Significance
Historically, Neisseria meningitidis serogroup A strains have caused large epidemics of meningitis across sub-Saharan Africa. Following mass vaccination from 2010, serogroup A outbreaks have been mostly eliminated. Starting in 2013 however, yearly epidemics of a previously unknown serogroup C strain have led to tens of thousands of cases in Nigeria and Niger. We show how this new strain evolved from a benign ancestor through the acquisition of virulence genes encoding the serogroup C capsule and a phage linked to invasiveness, illustrating that minor genetic changes in a microbe can have major public health consequences. Our reconstruction of the spatiotemporal outbreak dynamics in the Niger–Nigeria border region suggests direct epidemiological consequences of contrasting outbreak responses in the two countries.
Keywords: whole-genome sequencing, nanopore sequencing, horizontal gene transfer, serogroup C meningococci, virulence factors
Abstract
In the African meningitis belt, a region of sub-Saharan Africa comprising 22 countries from Senegal in the west to Ethiopia in the east, large epidemics of serogroup A meningococcal meningitis have occurred periodically. After gradual introduction from 2010 of mass vaccination with a monovalent meningococcal A conjugate vaccine, serogroup A epidemics have been eliminated. Starting in 2013, the northwestern part of Nigeria has been affected by yearly outbreaks of meningitis caused by a novel strain of serogroup C Neisseria meningitidis (NmC). In 2015, the strain spread to the neighboring country Niger, where it caused a severe epidemic. Following a relative calm in 2016, the largest ever recorded epidemic of NmC broke out in Nigeria in 2017. Here, we describe the recent evolution of this new outbreak strain and show how the acquisition of capsule genes and virulence factors by a strain previously circulating asymptomatically in the African population led to the emergence of a virulent pathogen. This study illustrates the power of long-read whole-genome sequencing, combined with Illumina sequencing, for high-resolution epidemiological investigations.
Invasive meningococcal disease caused by Neisseria meningitidis is one of the leading causes of death from infectious diseases globally, with a case-fatality rate of about 10% even with optimal treatment conditions (1). The normal habitat of the bacterium is the human oropharynx. N. meningitidis is carried asymptomatically by a large proportion of the human population and is transmitted between individuals through exchange of respiratory secretions (2). In a small proportion of colonized individuals, the bacteria may invade the bloodstream, causing septicaemia, and/or cross the blood–brain barrier, causing meningitis. A number of virulence factors are required for meningococci to survive in the bloodstream, the most important being the presence of a polysaccharide capsule (3). Meningococci may express 1 of 12 antigenically distinct capsule types that are used to classify the bacteria into serogroups (4). Invasive disease is most often caused by serogroups A, B, C, W, X, and Y, whereas noncapsulated meningococci are commonly found among carriers. The absence of capsule seems to enhance the bacteria’s ability for colonization, whereas a capsule is an advantage in invasive disease, helping the bacteria to evade complement-mediated and phagocytic killing by the host in the bloodstream. In addition to the capsule, a large number of genomic factors are involved in the ability of meningococci to adhere to epithelial cells of the oropharynx and/or to cause invasive disease (5).
In the African meningitis belt, a region of sub-Saharan Africa comprising 22 countries from Senegal in the west to Ethiopia in the east, large epidemics of meningococcal meningitis occur periodically in the dry season (6). Traditionally, these have been caused by N. meningitidis serogroup A (NmA), although more recently, serogroups W and X have been responsible for large outbreaks (7–9). A monovalent meningococcal A conjugate vaccine (MenAfriVac) was introduced gradually in the meningitis belt, starting in Burkina Faso, Mali, and Niger in 2010, with almost 300 million people having now been vaccinated. NmA cases have declined very rapidly in the region, and NmA epidemics have been all but eliminated (10). In the first years following the implementation of the vaccination program, meningitis cases were predominantly caused by Streptococcus pneumoniae, but outbreaks caused by N. meningitidis of serogroups W and X have persisted (11).
In 2013, Sokoto State in northwest Nigeria experienced an outbreak of meningitis caused by a strain of N. meningitidis serogroup C (NmC) (12). NmC cases have been extremely rare in the meningitis belt for many decades, with the last NmC outbreak in the region occurring in Burkina Faso in 1979, with 539 reported cases (13), whereas the last and only recorded NmC outbreak in northern Nigeria was in 1975 (14). In 2014, the adjacent state, Kebbi, was affected by a smaller outbreak. In contrast to the large NmA epidemics affecting many thousands of individuals, these NmC outbreaks were localized, and the number of cases was moderate (12). In 2015, however, yet another epidemic caused by NmC occurred, affecting a wider region of northern Nigeria (15) but also spreading to the neighboring country of Niger, where 8,500 cases including 573 deaths were reported (16). Cases were reported in the region in 2016 and 2017, and in 2017 the largest NmC epidemic ever globally recorded occurred in northern Nigeria, with more than 14,000 suspected cases (17). Because availability of serogroup C vaccines is severely limited (www.who.int/emergencies/nigeria/meningitis-c/en/), spread to additional countries in the region is a likely and alarming scenario.
The causative strain was assigned to a new sequence type (ST), ST-10217 (12), using multilocus sequence typing (MLST) (18). In addition, all these NmC isolates had the same rare major outer membrane porin, PorA type P1.21-15,16. This particular PorA type had been observed only once before 2013, in an N. meningitidis isolate recovered from a carriage study performed in Burkina Faso in 2012 (19).
Here we apply long- and short-read whole-genome sequencing to characterize the evolution and spread of this new outbreak clone in Nigeria and Niger. Our analyses suggest that acquisition of virulence genes by a nonencapsulated benign ancestor led to the emergence of this virulent strain, with dramatic consequence in the meningitis belt of Africa. Apart from two genomic islands encoding a functional capsule and a meningococcal disease-associated prophage (MDAΦ) (20), respectively, the genome of the virulent outbreak clone is nearly identical to that of the asymptomatically carried strain, highlighting the permeable boundary between commensals and pathogens.
Results
Origin of ST-10217.
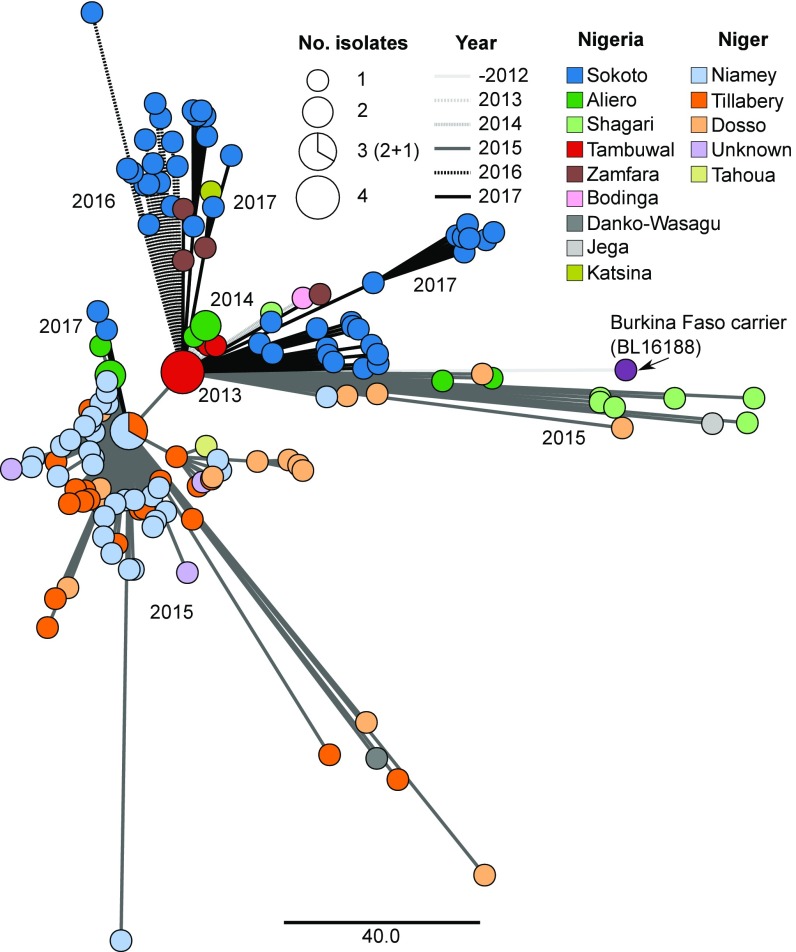
A total of 150 invasive isolates collected during the outbreaks in Nigeria and Niger from 2013 to 2017 were included in the analysis (SI Appendix, Table S1). All had identical PorA type, P1.21-15,16, and were classified as ST-10217 by MLST, except for a single isolate (SK99, from Nigeria, 2017), which had a variant abcZ allele and was thus classified as ST-11561. All outbreak isolates were serogroup C, except isolate Niga32_16, which was phenotypically nonserogroupable although it carried the gene cluster for serogroup C. The outbreak isolates represented a single monophyletic group with a high level of clonality: the outbreak-wide average pairwise allele distance (by cgMLST v1.0) was 54 allele differences out of 1,605 loci; that is, two randomly chosen isolates had identical allele profiles over 96.6% of loci. Within this cluster, two main subclusters could be found (Fig. 1): one chiefly containing isolates from Nigeria from 2013 to 2017 and the other one with isolates almost exclusively from Niger in 2015.
Fig. 1.
Minimum spanning tree of BL16188 and all sequenced outbreak isolates. Node colors correspond to region of origin, and edge pattern corresponds to sample year. Node size corresponds to the number of isolates with identical allelic profiles. Scale is in number of allelic differences in the cgMLST analysis.
Core genome MLST analyses comparing the outbreak isolates with the rare NmC isolates available from Africa revealed that the ST-10217 outbreak strain was distantly related to other invasive NmC strains, but very closely related to a noninvasive, nonserogroupable isolate from a carrier study in Burkina Faso. (SI Appendix, Fig. S1). The closest isolate to the outbreak strain was inferred to be BL16188, obtained from an asymptomatic carrier in Burkina Faso in 2012 (19). BL16188 had the same rare PorA type (P1.21-15,16) as the outbreak isolates and differed from those on average at only 130.8 alleles (i.e., identical at 91.8% of loci). This isolate was ST-9367, which differs from ST-10217 by a single nucleotide at position 25 of the fumC locus.
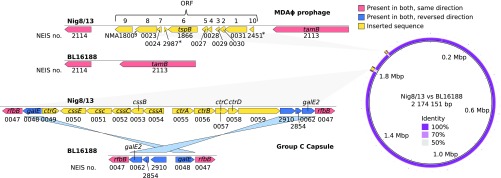
In-depth genome analyses, using both Illumina and Oxford Nanopore sequencing, revealed that BL16188 and the outbreak isolates were nearly identical, with the striking exception of two genomic islands present in the outbreak strain (Fig. 2). These were (i) a full serogroup C capsule locus, flanked by inverted galE/galE2 sequences, and (ii) an 8-kb prophage previously associated with hyperinvasiveness, termed Meningococcal Disease-Associated island (MDAΦ) (20–22), flanked by translocation/assembly module tamB (NEIS2113/NMA0295) and a peptide ABC transporter (NEIS2114/NMA0222). A characterization of these elements is shown in Table 1. A comparison of the MDAΦ phage to that of FAM18 (23), Z2491 (24), and MC58 (25) is shown in SI Appendix, Fig. S2 and Table S2. Both elements were found in all outbreak isolates. MDAΦ was identical in all outbreak isolates, except in the NEIS0023 locus, where the isolates in the Niger subcluster had allele 322, whereas the isolates in the Nigeria subcluster had allele 22. (This corresponds to a T/G polymorphism at position 373 of the reading frame, leading to a V124F mutation.)
Fig. 2.
(Left) Genomic architecture of the two putatively horizontally acquired virulence factors determined to be the major difference between carrier strain BL16188 and the outbreak strain. Acquired genes are colored yellow, and flanking regions which have been reversed are colored in blue. Identical regions are colored red. ORFs marked with an asterisk were called in PubMLST but not when using a separate gene caller. The gene with the section symbol was called by a separate gene caller but not in PubMLST. It has therefore been annotated with the corresponding NMA number rather than NEIS. ORF numbering is accordance with ref. 20. A description of these genes can be found in Table 1. NEIS2987 has not previously been described as part of the phage and is in the opposite direction of the other reading frames, and it might not represent an expressed gene. (Right) BRIG atlas showing the position in the chromosome where the two elements have been inserted. Nig8/13 is the reference genome, and BL16188 the comparison. Apart from these two inserts, only minor allele differences were observed between the outbreak and carrier strains. A near-100% nucleotide identity is seen elsewhere along the chromosome.
Table 1.
Annotation of horizontally acquired genes
| Context | NEIS no. | Gene | Product |
| Capsule | NEIS0049 | ctrG | Unknown |
| NEIS0050 | cssE | O-acetyl transferase | |
| NEIS0051 | csc | Polysialyltransferase | |
| NEIS0052 | cssC | N-acetylneuraminic acid synthetase | |
| NEIS0053 | cssB | CMP-N-acetylneuraminic acid synthase | |
| NEIS0054 | cssA | N-acetylglucosamine-6-P 2-epimerase | |
| NEIS0055 | ctrA | Capsule polysaccharide export outer membrane protein | |
| NEIS0056 | ctrB | Capsule polysaccharide export inner membrane protein | |
| NEIS0057 | ctrC | Capsule polysaccharide export inner membrane protein | |
| NEIS0058 | ctrD | Capsule polysaccharide export ATP-binding protein | |
| NEIS0059 | N/A | Putative transcriptional accessory protein | |
| Phage | NEIS2451 | orf10 | Hypothetical protein |
| NEIS0031 | orf1 | Putative phage replication initiation factor | |
| NEIS0030 | orf2 | Conserved hypothetical protein | |
| NEIS0029 | orf3 | Hypothetical protein | |
| NEIS0028 | orf4 | Major capsid protein | |
| NEIS0027 | orf5 | Hypothetical integral membrane protein | |
| NEIS1866 | orf6 | T- and B-cell stimulating protein B (tspB) | |
| NEIS2987 | orf7 | Hypothetical integral membrane protein | |
| NEIS0024 | orf8 | Conserved hypothetical integral membrane protein | |
| NEIS0023 | orf9 | Zonular occludens toxin-like protein / transposase |
N/A, not applicable.
As for the capsule, NEIS0052 (cssC) and NEIS0058 (ctrD) were identical in all strains, whereas the remaining loci contained minor differences. NEIS0050 (cssE), NEIS0051 (csc), and NEIS0053 (cssB) were assumed to be identical in all isolates. These three loci were incomplete in four, one, and one strains, respectively, but no polymorphism was seen in isolates with complete loci. NEIS0049 (ctrG), NEIS0055 (ctrA), and NEIS0059 each had a single synonymous SNP, at positions 286, 1089, and 702, respectively. NEIS0056 (ctrB) and NEIS0057 (ctrC) each had a single nonsynonymous SNP, at positions 1081 and 421, respectively. NEIS0054 (cssA) was the most variable locus with nonsynonymous SNPs at five positions: 28, 221, 233, 871, and 910. Thus, the SNP density across the capsule was 0.83 SNPs/kB, compared with 4.18 SNPs/kB across the entire core genome, or 1.84 SNPs/kB after removing putatively recombined regions of the alignment.
Apart from the capsule and the MDAΦ insertions, 11 recombination events were identified on the branch between the carrier and the outbreak strain, affecting a total of 35 genes (SI Appendix, Fig. S3 and Table S3). Two of the recombination events affected genes involved in iron acquisition: one affected the hypervariable tbpB, encoding transferrin-binding protein 1, and the other affected hmbR and hemO, which are involved in haem acquisition and utilization. The other recombination events appear to only have affected genes involved in housekeeping functions, particularly DNA replication and repair, as well as hypothetical genes. We also identified 13 instances of recombination on internal branches of the ST-10217 subtree (SI Appendix, Fig. S3 and Table S3). Unsurprisingly, tbpB appeared as a hot spot for recombination, with three additional events affecting this gene. One event affected lst, which is involved in sialylation of lipooligosaccharides. A particular branch of isolates from 2016 and 2017 had two independent recombination events affecting the gene penA, which encodes penicillin-binding protein 2. The effect, if any, of these recombination events is unclear.
Several lines of evidence strongly suggest that a noninvasive isolate, such as BL16188, an ancestor of ST-10217, acquired the capsule locus and the MDA phage: (i) the rarity of NmC in the meningitis belt in the past decades, (ii) the isolation of BL16188 a year before the first report of the first ST-10217 outbreak, and (iii) the basal phylogenetic position of BL16188 relative to the outbreak isolates (SI Appendix, Figs. S1, S3, and S4). Taken together, these data suggest that the capsule and the MDA phage were horizontally acquired by an ST-10217 ancestor. Abrupt expansion of this successful, highly transmissible clone resulted in massive outbreaks.
To estimate the time of acquisition of the capsule and the prophage, root-to-tip regressions were performed on a whole-genome SNP-based alignment. Because both the capsule locus and MDAΦ were absent from BL16188 but common to all outbreak isolates, the uptake of both elements likely occurred after the time of the most recent ancestor (TMRCA) for the whole group but before the TMRCA of the ST-10217 outbreak strain. This resulted in a likely temporal window of acquisition spanning from early 2007 to mid-2011 (SI Appendix, Fig. S4).
Origin of Acquired Virulence Genes.
An important question then is the origin of these two elements. By core genome MLST, ST-10217 showed some similarity to other African NmC (SI Appendix, Fig. S1). However, because the capsule locus was horizontally acquired, we created a separate phylogenetic tree for this locus alone (SI Appendix, Fig. S5). This revealed that the capsule locus of ST-10217 was actually more similar to that of ST-11 strains recovered from Europe and North America. A high identity could be seen in all ORFs except for NEIS0059, which has unknown function. For example, the entire capsule locus of Nig8/13 had only two allelic differences relative to M04_241156 (Neisseria PubMLST id: 29589), an invasive UK isolate from 2012. The two allelic differences corresponded to 2 SNPs in NEIS0057 (ctrC) and 54 in the more variable NEIS0059. In contrast, capsule loci in NmC isolates from Africa were more divergent relative to ST-10217. Among contemporary (after 2010) African isolates, the closest was the carrier isolate BF2880 from Burkina Faso, 2012, which had six allele differences across the capsule locus (NEIS0052, NEIS0053, NEIS0054, NEIS057, NEIS0058, and NEIS0059), corresponding to 55 single-nucleotide polymorphisms. The closest invasive African NmC was S13/98 (Neisseria PubMLST id: 38587), from Algeria, 1998, which had four allele differences from Nig8/13 across the capsule locus [NEIS0052 (cssC), NEIS0057 (ctrC), NEIS0058 (ctrD), and NEIS0059 (hypothetical gene), representing a total of 89 SNPs].
As for the MDAΦ island, there are currently more than 100 isolates with an identical allelic profile. The chronologically first identical match is the NM3683 isolate from Canada, 1970 (Neisseria PubMLST id: 31332). The MDAΦ prophage thus appears as too conserved to make meaningful predictions about potential donors to the ST-10217 clone.
Spatiotemporal Outbreak Dynamics.
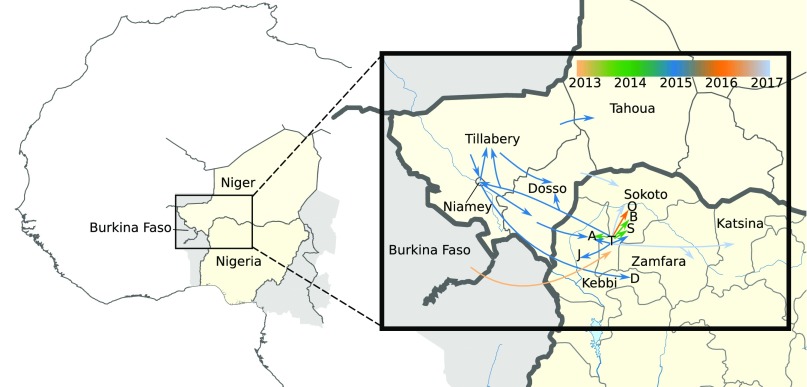
Extensive sampling across the full geographic and temporal range of the recent NmC outbreaks in Nigeria and Niger allowed a detailed reconstruction of the major chains of events driving these large outbreaks (Fig. 3). At some point between 2007 and 2011, a West African unencapsulated commensal strain became invasive after horizontally acquiring a group C capsule and an MDAΦ prophage. An outbreak was first reported from Tambuwal, a local government area in the state of Sokoto, northern Nigeria, in 2013. In 2014, new cases emerged in the nearby areas of Bodinga and Shagari, as well as in Aliero, which is located in the neighboring state Kebbi. Cases continued to occur in these areas in 2015 and in Jega, Kebbi state. Notably, new cases were more closely related to the original Tambuwal isolates than to local strains from the previous year, which could suggest effective handling of the disease in these sink locations, but with a steady stream of new cases coming from an uncontrolled reservoir around Tambuwal. Notably, in 2015 a secondary outbreak occurred in Niger, located immediately to the north of Sokoto state. The expansion of this strain led to massive outbreaks in the Niger states of Dosso and Tillabery and in the capital, Niamey. This strain subsequently spread to the state of Tahou and was also transmitted back across the border to Nigeria, with cases emerging in Aliero and Danko-Wasagu, both in Kebbi state. In 2016, new cases appeared in Sokoto state, and again these cases were most closely related to the original Tambuwal isolates. This pattern continued in 2017, with the majority of new cases in Sokoto being more closely related to Tambuwal–2013 than to Sokoto–2016. However, some new cases in Sokoto appeared to be of the Niger strain, suggesting some lingering reservoirs. Also in 2017, the Tambuwal strain spread to the eastern neighbor states of Zamfara and Katsina. The Niger strain, on the other hand, did not reestablish itself in Nigeria, with most new cases being of the Tambuwal type. By 2017, the geographical range of the outbreak appeared to be increasing.
Fig. 3.
(Left) Location of Niger and Nigeria within western Africa. (Right) Inferred spatiotemporal flow of the outbreak based on all sequenced data. Arrows are colored according to the inferred year of spread. Zoom coordinates: 10.5 −16.5 N, 0.0 − 8.8 E.
Discussion
This study illustrates the power of combining long-read and short-read technology of whole-genome sequencing for high-resolution epidemiological investigations. We sequenced the genomes of NmC isolates spanning the entire course of the outbreak across its full geographic range, which enabled us to reconstruct the evolution and expansion of a novel outbreak strain in unparalleled detail. Long-read sequencing allowed us to close and complete the genomes of both an early outbreak isolate and a benign strain closely related to the original recipient of the horizontally acquired virulence factors. We could thus fully characterize the horizontally acquired virulence genes and determine the exact location of their integration in the genome of the outbreak strain. When coupled with detailed epidemiological data, our genomic and phylogeographic analyses weave an unusually clear story of cause–effect relationships with respect to how a commensal becomes a pathogen by means of the stepwise acquisition of virulence factors.
Establishing causal relationships between pathogen genetic factors, virulence, and disease outbreaks is a complex task because pathogenic potential often relies on multiple genes (26) and even gene loss (27). An earlier rare example of a major outbreak driven by the introduction of a virulence gene in a novel genetic background is the 2011 outbreak of Escherichia coli O104:H4 in Germany, where strains had acquired the stx gene (28). A more indirect chain of events is exemplified by a highly unusual outbreak of tuberculosis caused by Mycobacterium bovis, which is generally attenuated in humans. In this case, comparative genome analyses revealed the presence of an insertion sequence immediately upstream of the phoPR two-component system in the outbreak strain, resulting in the up-regulation of the entire phoPR regulon, including a number of virulence genes (29). The access to closed and complete genome sequences from both a virulent outbreak strain and a closely related benign carrier strain, in addition to a large collection of genome sequences from an outbreak expanding rapidly in space, makes the current study unusual.
Speculation about the origins of the ST-10217 clone has centered around its possible relation to serogroup A, the serogroup most commonly involved in invasive disease in the region before the introduction of MenAfriVac. It is well known that hypervirulent clones can adapt to interventions or changing environment through capsule switches (30, 31). For example, serogroup W ST-2881, another invasive West African clone, appears to have evolved through capsule switching from serogroup Y ST-2881, also an invasive clone (32). However, we show here that ST-10217 did not emerge from invasive serogroup A but rather that this new, hypervirulent strain developed from a carrier strain through a limited number of recombination events. In N. meningitidis it is thought that many different genes are contributing to virulence and are necessary to development of invasive disease (33). Studies comparing the gene diversity in carriage and invasive strains that are genetically and epidemiologically closely related identified multiple differences in their genomes (34). Our results clearly showed that in fact, a previously unremarkable carrier strain circulating in the region acquired a serogroup C capsule and the filamentous bacteriophage MDAΦ, before the introduction of conjugate vaccine MenAfriVac. The capsule is an important virulence factor required by the bacteria to survive in the bloodstream (35). MDAΦ is overrepresented in disease isolates from young adults, and it has been suggested that it may significantly contribute to disease (20), possibly by allowing increased colonization on epithelial cells in the nasopharynx, which in turn leads to a higher bacterial load at the site of entry (22). In ORF6 it encodes TspB, a protein mediating Ig-binding, which may provide protection against immune response (36) and which allows adsorption through type IV pili (37). Except for these two horizontally transferred genetic elements, 11 recombination events, affecting 35 genes, were identified between the carrier and the outbreak strain. Remarkably, paired isolates collected 2 mo apart from the same asymptomatic carriers were found to differ from each other to the same extent, on average at 35 loci (38). The most notable of these 11 recombination events affected genes involved in iron acquisition and utilization which might have a role in invasiveness (39, 40). Other changes were predominantly in genes with housekeeping functions or hypothetical genes and were not deemed as important to the hyperinvasive phenotype of ST-10217.
The present study also underlines the importance of a prompt and determined response in the face of major public health threats. In 2015, the ST-10217 outbreak spread to Niger where it created a severe epidemic with ∼8,500 cases (16). By 2016 the outbreak appeared to be declining, with the WHO reporting ∼2,500 new cases in Niger and 1,000 in Nigeria (www.who.int/mediacentre/infographic/meningitis/meningitis-2018-2019.jpg). In 2015 in Niger, the WHO and its partners made available antibiotics for treatment of the patients, and massive vaccination campaigns were readily undertaken in the districts in epidemic. The response to the Nigerian outbreak, however, appeared to have been more fragmented and delayed and thus failed to prevent subsequent clonal outbreaks in 2016 and 2017, particularly in the Katsina and Zamfara states (17). In fact, the largest NmC outbreak ever reported in Africa is currently ongoing in Nigeria, with close to 16,000 suspected cases in 2017 and, at the start of this year’s meningitis season, cases are already reported in Nigeria and Niger (41). Prospects for the near future are grim, with a worst-case scenario from the WHO estimating up to 70,000 new cases in the region over the course of the next 2 y (2018–2019) (www.who.int/csr/disease/meningococcal/meningitis-c-epidemic-risk/en/). Clearly, a highly concerted and timely response, including vaccinations, ardent surveillance, and rapid medical care for those affected, is required to stamp out this as well as future outbreaks of N. meningitidis across Africa and beyond. The acute shortage of meningococcal serogroup C vaccine highlights the importance of accelerating the development of a multivalent conjugate vaccine affordable for the people and governments of Africa.
Materials and Methods
Bacterial Strains and Culture Conditions.
The invasive NmC isolates from 2013 to 2017 analyzed here were obtained from Médecins Sans Frontières in Niger and Nigeria and from the International Foundation Against Infectious Disease in Nigeria (SI Appendix, Table S1). For the years 2013–2015, cerebrospinal fluid from suspected meningitis patients in the outbreak areas in Nigeria was collected in Trans-Isolate media and sent to the WHO Collaborating Centre for Reference and Research on Meningococci, Oslo (WHO-CC, Oslo), for identification of the causative agent. The 2015 isolates from Niger and a large part of the 2016–2017 isolates from Nigeria were cultured in Niamey and Abuja before being sent to Oslo in various transport media. The carriage isolates were from a study performed in Burkina Faso in 2012 (19). NmC control isolates were from the collection of the WHO-CC, Oslo. Isolates were grown overnight on blood agar plates at 37 °C in an atmosphere of 5% CO2. The study was approved by the responsible scientific director at Norwegian Institute of Public Health, Oslo, and the WHO Collaborating Centre for Reference and Research on Meningococci, Oslo.
Illumina Sequencing.
Genomic DNA was extracted using an automated MagNAPure isolation station and MagNAPure 96 DNA and Viral NA Small Volume Kit (Roche), according to the manufacturer’s instructions. The sequencing libraries were made with KAPA HyperPlus Kit (KAPA Biosystems); sequencing was performed on the Illumina MiSeq platform with MiSeq Reagent Kits v2 500-cycles (Illumina Inc.) with 250 bp paired-end run modes; and the reads were subsequently trimmed, filtered, and assembled as previously described (40).
Oxford Nanopore Sequencing.
Long-read sequencing was performed on one encapsulated ST-10217 strain from 2013 (Nig8/13) and on the noncapsulated carrier strain from Burkina Faso with the same PorA type (BL16188). High-molecular weight DNA was extracted from cultured bacteria using the genomic-tip 20/G kit and buffer set (Qiagen). Sequencing libraries were prepared employing the rapid barcoding kit (SQK-RBK001) and 1D reads generated on the MinION platform using the R9.4 (FLO-MIN106) flow cell. The reads were demultiplexed and basecalled with Albacore version 2.1.2. Porechop version 0.2.3 (https://github.com/rrwick/Porechop) was used to remove adapters and split chimeric reads before assembly. This was done directly from Albacore’s fast5 output. For assembly we performed a hybrid approach with both MinION and Illumina reads using Unicycler version 0.4.3. The “normal” bridging mode was used. This resulted in a completely resolved circular genome of size 2,144,773 (carrier strain BL16188) and 2,174,151 (recipient strain Nig8/13). Both contigs were rotated to start with the dnaA (Uniprot Q9JXS7) gene.
Availability of Data.
Short reads were uploaded to the European Nucleotide Archive and are available under accession number PRJEB24294. Additionally, assembled genomes were uploaded to and are available from the PubMLST.org database (pubmlst.org/neisseria/) (SI Appendix, Table S1), which is served by the BIGS Database platform (42).
Genome Comparison and Phylogenetic Analyses.
Whole-genome alignments of all strains were created with the tool parsnp version 1.2 (43), using the closed genome of Nig8/13 as the reference. MUMi distance was set to be ignored, and PhiPack filtering (44) was enabled to remove SNPs identified in regions of recombination. We subsequently used Harvesttools (43) to extract variable sites (SNPs) only. The SNP alignment was used to build a maximum likelihood phylogenetic tree using the program IQ-tree version 1.5.5 (45). The BL16188 isolate was set to be the outgroup. Eighty-eight different DNA evolution models were tested, and the optimal [TVMe without gamma distributed rates; 96.8% weighted Bayesian information criterion (BIC) support] was chosen based on the BIC (46). Bootstrap iterations (n = 1,000) were performed, and bootstrap support was added to the final phylogenetic tree. The allele-based cgMSLT tree (SI Appendix, Fig. S1) was created using the Neisseria meningitidis cgMLST v1.0 scheme from PubMLST, and the minimum spanning tree was drawn using GrapeTree (47). The tree based on the capsule loci was created in a similar manner, but instead of the full cgMLST scheme, only sequences corresponding to NEIS0049 through NEIS0059 were used. The phage and capsule annotations were completed in a two-step procedure. In addition to annotation from PubMLST, they were also annotated using Prokka version 1.12 (48), with the option to first look for homologous proteins from the FAM18/Z2491/MC58 genomes.
Analyzing Recombination Blocks.
To investigate recombination hot spots, we ran Gubbins version 2.3.1 (49) on the full alignment of concatenated loci from PubMLST’s Neisseria meningitidis cgMLST v1.0 scheme, using BL16188 as an outgroup and the SNP-based tree described above as the starting tree. Putative recombination hot spots were visualized with Phandango (50), and the Gubbins tree was used for phylogenetic relationships. A recombination block was only reported if it occurred on an internal branch. Prokka version 1.12 (48) was used for annotation.
Dating the Acquisition of the Virulence Factors.
The time of the most recent common ancestor of all isolates was estimated by performing a simple regression of the root-to-tip distance in the ML SNP tree against each isolate’s date using Path-O-Gen (tree.bio.ed.ac.uk/software/pathogen). There was a clear correlation between root-to-tip genetic distance and sampling time, with a coefficient of determination (R2) of 0.43 (including the outbreak isolates and BL16188) and 0.23 (including only outbreak isolates).
Characterization and Visualization of Acquired Elements.
Annotation of ORFs on the putatively acquired elements (capsule locus and phage) was carried out by manually querying each predicted ORF against the Neisseria PubMLST database (https://pubmlst.org/neisseria/). The identification of the closest capsule locus relatives was performed by allele querying with mismatches allowed in PubMLST. Isolates with eight or more allelic matches to NEIS0049–NEIS0059 were included. Also, all other African NmC available in PubMLST were included (SI Appendix, Table S4). SI Appendix, Fig. S5, was drawn with GrapeTree (47). Global visual comparisons between BL16188 and Nig8/13 (Fig. 2) was created with BLAST Ring Image Generator (BRIG) version 0.95 (51). Detailed local visual comparisons between BL16188 and Nig8/13 were done with Artemis Comparison Tool version 16.0.11 (52). SI Appendix, Fig. S2, was created using EasyFig (53).
Supplementary Material
Acknowledgments
We are very grateful to Anne Witsø, Ingerid Ørjansen Kirkeleite, and Nadia Debech (all Norwegian Institute of Public Health) and Charity Kamau (Médecins sans Frontières laboratory advisor) for invaluable technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.P. is a guest editor invited by the Editorial Board.
Data deposition: All whole-genome sequencing (WGS) reads were uploaded to the European Nucleotide Archive (accession no. PRJEB24294).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802298115/-/DCSupplemental.
References
- 1.Brandtzaeg P, van Deuren M. Classification and pathogenesis of meningococcal infections. Methods Mol Biol. 2012;799:21–35. doi: 10.1007/978-1-61779-346-2_2. [DOI] [PubMed] [Google Scholar]
- 2.Yazdankhah SP, Caugant DA. Neisseria meningitidis: An overview of the carriage state. J Med Microbiol. 2004;53:821–832. doi: 10.1099/jmm.0.45529-0. [DOI] [PubMed] [Google Scholar]
- 3.Stephens DS. Conquering the meningococcus. FEMS Microbiol Rev. 2007;31:3–14. doi: 10.1111/j.1574-6976.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- 4.Harrison OB, et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis. 2013;19:566–573. doi: 10.3201/eid1904.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizza M, Rappuoli R. Neisseria meningitidis: Pathogenesis and immunity. Curr Opin Microbiol. 2015;23:68–72. doi: 10.1016/j.mib.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Greenwood B. Manson lecture. Meningococcal meningitis in Africa. Trans R Soc Trop Med Hyg. 1999;93:341–353. doi: 10.1016/s0035-9203(99)90106-2. [DOI] [PubMed] [Google Scholar]
- 7.Boisier P, et al. Meningococcal meningitis: Unprecedented incidence of serogroup X-related cases in 2006 in Niger. Clin Infect Dis. 2007;44:657–663. doi: 10.1086/511646. [DOI] [PubMed] [Google Scholar]
- 8.Nathan N, et al. Meningitis serogroup W135 outbreak, Burkina Faso, 2002. Emerg Infect Dis. 2007;13:920–923. doi: 10.3201/eid1306.060940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delrieu I, et al. Emergence of epidemic Neisseria meningitidis serogroup X meningitis in Togo and Burkina Faso. PLoS One. 2011;6:e19513. doi: 10.1371/journal.pone.0019513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diomandé FV, et al. Public health impact after the introduction of PsA-TT: The first 4 years. Clin Infect Dis. 2015;61:S467–S472. doi: 10.1093/cid/civ499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotter CL, et al. Impact of MenAfriVac in nine countries of the African meningitis belt, 2010-15: An analysis of surveillance data. Lancet Infect Dis. 2017;17:867–872. doi: 10.1016/S1473-3099(17)30301-8. [DOI] [PubMed] [Google Scholar]
- 12.Funk A, et al. Sequential outbreaks due to a new strain of Neisseria meningitidis serogroup C in northern Nigeria, 2013-14. PLoS Curr. 2014;6:ecurrents.outbreaks.b50c2aaf1032b3ccade0fca0b63ee518. doi: 10.1371/currents.outbreaks.b50c2aaf1032b3ccade0fca0b63ee518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broome CV, et al. Epidemic group C meningococcal meningitis in Upper Volta, 1979. Bull World Health Organ. 1983;61:325–330. [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammed I, Iliyasu G, Habib AG. Emergence and control of epidemic meningococcal meningitis in sub-Saharan Africa. Pathog Glob Health. 2017;111:1–6. doi: 10.1080/20477724.2016.1274068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow J, et al. Invasive meningococcal meningitis serogroup C outbreak in northwest Nigeria, 2015-third consecutive outbreak of a new strain. PLoS Curr. 2016;8:ecurrents.outbreaks.06d10b6b4e690917d8b0a04268906143. doi: 10.1371/currents.outbreaks.06d10b6b4e690917d8b0a04268906143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidikou F, et al. MenAfriNet consortium Emergence of epidemic Neisseria meningitidis serogroup C in Niger, 2015: An analysis of national surveillance data. Lancet Infect Dis. 2016;16:1288–1294. doi: 10.1016/S1473-3099(16)30253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nnadi C, et al. Large outbreak of Neisseria meningitidis serogroup C–Nigeria, December 2016-June 2017. MMWR Morb Mortal Wkly Rep. 2017;66:1352–1356. doi: 10.15585/mmwr.mm6649a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiden MC, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristiansen PA, et al. Persistent low carriage of serogroup A Neisseria meningitidis two years after mass vaccination with the meningococcal conjugate vaccine, MenAfriVac. BMC Infect Dis. 2014;14:663. doi: 10.1186/s12879-014-0663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bille E, et al. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med. 2005;201:1905–1913. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bille E, et al. Association of a bacteriophage with meningococcal disease in young adults. PLoS One. 2008;3:e3885. doi: 10.1371/journal.pone.0003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bille E, et al. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. 2017;13:e1006495. doi: 10.1371/journal.ppat.1006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentley SD, et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 2007;3:e23. doi: 10.1371/journal.pgen.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhill J, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 25.Tettelin H, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 26.Willemse N, et al. An emerging zoonotic clone in The Netherlands provides clues to virulence and zoonotic potential of Streptococcus suis. Sci Rep. 2016;6:28984. doi: 10.1038/srep28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson JS, Giogha C, Wong Fok Lung T, Hartland EL. The genetics of enteropathogenic Escherichia coli virulence. Annu Rev Genet. 2016;50:493–513. doi: 10.1146/annurev-genet-120215-035138. [DOI] [PubMed] [Google Scholar]
- 28.Frank C, et al. HUS Investigation Team Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalo-Asensio J, et al. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. Proc Natl Acad Sci USA. 2014;111:11491–11496. doi: 10.1073/pnas.1406693111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel U, Claus H, Frosch M. Rapid serogroup switching in Neisseria meningitidis. N Engl J Med. 2000;342:219–220. doi: 10.1056/NEJM200001203420319. [DOI] [PubMed] [Google Scholar]
- 31.Mustapha MM, et al. Genomic investigation reveals highly conserved, mosaic, recombination events associated with capsular switching among invasive Neisseria meningitidis serogroup W sequence type (ST)-11 strains. Genome Biol Evol. 2016;8:2065–2075. doi: 10.1093/gbe/evw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamelas A, et al. Emergence and genomic diversification of a virulent serogroup W:ST-2881(CC175) Neisseria meningitidis clone in the African meningitis belt. Microb Genom. 2017;3:e000120. doi: 10.1099/mgen.0.000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph B, et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS One. 2011;6:e18441. doi: 10.1371/journal.pone.0018441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren X, et al. Genomic, transcriptomic, and phenotypic analyses of Neisseria meningitidis isolates from disease patients and their household contacts. mSystems. 2017;2:e00127-17. doi: 10.1128/mSystems.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med. 2001;344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 36.Müller MG, Ing JY, Cheng MKW, Flitter BA, Moe GR. Identification of a phage-encoded Ig-binding protein from invasive Neisseria meningitidis. J Immunol. 2013;191:3287–3296. doi: 10.4049/jimmunol.1301153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer J, et al. Characterization of MDAΦ, a temperate filamentous bacteriophage of Neisseria meningitidis. Microbiology. 2016;162:268–282. doi: 10.1099/mic.0.000215. [DOI] [PubMed] [Google Scholar]
- 38.Bårnes GK, et al. Whole genome sequencing reveals within-host genetic changes in paired meningococcal carriage isolates from Ethiopia. BMC Genomics. 2017;18:407. doi: 10.1186/s12864-017-3806-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison OB, Bennett JS, Derrick JP, Maiden MC, Bayliss CD. Distribution and diversity of the haemoglobin-haptoglobin iron-acquisition systems in pathogenic and non-pathogenic Neisseria. Microbiology. 2013;159:1920–1930. doi: 10.1099/mic.0.068874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agnememel A, et al. Neisseria meningitidis serogroup X in sub-Saharan Africa. Emerg Infect Dis. 2016;22:698–702. doi: 10.3201/eid2204.150653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.WHO 2018. Meningitis Weekly Bulletin Week 2 (Inter Country Support Team - West Africa/WHO, Ouagadougou, Burkina Faso)
- 42.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruen T, Bruen T. PhiPack: PHI Test and Other Tests of Recombination. McGill University; Montreal: 2005. [Google Scholar]
- 45.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, et al. 2017. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. bioRxiv:10.1101/216788.
- 48.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadfield J, et al. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics. 2017;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver TJ, et al. ACT: The Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.