Significance
The heterodimeric cohesin SMC complex embraces duplex DNA and is associated with Rad21, which is cleaved in mitotic anaphase by a protease called separase/Cut1. Upon Rad21 cleavage, chromosomal DNAs are released from cohesin and segregated. We identified extragenic suppressors for separase and cohesin temperature-sensitive (ts) mutants using whole-genome sequencing and made the surprising discovery that cleavage of Rad21 is largely dispensable if suppressor causes physical disorders of cohesin interfaces among essential subunits. The predicted disorders provide insights into a DNA “hold-and-release” model in which hinge and head of SMC subunits are proximal to form arched coiled coils that close or open by their orientation. The model is distinct from the “ring” model and may promote further study.
Keywords: separase, securin, cohesin, cohesin loader Mis4, suppressor screen
Abstract
Cohesin is a fundamental protein complex that holds sister chromatids together. Separase protease cleaves a cohesin subunit Rad21/SCC1, causing the release of cohesin from DNA to allow chromosome segregation. To understand the functional organization of cohesin, we employed next-generation whole-genome sequencing and identified numerous extragenic suppressors that overcome either inactive separase/Cut1 or defective cohesin in the fission yeast Schizosaccharomyces pombe. Unexpectedly, Cut1 is dispensable if suppressor mutations cause disorders of interfaces among essential cohesin subunits Psm1/SMC1, Psm3/SMC3, Rad21/SCC1, and Mis4/SCC2, the crystal structures of which suggest physical and functional impairment at the interfaces of Psm1/3 hinge, Psm1 head–Rad21, or Psm3 coiled coil–Rad21. Molecular-dynamics analysis indicates that the intermolecular β-sheets in the cohesin hinge of cut1 suppressor mutants remain intact, but a large mobility change occurs at the coiled coil bound to the hinge. In contrast, suppressors of rad21-K1 occur in either the head ATPase domains or the Psm3 coiled coil that interacts with Rad21. Suppressors of mis4-G1326E reside in the head of Psm3/1 or the intragenic domain of Mis4. These may restore the binding of cohesin to DNA. Evidence is provided that the head and hinge of SMC subunits are proximal, and that they coordinate to form arched coils that can hold or release DNA by altering the angles made by the arched coiled coils. By combining molecular modeling with suppressor sequence analysis, we propose a cohesin structure designated the “hold-and-release” model, which may be considered as an alternative to the prevailing “ring” model.
New mutant alleles that affect important cellular processes may be identified using suppressor screening followed by identification of mutations. Extragenic suppressor analysis is a powerful tool to identify mutations that compensate for phenotypes of prior mutations. Identification of suppressor mutations has been a technical obstacle, but next-generation sequencing, especially when using a genomic DNA mixture as a template (1), has greatly facilitated identification of alterations caused by single-nucleotide changes throughout the genome (1–3).
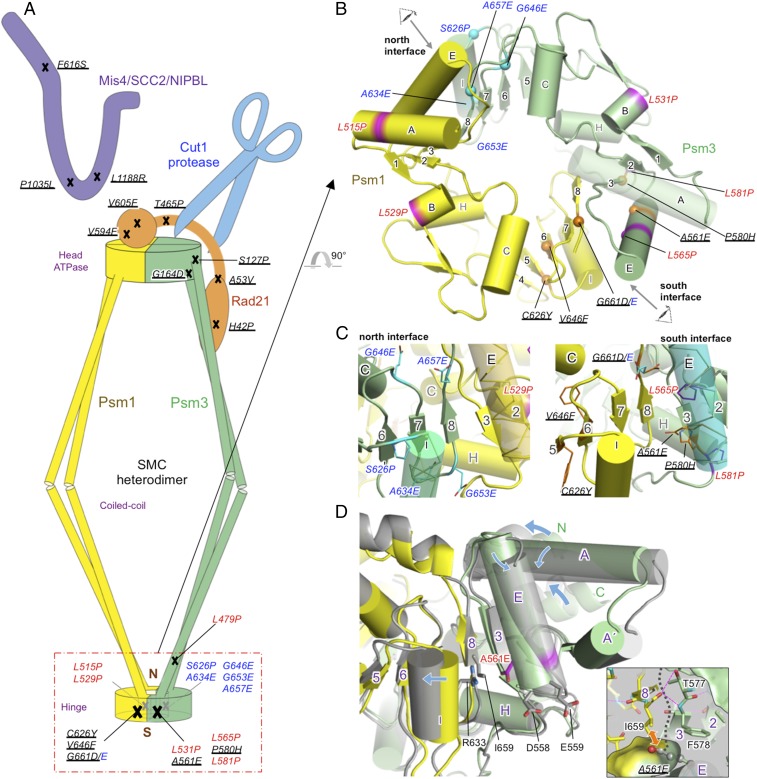
Cohesin forms a protein ring that topologically entraps DNA. It contains a heterodimer of Psm1/SMC1 and Psm3/SMC3, each of which comprises two head segments at the N and C termini, a hinge segment in the middle, and two 50-nm coiled coils linking the head and hinge segments (4, 5) (Fig. 1A). The cohesin head has ATPase activity (6). The hinge segments together form a doughnut-shaped structure with two (north and south) interfaces (7) (Fig. 1B). Rad21 associates with the Psm1 head and the Psm3 coiled coil adjacent to the head (8–10). Separase Cut1 is activated when securin Cut2 is ubiquitinated by the anaphase-promoting complex/cyclosome (APC/C) complex and degraded by the 26S proteasome (11, 12), and then Cut1 (illustrated as a pair of scissors) cleaves residues 179R and 231R of Rad21 (which bridges the head domains of Psm1 and Psm3) during the transition from mitotic metaphase to anaphase (13–15). The precise location of Mis4 in the cohesin complex is unknown, but cross-linking experiment suggested that its homolog Scc2 in Saccharomyces cerevisiae binds at or near the ATPase domains of the SMC dimer (16).
Fig. 1.
Destabilizing substitutions in cohesin subunits suppress cut1 and cut2. (A) Summary of 15 spontaneous suppressors in cohesin (underlined; SI Appendix, Table S1A), and other suppressors obtained by site-directed mutagenesis targeted to cohesin hinge (ts mutants, red; cs mutants, blue). Cut1 is represented as a pair of scissors. (B) Locations of the suppressors identified in the cohesin hinge. (C) Details of the hinge interfaces. Viewing directions are specified in B. (D) Structure at 100 ns in a MD simulation (Movie S1) of the hinge for Psm3-A561E suppressor, shows a change in orientation of the Psm3 coiled coil and preservation of the intermolecular β-sheet at the south interface. R633 (Psm1) and D558, E559 and E561 (Psm3) in hinge containing Psm3-A561E are shown with sticks. Blue arrows indicate structural changes arising from the A561E mutation. (Inset) A view of the south interface around A561 of Psm3, showing a steric clash between A561E (ball-and-stick) of Psm3 and I659 of Psm1. A black dotted line represents the boundary between Psm1 and Psm3, and magenta dashed lines represent hydrogen bonds. Yellow and green are used for wild type, and gray for the Psm3-A561E suppressor. β-Strands (1–8) and α-helices (A–H) are marked.
Here, we employed the comprehensive approach described in ref. 1 to isolate and analyze spontaneous extragenic suppressors for three classes of fission yeast temperature-sensitive (ts) or cold-sensitive (cs) mutants of the following: (i) caspase-like separase/separin Cut1 and its binding partner securin Cut2 (which acts as a chaperone and an inhibitor of Cut1) (11, 12, 17–21); (ii) Rad21, the target substrate of Cut1 (13–15); (iii) cohesin loader Mis4/Scc2/NIPBL, which is required for cohesin loading onto chromosomes in S phase (22, 23) [and its human homolog NIPBL is the causal gene of Cornelia de Lange syndrome (24, 25)]. When we employed molecular atomic modeling and dynamics analysis to analyze the suppressors identified, we were able to reveal a cohesin structural model designated the “hold-and-release” model, which may be presented as an alternative form of the prevailing “ring” model, explaining cohesin’s structure and genetic disorders, and suggesting the mechanism by which cohesin binds to and dissociates from DNA.
Results
Cut1 Suppressors Reside in Four Cohesin Subunits.
To identify suppressors that overcome inactive separase or securin, we reintegrated the ts mutation sites of cut1 and cut2 (refs. 13, 19, and 26; SI Appendix, Fig. S1A) into the “clean” wild-type genome, and obtained mutant phenotypes identical to those produced by the original cut1 and cut2 ts mutants (SI Appendix, Fig. S1 B and C). Various spontaneous revertants (frequencies, ∼10−6) produced at the semirestrictive or restrictive temperatures were then isolated and were subjected to determine suppressor mutation sites using an efficient next-generation whole-genome sequencing method, which employs genomic DNA mixtures (1). In total, we identified 15 extragenic suppressors in four cohesin-subunit genes: psm3, psm1, rad21, and mis4 (Fig. 1A and SI Appendix, Table S1A). Five of these mutations were identified in the hinge, two in the head, five in Rad21, and three in Mis4. No suppressor was located in other cohesin proteins, such as Wpl1 (WAPL), Pds5 (PDS5), or Psc3 (STAG1, 2, 3). This raised the question of how hinge mutations distant from Rad21-cleavage sites can compensate for inactive separase/Cut1.
Disruption of Cohesin Subunit Interfaces Alleviates the Cut1 Requirement.
Five spontaneous suppressors consisting of three psm1 and two psm3 mutations are located in the hinge south domain (Fig. 1A). Two psm3 mutations (S127P and G164D) are located in the head–coiled-coil junction region. They are close to the N terminus of Rad21, which contains another two suppressors (H42P and A53V). These mutations may perturb the interaction between the Psm3 coiled coil adjacent to head and the Rad21 N terminus, due to introduction of a helix-breaking proline or amino acids with larger side chains (G → D or A → V). Two other suppressors (V594F and V605F), located in the Rad21 C terminus, have similar substitution properties and should interfere with the proper interaction between the Rad21 C terminus and the Psm1 ATPase head. The precise location of the remaining T465P could not be determined, but its mutation (T → P) suggests the perturbation of helical structure. Thus, all these 12 suppressors weaken interfaces among cohesin subunits. Three remaining suppressors in Mis4 will be described below in light of the recently determined 3D structure of Scc2 (16, 27).
Hinge-Locating cut1 Suppressors Destabilize the Hinge Heterodimer Interface.
To examine the precise locations and natures of the five suppressor mutations in the hinge (psm3-A561E, -P580H, psm1-C626Y, -V646F, and -G661D), we constructed an atomic model for the cohesin hinge of Schizosaccharomyces pombe (Fig. 1B), adapted from the crystal structure determined for the mouse cohesin hinge (28). Psm1 (yellow) and Psm3 (green) are related by a pseudotwofold symmetry in the hinge, and its dimeric structure is stabilized by an intermolecular β-sheet and by salt bridges or hydrogen bonds between helices E and I at both the north and south interfaces (Fig. 1 B and C and SI Appendix, Fig. S2A). In addition, these helices stabilize the intermolecular β-sheet by hydrophobic interactions. Interestingly, all of the five separase/securin suppressor mutations are clustered at or near the south interface in the hinge and involve substitutions with bulkier residues. These would weaken the interface directly (A561E, P580H, and G661D) or indirectly (V646F and C626Y). Hence, impairment of the hinge interface seems to be key for suppression.
Three of the five suppressor sites are directly related to the hinge dimeric interaction. P580 of Psm3, facing helix E, is located on β3 (Fig. 1C), which forms an intermolecular β-sheet with β8 of Psm1 and makes van der Waals contacts with I659 on β8 (Fig. 1D). G661 of Psm1 is located at the side of β8, facing helix E of Psm3, and it contacts Y569 on helix E. A561 of Psm3 is located at the side of helix E, facing β3, and it contacts I659 on β8. Thus, replacements of these residues with bulkier ones (His, Asp, and Glu, respectively) would cause steric conflict at the south interface. The other two suppressor sites, C626 and V646 of Psm1, are located on β5 and β6, respectively, and are integrated into the intermolecular β-sheet. Their Tyr or Phe substitutions would also impair the south interface (Fig. 1 B and C). Such interference appears to significantly destabilize the hinge dimer interface, possibly resulting in hinge disorder.
To investigate structural changes in the hinge caused by the suppressing mutations, the mutations were introduced into the atomic model derived from the crystal structure of the mouse cohesin hinge [Protein Data Bank (PDB) ID code 2WD5] and all-atom molecular-dynamics (MD) simulations were performed for 100 ns. Interestingly, the intermolecular β-sheets at the south and north interfaces were preserved in all simulations, showing that full opening of the interfaces could not readily occur. However, some of the suppressors showed changes in orientation of the coiled coils caused by a small opening of the interface between helices E and I (Fig. 1D). In addition, normal mode analysis of the atomic models after MD simulations exhibited collision between helix E and helix A (which connected to the coiled coil), and then the collision produces large movements of helix A′, resulting in changes in orientation of the coiled coil (Movie S1). These observations suggest that increased movement of the coiled coils may play an important role in compensating for the cut1 and cut2 mutations.
Other Hinge-Locating Suppressors All Destabilize the Interface.
If such movement of the coiled coils suppresses the cut1 and cut2 mutations, destabilization of the north interface could conceivably also compensate for them, as its architecture is very similar to that of the south interface. Both hinge interfaces may be involved in stable association of cohesin with chromosomes (29). To examine this idea, we performed site-directed mutagenesis targeted to cohesin hinge regions. By introducing leucine (L) or serine (S) → proline (P), or glycine (G) or alanine (A) → glutamic acid (E), we isolated 11 ts or cs mutants with single-amino acid substitutions in the cohesin hinge and 1 ts mutant with a Psm3 L479P mutation at the hinge–coiled coil junction from 59 such single-amino acid substitutions selected (6 ts and 6 cs; Materials and Methods, Fig. 1 A and B, and SI Appendix, Fig. S3A). Double-mutant analysis indicated that all of these 12 mutants were able to rescue cut1-A1816T (SI Appendix, Fig. S3 B and C). Interestingly, all of the six ts mutants contain L → P helix-breaking substitutions. Five of the six cs mutants contained substitutions from small (G or A) to larger (E) substitutions. The other cs mutant S626P contained a S → P substitution so that physical constraints inevitably occurred in all of these cs mutants. Seven mutations were identified at or near the north interface and four at or near the south interface (Fig. 1 B and C). Taken together, in total, 16 hinge mutants with mutations in either the south or north part of the hinge were all able to suppress separase/cut1 mutants. Furthermore, all-atom MD simulations showed large movements of the coiled coils, while keeping the intermolecular β-sheet intact. Thus, disorder of the north or south interface is an effective way to suppress the cut1 and cut2 protease complex mutations by increasing the movement of the coiled coils.
The Mode of Suppression by psm3-A561E.
To investigate how suppression occurred, the single-suppressor segregant of the hinge cs mutant psm3-A561E was studied. cs psm3-A561E rescued the temperature sensitivity of cut1-L739S, -A1816T, cut2-R267Stop, and cut2-EA2 (SI Appendix, Fig. S4 A and B). Functionality of the A561 may require small neutral side chains, such as amino acids A, S, or G, which are found in other organisms (SI Appendix, Fig. S4C). The premature mitotic chromosome segregation phenotype of cs psm3-A561E at 20 °C (SI Appendix, Fig. S4D) resembles that of cohesin ts rad21-K1 at 36 °C (30). psm3-A561E is hypersensitive to DNA-damaging agents (SI Appendix, Fig. S4E) as in other cohesin mutants (31). Thus, the hinge destabilizing mutant, psm3-A561E, producing the authentic cohesin mutant phenotype, was able to overcome inactive separase or securin.
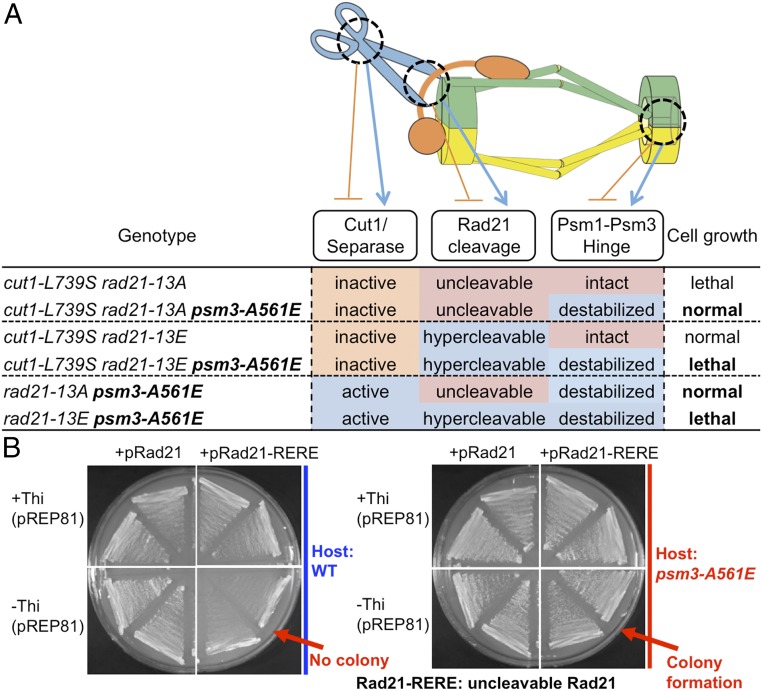
Dispensable Rad21 Cleavage in Hinge-Disordered Mutants.
The above results strongly suggested that Cut1 protease is dispensable when the cohesin complex is disordered, consistent with the fact that Cut1 disrupts the cohesin complex by cleaving Rad21 in mitotic anaphase (13–15). We further examined this issue using uncleavable Rad21. We used two rad21 mutants, rad21-13A and rad21-RERE, which were essentially uncleavable by Cut1 protease (31, 32). The Rad21-13A strain has alanine substitutions at 13 presumed polo kinase consensus sites. rad21-13A cut1-L739S double mutant could not be obtained; therefore, this unphosphorylatable (and uncleavable) rad21-13A was synthetic lethal in combination with separase-inactive cut1-L739S (Fig. 2A). In contrast, the phosphomimetic glutamate mutant rad21-13E (hypercleavable) was able to form colonies when combined with cut1-L739S. Interestingly, the hinge mutant psm3-A561E, described above, rescued the separase-inactive, Rad21-uncleavable lethality of cut1-L739S rad21-13A, indicating that Rad21 cleavage is unnecessary in the triple mutant with the separase inactivated and the cohesin hinge destabilized. In contrast, the hypercleavable, hinge-disrupted, and separase-defective triple mutant, cut1-L739S rad21-13E psm3-A561E, was lethal, perhaps because cohesin was too unstable due to hypercleavable rad21 and destabilized psm3. Conversely, the hypercleavable, separase-defective, double mutant, rad21-13E cut1-L739S, results in normal chromosome segregation at 26 °C. In addition, hinge-disrupted and Rad21 hypercleavable psm3-A561E rad21-13E is lethal, perhaps due to the nature of hyper-disordered and hypercleavable cohesin, while the hinge-disrupted and uncleavable-Rad21 double mutant, psm3-A561E rad21-13A, grew normally.
Fig. 2.
The psm3-A561E suppressor relieves noncleavable Rad21 toxicity. (A) Spot test results of six different strains containing the uncleavable rad21-13A or the hypercleavable rad21-13E mutation with or without cut1-L739S and hinge-destabilized psm3-A561E. (B) Streak test for colony formation of wild type or the psm3-A561E mutant carrying plasmid pRad21 wild type or the uncleavable pRad21-RERE mutant under the inducible promoter (+Thi off, −Thi on). psm3-A561E rescues the high toxicity of Rad21-RERE mutant protein overexpression.
Rad21-RERE is mutated in the two separase recognition sites (R179E and R231E) and is very toxic, even if mildly overproduced by plasmid pREP81 (32). This strong toxicity was completely abolished in the genetic background of hinge-destabilized mutant psm3-A561E (Fig. 2B). Taken together, these and the above results strongly suggest that the requirement of Rad21 cleavage can be alleviated by psm3-A561E, which induced a sterically disordered hinge interface, thus producing destabilized cohesin. Therefore, the strong toxicity of uncleavable Rad21 requires an intact cohesin hinge. For successful chromosome segregation, the cohesin hinge and head, which are well separated by the intervening coiled coils, may have to interact with each other.
Suppressors Disrupt the Interfaces Between Rad21 and Psm1/3.
We generated an atomic model of the Rad21-N, Rad21-C, head, and a part of the coiled-coil regions from Psm1 and Psm3, adapted according to the reported crystal structures of those from S. cerevisiae (8, 9) (PDB ID codes 4UX3 and 1W1W) and P. furiosus (33) (PDB ID code 4I99) (Fig. 3A). Rad21 (C) is bound to the head of Psm1, while Rad21 (N) contacts the coiled coil of Psm3 adjacent to the head (8–10). Recently, the crystal structure of DNA repair protein, Rad50, was determined (34–36). Rad50 binds to dsDNA, and its structure highly resembles the SMC Psm1–Psm3 head–coiled coil region (SI Appendix, Fig. S5 A and B; constructed from the structural data in ref. 36). Accordingly, presumed electrostatic potential in the Psm1–Psm3 head–coiled coil region might support DNA binding (SI Appendix, Fig. S5B). The orientations of coiled coils may affect the stability of DNA binding.
Fig. 3.
Suppressors of the cut1, cut2, and rad21 mutants in the head and coiled coil of cohesin. (A) Locations of the suppressors of the cut1, cut2, and rad21 mutants (magenta, suppressors of the cut1 and cut2 mutants; cyan, suppressors of rad21-K1) (SI Appendix, Table S1 A and B). I67, which is responsible for the rad21-K1 mutant, is also identified (boxed). (Inset) Steric clashes (double-headed arrows) caused by I67F mutation in Rad21. The I67 side chain appears in white stick, and that of Phe in ball-and-stick. (B) Details of the two ATP binding sites at the head interface. ATP and Mg2+ are shown with sticks and green spheres, respectively. In A and B, yellow, green, and orange are used for Psm1, Psm3, and Rad21, respectively. α-Helices (A–I for Psm1/3, AN–CC for Rad21) and β-strands (1–15 for Psm1/3, 1C–3C for Rad21) are marked. The suffix indicates the N- or C-terminal domain of Rad21. (C) Immunochemical detection of phosphorylation level of Rad21 (upper bands) using a polyclonal anti-Rad21 antibody.
Judging from the substituted amino acids, all six cut1 suppressors we identified in or near the head of cohesin (magenta characters in Fig. 3A) appear to destabilize the interfaces between Rad21 and Psm1/3, possibly affecting the capacity for DNA binding too. Mutant side chains of H42P and A53V (in Rad21-N), G164D and S127P (in Psm3), and V594F and V605F (in Rad21-C) are larger or helix breaking, so that the lack of Rad21 cleavage may be compensated due to unstable Rad21–Psm1 or Rad21–Psm3 interactions introduced by suppressors. The H42P substitution would disrupt the helix bundle formed by Rad21 and Psm3, because it would introduce a kink into the helix in which H42 is located.
Two suppressor mutations (S127P and G164D) identified in the head of Psm3 would also disrupt the interaction between Rad21 and Psm3 or between DNA and the head–coiled-coil domain, judging from its structural similarity to Rad50. S127 is located at the C terminus of helix B connected to the coiled coil via two short helices. The hydroxyl of S127 caps the helix and its Pro substitution would alter the path of the two short helices in the C-terminal side (SI Appendix, Fig. S6A). On the other hand, Psm3–G164 works as a pivot region, linking the head domain and the coiled-coil helix, and there are several charged and polar residues around G164 (Fig. 3A). Substitution of G164 with D164 may change the orientation of the coiled coil by interacting with charged or polar residues.
The C-terminal domain of Rad21, which contacts the head ATPase domain of Psm1, contains two suppressor sites: V594F and V605F (Fig. 3A). These two residues are present in the Winged helix-turn-helix domain (WHD) (37). It remains to be determined whether this Rad21 WHD really binds DNA. The side chain of V594 on the helix faces the interior of the C-terminal domain of Rad21, together with L542, N562, and F591, to form a hydrophobic core that would be disrupted by its substitution with Phe (F), leading to destabilization of the C-terminal domain. The other site, V605 on a β structure, contacts the aromatic ring of Y1208 in Psm1. The Phe substitution would clearly introduce steric interference with Y1208; thus, it is conceivable that suppression of separase/Cut1 inactivation occurred due to disordered Psm1–Rad21 interaction.
Suppressors of rad21-K1 in SMC ATPase Domains or Psm3 Coiled Coils.
To understand the role of Rad21 in cohesin, we then tried to isolate spontaneous suppressors for ts mutant rad21-K1 (30). We identified 17 distinct extragenic suppressors (Fig. 3A and SI Appendix, Table S1B). Interestingly, they reside exclusively in either the head ATPase or the Psm3 coiled coils adjacent to the head. The great majority (15/17) are psm3 mutations, and only two (R1127I and Q1168E) reside in the Psm1 head ATPase. Locations of Rad21 suppressors are schematized in Fig. 3A. There are nine mutations in the Psm3 coiled coil interacting with Rad21-N and eight in or near the ATPase head domains, respectively, suggesting that rad21 defects can be restored by alterations of the head ATPase or the Psm3 coiled coils. Coiled-coil mutations are modifying rather than destructive, as none of them involves proline. Hence, the alteration of the coiled coil seems to be critical for restoration of the rad21-K1 defect.
Three psm3 residues, S127 (ATPase), R199 (CC), and F1021 (CC), were twice isolated as different suppressor substitutions (Fig. 3A and SI Appendix, Table S1B). Hence, they may be critical residues for the structure of Psm3. Particularly distinct S127 substitutions (SI Appendix, Fig. S6B) were identified for two opposing ts mutants: separase/cut1 and rad21. The region around S127 is quite conserved among species (SI Appendix, Fig. S6C). Hence, S127 may have a pivotal role in controlling the Psm3–Rad21 interaction: S127 to P rescues cut1 by disrupting the Psm3–Rad21 interaction, and S127 to Y/F rescues rad21 by enhancing the Psm3–Rad21 interaction, respectively (Fig. 3A). Strikingly, all of the five ATPase mutations (K12E, G1099D, and A1133V in Psm3; R1127I, and Q1168E in Psm1) may affect ATP binding activity by the ATPase domains (Fig. 3 A and B). Judging from the exclusive location of rad21 suppressors in the coiled coil and ATPase head, these two distinct domains may be coordinated in restoring the defect of Rad21, perhaps by restoring the Psm3–Rad21 interaction.
I67F Is the Effective Mutation in rad21-K1 and Affects the Psm3–Rad21 Interaction.
To understand the nature of rad21-K1 suppressors, it was essential to determine the effective mutation site. Unfortunately, however, we found that the original ts strain (30) contained multiple mutations within the coding region. To determine which one is the responsible mutation for the ts phenotype of rad21-K1, we performed site-directed mutagenesis for each mutation: I67F, S89A, S396F, H453R, and E571G. Only rad21-I67F was found to be ts; therefore, I67F is the responsible mutation in rad21-K1. The crystal structure (Fig. 3A and SI Appendix, Fig. S7) reveals that residue I67 (α-helix CN) is located at the helix bundle, closely interacting with the Psm3 helix (Inset). Replacement with bulky F67 (I67 → F67) would interfere with surrounding residues (Fig. 3A, Inset). This rad21-I67F mutation probably disorders the Psm3–Rad21 helix bundle interaction, leading to the loss of cohesin at the restrictive temperature and readily explaining the cohesin-defect phenotype.
Phosphorylation of Rad21 Is Diminished in the rad21-I67F Mutant.
Rad21/Scc1 is hyperphosphorylated (31, 38, 39), and this can be monitored by immunoblot using an anti-Rad21 polyclonal antibody. Besides the typical cohesionless cell phenotype, a striking feature of rad21-K1 was the great reduction of Rad21 phosphorylation (Fig. 3C; detailed description in ref. 31). Therefore, Rad21 hyperphosphorylation might serve as an indicator of functional cohesin. If cohesin’s DNA binding activity decreases, Rad21 phosphorylation level may decrease. We found that single segregant rad21-I67F mutant cells showed the same great loss of Rad21 phosphorylation as in rad21-K1. The result is consistent with the finding that a single I67F mutation caused the same phenotype as the rad21-K1 mutant. Thus, suppressor analysis is consistent with the crystal structure, indicating that defects of Rad21-I67F in the N-terminal CN helix can be rescued by suppressors in the ATPase or Psm3 coiled coil interacting with Rad21, which may directly or indirectly enhance DNA-dependent ATPase via the Psm3–Rad21 interaction.
cut1 Suppressors in mis4 Cause Loss of Cohesion.
How can suppressors in mis4 (F616S, P1035L and L1188R; Fig. 1A and SI Appendix, Table S1A) overcome inactive cut1/separase? We were able to assign the mutation sites in the atomic model of Mis4 (Fig. 4 A and B) adapted from the recently solved 3D crystal structure of the Mis4 homolog, Scc2/NIPBL (16, 27). In addition, the responsible mutation of the ts mutant mis4-242, G1326E, was indicated too (22) (Fig. 4B). Basic regions are candidates for DNA binding (40) (Fig. 4A). P1035L would cause steric hindrance with neighboring residues (V1072 and F1075; Fig. 4C), suggesting that cohesin DNA binding might be impaired in the mis4-P1035L suppressor mutant.
Fig. 4.
The cut1 suppressors in Mis4/Scc2/NIPBL. (A) Surface potential of Mis4, based on the crystal structure of Scc2. Large positively charged (blue) areas that are likely to form a binding site for double-stranded DNA (schematically shown with magenta helices) are indicated by two green arrows. Inset shows an end view of DNA drawn to scale. (B) The architecture of the central part of Mis4/Scc2 and locations of the cut1 suppressors. Mutations are shown in green, and substitutions are shown with orange spheres. Heat-repeat motifs in which the mutations are identified are in magenta. (C) Boxed area in B, showing steric clashes caused by the P1035L mutation. (D) Spot test showing that all of the three cut1 suppressors in mis4 are UV damage sensitive. (E) Rad21 phosphorylation level (upper bands) detected using a polyclonal anti-Rad21 antibody. (F) Location of mis4-G1326E intragenic suppressors. (G) Locations of psm1 and psm3 head mutations that suppress mis4-G1326E or rad21-K1. rad21 suppressors are in blue, and mis4 suppressors are in magenta.
Consistently, all of the three single suppressor segregants were DNA damage sensitive (Fig. 4D). Furthermore, hyperphosphorylation of Rad21 was greatly diminished in these three mis4 suppressor mutants as well as in mis4-G1326E (Fig. 4E), which supports that cohesion might be impaired in these mis4 mutants.
Mis4/Scc2/NIPBL Suppressors May Restore Its Interaction with Psm1/3 Heads.
To identify mutations that can compensate for mis4, we isolated a number of spontaneous suppressors for ts mutant mis4-G1326E (22), which had been integrated into the clean genome of wild-type fission yeast by homologous recombination. A number of Ts+ intragenic suppressors (listed in SI Appendix, Table S1C) were obtained within the mis4 gene, the majority of which were located in two narrow domains (amino acids 803–844 and 1,332–1,387). Mis4/Scc2/NIPBL has the shape of a handle and a hook (16, 27). The intragenic suppressor mutations sites are largely concentrated on the hook and the boundary between the hook and handle (Fig. 4F). Since the two narrow regions may be directly involved in DNA binding (Fig. 4A), a possible explanation is that these suppressors may restore the binding of Mis4 to DNA.
mis4 intergenic suppressors were concentrated on the Psm1 and Psm3 head domains. Twenty-one and two suppressors were identified in the head domains of Psm3 and Psm1, respectively (Fig. 4G). They are broadly distributed in the head domains, in sharp contrast to Rad21 suppressors, which are restricted to ATPase domains nearby its ATP-binding sites. Our finding of mis4 suppressors in the Psm1 and Psm3 head domains is consistent with the recent cross-linking and electron microscopy results, showing that Scc2 (the Mis4 homolog) binds cohesin at or near its heads (16).
Pds5 Is Not Required for Suppression of the cut1 Mutation.
A question, whether Pds5–Wpl1 heterodimeric complex is involved in suppression of cut1 mutants by cohesin mutations, is addressed. Pds5 and Wpl1 are nonessential but act as cohesin interactors by disordering the interface between the SMC (Psm3) coiled coil and Rad21’s N terminus in a nonproteolytic way (41–43). Therefore, we examined whether suppression of the cut1 mutant by psm3-A561E is dependent on the presence of Pds5. If suppression requires Pds5, the triple-mutant cut1-A1816T psm3-A561E Δpds5 should abolish the suppression. As shown in SI Appendix, Fig. S8, however, Δpds5 had no effect on the cut1-A1816T psm3-A561E double-mutant phenotype, demonstrating that Pds5-dependent release of cohesin does not participate in the cohesin release in the cut1-A1816T psm3-A561E separase-cohesin hinge double mutant. We thus concluded that Pds5 is not required for the suppression of separase mutants.
Discussion
At the onset of anaphase, activated Cut1 separase cleaves Rad21 in the loop connecting the Psm1 head and Psm3 coiled coil, thereby releasing DNA from cohesin. In suppressors that overcome the inactive separase, we expected some kind of disorder to occur in the vicinity of the cleavage site due to suppressor mutations. Indeed, certain cut1 suppressors suggested destabilization in Psm1–Rad21 and Psm3–Rad21 interfaces. However, many other suppressors identified are actually at the interface within the hinge. Destabilization of the hinge might greatly contribute to the release of DNA. How then does the release occur in the hinge mutants? Currently, the ring model of cohesin (40, 41, 44–46) is generally accepted (Fig. 5A). One (or two) duplex DNAs may be embraced within the ring (SI Appendix, Fig. S9A) that is composed of two distinct SMC and Rad21 subunits. Rad21 is the linker in the head. Our work is based on suppressor analysis, which apparently dispenses with the separase-induced cleavage or overcomes destabilized cohesin subunits. Thus, an explanation of cut1 and rad21 suppressor results is possible through this ring model: the destabilized hinge or the head interfaces are open in cut1/cut2 suppressors; therefore, the cohesin ring may be fully open (or somehow unfolded) and can release DNAs (SI Appendix, Fig. S9B), while the cohesin ring might be disrupted in rad21-K1 but restored by rad21-K1 suppressor mutations (SI Appendix, Fig. S9C). Although our MD analysis suggests a tight association in the hinge interface in suppressors, the release of DNA might still be possible through infrequent opening of the hinge, etc.
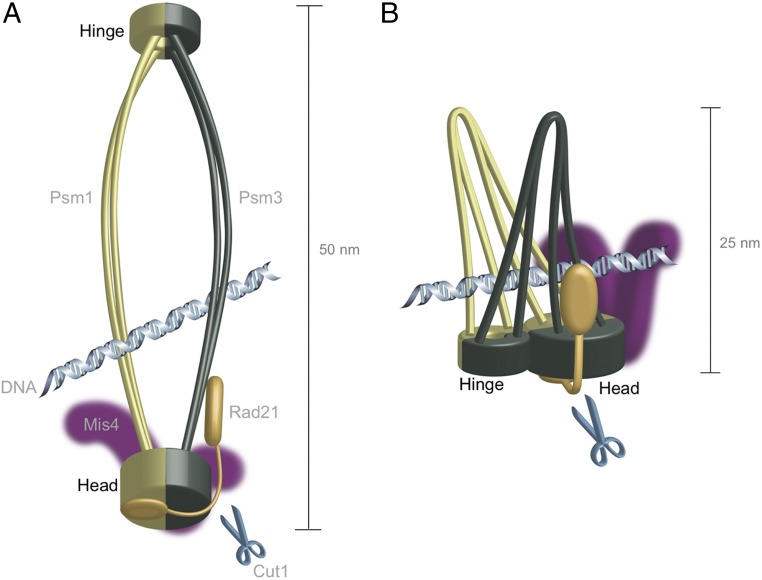
Fig. 5.
The prevailing ring model and the hold-and-release model proposed in this study. (A) Cohesin protein complex forms a ring that topologically entraps chromosomal DNAs. In anaphase, Cut1/separase cleaves Rad21, resulting in the release of DNAs. (B) The hold-and-release model proposes to explain how cohesin hinge interface mutations dispense with Rad21 cleavage. In this model, SMC subunits’ head and hinge are proximal to form arched coiled coils, which can hold or release DNA.
Structural changes other than hinge opening might also be able to suppress the absence of loop cleavage. We are interested in the large movements of the coiled-coil portions in suppressors and want to propose an alternative hold-and-release model (described below). The hold-and-release model requires the interaction of DNA with arched coiled coils. The model does not negate the embrace models, including handcuff model (47), but explains the binding and release of DNA from cohesin without protein cleavage. Sister chromatids may be held by coiled-coil interactions rather than by the loop (SI Appendix, Fig. S10 A and B).
The previous studies of cohesin using atomic force microscopy (48, 49) or electron microscopy (16) showed that cohesin forms a tadpole-like structure and that the length of the tadpole tail (∼25 nm) is about one-half of the planar head–hinge distance. The cohesin coiled coils might thus fold so that the hinge and head can interact directly, forming the arched coiled coils (shown in Fig. 5B and SI Appendix, Fig. S10A). In fact, the possibility of such interaction between hinge and head in SMCs has already been suggested in several biochemical studies. The condensin hinge interface was previously shown to undergo in vitro phosphorylation, which resulted in dissociation from DNA, and this phosphorylation was abolished by a mutation in the condensin head ATPase (50). Binding between DNA and the head might be diminished by hinge interface phosphorylation driven by the head ATPase. Since condensin and cohesin adopt similar structures, the hinge and head might thus directly interact with DNA in cohesin too. Acetylation and ATPase activity of head have been reported to be modulated by hinge (28, 51, 52). Furthermore, isolated SMC3 hinge and SMC1 head were immuno-coprecipitated with SMC1 hinge, suggesting that hinge and head are possibly directly associated (53). This report also suggested that head-interacting subunit Pds5 is closely situated on hinge. More recently, the direct binding of cohesin hinge to Psc3, which binds to Rad21 and is located near cohesin head, has been shown by a coimmunoprecipitation assay (41). Waldman et al. (54) identified the presence of breaks in the coiled-coil structure and suggested that such breaks would induce “folded” coiled coils. Hence, we propose that arched coiled coils might exist and can hold DNA, and the angle made by the arched coils is the determining factor of holding and releasing DNA (Fig. 5B). The suppressing mutations identified in the hinge might actually widen the angle of the arched coils so that DNA is less strongly held and more easily released.
The crystal structure of Rad50 with bound DNA was recently solved (34–36) and is helpful in interpreting our results. Rad50, a subunit of the Mre11 DNA damage repair complex (55, 56), contains an SMC head-like ATPase and coiled coils, but lacks the hinge. Its structure and electrostatic surface potential are very similar to the S. pombe cohesin head (SI Appendix, Fig. S5 A and B). Therefore, we assume putative DNA-binding sites in the cohesin head. Two bands of positive charges are present on the surface of the cohesin head, while a cluster of positive charges exists near the root of coiled coil. These positively charged patches might be used for DNA binding, as proposed for Rad50 (34, 36), when the two strands of the coiled coil are open by unwinding, but the space for DNA may become narrowed if the coiled coils adopt a more upright orientation. Consistently, reports (57–59) have shown that the SMC hinge acquires higher affinity for DNA when coiled coils are present. Taken together, the head–hinge interaction of cohesin may affect the opening angle of the coiled coil arch.
The hold-and-release model requires dynamic interactions of DNA with the coiled coil, hinge, and head (Fig. 5B and SI Appendix, Fig. S10 A and B). The hinge and the head are brought into proximity by the folding of the coiled coils so that DNA is sandwiched between the arched coiled coils. Extensive opening of the “arched” (folded) coiled coils leads to the release of DNA, so that coiled-coil movements may play a central role in DNA binding and dissociation (Movie S1 shows larger movements of the coiled coil in a hinge mutant). Accordingly, opening/closing of coiled coils may be indirectly controlled by the head ATPase. Note that some rad21 suppressors are located closely to the head ATP sites possibly controlling the orientation of the coiled coil. This hold-and-release model does not interfere with the gliding movements of cohesin along DNA (60, 61). Chemical cross-linking (10, 16) indicated that cohesin coiled coils closely interact with each other. The hinge and head may thus indirectly affect the force of DNA binding via a change in orientation or the opening/closing of the folded coiled coils. DNA may simultaneously fall out of cohesin through the Rad21-cleavage independent signal, leading to chromosome segregation. This may explain many suppressors we isolated in this study of inactive separase. Conversely, Psm3 coiled coils and Psm1/Psm3 head mutations that suppress ts rad21 may restore DNA holding. The hold-and-release model offers potentially a unique perspective on the dynamic association of cohesin proteins with chromosomal DNA. Note that this model can also be applied to the wild-type situation. Further investigation is needed to understand how cohesin functions to achieve sister chromatid cohesion and separation. In particular, the model does not explain how two duplex DNAs are held together. If only one duplex DNA can bind to the arched coiled coil, two sets of them would be needed for sister DNAs’ cohesion. To this end, Mis4 may play an essential role in tethering two sister DNAs together. In SI Appendix, Fig. S10B, two postreplicative duplex DNAs, each of which binds to cohesin complex, is illustrated for wild type and for cut1 suppressor.
In conclusion, our suppressor analysis identifies the functional interfaces in cohesin. Since the method is based on obtaining spontaneous suppressors, this unbiased way of identifying essential functional interfaces may be further exploited not only in cohesin but also in other chromosomal or even protein complexes in general, when whole-genome sequencing can be applied to identify suppressor sites (1) and 3D structures have been previously determined.
Materials and Methods
S. pombe cut1 and cut2 Mutants Used for Suppressor Screening.
Two previously characterized cut1 ts mutants (cut1-206, cut1-693) and cut2 ts mutants (cut2-364, cut2-EA2) were selected for suppressor screening (13, 19). Responsible mutations (SI Appendix, Fig. S1A) were reintegrated into the 972h− wild-type strain using site-directed mutagenesis. Newly constructed mutants were ts and exhibited the same phenotypes as the original ts mutants (SI Appendix, Fig. S1 B and C).
Isolation of Cohesin Hinge Ts/Cs Mutants.
Beside spontaneous suppressor mutations, we isolated cohesin hinge ts/cs mutants using site-directed mutagenesis targeted to cohesin hinge regions. Leucine (L) or serine (S) → proline (P), and glycine (G) or alanine (A) → glutamic acid (E) often cause ts or cs. The residues change from L to P was successful in isolating condensin ts mutants (62). While mutant psm3-A561E produced the cs phenotype, mutants mis4-242, mis6-302, and mis12-537 containing G1326E, G135E, and G52E substitutions, respectively (22, 63, 64), produced the ts phenotype. Thus, we made 59 new hinge mutants (33 psm3 and 26 psm1, respectively) using the above substitutions (SI Appendix, Fig. S3A), and found 6 ts and 6 cs mutants. Detailed procedures for PCR mutagenesis and chromosomal integration followed by selection have been previously described (62).
Suppressor Screening, Next-Generation Sequencing, and Mutation Identification.
Suppressor screening, next-generation sequencing, and mutation identification follow the procedures described in ref. 1. Briefly, appropriate restrictive temperatures were then selected for each ts mutant to ensure revertant frequencies between 1×10−8 and 1×10−6. Mutant cells were plated on YPD plates and cultured at selected restrictive temperatures for 4 d. Colonies (revertants that contained suppressor mutations in addition to the original ts mutation) were selected. In total, we isolated and sequenced 888 revertants (103 for cut1-L739S; 245 for cut1-A1816T; 84 for cut2-R267Stop; 17 for cut2-EA2; 93 for rad21-K1; and 446 for mis4-G1326E) and identified 100 suppressor mutations in cohesin subunits or its loader Mis4 (SI Appendix, Table S1). Genomic DNAs were extracted, and then genomic DNA of each ∼10 revertants of the same ts mutant were mixed together in equal amounts and used for library construction. Each 10 DNA mixtures were barcoded for one lane of pooled sequencing. DNA libraries for Illumina sequencing were generated using standard protocols (Illumina). The libraries were sequenced with paired-end (2 × 150-bp) runs using Illumina HiSeq 2000 sequencers. Sequence reads were mapped against the S. pombe reference genome using the Novoalign mapping tool with default settings. Mutations were called with SNVer (65) (version 0.5.3). Genes with two or more independent mutations or two or more independent mutations in genes involved in same complex/pathway were manually selected as probable suppressors for further analysis.
Immunochemistry.
Protein extracts were prepared by cell breakage using glass beads in extraction buffer (25 mM Tris⋅HCl at pH 7.5, 15 mM EDTA, 85 mM β-glycerophosphate, 0.1% Nonidet P-40, 10% glycerol, 1 mM DTT, 1 mM PMSF) supplied with protease inhibitor tablets, EDTA-free (Roche). Extracts were boiled with LDS sample buffer and loaded onto a custom-made 3–8% gradient Tris-acetate gel (NuPAGE; Invitrogen). Immunoblotting was performed using the anti-Rad21 (13, 31, 39) and anti-PSTAIR (a gift from Dr. Yoshitaka Nagahama, National Institute for Basic Biology, Okazaki, Japan) antibodies with Ponceau S staining.
Generating Atomic Models of S. pombe Cohesin Hinge and Head.
For homology modeling, sequences of S. pombe Psm1 and Psm3 were aligned with those of SMC1 and SMC3 from mammals and S. cerevisiae, and condensin from Pyrococcus furiosus. Then, an atomic model of S. pombe cohesin hinge was generated from the crystal structure of the murine cohesin hinge (PDB ID code 2WD5; ref. 28) based on the sequence alignment using Modeler (66). An atomic model of S. pombe cohesin head was also generated from the crystal structures of SMC heads from S. cerevisiae [PDB ID codes 1W1W (9) and 4UX3 (8)] and from P. furiosus [PDB ID code 4I99 (33)] with the same procedure.
MD Simulations of the Mouse Cohesin Hinge.
To evaluate structural changes of the hinge due to suppressing mutations, MD simulations were carried out with the atomic models generated from the crystal structure of the mouse cohesin hinge [PDB ID code 2WD5 (28)]. The mutations were manually introduced using Coot (67). Potential protonation in the models was examined using PDB2PQR server (68). K+ and Cl− ions were added at a concentration of ∼0.15 M into the systems using CHARMM-GUI (69). The systems solvated using water molecules (TIP3P) are 88 × 88 × 88 Å3 in size and contain ∼63,400 atoms. All simulations were carried out with NAMD using the CHARMM36 force field (70). The systems were preequilibrated using energy minimization and 50-ps MD simulation with harmonic restraints on the heavy atoms of the protein, and then productive runs were performed for 100 ns. Normal-mode analyses of the models generated from the MD simulations were carried out with ANM 2.1 (71).
Supplementary Material
Acknowledgments
We thank Prof. Hongtao Yu for his valuable comments to improve our manuscript; Dr. Kenichi Sajiki for his help in next-generation sequencing; Ms. Orie Arakawa and Ms. Kaori Serakaki for technical assistance and illustration, respectively; and Dr. Steven D. Aird for technical editing. Generous support from the Okinawa Institute of Science and Technology Graduate University is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database (accession number PRJNA450289).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803564115/-/DCSupplemental.
References
- 1.Xu X, Wang L, Yanagida M. Whole-genome sequencing of suppressor DNA mixtures identifies pathways that compensate for chromosome segregation defects in Schizosaccharomyces pombe. G3 (Bethesda) 2018;8:1031–1038. doi: 10.1534/g3.118.200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbatsh AMO, et al. Cohesin releases DNA through asymmetric ATPase-driven ring opening. Mol Cell. 2016;61:575–588. doi: 10.1016/j.molcel.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marinova IN, et al. Single site suppressors of a fission yeast temperature-sensitive mutant in cdc48 identified by whole genome sequencing. PLoS One. 2015;10:e0117779. doi: 10.1371/journal.pone.0117779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 5.Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: Long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 8.Gligoris TG, et al. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haering CH, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Huis in ’t Veld PJ, et al. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- 11.Funabiki H, et al. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 12.Funabiki H, et al. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 14.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 15.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 16.Chao WC, et al. Structure of the cohesin loader Scc2. Nat Commun. 2017;8:13952. doi: 10.1038/ncomms13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. Classification of the caspase-hemoglobinase fold: Detection of new families and implications for the origin of the eukaryotic separins. Proteins. 2002;46:355–367. doi: 10.1002/prot.10060. [DOI] [PubMed] [Google Scholar]
- 18.Boland A, et al. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat Struct Mol Biol. 2017;24:414–418. doi: 10.1038/nsmb.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z, Luo X, Yu H. Structural basis of cohesin cleavage by separase. Nature. 2016;532:131–134. doi: 10.1038/nature17402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo S, Tong L. Molecular mechanism for the regulation of yeast separase by securin. Nature. 2017;542:255–259. doi: 10.1038/nature21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 26.Hirano T, Funahashi S, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikuchi S, Borek DM, Otwinowski Z, Tomchick DR, Yu H. Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc Natl Acad Sci USA. 2016;113:12444–12449. doi: 10.1073/pnas.1611333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurze A, et al. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J. 2011;30:364–378. doi: 10.1038/emboj.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra A, et al. Both interaction surfaces within cohesin’s hinge domain are essential for its stable chromosomal association. Curr Biol. 2010;20:279–289. doi: 10.1016/j.cub.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatebayashi K, Kato J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: Possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–57. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adachi Y, Kokubu A, Ebe M, Nagao K, Yanagida M. Cut1/separase-dependent roles of multiple phosphorylation of fission yeast cohesion subunit Rad21 in post-replicative damage repair and mitosis. Cell Cycle. 2008;7:765–776. doi: 10.4161/cc.7.6.5530. [DOI] [PubMed] [Google Scholar]
- 32.Toyoda Y, et al. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr Biol. 2002;12:347–358. doi: 10.1016/s0960-9822(02)00692-9. [DOI] [PubMed] [Google Scholar]
- 33.Bürmann F, et al. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016;35:743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojowska A, et al. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J. 2014;33:2847–2859. doi: 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleiffer A, et al. Kleisins: A superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 38.Birkenbihl RP, Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- 39.Tomonaga T, et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murayama Y, Uhlmann F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015;163:1628–1640. doi: 10.1016/j.cell.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang Z, Yu H. Releasing the cohesin ring: A rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. BioEssays. 2017;39:1600207. doi: 10.1002/bies.201600207. [DOI] [PubMed] [Google Scholar]
- 43.Ouyang Z, et al. Structure of the human cohesin inhibitor Wapl. Proc Natl Acad Sci USA. 2013;110:11355–11360. doi: 10.1073/pnas.1304594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 45.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 46.Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. Establishment of DNA-DNA interactions by the cohesin ring. Cell. 2018;172:465–477.e15. doi: 10.1016/j.cell.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eng T, Guacci V, Koshland D. Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol Biol Cell. 2015;26:4224–4235. doi: 10.1091/mbc.E15-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai A, Hizume K, Sutani T, Takeyasu K, Yanagida M. Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein-protein assembly. EMBO J. 2003;22:2764–2775. doi: 10.1093/emboj/cdg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimura SH, et al. Condensin architecture and interaction with DNA: Regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 50.Akai Y, et al. ATPase-dependent auto-phosphorylation of the open condensin hinge diminishes DNA binding. Open Biol. 2014;4:140193. doi: 10.1098/rsob.140193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirano M, Hirano T. Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 53.Mc Intyre J, et al. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldman VM, Stanage TH, Mims A, Norden IS, Oakley MG. Structural mapping of the coiled-coil domain of a bacterial condensin and comparative analyses across all domains of life suggest conserved features of SMC proteins. Proteins. 2015;83:1027–1045. doi: 10.1002/prot.24778. [DOI] [PubMed] [Google Scholar]
- 55.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 56.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu A, Revenkova E, Jessberger R. DNA interaction and dimerization of eukaryotic SMC hinge domains. J Biol Chem. 2004;279:26233–26242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- 58.Roy MA, Dhanaraman T, D’Amours D. The Smc5-Smc6 heterodimer associates with DNA through several independent binding domains. Sci Rep. 2015;5:9797. doi: 10.1038/srep09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soh YM, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson IF, et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 2016;35:2671–2685. doi: 10.15252/embj.201695402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stigler J, Çamdere GO, Koshland DE, Greene EC. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 2016;15:988–998. doi: 10.1016/j.celrep.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X, Nakazawa N, Yanagida M. Condensin HEAT subunits required for DNA repair, kinetochore/centromere function and ploidy maintenance in fission yeast. PLoS One. 2015;10:e0119347. doi: 10.1371/journal.pone.0119347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 64.Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: A statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39:e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 70.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eyal E, Lum G, Bahar I. The anisotropic network model web server at 2015 (ANM 2.0) Bioinformatics. 2015;31:1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.