al. (8) built a coexpression network model for senescence- or stress-related genes (salt and oxidative stress) and predicted a regulatory module as a set of densely interacting nodes in the network model. In these approaches, network models delineate static views of interactions among candidate senescence regulators. However, senescence is determined by time-dependent interactions among senescence regulators during leaf aging. Thus, there is a need for a method based on time-evolving networks that describe the temporal transition of regulatory interactions among senescence regulators during leaf aging.
Several types of senescence regulators have been reported in plants, including receptors, kinases/phosphatases, TFs, epigenetic regulators (e.g., methyltransferases and demethylases), and regulatory RNAs (e.g., microRNAs) (3). Of these, the TFs, such as NACs (NAM/ATAF/CUC), WRKYs, and MYBs, are important for regulating the temporal expression of senescence-associated genes during leaf aging (3, 9–16). Among the TF families, the NAC family is one of the largest (17, 18) and is a central regulator of leaf senescence. Many NACs (57.5% or 65/113 NAC genes) show expression changes during leaf aging in Arabidopsis, and genetic studies identified many NACs as positive (ANAC016, ANAC029/AtNAP, ANAC059/ORS1, and ANAC092/ORE1) and negative (ANAC042/JUB1 and ANAC083/VNI2) regulators of leaf senescence (4, 19–24). Moreover, the NACs are reported to regulate their own and each other’s transcription (25). Thus, time-evolving regulatory networks for NACs likely provide sufficient information regarding the temporal transition of regulatory relationships, thereby enabling the prioritization of NACs or NAC-regulatory modules involved in leaf senescence.
Here, we present time-evolving networks that delineate causal regulatory relationships among 49 senescence-associated NACs, revealing a shift from positive to negative regulation among NACs before senescence. Three NACs, ANAC017, ANAC082, and ANAC090, referred to as a “NAC troika” (regulatory module), governed this regulatory shift. We found that the NAC troika negatively regulates leaf senescence at presenescent stages by inhibiting senescence-promoting pathways, including salicylic acid (SA) and reactive oxygen species (ROS) pathways.
Results
Time-Evolving NAC Networks Reveal a Regulatory Shift at a Presenescent Stage.
In our network-based approach, we first selected a set of senescence regulators to construct time-evolving networks. Among 113 NACs in Arabidopsis (26), we selected the 65 NACs that show gene-expression changes over the lifespan of leaves (SI Appendix, Fig. S1A) (5). To infer the temporal evolution of regulatory relationships among the selected NACs, we obtained knockout mutants for 49 of the 65 NACs from the Arabidopsis Biological Resource Center (SI Appendix, Table S1). We sampled the third and fourth true leaves of wild-type (Col) and mutant plants at 4-d intervals from a mature leaf stage (14 d of leaf age) to a middle-senescent stage (26 d of leaf age) that displays more than 40% loss of chlorophyll (SI Appendix, Fig. S1 B–E). Previously, transcript levels of SEN4, a senescence-inducible marker, were reported to be significantly (twofold or more) increased in leaves at 22 d of leaf age, compared with those at the mature stage (14 d), with about a 20% loss of chlorophyll (27). We thus defined 22 d of leaf age as an early senescent stage and 18 d of leaf age, between the mature (14 d) and early-senescent (22 d) stages, as a presenescent stage.
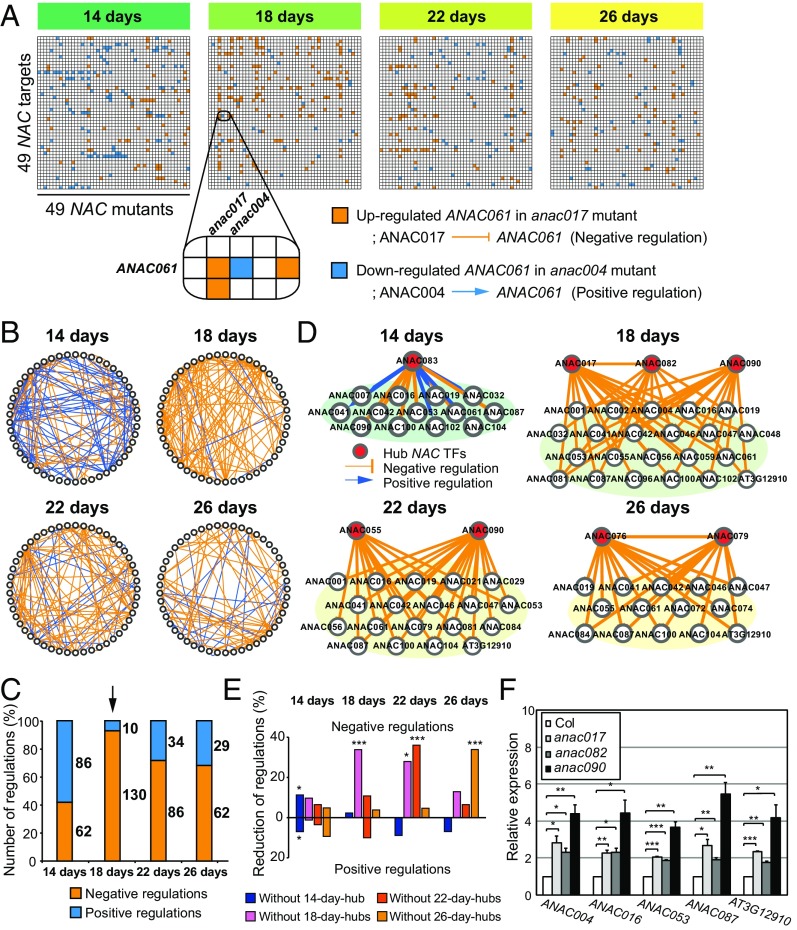
We measured mRNA-expression levels in wild-type and mutant leaves at these four stages (mature and pre-, early-, and middle-senescent stages) by NanoString nCounter analysis (28). For each stage, we identified differentially expressed NACs in individual mutants relative to the wild type, which were considered targets of the mutated NAC. Up- and down-regulation of the target NACs in mutants reflect negative (repressive) and positive (activating) regulation, respectively, in wild-type plants (Fig. 1A). In total, 148, 140, 120, and 91 differentially expressed NACs were identified from the 49 NAC mutants at 14, 18, 22, and 26 d of leaf age, respectively (Dataset S1). For each stage, we built a genetic regulatory network that describes positive and negative regulatory relationships between the mutated NACs and their target NACs (Fig. 1B).
Fig. 1.
Time-evolving NAC networks reveal a drastic inversion in their regulatory patterns at the presenescent stage. (A) Causal regulatory relationships among 49 aging-associated NACs at four stages (14, 18, 22, and 26 d of leaf age). The heat map at each stage shows the relationships inferred from differentially expressed NACs between NAC-knockout mutants and wild-type plants at 14, 18, 22, and 26 d. For example, ANAC061 was up-regulated (orange in the heat map) in the anac017 mutant, indicating negative (repressive) regulation on ANAC061 by ANAC017. In contrast, ANAC061 was down-regulated (blue in the heat map) in the anac004 mutant, indicating positive (activating) regulation on ANAC061 by ANAC004. (B) Genetic regulatory networks that show causal regulatory relationships among 49 NACs at the four stages. In each network, circles represent 49 NACs, and the blue and orange lines indicate positive and negative regulations inferred from NACs differentially expressed in the mutants and the wild type. (C) Relative proportions of positive (blue) and negative (orange) regulations at each stage. The arrow indicates the presenescent stage (18 d) in which the number of negative regulations exceeded that of positive regulations, referred to as the “positive-to-negative regulatory inversion.” (D) Subnetworks showing negative (orange) and positive (blue) regulatory relationships between hub NACs and their target NACs at four stages (14, 18, 22, and 26 d of leaf age). In each subnetwork, circles represent target NACs regulated by the corresponding hub NACs, and blue and orange lines indicate positive and negative regulations of target NACs by the hub NACs. (E) Reduced percentages of negative and positive regulatory relationships at each stage after removing the hub NACs identified at individual stages from the time-evolving networks. Colors represent the percentages after removal of the indicated hub NACs. The significance (P) of the reduction percentage was evaluated using an empirical distribution of the reduction percentage obtained from random removal experiments. *P < 0.05; ***P < 0.001. (F) Validation of the up-regulation of five representative NAC targets in anac017, anac082, and anac090 mutants at 18 d. mRNA expression levels were analyzed by qRT-PCR and then were normalized by those of ACT2. For each gene, the normalized expression levels were further normalized by those in wild-type plants. The normalized expression levels were compared with those in wild-type plants using ANOVA with Dunnett’s correction as a post hoc test. *P < 0.1; **P < 0.05; ***P < 0.01. Values are means ± SE (n = 3).
Intriguingly, the time-evolving NAC networks revealed a drastic inversion in the dominant regulatory patterns from positive regulation at 14 d (mature stage) to negative regulation at 18 d (presenescent stage) (Fig. 1B). The number of positive regulations decreased from 86 at 14 d to 10 at 18 d (8.6-fold), whereas the number of negative regulations increased from 62 to 130 (2.1-fold) (Fig. 1C). Negative regulations remained dominant after 18 d (Fig. 1C and SI Appendix, Fig. S1 F and G). The positive-to-negative regulatory inversion in the time-evolving NAC networks suggests the suppression or maintenance of NAC expression, possibly as a strategy to prevent untimely senescence at the presenescent stage.
We next attempted to identify early-senescence regulators or regulatory modules from the time-evolving network. Many biological networks have been shown to be scale-free with the hubs that govern functions of the networks (29). Our NAC networks were also found to be scale-free at individual stages based on out-degree distributions (29), suggesting the presence of hub NACs at individual stages (SI Appendix, Fig. S2A). This observation led us to hypothesize that such hub NACs can govern the positive-to-negative regulatory inversion at the presenescent stage. To test this hypothesis, we identified seven hub NACs that have a significantly (P < 1.0 × 10−4) large number of target NACs (i.e., significant out-degree centrality) in the time-evolving networks, as described in SI Appendix, SI Materials and Methods [SI Appendix, Fig. S2B: ANAC083 at 14 d (mature); ANAC017, ANAC082, and ANAC090 at 18 d (presenescent); ANAC055 and ANAC090 at 22 d (early senescent); and ANAC076 and ANAC079 at 26 d (middle senescent)]. The subnetworks for the hub NACs and their targets showed that the hub NAC at 14 d (ANAC083) regulated its targets positively and negatively, whereas the hub NACs from 18 to 26 d regulated their targets only negatively (Fig. 1D), indicating the importance of the hub NACs at 18 d in the establishment of the regulatory inversion. To further assess such importance, we evaluated whether removing the hub NACs from the time-evolving networks affected the positive-to-negative regulatory inversion. Among the hub NACs, the three hub NACs ANAC017, ANAC082, and ANAC090 at 18 d, as a NAC troika, showed the most significant reduction (P < 1.010−4) in negative regulations, suggesting that the NAC troika has the largest effect on the regulatory inversion (Fig. 1E).
We confirmed target NACs of the NAC troika at 18 d by quantitative real-time PCR (qRT-PCR) (Fig. 1F). Finally, we examined whether the perturbation of the NAC troika could affect the induction kinetics of their target NACs at the presenescent stage. The induction of target NACs was accelerated in single and double mutants of the NAC troika compared with wild-type plants, whereas plants overexpressing the NAC troika displayed delayed the induction of target NACs (SI Appendix, Fig. S3), suggesting that the NAC troika critically contributes to the regulatory inversion kinetics in time-evolving NAC networks. Taken together, these data suggest that the NAC troika, as a regulatory module of early-senescence regulators, controls the positive-to-negative regulatory inversion at the presenescent stage in the time-evolving NAC networks.
The NAC Troika Negatively Regulates Various Leaf-Senescence Phenotypes.
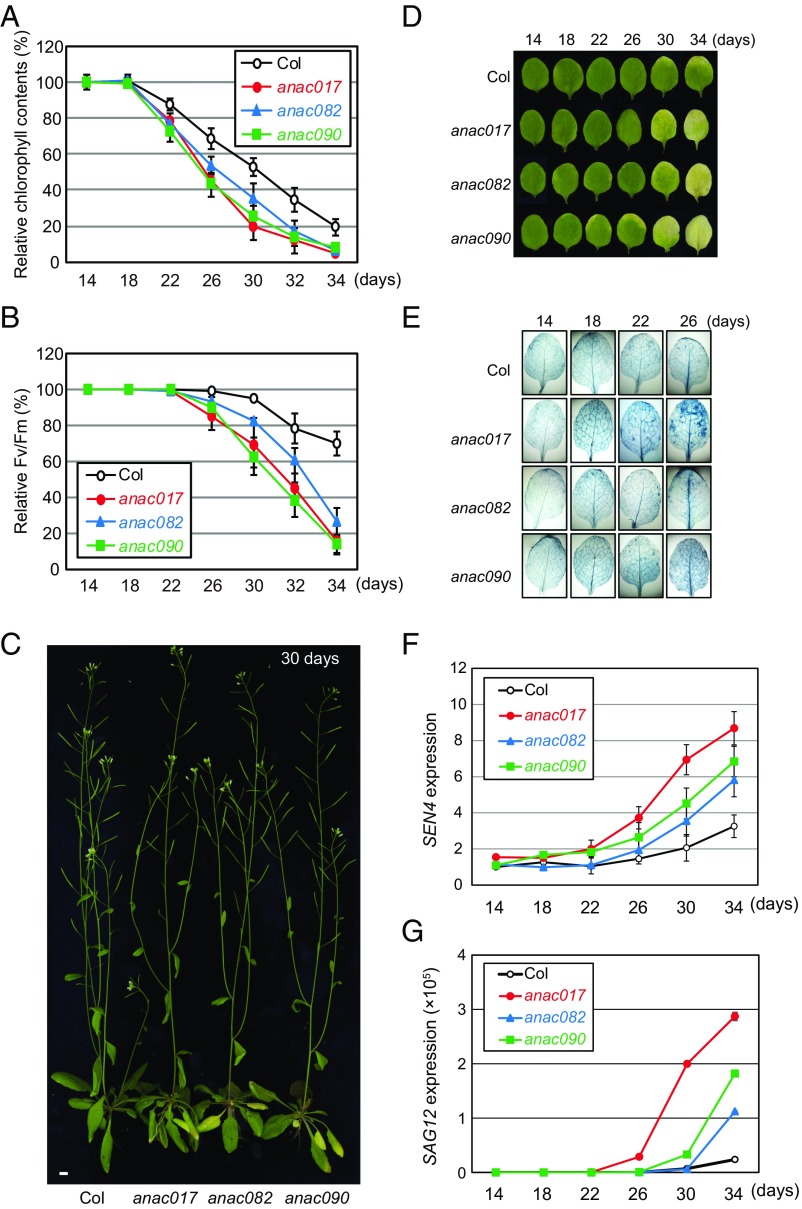
Next, we investigated the physiological relevance of the presenescent regulatory inversion in the time-evolving NAC networks. To this end, we examined whether the NAC troika that governs this inversion was associated with leaf senescence. First, we measured two physiological features of leaf senescence, chlorophyll content and photochemical efficiency (Fv/Fm), in the mutants of the seven hub NACs during leaf aging. Consistent with the findings in Fig. 1E, among the seven identified hub NACs, the mutants of the NAC troika showed the most substantial acceleration in the loss of chlorophyll content and photochemical efficiency compared with the wild type (Fig. 2 A and B and SI Appendix, Fig. S4).
Fig. 2.
Mutants of the NAC troika exhibit strong associations with leaf-senescence phenotypes. (A and B) Relative changes in chlorophyll content (A) and photochemical efficiency (Fv/Fm) (B) in single mutants of the NAC troika (anac017, anac082, and anac090 mutants) compared with wild-type (Col) plants during leaf aging. Values are means ± SD (n > 20) and are normalized to those at 14 d in individual mutants. Colors represent the data measured from the indicated single mutants. (C) Representative photographs of a wild-type plant (Col) and single mutants at 30 d after the emergence of the third or fourth leaves. (Scale bar, 1 cm.) (D) Leaf yellowing in wild-type plants and single mutants at the indicated leaf age (in days). (E) Trypan blue staining of leaves at the indicated leaf age. In each plant, blue-colored patches of cells were developed by Trypan blue staining, indicating areas of dead or dying cells. (F and G) mRNA expression levels of SEN4 (F) and SAG12 (G) in wild-type plants and single mutants at the indicated leaf age. The mRNA expression level of each gene was measured by qRT-PCR and normalized by that of ACT2. Values are means ± SD (n = 2).
Next, we examined the role of the NAC troika in leaf-yellowing phenotypes, cell death, and the expression of molecular senescence markers during leaf aging. The overall growth and development of anac017, anac082, and anac090 mutants were not significantly distinguishable from those of wild-type plants (Fig. 2C). However, rosette leaves (the third or fourth leaf) of anac017, anac082, and anac090 mutants began to yellow at 30 d of leaf age and were completely yellow at 34 d, whereas wild-type leaves retained their integrity and remained green even at 34 d (Fig. 2D). In addition, Trypan blue staining revealed increased cell death in the third or fourth leaves of anac017, anac082, and anac090 mutants relative to the wild type (Fig. 2E). Consistent with these findings, these mutants exhibited accelerated induction of two senescence marker genes, SEN4 and SAG12, during leaf aging, compared with the wild type (Fig. 2 F and G). Further, double mutants of the NAC troika exhibited accelerated leaf yellowing compared with the single mutants, whereas overexpression plants showed delayed leaf yellowing (SI Appendix, Fig. S5A). Similar patterns were observed for chlorophyll loss (SI Appendix, Fig. S5B), which was measured as the median relative chlorophyll content during leaf aging, and for the expression of SAG12 (SI Appendix, Fig. S5C). Taken together, these data show that the NAC troika is strongly associated with leaf senescence, suggesting that the presenescent regulatory shift governed by the NAC troika in the time-evolving NAC networks is linked to leaf senescence.
The NAC Troika Suppresses Genes Involved in Senescence-Promoting Processes.
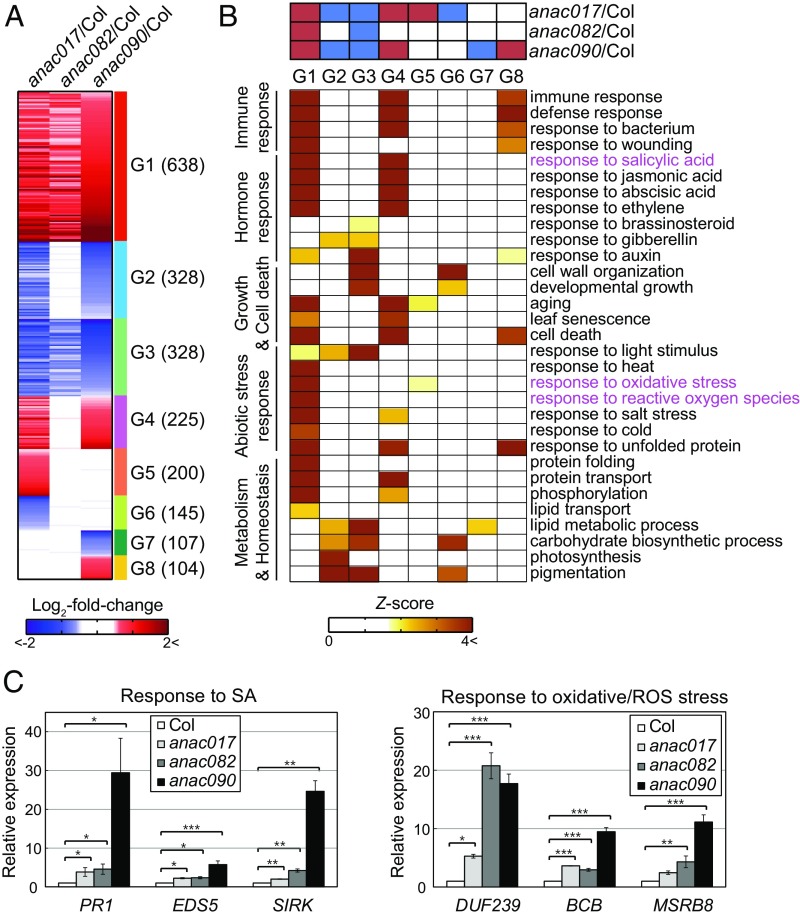
To investigate senescence-associated genes regulated by the NAC troika, we performed genome-wide mRNA expression analysis of wild-type and mutant leaves at the presenescent stage (18 d) (Fig. 3A). We compared the transcriptomes of wild-type leaves with those of anac017, anac082, and anac090 mutants and identified 1,977, 1,355, and 1,907 differentially expressed genes (DEGs), respectively (Dataset S2) (30). We found that 1,796 (72.6%) of the union of these DEGs (2,473 genes) were differentially expressed in more than one mutant (SI Appendix, Fig. S6A), suggesting substantial coregulation by the NAC troika.
Fig. 3.
The NAC troika negatively regulates genes involved in senescence-promoting processes. (A) Heat map showing eight major groups (G1–8) of genes up-regulated (red) and down-regulated (blue) in anac017, anac082, and anac090 mutants compared with the wild type. The numbers of target genes in each group are denoted in parentheses. Red and blue colors represent up- and down-regulation in the mutants, respectively, compared with the wild type. The color bar shows the gradient of log2 fold changes between the mutants and the wild type. (B) GOBPs represented by the genes in G1–8. (Upper) The heat map shows up-regulation (red) and down-regulation (blue) patterns of the genes in G1–8 from the three comparisons: anac017 vs. wild type (anac017/Col), anac082 vs. wild type (anac082/Col), and anac090 vs. wild type (anac090/Col). (Lower) The heat map shows the significance (Z-score) of GOBPs being enriched by the genes in G1–8. The enriched GOBPs were categorized into the following functional groups: immune response, hormone response, growth and cell death, abiotic stress response, and metabolism and homeostasis. Two top-ranked GOBPs according to GOBP-association analysis (SI Appendix, Fig. S5 B and C), responses to SA, and oxidative/ROS stress, are labeled in magenta. The color bar shows the Z-score gradient that represents the enrichment significance (P value): Z = N−1(1 − P) where N−1 is the inverse standard normal distribution and P is the enrichment P value. (C) Confirmation of up-regulation of SA- and oxidative/ROS stress-inducible genes in the mutants, compared with wild-type plants, at 18 d using qRT-PCR analysis. mRNA expression levels of SA-inducible genes (PR1, EDS5, and SIRK) and oxidative/ROS stress-inducible genes (DUF239, BCB, and MSRB8) were measured by qRT-PCR and then were normalized by those of ACT2. For each gene, the normalized expression level was further normalized by that in wild-type plants. The normalized expression levels were compared with those in wild-type plants using one-way ANOVA with Dunnett's correction as a post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001. Values are means ± SE (n = 3 or 4).
We categorized the 2,473 DEGs into 21 groups based on their patterns of up- and down-regulation in the three mutants relative to the wild type (SI Appendix, Table S2). We focused on eight major groups (G1–8), each of which includes more than 100 DEGs (Fig. 3A). To determine the cellular processes associated with the genes in G1–8, we performed enrichment analysis of Gene Ontology (GO) biological processes (GOBPs) (Dataset S3) (31). Groups G1 and G4 were significantly (P < 0.01) associated with aging and leaf senescence (Fig. 3B); groups G2 and G3 were mainly associated with developmental growth (photosynthesis and carbohydrate/lipid metabolism) and responses to development and growth hormones (brassinosteroid and gibberellin); and groups G5–8 were associated with subsets of these processes. Based on these results, we focused on groups G1 and G4 that were significantly associated with aging and leaf senescence (Fig. 3B). Moreover, G1 was more strongly associated than G4 with senescence-promoting processes such as immune responses, responses to abiotic stresses (oxidative/ROS and salt stresses), and responses to senescence-promoting hormones [SA, jasmonic acid (JA), abscisic acid (ABA), and ethylene]. Furthermore, among these processes, the GOBP-association analysis revealed that responses to SA and oxidative/ROS stress are the top senescence-promoting processes regulated by the NAC troika (SI Appendix, Fig. S6 B and C). Finally, using qRT-PCR analysis, we confirmed that genes involved in responses to SA (PR1, EDS5, and SIRK) and ROS (DUF239, BCB, and MSRB8) were up-regulated in presenescent-stage leaves from the mutants compared with the wild type (Fig. 3C). Collectively, these data suggest that the NAC troika negatively regulates senescence-promoting processes, including responses to SA and oxidative/ROS stress, at the presenescent stage.
ANAC090 and ANAC017 Predominantly Suppress SA and ROS Responses, Respectively, at the Presenescent Stage.
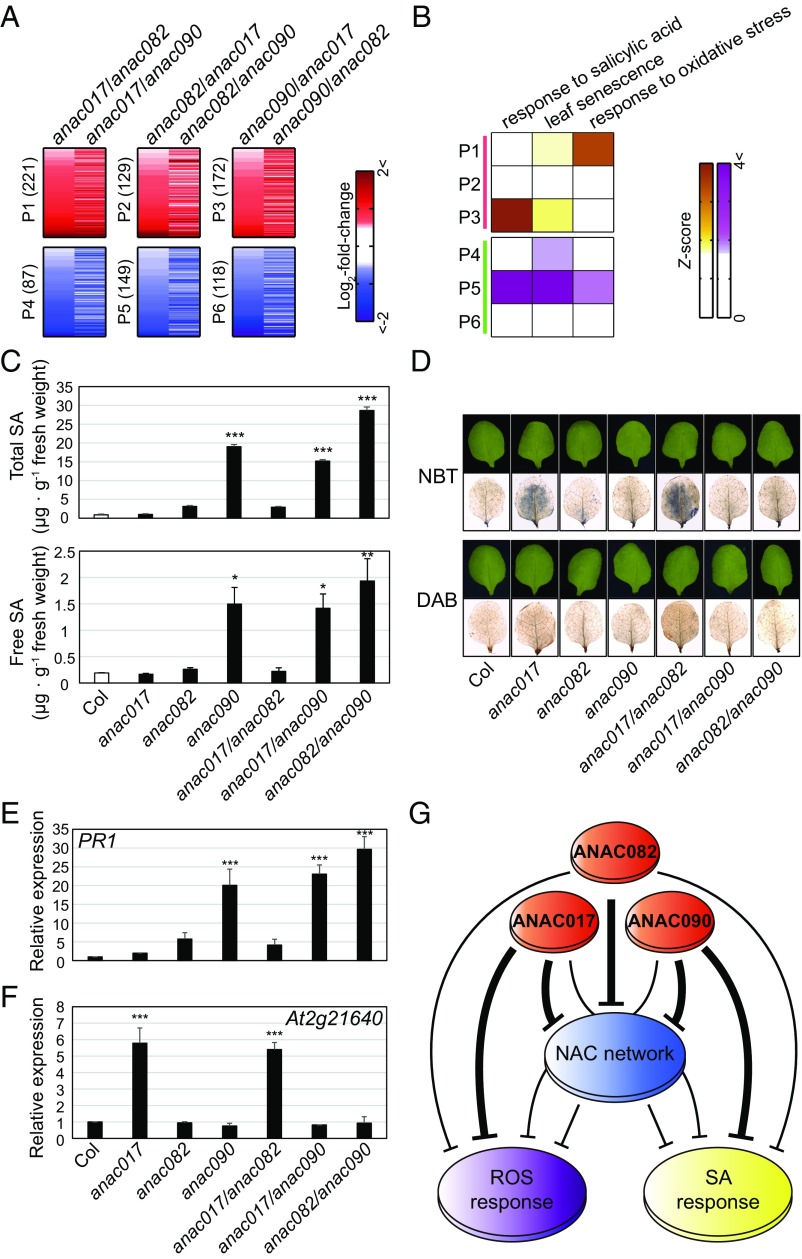
Next, to identify genes that are differentially regulated by each component of the NAC troika, we compared expression profiles among single mutants. We identified 221 genes that were up-regulated in anac017 relative to anac082 and anac090 (P1), 129 genes that were up-regulated in anac082 relative to anac017 and anac90 (P2), and 172 genes that were up-regulated in anac90 relative to anac017 and anac082 (P3) (Fig. 4A). Similarly, we identified 87, 149, and 118 genes that were specifically down-regulated in anac017, anac082, and anac090 mutants, respectively, compared with the other single mutants (P4–6 in Fig. 4A; also see SI Appendix, SI Materials and Methods and Dataset S4). GOBP enrichment analysis for the genes in P1–6 revealed that responses to SA and oxidative/ROS stress were more strongly represented by the up-regulated genes in anac090 (P3) and anac017 (P1) mutants, respectively (Fig. 4B and Dataset S5). Consistently, ANAC017 was reported to regulate more than 80% of genes involved in the response to hydrogen peroxide (H2O2) in Arabidopsis, thereby conferring the hypersensitivity to H2O2 in anac017 mutants (32). However, little is known about SA-associated functions of ANAC090 in leaf senescence. To further investigate this differential regulation of the SA response by ANAC090, we measured endogenous steady-state levels of SA in the leaves of wild-type plants and in single and double mutants at 18 d of leaf age. Total and free SA levels were elevated in leaves with an anac090 mutation (anac090, anac017/090, and anac082/090) relative to those without this mutation (Fig. 4C), suggesting a predominant role for ANAC090 in suppressing SA accumulation at the presenescent stage.
Fig. 4.
ANAC017 and ANAC090 differentially regulate ROS and SA responses. (A) Heat maps showing up-regulation (red) and down-regulation (blue) of genes predominantly in anac017 (P1 and P4), anac082 (P2 and P5), or anac090 (P3 and P6) mutants. These genes were identified from the following comparisons: for P1 and P4, anac017 vs. anac082 (anac017/anac082) and anac017 vs. anac090 (anac017/anac090); for P2 and P5, anac082 vs. anac017 (anac082/anac017) and anac082 vs. anac090 (anac082/anac090); and for P3 and P6, anac090 vs. anac017 (anac090/anac017) and anac090 vs. anac082 (anac090/anac082). The number of genes in each pattern is denoted in parenthesis. The color bar represents the gradient of log2 fold changes from each comparison. (B) Representative aging-associated GOBPs enriched by genes in P1–6. The color bar represents the Z-score gradient representing enrichment significance (P value): Z = N−1(1 − P) where N−1 is the inverse standard normal distribution and P is the enrichment P value. (C) Endogenous levels of free SA and total SA (free SA + SA glucosides) measured from the third or fourth leaves of wild-type (Col) plants, single mutants, and double mutants at 18 d of leaf age. Values are means ± SE (n = 3). (D) Staining of O2− and H2O2 in the third or fourth leaves of wild-type plants, single mutants, and double mutants at 18 d of leaf age using nitroblue tetrazolium (NBT) and diaminobenzidine tetrahydrochloride (DAB), respectively. (E and F) mRNA expression levels of PR1 (E) and At2g21640 (F) in leaves from wild-type (Col) plants, single mutants, and double mutants at 18 d of leaf age. mRNA expression levels were analyzed by qRT-PCR and then were normalized by those of ACT2. For each gene, the normalized expression levels were further normalized by those in wild-type plants. Values are means ± SD (n = 4). (G) A proposed model for shared and differential regulation by the NAC troika of SA and ROS responses. SA levels and the normalized expression levels were compared using one-way ANOVA with Dunnett's correction as a post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, to investigate the differential regulation of the ROS response by ANAC017, we measured ROS, superoxide (O2−), and H2O2 content in leaves from wild-type plants and single and double mutants at 18 d of leaf age. O2− and H2O2 levels were much higher in leaves containing an anac017 mutation (anac017 and anac017/082 mutants) than in those without this mutation (Fig. 4D), suggesting a prominent role for ANAC017 in regulating ROS levels at the presenescent stage. Interestingly, despite the presence of an anac017 mutation, O2− and H2O2 levels in the anac017/090 mutant did not differ from wild-type levels, suggesting potential crosstalk between anac017 and anac090 on the regulation of oxidative stress (Discussion). Consistent with these observations, the mRNA levels of PR1, a marker of SA induction, were increased significantly (P < 0.01) in anac090, anac017/090, and anac082/090 mutants (Fig. 4E), whereas the mRNA levels of At2g21640, previously reported as a marker of oxidative stress (33), were significantly (P < 0.001) increased in anac017 and anac017/082 mutants but not in anac017/090 mutants (Fig. 4F). On the other hand, the GOBP enrichment analysis above showed the shared regulation of SA and ROS responses by the NAC troika (Fig. 3B), suggesting that a subset of genes or additional genes involved in SA and ROS responses were more strongly regulated by ANAC090 and ANAC017, respectively (P3 and P1, respectively, in Fig. 4A) (Discussion). Taken together, these results suggest that the NAC troika collectively suppresses the induction of genes involved in senescence-promoting processes (Fig. 3B) but with additional predominant regulation of genes involved in SA and ROS responses by ANAC090 and ANAC017, respectively (Fig. 4G).
ANAC090 Directly Binds and Suppresses Promoters of Target Genes Involved in the SA Response.
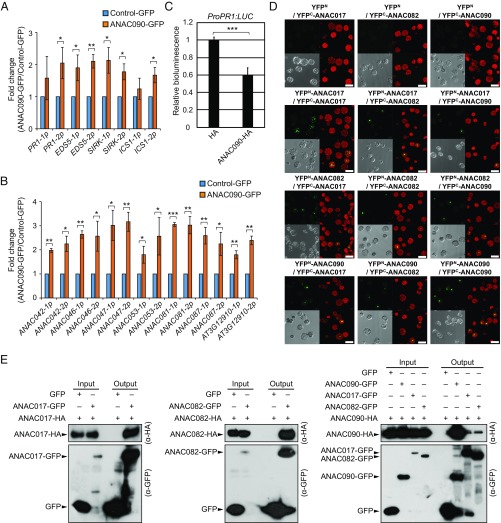
Having established that the NAC troika influences leaf-senescence phenotypes via negative regulation of target genes that promote senescence, we next examined whether the NAC troika binds directly to promoters of their target genes. We generated plants overexpressing GFP-tagged ANAC090 (SI Appendix, Fig. S7 A and B) and performed ChIP-PCR for SA-inducible genes (PR1, EDS5, and SIRK) (Fig. 3C) and for ICS1, a key SA synthetic enzyme gene. We found that ANAC090 bound to the promoters of all tested SA-inducible genes (Fig. 5A and SI Appendix, Fig. S7C) as well as to the promoters of the previously tested seven target NACs of the NAC troika (ANAC0042, ANAC046, ANAC047, ANAC053, ANAC081, ANAC087, and AT3G12910) (Fig. 5B and SI Appendix, Fig. S7C). Furthermore, we measured luciferase activities in protoplasts cotransfected with a proPR1:LUC reporter and ANAC090-HA and observed decreased activity of PR1 promoter, indicating negative regulation of the PR1 gene by ANAC090 (Fig. 5C). These data suggest that ANAC090 directly suppresses the expression of target genes, including PR1. We were unable to perform ChIP-PCR experiments with ANAC017 and ANAC082 due to the instability of these proteins after protein extraction.
Fig. 5.
Components of the NAC troika interact among themselves, and ANAC090 binds to the promoter of SA-inducible genes and target NACs. (A and B) ChIP-qPCR assays of selected DNA fragments in the promoters of SA-inducible genes (PR1, EDS5, SIRK, and ICS1) (A) and in the promoters of downstream target NACs (ANAC042, ANAC046, ANAC047, ANAC053, ANAC081, ANAC087, and At3g12910) (B). Chromatin from aerial parts of 2-wk-old pCsVMV:ANAC090-GFP plants was immunoprecipitated with an anti-GFP antibody. Enrichment fold changes were normalized to those of TA3. Values are means ± SD (n = 3). The normalized enrichment fold changes were compared between control-GFP and ANAC090-GFP samples using the paired t test (right-sided). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Transient expression assays in Arabidopsis protoplasts showing the transrepression of the PR1 promoter by ANAC090. Protoplasts were cotransfected with the proPR1:LUC reporter and an effector plasmid expressing ANAC090-HA. The expression of proPR1:LUC was normalized to that of 35Spro:RLuc (internal control). The resulting expression in ANAC090-HA was normalized to that in protoplasts that were transfected with the reporter plasmid and an effector plasmid expressing HA only. Values are means ± SD (n = 3). The relative expression levels in ANAC090-HA and HA samples were compared using a t test. ***P < 0.001. (D) BiFC analysis of protein interactions among ANAC017, ANAC082, and ANAC090. Fluorescence was detected using a confocal laser-scanning microscope: BiFC fluorescence (green signal, Upper Left), chlorophyll fluorescence (red signal, Upper Right), bright-field images (Lower Left), and merged images (Lower Right) are shown. (Scale bars, 50 μm.) (E) Coimmunoprecipitation analysis of interactions among ANAC017, ANAC082, and ANAC090. Protoplast cells were cotransfected with the indicated combinations of HA- and GFP-tagged proteins (ANAC017, ANAC082, or ANAC090) along with the GFP control. GFP-tagged proteins and GFP control proteins were immunoprecipitated with an anti-GFP antibody (α-GFP), and the immunoblots were probed with anti-HA (α-HA) and anti-GFP antibodies.
NAC TFs have been shown to form homodimers or heterodimers that bind the promoters of target genes (34, 35). To determine whether the components of the NAC troika interact, we performed bimolecular fluorescence complementation (BiFC) experiments. BiFC signals were detected in the nucleus (puncta in images) for all pairwise combinations of ANAC017, ANAC082, and ANAC090 (Fig. 5D), revealing that they form homodimers and heterodimers. We performed coimmunoprecipitation experiments and confirmed the formation of homodimers of ANAC017 and ANAC082 (Fig. 5E, Left and Middle, respectively), as well as homodimers of ANAC090 and heterodimers of ANAC090 with ANAC017 and ANAC082 in protoplasts (Fig. 5E, Right). Collectively, our results suggest that the components of the NAC troika interact and directly regulate the promoters of target genes, which may contribute to the positive-to-negative regulatory inversion and/or to the inhibition of senescence-promoting processes before leaf senescence.
Discussion
We show that the NAC troika, consisting of ANAC017, ANAC082, and ANAC090, is a negative regulator of leaf senescence that increases in mRNA expression as leaves approach senescence (SI Appendix, Fig. S8A) (5). Our findings suggest that senescence-promoting genes as well as anti-senescence regulators are up-regulated during leaf senescence, possibly to control the initiation and progression of leaf senescence. The mRNA levels of all three NACs began to increase at the presenescent stage, which might underlie the positive-to-negative regulatory shift at the presenescent stage.
We showed shared regulation of SA and ROS responses by the NAC troika (Fig. 3) but also observed differential regulation of SA and ROS responses by ANAC090 and ANAC017, respectively (Fig. 4). To clarify shared and differential regulation of the NAC troika on SA and ROS responses, we built a network model that describes the interactions among the genes involved in the shared (G1 and G4 in Fig. 3A) and differential (P1 and P3 in Fig. 4A) regulation of SA and ROS responses by the NAC troika (SI Appendix, Fig. S8B and Dataset S6). For the SA response, the network model showed that the genes involved in SA synthesis (ICS1, EDS5, CBP60G, SARD1, and MES9) and downstream TFs of SA signaling (MYB2, MYB50, MYB51, WRKY18, WRKY30, WRKY38, WRKY40, WRKY53, WRKY54, WRKY60, WRKY62, WRKY70, and ANAC081) belonged to G1 or G4, suggesting that these central processes in SA response are commonly regulated by the NAC troika. However, a significant portion of these genes (CBP60G for SA synthesis and WRKY18, WRKY38, WRKY40, WRKY53, WRKY54, and WRKY70 for SA signaling) belonged to P3, suggesting that the central processes in SA response were more strongly regulated by ANAC090 beyond their basal shared regulations with ANAC017 and ANAC082. Similarly, the network model showed that the genes involved in ROS production (PER34 and PER71) and reduction (PER4, PER52, DUF239, GRXC9, GRXS13, MSRB8, MSRB9, AOX1D, and ANAC042), downstream processes of ROS signaling (CRT1 and CRT2 for mitochondrial dysfunction and PLP2, GSTF6, CYP71A12, CYP71A13, CYP71B15, and PAD4 for defense response), and NAD/NAD(P)H, FAD/FADH2 regulation (NUDT7, AT1G30720, and AT5G48440) belonged to G1 or G4, but additional genes [PER71 for ROS production, PER52 for ROS reduction, DOX1, CYP71A12, CYP79B3, and PAD4 for defense response, ANAC013 for mitochondrial retrograde signaling, and AT1G30720, AT5G01670, AT5G16980, AT5G44380, and AT5G48440 for NAD(P)/NAD(P)H and FAD/FADH2 regulation] involved in the same processes belonged to P1, suggesting the basal shared regulation of ROS response and its additional regulation by ANAC017. Thus, the network model can explain how the NAC troika can commonly and differentially regulate SA and ROS responses.
In this study, we found that O2− and H2O2 levels in anac017/090 mutants, unlike anac017 and anac017/082 mutants, did not differ from wild-type levels (Fig. 4D). Similar to anac017/090 mutants, anac090 mutants showed no significant difference in the levels of O2− and H2O2 from the wild type. On the other hand, all mutants containing the anac090 mutation showed increased levels of SA in leaves. Previously, SA was shown to reduce ROS through the antioxidant defense system, although SA was also shown to contribute to the accumulation of ROS (36). Based on these observations, we can develop a model for the negative regulation of ROS levels by SA (SI Appendix, Fig. S8C). According to this model, SA levels were increased in the mutants containing the anac090 mutation, which activates an antioxidant defense system by increasing the levels of SA-inducible redoxins (e.g., GRXC9 in SI Appendix, Fig. S8B) and glutathione, in turn leading to a reduction in ROS levels (SI Appendix, Fig. S8C). In anac017/090 mutants, ROS levels are expected to increase due to the loss of ANAC017 but also to decrease due to the loss of ANAC090 and increased SA. This opposing regulation of ROS levels might explain why ROS levels do not increase in the anac017/090 mutant. This crosstalk between SA and ROS responses can maintain the levels of antioxidants sufficiently to protect the senescing leaf cells from premature death, ensuring the slow degeneration of cells during leaf senescence. In this regard, the SA–ROS crosstalk at the early stage of leaf senescence might be critical to prevent an oxidative burst and acute cell death in response to environmental stresses.
A number of studies have previously reported the associations of the NAC TF family with ROS and SA responses. First, increased ROS levels, owing to a decline in antioxidant capacity, are highly associated with the progression of leaf senescence (36). H2O2 treatment was reported to induce the expression of several senescence-associated NACs, such as ANAC029/AtNAP, ANAC002/ATAF1, ANAC042/JUB1, and ANAC059/ORS1 (20, 23, 37). Among them, ANAC042 was reported as the only negative regulator of leaf senescence that modulates cellular H2O2 levels (23). Interestingly, however, ANAC017 is not induced by H2O2, although it mediates H2O2 responses in plants (32) and, according to our data, loss of ANAC017 triggers increased ROS levels (Fig. 4D) and accelerates leaf senescence (Fig. 2). These data suggest that ANAC017, compared with the other ROS-induced NACs, acts as an upstream master negative regulator of ROS response with no feedback regulatory loop from ROS on its expression. Second, SA levels were previously shown to be low in young leaves but high in senescing leaves (38, 39), and overexpression of the S3H gene encoding an SA 3-hydroxylase reduced SA levels and delayed leaf senescence (40), suggesting that SA contributes to the promotion of leaf senescence. However, little is known about NACs that regulate SA-mediated leaf senescence. CBNAC/NTL9 and ANAC081/ATAF2 were increased in their expression levels after exposure to SA (41, 42), but they played no significant roles in leaf senescence according to our data (SI Appendix, Fig. S8D), suggesting that they contribute to SA-mediated pathogen resistance (41, 43) rather than to SA-mediated leaf senescence. In this study, we identified ANAC090 as a negative regulator of SA-mediated leaf senescence; loss of ANAC090 led to increased SA levels (Fig. 4C) and accelerated leaf senescence (Fig. 2). Of note, previous genetic studies have not identified ANAC090 in association with leaf senescence. Senescence-associated NACs have been identified mostly based on the alteration in their expression levels during leaf senescence. However, ANAC090 is not induced by SA treatment, and its expression drops to the basal level at the late-senescence stage, unlike known senescence-associated NACs (SI Appendix, Fig. S8A). Also, dysfunction of ANAC090 causes no severe alteration in morphological phenotype during leaf senescence. These data demonstrate the unique value of our time-evolving network-based approach for identifying senescence regulators such as ANAC090. Furthermore, our approach can be applied to other TF families, such as the WRKY and MYB families, to identify key presenescence regulators in these TF families.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana ecotype Columbia (Col) was the parent strain for all mutants used in this study. The transfer DNA (T-DNA) insertion lines of NAC genes were obtained from the SALK T-DNA insertion collection, SAIL collection, or GABI-KAT collection (44–46). The genotype of each single- or double-mutant line was confirmed by PCR-based genotyping (SI Appendix, Table S1). For the ANAC017-HA, ANAC082-HA, and ANAC090-HA or ANAC090-GFP overexpression lines, cDNAs covering the complete coding sequence were cloned into pCR-CCD_F (47) and were recombined into the gateway version of the pCsVMV-HA3-N-1300 or pCsVMV-GFP-N-1300 vector. Plants used for most experiments were grown in an environmentally controlled growth room under a 16-h/8-h light/dark cycle (photon flux density 100 μmol⋅m−2⋅s−1) at 22 °C.
Data Availability.
The raw and normalized data of the NanoString nCounter assay and RNA-sequencing have been deposited in the National Center for Biotechnology Gene Expression Omnibus database (48) with the accession numbers of GSE92369 and GSE92314, respectively, under the GEO SuperSeries accession no. GSE92371.
More detailed information regarding the experimental and computational procedures used in this study is provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wonhee Lee and Sukjoon Jung for insightful discussions. This research was supported by Institute for Basic Science Grants IBS-R013-D1 (to H.J.K., J.G.K., J.J.K., S.H., J.K., J.H.K., and H.G.N.) and IBS-R013-A1 (to J.-H.P. and D.H.) and by the Mid-career Researcher Program through National Research Foundation of Korea (funded by the Ministry of Science, ICT and Future Planning) Grants 2015R1A2A2A01005820 (to H.R.W.) and 2017R1A2B4012937 (to P.O.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw and normalized data of the Nanostring nCounter assay and RNA sequencing have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession nos. GSE92369 and GSE92314, respectively) under the GEO SuperSeries accession no. GSE92371.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721523115/-/DCSupplemental.
References
- 1.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 2.Oh SA, et al. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12:527–535. doi: 10.1046/j.1365-313x.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- 3.Woo HR, Kim HJ, Nam HG, Lim PO. Plant leaf senescence and death–Regulation by multiple layers of control and implications for aging in general. J Cell Sci. 2013;126:4823–4833. doi: 10.1242/jcs.109116. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016;33:48–56. doi: 10.1016/j.pbi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Woo HR, et al. Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 2016;171:452–467. doi: 10.1104/pp.15.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breeze E, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penfold CA, Buchanan-Wollaston V. Modelling transcriptional networks in leaf senescence. J Exp Bot. 2014;65:3859–3873. doi: 10.1093/jxb/eru054. [DOI] [PubMed] [Google Scholar]
- 8.Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S. Salt stress and senescence: Identification of cross-talk regulatory components. J Exp Bot. 2014;65:3993–4008. doi: 10.1093/jxb/eru173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013;13:192. doi: 10.1186/1471-2229-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, et al. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011;52:138–148. doi: 10.1093/pcp/pcq180. [DOI] [PubMed] [Google Scholar]
- 11.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zentgraf U, Laun T, Miao Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol. 2010;89:133–137. doi: 10.1016/j.ejcb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Jiang Y, Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Gan S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011;156:1612–1619. doi: 10.1104/pp.111.177022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CK, et al. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol Biol. 2015;88:269–286. doi: 10.1007/s11103-015-0321-2. [DOI] [PubMed] [Google Scholar]
- 17.Jensen MK, et al. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 18.Podzimska-Sroka D, O’Shea C, Gregersen PL, Skriver K. NAC transcription factors in senescence: From molecular structure to function in crops. Plants (Basel) 2015;4:412–448. doi: 10.3390/plants4030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- 20.Balazadeh S, et al. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant. 2011;4:346–360. doi: 10.1093/mp/ssq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 2013;54:1660–1672. doi: 10.1093/pcp/pct113. [DOI] [PubMed] [Google Scholar]
- 22.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 23.Wu A, et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24:482–506. doi: 10.1105/tpc.111.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang SD, Seo PJ, Yoon HK, Park CM. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell. 2011;23:2155–2168. doi: 10.1105/tpc.111.084913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot. 2014;65:4023–4036. doi: 10.1093/jxb/eru112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin J, et al. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo HR, et al. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot. 2010;61:3947–3957. doi: 10.1093/jxb/erq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Barabási AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 30.Lee HJ, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285:9262–9272. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Ng S, et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gadjev I, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst HA, Olsen AN, Larsen S, Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen ANEH, Lo Leggio L, Skriver K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005;169:785–797. [Google Scholar]
- 36.Rogers H, Munné-Bosch S. Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: Similar but different. Plant Physiol. 2016;171:1560–1568. doi: 10.1104/pp.16.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balazadeh S, Wu A, Mueller-Roeber B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behav. 2010;5:733–735. doi: 10.4161/psb.5.6.11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris K, et al. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 39.Khan M, Rozhon W, Poppenberger B. The role of hormones in the aging of plants–A mini-review. Gerontology. 2014;60:49–55. doi: 10.1159/000354334. [DOI] [PubMed] [Google Scholar]
- 40.Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA. 2013;110:14807–14812. doi: 10.1073/pnas.1302702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HS, et al. A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 2012;40:9182–9192. doi: 10.1093/nar/gks683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delessert C, et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43:745–757. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 43.Alves MS, et al. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes. 2014;2:85–106. doi: 10.3390/proteomes2010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012;40:D1211–D1215. doi: 10.1093/nar/gkr1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alonso JM, Stepanova AN. T-DNA mutagenesis in Arabidopsis. Methods Mol Biol. 2003;236:177–188. doi: 10.1385/1-59259-413-1:177. [DOI] [PubMed] [Google Scholar]
- 46.Sessions A, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Somers DE. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 2010;154:611–621. doi: 10.1104/pp.110.162271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and normalized data of the NanoString nCounter assay and RNA-sequencing have been deposited in the National Center for Biotechnology Gene Expression Omnibus database (48) with the accession numbers of GSE92369 and GSE92314, respectively, under the GEO SuperSeries accession no. GSE92371.
More detailed information regarding the experimental and computational procedures used in this study is provided in SI Appendix, SI Materials and Methods.