Significance
Renal epithelial cells (RECs) contain abundant mitochondria that are essential to support renal reabsorption of electrolytes, glucose, and amino acids. However, it remains poorly understood how mitochondrial metabolism is coordinated with kidney reabsorptive functions. Here we show that deletion of estrogen-related receptor gamma (ERRγ) in RECs results in severe renal mitochondrial and reabsorptive dysfunction with fluid-filled cysts. ERRγ directly regulates mitochondrial metabolism and cooperates in regulating renal reabsorption genes with hepatic nuclear factor 1 beta (HNF1β), mutations of which cause strikingly similar renal dysfunction and cysts in animals and humans. These findings reveal a role for ERRγ in simultaneously coordinating a transcriptional program of renal energy-generating mitochondrial and energy-consuming reabsorptive functions relevant to kidney disease.
Keywords: nuclear receptor, ERRγ, mitochondria, kidney, renal reabsorption
Abstract
Mitochondrial dysfunction is increasingly recognized as a critical determinant of both hereditary and acquired kidney diseases. However, it remains poorly understood how mitochondrial metabolism is regulated to support normal kidney function and how its dysregulation contributes to kidney disease. Here, we show that the nuclear receptor estrogen-related receptor gamma (ERRγ) and hepatocyte nuclear factor 1 beta (HNF1β) link renal mitochondrial and reabsorptive functions through coordinated epigenomic programs. ERRγ directly regulates mitochondrial metabolism but cooperatively controls renal reabsorption via convergent binding with HNF1β. Deletion of ERRγ in renal epithelial cells (RECs), in which it is highly and specifically expressed, results in severe renal energetic and reabsorptive dysfunction and progressive renal failure that recapitulates phenotypes of animals and patients with HNF1β loss-of-function gene mutations. Moreover, ERRγ expression positively correlates with renal function and is decreased in patients with chronic kidney disease (CKD). REC-ERRγ KO mice share highly overlapping renal transcriptional signatures with human patients with CKD. Together these findings reveal a role for ERRγ in directing independent and HNF1β-integrated programs for energy production and use essential for normal renal function and the prevention of kidney disease.
Mitochondria are organelles that generate the majority of cellular energy through oxidative phosphorylation (OxPhos) and fatty acid oxidation (FAO). Their optimal function is central to health. Mutations of mitochondrial DNA and proteins directly cause mitochondrial disease, with severe defects often observed in organs of high mitochondrial content and energetic demand, including the kidney (1–3). Mitochondrial dysfunction in general has been recognized to broadly contribute to heart disease, obesity, diabetes, neurodegeneration, aging, and many kidney diseases including acute kidney injury, polycystic kidney disease, and chronic kidney disease (CKD) (1, 4–8). CKD is characterized by gradual loss of kidney function with various etiologies, incompletely understood pathophysiology, and no cure. CKD features both glomerular and tubular cell dysfunction as well as metabolic dysregulation (9). Transcriptomic and metabolomic studies have implicated impaired mitochondrial OxPhos in many kidney diseases including CKD (10, 11). A recent study also shows that renal mitochondrial OxPhos and FAO are among the top dysregulated cellular pathways in both patients with CKD and animal models (12).
Among all kidney cells, the tubule and collecting duct renal epithelial cells (RECs) have a very high density of mitochondria. The main physiological function of RECs is to maintain whole-body homeostasis of osmolality, acid-base balance, and extracellular fluid volume through reabsorption of water, critical nutrients, and electrolytes. Mechanistically, these reabsorptive functions are achieved by the work of many membrane transporters and channels specific for water, glucose, amino acids, sodium, chloride, bicarbonate, and other biomolecules on the REC apical and basolateral surfaces, using cellular ATP as the ultimate energy source. Accordingly, RECs are densely packed with mitochondria and depend on mitochondrial OxPhos and FAO to generate energy to support their reabsorptive functions. However, it remains poorly understood how mitochondrial metabolism is coordinated with kidney reabsorptive functions, and how mitochondrial dysfunction contributes to kidney disease.
Recent work has revealed nuclear receptor estrogen-related receptor gamma (ERRγ) as a critical transcriptional regulator of mitochondrial OxPhos and FAO (13–15). Studies using cell type-specific ERRγ KO mice bypassed the perinatal lethality of whole-body ERRγ KO mice and provided definitive evidence for an essential role of ERRγ in neuronal metabolism and learning/memory, cardiac metabolism and contraction/conduction, functional maturation of pancreatic β cells, brown adipocyte thermogenic function, and skeletal muscle fiber type determination and function (16–22). Genomic studies show that ERRγ directly binds to and activates the transcription of hundreds of genes important in mitochondrial OxPhos in neurons, and OxPhos, FAO, and cardiac contraction genes in heart cells (17, 23). In addition, ERRγ is highly expressed in the kidney and is essential for normal embryonic kidney development (24). Notably, human genetic studies reveal that a de novo reciprocal translocation at t(1, 2)(q41; p25.3) involving the ERRγ locus is associated with bilateral renal agenesis/hypoplasia/dysplasia (25).
Here we generated REC-ERRγ KO mice and show that they develop renal mitochondrial and reabsorption dysfunction. Genomic studies suggest that ERRγ regulates mitochondrial metabolism and renal reabsorption through distinct mechanisms: although ERRγ alone regulates mitochondrial OxPhos/FAO functions, ERRγ cooperates with HNF1β to activate the expression of renal reabsorption genes. In addition, human patients with CKD show decreased kidney ERRγ expression and share highly overlapping renal transcriptional signatures with REC-ERRγ KO mice. Together, these results uncover a mechanism for coordinated regulation of renal mitochondrial and reabsorption functions and identify ERRγ signaling as an important link to mitochondrial dysfunction associated with kidney disease.
Results
Loss of ERRγ in RECs Results in Kidney Disease with Cysts.
We first determined where ERRγ is expressed in postnatal kidneys by analyzing an ERRγ heterozygous mouse strain in which LacZ was knocked into the ERRγ locus (16). X-gal staining revealed that ERRγ was highly and specifically expressed in the RECs of all of the segments of tubules and collecting ducts, but barely detectable in any other cells, including those in the glomeruli (SI Appendix, Fig. S1A). Furthermore, ERRγ protein was found exclusively in the nucleus but not in mitochondria or cytosol, in line with its function as a nuclear transcription factor (SI Appendix, Fig. S1B).
To determine the importance of ERRγ in postnatal kidney biology and disease, we generated REC-ERRγ KO mice by crossing mice harboring a floxed allele of ERRγ (17) with Sim1-Cre strain. Strong renal Cre expression occurred at birth and resulted in rapid postnatal loss of kidney ERRγ expression with only 15% ERRγ mRNA remaining by 1 wk of age (SI Appendix, Fig. S1C). Using a double fluorescent Cre reporter mouse strain (26), we further found that this Sim1-Cre line mediated recombination specifically in all segments of the tubule and collecting duct epithelium within the kidney (SI Appendix, Fig. S1D), exactly where ERRγ is normally expressed (SI Appendix, Fig. S1A). We used both male and female REC-ERRγ KO mice for all subsequent studies.
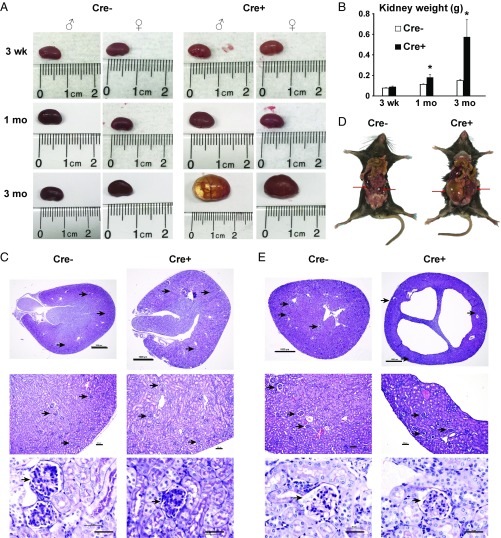
Both male and female REC-ERRγ KO mice (Cre+) were born and survived perinatally at the expected Mendelian ratio. Their kidneys were of normal weight and histological appearance at birth and at 3 wk of age, suggesting no developmental defects (Fig. 1 A–C). Consistent with the histological observations, electron microscopy showed that renal ultrastructure, including mitochondria in the glomeruli, proximal tubules, and distal tubules, was comparable to controls at 3 wk of age (SI Appendix, Fig. S2). Subsequently, however, both male and female REC-ERRγ KO mice developed progressive renal abnormalities (Fig. 1A). Kidneys of REC-ERRγ KO mice were significantly heavier than controls by 1 mo of age, and threefold heavier by 3 mo of age (Fig. 1 B and D). Histological examination showed that REC-ERRγ KO mouse kidneys contained multiple fluid-filled cysts of variable sizes (Fig. 1E), resulting in the disruption of overall kidney organization and structure. Glomeruli were present and appeared histologically normal based on H&E staining. The carbohydrate-rich basement membranes of glomeruli including capillary loops responsible for renal filtration function also remained intact, as evaluated by periodic acid-Schiff staining (Fig. 1E). These findings suggest that ERRγ is critical for maintaining normal kidney structure and function in vivo.
Fig. 1.
REC-ERRγ KO mice develop kidney disease with fluid-filled cysts. (A and B) Representative pictures (A) and weight (B) of control and REC-ERRγ KO kidneys at different ages (n = 12–19). (C) Three-week-old control and REC-ERRγ KO kidney morphology. (Top) H&E stain. (Scale bar, 1,000 µm.) (Middle) H&E stain. (Scale bar, 100 µm.) (Bottom) Periodic acid-Schiff stain (Scale bar, 100 µm.) Arrowheads point to glomeruli. (D) Kidneys (red arrows) of 3-mo-old control and REC-ERRγ KO mice, illustrating their relative sizes to the body. (E) Three-month-old control and REC-ERRγ KO kidney morphology. Staining and scale bars are the same as in C. Error bar indicates SEM. *P < 0.05 between Cre− and Cre+ by t test. Both male and female mice were included.
ERRγ Directly Regulates Mitochondrial Metabolic and Renal Reabsorptive Function.
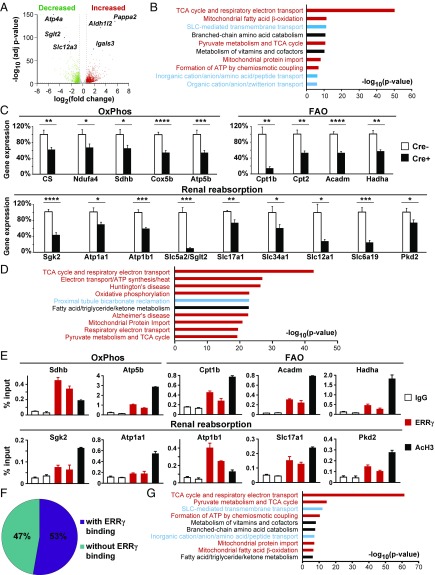
To further understand the molecular mechanisms underlying the observed kidney disease phenotype, we used RNA-Seq to quantify the transcriptomic changes between control and REC-ERRγ KO mouse kidneys (n = 4 each) at 3 wk of age, just before morphological abnormalities became observable. We found that expression levels of 1,018 genes were significantly decreased and 1,406 genes were significantly increased in REC-ERRγ KO mouse kidneys (Fig. 2A; P < 0.01; fold change > ±1.5). Pathway analysis revealed that the down-regulated 1,018 genes were enriched for two cellular functions: mitochondrial metabolism (OxPhos, FAO, etc.) and transmembrane transport associated with renal reabsorption (Fig. 2B). Decreased mitochondrial metabolism (OxPhos, FAO and amino acid metabolism) was recently identified as a genetic signature of human CKD (12). Pathways enriched in the up-regulated 1,406 genes included inflammatory response (SI Appendix, Fig. S3A), which has also been implicated in human CKD (12). We validated expression changes for many of these genes, using qRT-PCR. For example, renal expression of genes important in OxPhos (CS, Ndufa4, Sdhb, Cox5b, and Atp5b) and FAO (Cpt1b, Cpt2, Acadm, and Hadha) were reduced in REC-ERRγ KO mice (Fig. 2C). Further, transcripts implicated in renal reabsorption were significantly decreased in REC-ERRγ KO kidneys, including Slc5a2/Sglt2 (Na+/glucose cotransporter), Atp1a1, and Atp1b1 (Na+/K+ ATPases, genetic variations of which are associated with hypertension), Slc34a1 (Na+/phosphate cotransporter, mutations of which cause Fanconi Renotubular Syndrome 2 in humans), Slc12a1 (Na+/K+/Cl− cotransporter, mutations of which cause Bartter syndrome type 1 featuring renal tubular hypokalemic alkalosis), Slc6a19 (neutral amino acid transporter, mutations of which cause Hartnup Disease because of blocked reabsorption of neutral amino acids in humans), Sgk2 (a regulator of renal transport), and Pkd2 (mutations of which cause autosomal dominant polycystic kidney disease in humans; Fig. 2C).
Fig. 2.
ERRγ is essential for maintaining normal expression of REC mitochondrial metabolic and renal reabsorptive genes. (A) Volcano plot showing genes differentially expressed in 3-wk-old control and REC-ERRγ KO kidneys by RNA-Seq (n = 4). Filtering criteria is fold change >±1.5 and P < 0.01 (dotted lines). (B) Top 10 enriched pathways of down-regulated genes. (C) Gene expression in 3-wk-old control (n = 7) and REC-ERRγ KO (n = 10) kidneys by qPCR. (D) Top 10 ranked pathways of genes with nearby ERRγ ChIP-Seq peaks. (E) ChIP-qPCR shows that ERRγ binds to OxPhos, FAO, and renal reabsorption genes. (F) Pie graph of down-regulated genes with or without annotated ERRγ binding peaks. (G) Top enriched pathways of down-regulated genes with annotated ERRγ binding peaks. Error bar indicates SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 between Cre− and Cre+ by t test. Both male and female mice were included.
We next investigate whether the renal transcriptome changes in REC-ERRγ KO mice are gender-dependent. Both male and female REC-ERRγ KO mice exhibited similar pathway changes for both down- and up-regulated genes (SI Appendix, Fig. S3B). This is consistent with the fact that both sexes of REC-ERRγ KO mice develop kidney disease. To determine whether these molecular defects of mitochondrial and renal functions persisted in the later stage of kidney disease, we performed RNA-Seq analysis in 3-mo-old male control and REC-ERRγ KO mice and compared it with RNA-Seq data of 3-wk-old male mice. All the down-regulated and most of the up-regulated cellular pathways were maintained in 3-mo-old REC-ERRγ KO mice (SI Appendix, Fig. S3C). A few additional up-regulated cellular pathways were found at 3 mo of age, likely reflecting secondary changes at the later stage of kidney disease.
To determine whether expression of these genes was directly regulated by ERRγ, we performed chromatin immunoprecipitation sequencing (ChIP-Seq) of ERRγ, using two biological replicates of WT mouse kidney samples. Because ERRγ is specifically expressed in RECs, this effectively mapped the ERRγ cistrome in RECs. ERRγ binding peaks were distributed mainly in introns, intergenic and promoter regions (SI Appendix, Fig. S4A). Eighty-six percent (7,463/8,702) of the ERRγ peaks contain the consensus ERR binding motif (AAGGTCA). Pathway analysis revealed that mitochondrial metabolism, including OxPhos pathway genes that comprise most of the lists for Huntington’s disease and Alzheimer disease, and renal reabsorption are among the most enriched pathways of genes annotated to the kidney ERRγ cistrome (Fig. 2D). This is nearly the same pattern seen among down-regulated RNA-Seq pathways in REC-ERRγ KO kidneys (Fig. 2B). We validated these ERRγ ChIP-Seq results using ChIP-qPCR which showed that ERRγ directly bound to the same set of OxPhos, FAO, and renal reabsorption genes (Fig. 2E). By comparing the ChIP-Seq and RNA-Seq datasets, we found that 53% (537 of 1,018) of the genes down-regulated in REC-ERRγ KO mice were bound by ERRγ (Fig. 2F). These genes are again highly enriched for mitochondrial metabolism (OxPhos, FAO, etc.) and renal reabsorption pathways (Fig. 2G). In contrast, only 19% (271 of 1,406) of the up-regulated genes were bound by ERRγ (SI Appendix, Fig. S4B). Inflammatory pathway was again enriched in this list, but to a lesser extent than was observed in the ERRγ transcriptome as a whole (SI Appendix, Fig. S4C). These results suggest that the major biological function of ERRγ in RECs is transcriptional activation of mitochondrial metabolism and renal reabsorption.
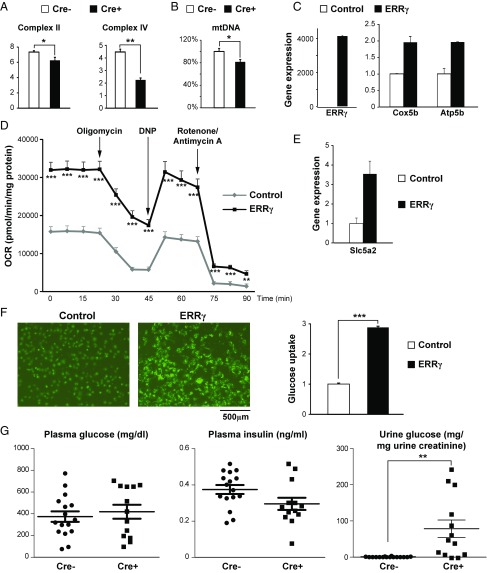
Next we investigated whether mitochondrial metabolism and renal reabsorption function were impaired in REC-ERRγ KO mice. Enzymatic activities of multiple mitochondrial electron transport chain complexes were found decreased in 3-wk-old REC-ERRγ KO kidneys (Fig. 3A), consistent with their reduced RNA expression. In addition, mtDNA content was significantly reduced (Fig. 3B), despite normal mitochondrial ultrastructures, indicating mitochondrial dysfunction. Importantly, all these mitochondrial metabolic defects were present at 3 wk of age, before any morphological or functional renal defects were observed, suggesting that mitochondrial dysfunction is a genesis for renal defects in REC-ERRγ KO mice.
Fig. 3.
ERRγ regulates REC mitochondrial and reabsorptive functions. (A) Enzymatic activities (µmol/min/mg tissue) of mitochondrial electron transport chain complexes in 3-wk-old control (n = 3) and REC-ERRγ KO (n = 6) kidneys. (B) Relative mitochondrial DNA content in 3-wk-old control (n = 13) and REC-ERRγ KO (n = 12) kidneys. (C) ERRγ and mitochondrial OxPhos genes expression in HKC-8 cells by qPCR. (D) Oxygen consumption rate (OCR) in HKC-8 cells measured by Seahorse XF24 analyzer. n = 7 for control and n = 8 for ERRγ. (E) Slc5a2/Sglt2 expression in HKC-8 cells by qPCR. (F) Glucose transport into HKC-8 cells. (F, Left) Representative pictures visualizing fluorescent 2-NBDG uptake. (F, Right) Quantification (n = 10). (G) Plasma and urine glucose and plasma insulin levels in 10–13-wk-old control (n = 16) and REC-ERRγ KO (n = 13) mice 2 wk after STZ treatment. Error bar indicates SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by t test. Both male and female mice were included.
The Sim1-Cre line also mediates recombination in nonrenal tissues such as the hypothalamus (27). We next studied whether ERRγ regulates REC mitochondrial functions in a cell-autonomous manner. We found that adenoviral-mediated ERRγ overexpression increased expression of mitochondrial metabolic genes (Fig. 3C), leading to increase in both basic and stimulated mitochondrial respiration in human tubular REC HKC-8 cells (Fig. 3D). In addition, siRNA-mediated knockdown of ERRγ resulted in decreased mitochondrial respiration, which was rescued by reintroduction of ERRγ (SI Appendix, Fig. S4D). These results indicate that the mitochondrial defects in REC-ERRγ KO mice occur as a result of kidney intrinsic functions of ERRγ.
A central function of the kidney is the reabsorption of water, essential nutrients, and electrolytes. For example, in normal health, almost all urine glucose is reabsorbed, mostly through the high-capacity sodium-glucose cotransporter 2 (SGLT2/Slc5a2) on the apical membrane of the proximal tubule RECs. Loss or inhibition of SGLT2 in humans leads to significant loss of glucose reabsorption and glucosuria (28). The almost complete dependence of renal Sglt2 expression on ERRγ is particularly interesting because a new class of SGLT2 inhibitors (Gliflozins) have become the latest class of potent, FDA-approved medications to treat type 2 diabetes (29, 30). We found that ERRγ increased Sglt2 expression (Fig. 3E) and significantly elevated cellular glucose uptake in human RECs (Fig. 3F).
SGLT2 inhibitors are clinically used to treat diabetes through inhibiting renal glucose reabsorption, resulting in glucosuria. Although REC-ERRγ KO mice showed similar, very low urine glucose levels compared with controls under normal conditions (SI Appendix, Fig. S4E), likely because of a small amount of remaining SGLT2, they decompensated in the setting of streptozotocin (STZ)-induced diabetes. In this model, STZ induced acute diabetes with very high plasma glucose and low plasma insulin levels in both control and REC-ERRγ KO mice (Fig. 3G). However, as seen in patients with diabetes treated with SGLT2 inhibitors, REC-ERRγ KO mice exhibited strong glucosuria with urine glucose levels soaring more than 100-fold above controls because of the loss of SGLT2 and renal glucose reabsorption function. Together, these results demonstrate that ERRγ is an essential regulator of renal glucose reabsorption through control of Sglt2 transcription.
ERRγ Regulates Mitochondrial and Renal Reabsorption Genes Through Distinct Mechanisms.
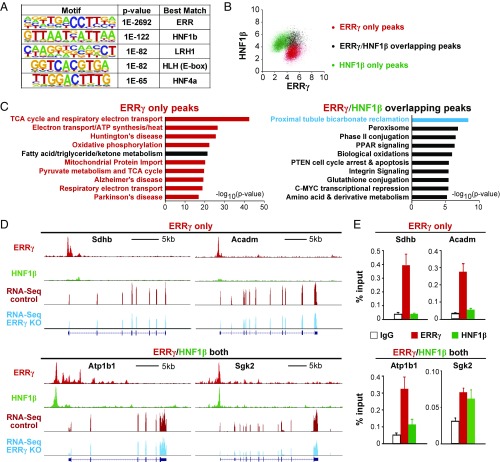
Genomic approaches including ChIP-Seq and ChIP-on-chip have been used to unambiguously demonstrate that ERRγ activates mitochondrial metabolic genes by directly binding to their promoters or enhancers in neurons and hearts (17, 23). It is therefore not surprising that ERRγ binds to and regulates mitochondrial metabolic genes in the kidney, another organ with abundant mitochondria and high energetic demand. It is, however, unexpected that ERRγ also directly binds to and activates the transcription of reabsorption genes, which was not the case in neurons or heart cells (17, 23). To understand whether these two sets of molecular signatures are regulated by ERRγ via similar mechanisms, we further examined the ERRγ binding patterns around mitochondrial and renal reabsorption genes. Notably, de novo motif analysis of kidney ERRγ binding peaks revealed that the second most enriched motif after ERR is HNF1β (Fig. 4A), a homeodomain-containing transcription factor known to cause maturity onset diabetes of the young (MODY5) (31). HNF1β is highly expressed in both tubule and collecting duct RECs and is critical for kidney development and function (32–34). Mutations of HNF1β are the most frequent monogenic cause of developmental kidney disease. The most common features of HNF1β-associated kidney disease include fluid-filled renal cysts, renal hypoplasia, and electrolyte abnormalities (34), the very same symptoms seen in humans with chromosome translocation mapped to ERRγ locus (25) or in our REC-ERRγ KO mice. In addition, perinatal deletion of HNF1β in mouse kidneys results in similar cystic renal disease (33, 35). The common pathological features of HNF1β and ERRγ loss-of-function prompted us to investigate further a possible role for HNF1β in ERRγ’s regulation of kidney metabolism and function.
Fig. 4.
ERRγ cooperates with HNF1β to regulate the transcription of renal reabsorptive genes. (A) Top 5 de novo motifs near ERRγ ChIP-Seq peaks. (B) Overlap of renal ERRγ and HNF1β ChIP-Seq peaks. Axis is log2 tags/107 reads. (C) Top 10 pathways of genes annotated to ERRγ and/or HNF1β ChIP-Seq peaks. (D) Representative ERRγ and HNF1β ChIP-Seq peaks. y axis is peak reads. Same scale for control and ERRγ KO RNA-Seq tracks. (E) Binding of ERRγ and/or HNF1β near OxPhos, FAO and renal reabsorption genes by ChIP.
We performed HNF1β ChIP-Seq in biological duplicate WT mouse kidney samples and compared these results with our ERRγ genome-wide binding datasets. The renal ERRγ and HNF1β cistromes shared 1,109 common peaks, representing about 13% of the total ERRγ peaks and 9% of the total HNF1β peaks (Fig. 4B). Pathway analysis showed that ERRγ-only peaks annotate to mitochondrial metabolic pathways (OxPhos, FAO, and neurodegenerative diseases in which mitochondrial dysfunction is known to contribute), but are not associated with any renal reabsorption pathway (Fig. 4C). In sharp contrast, ERRγ/HNF1β overlapping peaks had stronger representation of the renal absorption pathway compared with ERRγ- or HNF1β-only peaks (Fig. 4C and SI Appendix, Fig. S5A), and importantly, lacked association with mitochondrial metabolic pathways (Fig. 4C). This complete separation of mitochondrial metabolism and renal reabsorption through pathway analysis was further confirmed by visualization of ChIP-Seq peaks near these genes (Fig. 4D and SI Appendix, Fig. S5B) and validated by ChIP-qPCR (Fig. 4E and SI Appendix, Fig. S5C). These results suggest that ERRγ regulates the transcription of mitochondrial and renal reabsorption genes through distinct mechanisms: mitochondrial metabolic genes are bound by ERRγ, as seen in neurons and heart cells (17, 23), whereas renal reabsorption genes are bound by both ERRγ and HNF1β.
ERRγ and HNF1β Functionally Cooperate to Regulate Renal Reabsorption Genes.
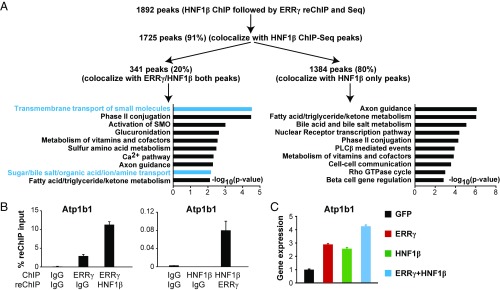
Our ChIP-Seq results clearly suggest that ERRγ and HNF1β bind to a common set of genes important in renal reabsorption. However, it could not yet be excluded that ERRγ and HNF1β may bind to these genes in different cells or, rarely, occupy their common binding regions at the same time in the same cell because ChIP-Seq results represent an average binding signal from millions of cells. To address this, we next used HNF1β ChIP-ERRγ reChIP-Seq to determine whether ERRγ and HNF1β bind to any of these common peaks simultaneously in the same cells. By applying similar statistical and filtering parameters used in our ChIP-Seq studies, ChIP-reChIP-Seq identified a total of 1,892 peaks (Fig. 5A). Of these peaks, 1,725 (91%) colocalize with HNF1β ChIP-Seq peaks (related to the first ChIP step of the ChIP-reChIP-Seq), validating the reproducibility of our ChIP and sequencing assay. Importantly, 20% (341) of these peaks colocalize to previously found ERRγ/HNF1β overlapping peaks. This 20% is a significantly higher percentage than would be expected by the overlap of single ERRγ and HNF1β ChIP-seq results (9%, P = 5 × 10−56 by hypergeometric test). These 341 peaks are highly likely to be simultaneously bound by both ERRγ and HNF1β in the same cells, and they annotate to many renal reabsorption genes, including Atp1b1. These peaks are enriched for renal reabsorption-related pathways compared with the other 1,384 peaks, but not for any mitochondrial metabolic pathways (Fig. 5A). We validated this result on Atp1b1 using reciprocal ERRγ and HNF1β ChIP-reChIP-qPCR (Fig. 5B).
Fig. 5.
Functional cooperation between ERRγ and HNF1β in regulating renal function. (A) HNF1β ChIP-ERRγ reChIP-Seq and data analysis. (B) Atp1b1 is cobound by ERRγ and HNF1β in the same cells by ChIP-reChIP-qPCR. (C) Enhanced activation of Atp1b1 expression by both ERRγ and HNF1β.
We next investigated whether ERRγ and HNF1β functionally cooperate to regulate the transcription of renal reabsorption genes. We were unable to detect a physical interaction between ERRγ and HNF1β in lysates from kidney cells or mouse kidneys, using coimmunoprecipitation followed by Western blot (SI Appendix, Fig. S5D). This suggests that they either do not interact directly or their interaction is not strong enough to be detected by this method, as previously reported in cases of transcription factor tethering (36). The expression of ERRγ and HNF1β cobound genes such as Atp1b1 was activated by either ERRγ or HNF1β, and further increased with both ERRγ and HNF1β (Fig. 5C). Together, these results suggest that ERRγ and HNF1β functionally cooperate to regulate renal reabsorption genes.
Human Patients with CKD Show Decreased Kidney ERRγ Expression and Share Highly Overlapping Renal Transcriptional Signatures with REC-ERRγ KO Mice.
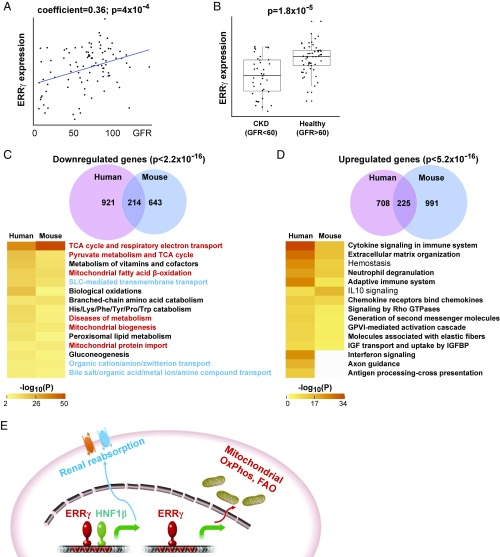
To relate these findings of ERRγ to human kidney diseases beyond congenital disorders (25), we measured renal ERRγ expression in 95 human kidney tubule samples from patients with CKD and control cohorts. We found that ERRγ expression positively correlated with glomerular filtration rate (GFR), a well-established clinical index for kidney function (Fig. 6A). In addition, kidney ERRγ expression was significantly reduced in patients with CKD (GFR < 60) compared with healthy controls (GFR > 60; Fig. 6B). We next compared the renal transcriptomes between REC-ERRγ KO mice and human patients with CKD (12). About 25% of down-regulated and 19% of up-regulated genes in REC-ERRγ KO mice were shared in human patients with CKD (Fig. 6 C and D). There is a significant overlap of affected pathways between REC-ERRγ KO mice and human patients with CKD. In particular, pathways down-regulated are nearly identical in mouse and human and include many mitochondrial function and renal reabsorption pathways that are regulated by ERRγ. These results suggest a broad and critical role for ERRγ in human kidney disease.
Fig. 6.
Human patients with CKD show decreased kidney ERRγ expression and share highly overlapping renal transcriptional signatures with REC-ERRγ KO mice. (A) ERRγ expression positively correlates with GFR across 95 human kidney tubule samples, by Pearson correlation. (B) ERRγ expression is significantly reduced in human CKD kidney tubules, by t test. (C and D) Venn diagram and heat map comparison of top enriched pathways of down-regulated (C) or up-regulated (D) kidney genes between 3-wk-old REC-ERRγ KO mice and human patients with CKD. Statistical analysis was performed using Fisher’s exact test. (E) Model illustrating that ERRγ coordinates energy-generating mitochondrial metabolism and energy-consuming renal reabsorption functions through distinct mechanisms.
Discussion
Mitochondrial dysfunction has been increasingly recognized as an important determinant of kidney diseases including CKD (4–7, 10, 11). ERRγ has recently emerged as a key transcriptional regulator of mitochondrial biogenesis and function, and human genetic studies have linked ERRγ to kidney disease (13–15, 25). However, definitive evidence linking regulation of mitochondrial function and kidney biology/disease is still missing. Here we show that ERRγ is an essential transcriptional coordinator of kidney mitochondrial and reabsorptive functions relevant to human kidney disease. Renal ERRγ expression correlates with kidney function in humans. Mice lacking REC ERRγ develop kidney mitochondrial and reabsorptive dysfunction with similar signs and molecular signatures as seen in human CKD. Mechanistically, we demonstrate that ERRγ directly regulates mitochondrial metabolism and renal reabsorption, but through distinct mechanisms. The functional cooperation between ERRγ and HNF1β, another transcriptional regulator important in kidney biology and disease, further uncovers the transcriptional network optimized to coordinate kidney mitochondrial and reabsorptive functions. Together, these results improve our understanding of how mitochondrial function is coordinately regulated to support normal kidney function and how its dysregulation contributes to kidney disease (Fig. 6E).
The near complete dependence of SGLT2 expression and renal glucose reabsorption on ERRγ is particular noteworthy because of the recent clinical use and efficacy of SGLT2 inhibitors in treating type 2 diabetes, and their therapeutic potential in treating obesity, kidney disease, and cardiovascular disease. We further demonstrated that ERRγ can increase Sglt2 expression and REC glucose uptake in a cell-autonomous manner. Under STZ-induced diabetic conditions, REC-ERRγ KO mice faithfully recapitulated glucosuria seen in patients treated with SGLT2 inhibitors. However, this did not result in lower blood glucose, as observed clinically. One possible explanation for this is there remains a small amount of SGLT2 and intact SGLT1 in the REC-ERRγ KO animals. This incomplete glucose excretion phenomenon has also been observed in Sglt2 KO mice (37), as well as in SGLT2 inhibitor-treated patients with diabetes in whom only 40–50% of the expected glucose reabsorption inhibition can be achieved with a full dose (30). Another possible explanation is that STZ destroys pancreatic β cells, causing type 1 diabetes and very high plasma glucose levels. Loss of insulin production and incomplete glucose excretion may explain the lack of blood glucose normalization in REC-ERRγ mice, similar to what was reported in Sglt2 KO mice (37). Notably, SGLT2 inhibitors are not currently approved for type 1 diabetes.
ERRγ has been shown to promote energy-generating mitochondrial functions in several energy-demanding cell types, including neurons, cardiomyocytes, skeletal myocytes, brown adipocytes, and now in RECs (17–23, 38). In three of these cell types (neurons, cardiomyocytes, and RECs) ChIP-Seq or ChIP-on-chip has been used to demonstrate that ERRγ directly binds near mitochondrial OxPhos genes (17, 23). However, ERRγ cistromes also exhibit cell-type-specific features that match the metabolic profile and energy-intensive cellular functions of individual cell types. Neurons depend on aerobic catabolism of glucose, including OxPhos as an energy source, and they cannot metabolize fat. Accordingly, the neuronal ERRγ cistrome includes many glycolysis and OxPhos genes but is depleted of FAO genes (17). Cardiomyocytes, in contrast, use both glucose and fat as fuel. As a consequence, cardiac ERRγ binds to and activates the transcription of FAO genes in addition to OxPhos genes (23). Here in RECs, we demonstrate that ERRγ directly regulates both OxPhos and FAO, key metabolic pathways used by these cells (12). Therefore, ERRγ is a key transcriptional regulator of cell-type specific metabolic functions. In addition, ERRγ has been shown to directly bind to and activate genes for cardiac contraction and conduction, two major cellular energy utilization processes specific to the heart (18, 23). Here, we show that ERRγ directly regulates the major energy consumption process in another cell type; namely, reabsorption in RECs, via functional cooperation with the REC-related transcription factor HNF1β. Together these studies reveal a common theme of cellular metabolic regulation: cellular energy production and consumption must be coordinated because most cells do not have an extensive capacity to fully store energy. Our studies thus demonstrate that ERRγ is a central transcriptional coordinator of cell-type specific energy production and utilization.
Materials and Methods
All mouse studies were approved by and performed under the guidelines of the Institutional Animal Care and Use Committee of the Children’s Hospital of Philadelphia. Adenoviruses were generated as previously described (39, 40). Transmission electron microscopy, gene expression, Western blot, cellular oxygen consumption measurement, mitochondrial DNA, and enzyme activity analysis were performed as previously described (18, 41). Please see SI Appendix, SI Materials and Methods for detailed description of mouse studies, cell culture, glucose uptake, histological analysis, RNA-Seq, ChIP-Seq, and ChIP-reChIP-Seq experiments. Statistical analysis was performed using Student’s t test, Pearson correlation, or Fisher’s exact test. Microarray data were deposited in ArrayExpress database (E-MTAB-2502). RNA-Seq, ChIP-Seq, and ChIP-reChIP-Seq data were deposited in the GEO database (GSE104907).
Supplementary Material
Acknowledgments
We thank Dr. Douglas Wallace, Dr. Mitchell Lazar, Dr. Matthew Weitzman, Dr. Amita Sehgal, Dr. Michael Marks, and Dr. Mark Kahn for critical discussion of the project. We thank Dr. Biao Zuo, Dr. Ray Meade, and the UPenn Electron Microscopy Resource Laboratory core facility for technical support. The authors and this work were supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program under Award W81XWH-16-1-0400, and pilot awards from the Diabetes Research Center at the University of Pennsylvania from a grant sponsored by the NIH (DK19525; DK111495 to L.P.; DK099379 to B.J.W.; DK108987 to G.D.B.; and DK087635 and DP3 DK108220 to K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data were deposited in ArrayExpress database (E-MTAB-2502). RNA-Seq, ChIP-Seq, and ChIP-reChIP-Seq data were deposited in the GEO database (GSE104907).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804965115/-/DCSupplemental.
References
- 1.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Ouweland JM, Maechler P, Wollheim CB, Attardi G, Maassen JA. Functional and morphological abnormalities of mitochondria harbouring the tRNA(Leu)(UUR) mutation in mitochondrial DNA derived from patients with maternally inherited diabetes and deafness (MIDD) and progressive kidney disease. Diabetologia. 1999;42:485–492. doi: 10.1007/s001250051183. [DOI] [PubMed] [Google Scholar]
- 3.Dinour D, Mini S, Polak-Charcon S, Lotan D, Holtzman EJ. Progressive nephropathy associated with mitochondrial tRNA gene mutation. Clin Nephrol. 2004;62:149–154. doi: 10.5414/cnp62149. [DOI] [PubMed] [Google Scholar]
- 4.Che R, Yuan Y, Huang S, Zhang A. Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2014;306:F367–F378. doi: 10.1152/ajprenal.00571.2013. [DOI] [PubMed] [Google Scholar]
- 5.Emma F, Montini G, Parikh SM, Salviati L. Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nat Rev Nephrol. 2016;12:267–280. doi: 10.1038/nrneph.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhargava P, Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol. 2017;13:629–646. doi: 10.1038/nrneph.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishimoto Y, et al. Mitochondrial abnormality facilitates cyst formation in autosomal dominant polycystic kidney disease. Mol Cell Biol. 2017:MCB.00337-17. doi: 10.1128/MCB.00337-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei L, Wallace DC. Mitochondrial etiology of neuropsychiatric disorders. Biol Psychiatry. 2018;83:722–730. doi: 10.1016/j.biopsych.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breyer MD, Susztak K. The next generation of therapeutics for chronic kidney disease. Nat Rev Drug Discov. 2016;15:568–588. doi: 10.1038/nrd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granata S, et al. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genomics. 2009;10:388. doi: 10.1186/1471-2164-10-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma K, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichner LJ, Giguère V. Estrogen related receptors (ERRs): A new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 14.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 15.Misra J, Kim DK, Choi HS. ERRγ: A junior orphan with a senior role in metabolism. Trends Endocrinol Metab. 2017;28:261–272. doi: 10.1016/j.tem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Alaynick WA, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Pei L, et al. Dependence of hippocampal function on ERRγ-regulated mitochondrial metabolism. Cell Metab. 2015;21:628–636. doi: 10.1016/j.cmet.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, et al. Estrogen-related receptor α (ERRα) and ERRγ are essential coordinators of cardiac metabolism and function. Mol Cell Biol. 2015;35:1281–1298. doi: 10.1128/MCB.01156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihara E, et al. ERRγ is required for the metabolic maturation of therapeutically functional glucose-responsive β cells. Cell Metab. 2016;23:622–634. doi: 10.1016/j.cmet.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan Z, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J Clin Invest. 2013;123:2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantner ML, Hazen BC, Eury E, Brown EL, Kralli A. Complementary roles of estrogen-related receptors in brown adipocyte thermogenic function. Endocrinology. 2016;157:4770–4781. doi: 10.1210/en.2016-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmadian M, et al. ERRγ preserves brown fat innate thermogenic activity. Cell Rep. 2018;22:2849–2859. doi: 10.1016/j.celrep.2018.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Berry R, et al. Esrrg functions in early branch generation of the ureteric bud and is essential for normal development of the renal papilla. Hum Mol Genet. 2011;20:917–926. doi: 10.1093/hmg/ddq530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harewood L, et al. Bilateral renal agenesis/hypoplasia/dysplasia (BRAHD): Postmortem analysis of 45 cases with breakpoint mapping of two de novo translocations. PLoS One. 2010;5:e12375. doi: 10.1371/journal.pone.0012375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 27.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 28.van den Heuvel LP, Assink K, Willemsen M, Monnens L. Autosomal recessive renal glucosuria attributable to a mutation in the sodium glucose cotransporter (SGLT2) Hum Genet. 2002;111:544–547. doi: 10.1007/s00439-002-0820-5. [DOI] [PubMed] [Google Scholar]
- 29.Chao EC, Henry RR. SGLT2 inhibition–A novel strategy for diabetes treatment. Nat Rev Drug Discov. 2010;9:551–559. doi: 10.1038/nrd3180. [DOI] [PubMed] [Google Scholar]
- 30.Vallon V. The mechanisms and therapeutic potential of SGLT2 inhibitors in diabetes mellitus. Annu Rev Med. 2015;66:255–270. doi: 10.1146/annurev-med-051013-110046. [DOI] [PubMed] [Google Scholar]
- 31.Anık A, Çatlı G, Abacı A, Böber E. Maturity-onset diabetes of the young (MODY): An update. J Pediatr Endocrinol Metab. 2015;28:251–263. doi: 10.1515/jpem-2014-0384. [DOI] [PubMed] [Google Scholar]
- 32.Lazzaro D, De Simone V, De Magistris L, Lehtonen E, Cortese R. LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development. 1992;114:469–479. doi: 10.1242/dev.114.2.469. [DOI] [PubMed] [Google Scholar]
- 33.Gresh L, et al. A transcriptional network in polycystic kidney disease. EMBO J. 2004;23:1657–1668. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clissold RL, Hamilton AJ, Hattersley AT, Ellard S, Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat Rev Nephrol. 2015;11:102–112. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 35.Verdeguer F, et al. A mitotic transcriptional switch in polycystic kidney disease. Nat Med. 2010;16:106–110. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. HNF6 and Rev-erbα integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 2016;30:1636–1644. doi: 10.1101/gad.281972.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallon V, et al. SGLT2 mediates glucose reabsorption in the early proximal tubule. J Am Soc Nephrol. 2011;22:104–112. doi: 10.1681/ASN.2010030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narkar VA, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pei L, et al. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 40.Pei L, et al. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat Med. 2011;17:1466–1472. doi: 10.1038/nm.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, et al. GDF15 is a heart-derived hormone that regulates body growth. EMBO Mol Med. 2017;9:1150–1164. doi: 10.15252/emmm.201707604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.