Significance
Nosocomial infections and infections of indwelling devices are major healthcare problems worldwide. These infections are strongly associated with the ability of pathogens to form biofilms on biotic and abiotic surfaces. Panantibiotic-resistant Acinetobacter baumannii is one of the most troublesome pathogens, capable of colonizing medical devices by means of Csu pili, an adhesive organelle that belongs to the widespread class of archaic chaperone–usher pili. Here, we report an atomic-resolution insight into the mechanism of bacterial attachment to abiotic surfaces. We show that archaic pili use a binding mechanism that enables bacterial adhesion to structurally variable substrates. The results suggest a simple and cheap solution to reduce infections of A. baumannii and related pathogens.
Keywords: archaic pili, chaperone–usher pathway, bacterial adhesion, biofilm, Acinetobacter baumannii
Abstract
Acinetobacter baumannii—a leading cause of nosocomial infections—has a remarkable capacity to persist in hospital environments and medical devices due to its ability to form biofilms. Biofilm formation is mediated by Csu pili, assembled via the “archaic” chaperone–usher pathway. The X-ray structure of the CsuC-CsuE chaperone–adhesin preassembly complex reveals the basis for bacterial attachment to abiotic surfaces. CsuE exposes three hydrophobic finger-like loops at the tip of the pilus. Decreasing the hydrophobicity of these abolishes bacterial attachment, suggesting that archaic pili use tip-fingers to detect and bind to hydrophobic cavities in substrates. Antitip antibody completely blocks biofilm formation, presenting a means to prevent the spread of the pathogen. The use of hydrophilic materials instead of hydrophobic plastics in medical devices may represent another simple and cheap solution to reduce pathogen spread. Phylogenetic analysis suggests that the tip-fingers binding mechanism is shared by all archaic pili carrying two-domain adhesins. The use of flexible fingers instead of classical receptor-binding cavities is presumably more advantageous for attachment to structurally variable substrates, such as abiotic surfaces.
The Gram-negative bacterium Acinetobacter baumannii, also known as “Iraqibacter” due to its emergence in US military treatment facilities in Iraq, has quickly become one of the most troublesome pathogens for healthcare institutions globally and currently tops the priority pathogens list for development of new antibiotics (1, 2). The outstanding survival properties and antibiotic resistance of A. baumannii are strongly associated with its ability to form biofilms (3). The pathogen was shown to colonize various objects, including medical equipment and tools, hospital furniture, and even gowns and gloves of healthcare providers (4, 5). A. baumannii biofilm formation on such abiotic surfaces is mediated by Csu pili (6) (Fig. 1).
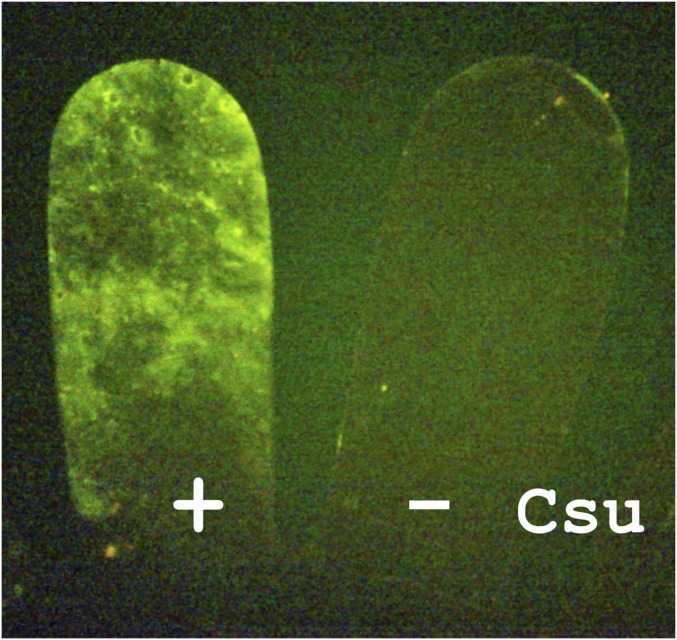
Fig. 1.
Csu pili-mediate bacterial attachment to abiotic surfaces. Two fingers of a polyethylene glove, Left and Right in the image, were dipped into culture medium of Escherichia coli expressing and not expressing Csu pili, respectively. To visualize bacteria, expression of yellow fluorescence protein (YFP) was induced.
The Csu pilus is elaborated from four protein subunits, CsuA/B, CsuA, CsuB, and CsuE (6), via the archaic (σ) chaperone–usher (CU) pathway. Archaic CU pili constitute the largest family of CU systems and together with the alternative CU family form the “nonclassical” branch of the CU superfamily (7, 8). Although archaic systems have a far wider phylogenetic distribution and are associated with a broader range of diseases than their classical equivalents, they were discovered relatively recently (8). The crystal structure of the preassembly CsuC-CsuA/B chaperone–subunit complex provided the first high-resolution insight into the assembly mechanism of archaic pili (7). The chaperone-bound CsuA/B has a highly flexible incomplete Ig-like fold in a six-stranded β-sandwich, in which the absent seventh strand (G) leaves a large hydrophobic cleft (7). In the polymer, CsuA/B subunits are linked by donor strand complementation (DSC) (9–12) with the N-terminal sequence of one subunit inserted into the hydrophobic cleft of a neighboring subunit (7). In contrast to CsuA/B, CsuE is not capable of self-assembly (7). Instead of the donor sequence, this subunit is predicted to contain an additional domain (7). This implies that CsuE is located at the pilus tip. Since many two-domain tip subunits in classical systems have been shown to act as host cell binding adhesins (TDAs) (13–16), CsuE could also play a role in bacterial attachment to biotic and abiotic substrates. However, adhesion properties of Csu subunits are not known, and the mechanism of archaic pili-mediated biofilm formation remains enigmatic. Here, we report the crystal structure of the CsuE subunit complexed with the CsuC chaperone. Our structural study together with site-directed mutagenesis and site-specific antibody biofilm inhibition reveals a binding mechanism that enables bacterial adhesion to structurally variable substrates and suggests approaches to restrict the spread of A. baumannii and other pathogens in hospital setting as well as colonization of indwelling devices.
Results and Discussion
To produce a convenient model to study the function of Csu pili, the Csu gene cluster was cloned in Escherichia coli. Expression of pili resulted in strong adherence of the recombinant strain to hydrophobic plastics, in particular polystyrene, polypropylene, and polyethylene (Fig. 1 and SI Appendix, Fig. S1). These materials are widely used in medical equipment implicated in the spread of A. baumannii (3). As in the case of the A. baumannii 19606 prototype strain (6), recombinant E. coli biofilms were particularly pronounced at the liquid–air interface and were visualized as a fluorescent ring of bacteria coexpressing Csu pili with yellow fluorescent protein (YFP) or a blue ring after staining cells with crystal violet (SI Appendix, Fig. S1). Adhesion of the strain to hydrophilic surfaces, such as glass and cellophane, was also observed (SI Appendix, Fig. S1). However, such biofilms were easily washed away, suggesting that Csu pili mediate tight bacterial attachment only to hydrophobic substrates.
Transmission electron microscopy (TEM) of the induced strain revealed thin (∼3.5 nm), compared with the classical type 1 and P pili (17, 18), but unusually long pili (SI Appendix, Fig. S2). Deletions of CsuA/B and CsuE completely abrogated pilus assembly (SI Appendix, Fig. S3 A and E). Deletions of CsuA and CsuB led to assembly of a few abnormal fibers (SI Appendix, Fig. S3 B–D). However, none of the deletion mutants were capable of biofilm formation on plastics, suggesting that all four subunits are required to form functional pili (SI Appendix, Fig. S4). Whereas CsuA/B, CsuA, and CsuB may have structural roles as building blocks of the pilus stalk (6, 7, 19), CsuE must be critical for initiation of assembly, similarly to tip-located TDAs of type 1 and P pili (20, 21). To gain insight into the role of CsuE, we determined the X-ray crystal structure of CsuE complexed with the CsuC chaperone (Fig. 2).
Fig. 2.
Three finger-like loops form the hydrophobic tip of the Csu pilus. (A) Stereoview of the CsuC-CsuE complex. CsuE is colored cyan, except for the hydrophobic binding motifs in loops B12-C12 and D11″-α2 (yellow, 1), F12-G11 (orange, 2), and G11-G12 (light orange, 3). CsuC is colored magenta, except for the G1 “donor” strand, which is violet. (B) Topology diagram of CsuE. (C) Cartoon diagram and molecular surface of CsuE. Residues comprising the hydrophobic sequences are shown as ball-and-stick. (D) Cartoon diagram and semitransparent surface of CsuE viewed from the pilus tip. (E) Alignment of TDAs of archaic pili from Acinetobacter baumannii, Burkholderia pseudomallei, Pseudomonas aeruginosa, and Yersinia pestis. Amino acid identities and similar residues are indicated by background shading in cyan and gray, respectively. Hydrophobic binding motifs are framed. Within motifs, hydrophobic residues are shown on a yellow background. Cysteines are colored red. Disulfide bonds are indicated with connecting lines. Limits and nomenclature for secondary structure elements are shown above the sequence. Dashed and wavy lines indicate unstructured and domain-linking sequences, respectively. Alignment of a large set of archaic TDAs is shown in SI Appendix, Fig. S11.
The structure of the CsuC-CsuE complex was solved using selenomethionine single anomalous dispersion (Se-SAD) phasing to a resolution of 2.3 Å (22). Refinement statistics are shown in SI Appendix, Table S3. Two copies of the CsuC-CsuE heterodimer are present in the P1 unit cell. The structure of CsuC is virtually the same in both copies, with an RMSD deviation between Cα atoms of 0.5 Å, and is similar to the structure of CsuC in the CsuC-CsuA/B complex (7). Comparison of CsuE molecules revealed much larger differences (RMSD 1.4 Å) (SI Appendix, Fig. S5). CsuE folds into two β-barrel domains (Fig. 2 A–C). The angle between the domains gives the molecule an overall C-like shape, bending over domain 1 of CsuC (SI Appendix, Fig. S6).
The C-terminal domain of CsuE binds in the cleft between the CsuC domains (Fig. 2A). It has an incomplete Ig-like fold that lacks the canonical C-terminal strand G (Fig. 2 A and B). The missing strand is provided by CsuC, which inserts its strand G1 into a hydrophobic groove at the edge of β-sheet DCC′D′. Since the groove is likely to accept a donor strand during pilus assembly to connect CsuE to the rest of the fiber, we refer to this domain as the pilin domain (CsuEpd). Indeed, CsuEpd shows significant structural similarity to CsuA/B (Z-score = 9.9, SI Appendix, Fig. S7), which currently serves as a prototype of stalk pilins in the archaic CU pathway (7). Like CsuC-bound CsuA/B, CsuEpd has a large fraction of disordered or poorly ordered sequence (Fig. 2A and SI Appendix, Fig. S8A), further supporting our previous conclusion that nonclassical chaperones, unlike their classical counterparts, maintain subunits in a substantially disordered conformational state (7).
The N-terminal domain is connected to CsuEpd by a three-residue linker with a proline in the middle (Fig. 2 A and C and SI Appendix, Fig. S9). Hydrophobic interactions and a network of hydrogen and ionic bonds dominate the interface between the domains (SI Appendix, Fig. S9). The CsuE N-terminal domain (CsuENTD) has a mixed α/β structure. The core of the domain is formed by an Ig-like sandwich that adopts a β-barrel shape due to switches of edge strands A and D between the β-sheets (Fig. 2B). Association of strand A1′ with strand G12 results in a mixed parallel–antiparallel structure of the second β-sheet (Fig. 2B), which is also a common feature of lectin domains of TDAs in both classical and alternative systems (SI Appendix, Fig. S10). Indeed, CsuENTD shares considerable structural similarity with lectin domains of CfaE and FimH (Z-scores of 6.9 and 5.5, respectively), again pointing to a possible role of CsuENTD in bacterial adhesion. At the same time, comparison of these structures (SI Appendix, Fig. S10) uncovered a major topological difference. In CsuENTD, both β-sheets are split due to loop insertions in all major β-strands. Hence, the structure consists not of one, but rather two β-barrels, each of which is partially sealed by an α-helix. This unique arrangement makes CsuENTD longer than other TDAs of known structure. Its length is further increased by the presence of three large loops forming a three-fingered structure at the tip of CsuENTD (Fig. 2 C and D). Strikingly, the finger loops are very hydrophobic, with hydrophobic motifs consisting of 24 surface-exposed residues, only one of which has a polar side chain (Asn45) (Fig. 2E). The first two hydrophobic fingers are conserved among archaic TDAs, whereas the third loop is specific for Csu pili of A. baumannii (Fig. 2E and SI Appendix, Fig. S11).
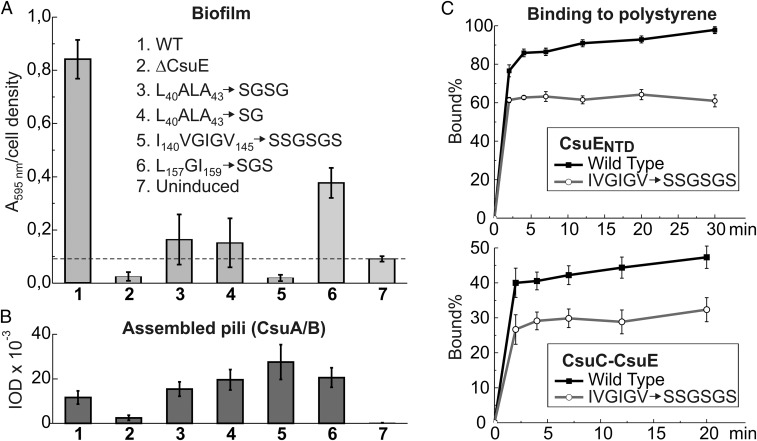
The hydrophobic nature of the finger loops prompted us to examine their possible involvement in A. baumannii biofilm formation on hydrophobic substrates (Fig. 3A). We introduced four mutations decreasing the hydrophobicity of the fingers. Replacement of residues L40ALA43 with SGSG or SG, residues I140VGIGV145 with SSGSGS, and residues L157GI159 with SGS disrupted hydrophobic fingers 1–3, respectively. In contrast to deletion of the entire CsuE subunit, these mutations neither decreased the level of assembly of Csu pili (Fig. 3B) nor affected their morphology (SI Appendix, Fig. S3F). However, mutations in finger 3 dramatically decreased, and mutations in fingers 1 and 2 practically abolished biofilm formation (Fig. 3A). These results provide conclusive evidence that the three-finger structure indeed plays an essential role in biofilm formation.
Fig. 3.
Hydrophobic loops in CsuE are essential for biofilm formation. (A) Quantification of biofilms of E. coli harboring the wild-type (1, 7) and mutant (2–6) Csu gene cluster (2). Deletion of the entire gene of CsuE (3–6); mutations in hydrophobic loops of CsuE. Uninduced cells were used as a negative control (7). Biofilms were stained with crystal violet and quantified by measuring absorbance at 595 nm. (B) Quantification of pilus assembly with Western blotting using anti-CsuA/B antibody. Integral optical density (IOD) of the CsuA/B band is proportional to the amount of CsuA/B assembled into the pili. (C) Effect of the IVGIGV → SSGSGS mutation on binding of CsuENTD (Top) and CsuC-CsuE (Bottom) to polystyrene plates. The results in A–C are representative of at least three independent experiments. Error bars represent one SD.
To investigate the role of the finger loops in binding to hydrophobic plastics, we examined the effect of mutation I140VGIGV145 → SSGSGS on binding of CsuE complexed to CsuC, and of CsuENTD (SI Appendix, Fig. S12), to polystyrene surfaces. To enable quantitative analysis of protein binding to plastics, wild-type and mutant CsuC-CsuE and CsuENTD were labeled with Eu3+. Identical amounts of equally labeled wild-type and mutant proteins were incubated in polystyrene plates. Although both wild-type and mutant proteins bound to polystyrene plates in our experimental setup, measurements of Eu fluorescence in the samples revealed that wild-type proteins consistently bound more efficiently to plastics than the corresponding mutants (Fig. 3C). This result strongly suggests that the hydrophobic fingers directly mediate binding of Csu pili to hydrophobic plastics.
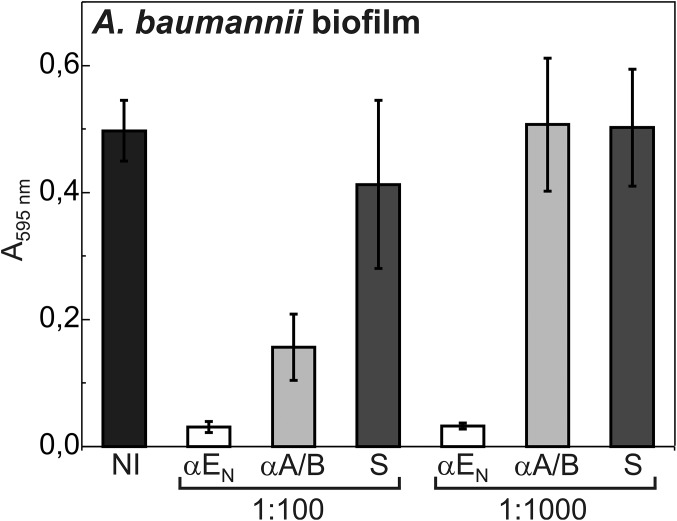
To further investigate Csu pili-mediated biofilm formation, we raised polyclonal antibodies against self-complemented CsuA/B subunit (αA/B) and CsuENTD (αEN) and examined the effect of these antibodies on biofilm formation of A. baumannii (Fig. 4) and Csu pili-expressing E. coli (SI Appendix, Fig. S13). Bacterial cultures were incubated with different dilutions of αA/B and αEN antibodies and preimmune serum (negative control) and then assayed for biofilm formation in wells of polystyrene microtiter plates. αEN diluted up to several thousand times completely blocked biofilm formation of A. baumannii (Fig. 4). αA/B also inhibited biofilm formation, but inhibition was observed only at high concentration. This result further underlines the critical role of CsuENTD in bacterial adhesion to hydrophobic plastics.
Fig. 4.
Antitip antibody blocks A. baumannii biofilm formation. A. baumannii 19606 cells were preincubated with PBS buffer, 1:100 and 1:1,000 dilutions of antibodies raised against the N-terminal domain of CsuE (αEN) and CsuA/B (αA/B) or the preimmune serum (S) and then assayed for biofilm formation in polystyrene microtiter plates. Similar results were obtained for other A. baumannii strains from the CCUG collection (SI Appendix, Table S1). The results are representative of three independent experiments. Error bars represent one SD.
The Csu gene cluster is highly conserved and is present in nearly all clinical isolates of A. baumannii (23). To study Csu pili expression in different A. baumannii strains, we analyzed bacterial surface extracts of five unrelated isolates of A. baumannii from the Culture Collection of the University of Gothenburg (SI Appendix, Table S1), including type strain 19096T [known to express large amounts of Csu pili (6)], with Western blotting using αA/B serum (SI Appendix, Fig. S14). Csu pili were detected in all five isolates. In three isolates, the expression level was about two to three times lower than in 19096T, and in one isolate it was notably higher. αEN antibody completely blocked biofilm formation of all five isolates. Hence, Csu pili are constitutively expressed in a wide variety of A. baumannii isolates and are responsible for A. baumannii biofilm formation on hydrophobic plastics. This finding further highlights the role of Csu pili in the spread of A. baumannii in medical settings. The strong inhibitory effect of αEN antibody presents a means to prevent the spread of the pathogen. The use of hydrophilic materials instead of hydrophobic plastics in medical devices represents another simple solution to reduce pathogen outbreaks. For example, we observed much weaker biofilms on gloves made of polyvinyl chloride than on polyethylene gloves.
Another multidrug-resistant pathogen, Pseudomonas aeruginosa, assembles phylogenetically related CupE pili, which have recently been shown to mediate morphologically similar biofilms (24). Considerable sequence similarity between two-domain tip subunits CsuE and CupE6 and the presence of two hydrophobic loop motifs in CupE6 corresponding to hydrophobic fingers 1 and 2 in CsuE (Fig. 2E) suggest that CupE pili may use a similar attachment mechanism. Hence, we predict that the means suggested for A. baumannii above might also be applied to control the spread of P. aeruginosa infections.
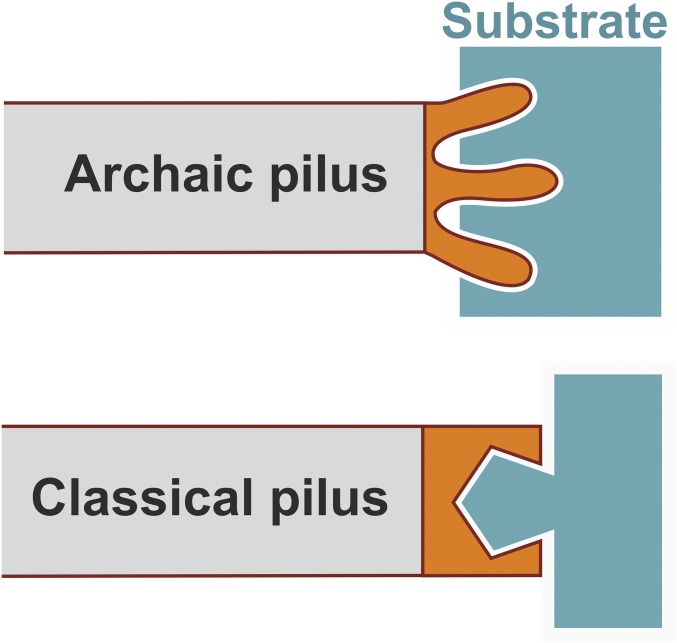
Comparison of binding mechanisms of archaic and classical pili reveals an important conceptual difference (Fig. 5). Like many other proteins, which function relies on substrate or receptor recognition, classical pili use a depression or a binding cavity, which is complementary in shape and chemical properties to the minimal binding determinant of the host cell receptor (13–15, 25, 26). Our study shows that archaic pili have no such binding cavity. Instead, they use protruding finger-like loops that may insert into cavities in substrates to establish tight attachment. The use of flexible fingers (SI Appendix, Fig. S5) instead of preformed receptor-binding cavities may have an important advantage when bacteria need to attach to structurally variable substrates, such as abiotic surfaces. Although being less specific, this flexible mode of binding may promote an equally tight attachment because, in the process of surface scanning, the fingers would likely find a suitable cavity in the substrate. Fingers 1 and 2 are predicted in all TDA subunits of archaic pili (SI Appendix, Fig. S11), suggesting that the tip-fingers binding mechanism is shared by all archaic systems and has likely evolved early in nonpathogenic bacteria to mediate adhesion to abiotic substrates.
Fig. 5.
Conceptual difference in substrate recognition between archaic and classical pili.
Our first atomic-resolution insight into bacterial colonization of human-made substrates may be very useful in addressing another very important medical problem—biofilm-associated infections of indwelling medical devices (IMDs). IMDs are often colonized by bacterial and fungal pathogens, increasing mortality rates by more than 25% in some cases (27). The knowledge of the nature of microbe–substrate interactions presented here will help development of device coatings preventing colonization.
Materials and Methods
Bacterial Strains and Plasmids.
Characteristics and source of the bacterial strains used in this study are given in SI Appendix, Table S1. Oligonucleotides are listed in SI Appendix, Table S2. Expression plasmids were constructed based on the pBAD-ENSPA plasmid (25) or pET101D expression vector (Invitrogen) as described in SI Appendix. The pBAD-Csu plasmid and its derivatives were used to express wild-type and mutant Csu pili, and the pET101-CsuENPD6H plasmid and its derivative were used to express wild-type and mutant CsuENTD. Deletions and substitutions were generated using reverse PCR as described in SI Appendix.
Protein Purification.
CsuC-CsuE expression and purification is described in ref. 22. To express CsuENTD, E. coli BL21-AI was transformed with pET101-CsuENPD6H. Bacterial cells were cultivated in Luria Broth (LB) medium containing ampicillin (80–100 mg/L) at 37 °C. The cells were grown at 110 rpm in baffled flasks to an optical density (OD) of 0.8–1 at 600 nm and were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and 0.2% arabinose for protein expression. The culture was grown for a further 2.5 h. The cells were harvested by centrifugation, and CsuENTD was purified from the periplasm by metal–ion affinity chromatography at 4 °C using a 5-mL HiTrap Ni-IMAC column (GE Healthcare) essentially as the CsuC-CsuA/B complex (28). Sample of CsuENTD was used to generate polyclonal rabbit antibodies (Innovagen AB).
Structure Determination.
Details of crystallization, X-ray diffraction data collection and phasing were previously published (22). Model building and refinements were performed by the PHENIX refinement module. Manual corrections were done with the molecular modeling program COOT (29). Refinement statistics are given in SI Appendix, Table S3.
Biofilm Assays.
E. coli strain BL21 harboring pBAD-Csu or its derivatives was cultured overnight in LB medium in the presence of 100 mg/L ampicillin. One milliliter of the fresh medium in a 12-mL polypropylene tube was inoculated with 20 μL of the overnight culture and then grown at 37 °C with vigorous shaking for 2 h. Arabinose was added to 0.02% final concentration to induce expression of the Csu pili, and the cells were further grown with shaking at 200 rpm for 2.5 h. The cell density was determined by measuring OD at 600 nm. The tubes were emptied and rinsed briefly with PBS. The biofilms were stained with 1% crystal violet for 15 min, rinsed with water, and dissolved in 1.5 mL of 0.2% Triton X-100. Three hundred microliters of this solution was transferred into microtiter wells, and OD was measured with a 96-well plate spectrometer reader at 595 nm.
To study A. baumannii biofilms, strains from the Culture Collection, University of Gothenburg (CCUG), were transferred with an inoculation loop from an agar slant into 5 mL of Super Broth (SB) medium in a 12-mL polypropylene or polystyrene tube and grown overnight at 37 °C without shaking. On the following day, the tubes were emptied, rinsed with PBS, and remaining biofilms were analyzed as described above.
To study biofilms of living cells, E. coli strain BL21 was cotransformed with plasmids pBAD-Csu and pYPet-His (gift from Patrick Daugherty, University of California, Santa Barbara, CA, Addgene plasmid #14031), overexpressing Csu pili and YFP, respectively. Bacteria were cultured overnight in LB medium in the presence of 100 mg/L ampicillin and 25 mg/L chloramphenicol. On the following day, 20 mL LB medium was inoculated with 1 mL of the overnight culture, grown for 2 h, and expression of pili and YFP was induced for 2.5 h with 0.04% arabinose. The induced cells were used to examine biofilm formation on various surfaces (plastic and glass tubes, gloves, and straps of cellophane). Uninduced cells were used as a negative control. After incubation, biofilms were rinsed gently (hydrophilic surfaces) or thoroughly (hydrophobic surfaces) with water, and YFP fluorescence was observed on a Dark Reader blue transilluminator (Clare Chemical Research) and photographed.
Biofilm Inhibition.
Cell culture samples were mixed with antibody or serum dilution in an Eppendorf tube for 30 min at 22 °C with gentle shaking. Two 150-µL aliquots from each mixture were transferred with a micropipette to wells of a microtiter plate. The plate was incubated at 22 °C for 2 h without shaking. Wells were then emptied and washed three times with 300 µL of distilled water. Any remaining biofilm was stained with 1% crystal violet for 30 min, washed with water again, and dissolved in 250 µL of 0.2% Triton X-100. Optical density at 595 nm was determined with a 96-well plate spectrometer reader.
Purification of Csu pili.
E. coli strain BL21 harboring pBAD-Csu or A. baumannii strains from CCUG were cultured in appropriate medium as described above, centrifuged at 10,000 g for 15 min, washed with 0.5 mM Tris, 75 mM NaCl, pH 7.3, and suspended in the same buffer. Samples were incubated at 65 °C for 30 min and then centrifuged at 4,000 g for 15 min. Supernatants containing detached pili were carefully removed and stored at 4 °C before analysis.
Electron Microscopy.
Bacterial cells or purified pili were sampled with Formvar-coated gold grids, negative-stained with 2% uranyl acetate, and visualized using a JEOL JEM-1400 Plus transmission electron microscope operated at 80-kV acceleration voltage.
Western Blotting.
Cell cultures grown for biofilm assay were mixed with Laemmli buffer, and the samples were incubated at 22 °C or boiled. The proteins were separated by electrophoresis in 18% SDS polyacrylamide gels and transferred onto an Immunoblot polyvinylidene difluoride membrane (Bio-Rad Laboratories) in Bio-Rad A-buffer (25 mM Tris, pH 8.3, 192 mM glycine, with 20% methanol and 0.1% SDS) at 100 V for 1 h. Membrane was blocked with 5% skim milk in PBS/Tween, incubated with primary anti-CsuA/B rabbit polyclonal antibody (Innovagen AB), followed by incubation with secondary IRDye 68RD-conjugated anti-rabbit goat antibody (Li-Cor Biosciences). Protein bands were detected and quantified with the Odyssey system (Li-Cor Biosciences).
Analysis of CsuE Binding to Plastics.
Eu+3 labeled proteins were incubated in 5.5-cm polystyrene Petri dishes (Sarstedt). Aliquots were withdrawn at different time points to measure delayed Eu+3 fluorescence and generate binding curves. See SI Appendix for detailed experimental protocols.
Supplementary Material
Acknowledgments
This work was supported by grants from the Academy of Finland (273075) and S. Juselius Foundation (to A.V.Z.). N.P. was supported by a stipend from the Finnish Cultural Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6FJY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800961115/-/DCSupplemental.
References
- 1.Maragakis LL, Perl TM. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 2.WHO . Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO; Geneva: 2017. pp. 1–7. [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilks M, et al. Control of an outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus colonization and infection in an intensive care unit (ICU) without closing the ICU or placing patients in isolation. Infect Control Hosp Epidemiol. 2006;27:654–658. doi: 10.1086/507011. [DOI] [PubMed] [Google Scholar]
- 5.Morgan DJ, et al. Frequent multidrug-resistant Acinetobacter baumannii contamination of gloves, gowns, and hands of healthcare workers. Infect Control Hosp Epidemiol. 2010;31:716–721. doi: 10.1086/653201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 7.Pakharukova N, et al. Structural insight into archaic and alternative chaperone-usher pathways reveals a novel mechanism of pilus biogenesis. PLoS Pathog. 2015;11:e1005269. doi: 10.1371/journal.ppat.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuccio SP, Bäumler AJ. Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhury D, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 10.Sauer FG, et al. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 11.Zavialov AV, et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: Preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 12.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 13.Dodson KW, et al. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 14.Moonens K, et al. Structural insight in histo-blood group binding by the F18 fimbrial adhesin FedF. Mol Microbiol. 2012;86:82–95. doi: 10.1111/j.1365-2958.2012.08174.x. [DOI] [PubMed] [Google Scholar]
- 15.Bouckaert J, et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol Microbiol. 2005;55:441–455. doi: 10.1111/j.1365-2958.2004.04415.x. [DOI] [PubMed] [Google Scholar]
- 16.Le Trong I, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hospenthal MK, et al. Structure of a chaperone-usher pilus reveals the molecular basis of rod uncoiling. Cell. 2016;164:269–278. doi: 10.1016/j.cell.2015.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hospenthal MK, et al. The cryoelectron microscopy structure of the type 1 chaperone-usher pilus rod. Structure. 2017;25:1829–1838.e4. doi: 10.1016/j.str.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 20.Phan G, et al. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pakharukova N, Tuittila M, Paavilainen S, Zavialov A. Methylation, crystallization and SAD phasing of the Csu pilus CsuC-CsuE chaperone-adhesin subunit pre-assembly complex from Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun. 2017;73:450–454. doi: 10.1107/S2053230X17009566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriel DG, et al. Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS One. 2013;8:e77631. doi: 10.1371/journal.pone.0077631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraud C, et al. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol. 2011;13:666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- 25.Pakharukova N, et al. Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues. Mol Microbiol. 2016;102:593–610. doi: 10.1111/mmi.13481. [DOI] [PubMed] [Google Scholar]
- 26.Bao R, et al. Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis. Proc Natl Acad Sci USA. 2013;110:1065–1070. doi: 10.1073/pnas.1212431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008;59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- 28.Pakharukova N, Tuittila M, Paavilainen S, Zavialov A. Crystallization and preliminary X-ray diffraction analysis of the Csu pili CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun. 2015;71:770–774. doi: 10.1107/S2053230X15007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.