Significance
Sex determination involves antagonistic interactions between the testis-determining (SRY-SOX9-FGF9) and ovary-promoting (RSPO1-WNT/β-catenin-FOXL2) pathways, but the underlying molecular mechanisms remain unclear. We show that ZNRF3, an E3 ubiquitin ligase that inhibits WNT signaling and is a direct target of RSPO1-mediated membrane clearance, is testis-determining in mice. Testis determination defects in the absence of ZNRF3 arise due to ectopic canonical WNT signaling in XY gonads at the sex-determining stage. We identify human ZNRF3 sequence variants in cases of 46,XY disorders of sex development with XY female presentation. In vitro functional assays show that these variants disrupt ZNRF3 function. Our data reveal a sex-determining role for ZNRF3 and indicate that interactions between ZNRF3 and RSPO1 regulate mammalian sex determination.
Keywords: sex determination, ZNRF3, DSD, WNT signaling, organogenesis
Abstract
Mammalian sex determination is controlled by the antagonistic interactions of two genetic pathways: The SRY-SOX9-FGF9 network promotes testis determination partly by opposing proovarian pathways, while RSPO1/WNT-β-catenin/FOXL2 signals control ovary development by inhibiting SRY-SOX9-FGF9. The molecular basis of this mutual antagonism is unclear. Here we show that ZNRF3, a WNT signaling antagonist and direct target of RSPO1-mediated inhibition, is required for sex determination in mice. XY mice lacking ZNRF3 exhibit complete or partial gonadal sex reversal, or related defects. These abnormalities are associated with ectopic WNT/β-catenin activity and reduced Sox9 expression during fetal sex determination. Using exome sequencing of individuals with 46,XY disorders of sex development, we identified three human ZNRF3 variants in very rare cases of XY female presentation. We tested two missense variants and show that these disrupt ZNRF3 activity in both human cell lines and zebrafish embryo assays. Our data identify a testis-determining function for ZNRF3 and indicate a mechanism of direct molecular interaction between two mutually antagonistic organogenetic pathways.
Mammalian sex determination involves the sexually dimorphic development of a gonadal primordium. In the presence of SRY on the Y chromosome, supporting cell precursors of the developing gonad differentiate into Sertoli cells, and this somatic lineage orchestrates morphological events required for testis determination (reviewed in refs. 1 and 2). Thus, the “decision” as to whether supporting cells develop as Sertoli (testicular) or granulosa (ovarian) cells is pivotal to sex determination, and understanding the molecular events that result in fate specification of this lineage remains critical to our understanding of gonadogenesis.
SRY acts to up-regulate the expression of the testis-determining gene SOX9 (3), a transcription factor that initiates a program of gene activity that directs Sertoli cell differentiation (4, 5). The timing of these protestis events is crucial: Any delay in the expression of Sry can result in sex reversal or ovotestis development in XY mice (6). Studies have shown that the testis-determining genetic pathway is important for the inhibition of the equivalent ovarian-determining pathway. Indeed, the two pathways, most notably FGF signaling in the testis and canonical WNT signaling in the ovary, act in a mutually antagonistic fashion (7). This mutual antagonism persists in the adult gonad: Postnatal deletion of genes such as DMRT1 (8) and FOXL2 (9) can result in reprogramming of cells of the adult testis and ovary, respectively, to the alternative sexual fate.
Canonical WNT/β-catenin signals are required for normal ovarian development from the embryonic XX gonad: Loss of WNT4 or Rspondin-1 (RSPO1), which effect such signals through stabilization of β-catenin, can result in partial XX gonadal sex reversal in mice and 46,XX testicular disorders of sex development (DSD) or virilization in humans (10–14). Mechanistically, R-spondins, in association with LGR4/5 cell surface receptors, promote WNT signaling by binding to and sequestering the transmembrane E3 ubiquitin ligases ZNRF3 and RNF43; these two molecules, in turn, inhibit WNT signaling by targeting Frizzled receptor for degradation by ubiquitination and increased membrane turnover (15–19). Loss of function genetic studies show that testis determination requires the inhibition of proovarian canonical WNT/β-catenin signals (7, 20), and these observations are consistent with the report that ectopic stabilization of β-catenin in transgenic XY mice can disrupt testis development (21). However, the molecular effectors of this inhibition of WNT/β-catenin during testis determination have not been identified. Components of the core sex-determining pathways are only very rarely identified, but, given their inhibition by RSPO1 in other contexts, ZNRF3 and RNF43 are excellent candidates for gene products that act to inhibit canonical WNT signaling during sex determination to tilt the balance toward the testicular fate.
Here, we report testis determination defects, including gonadal sex reversal, in mice lacking ZNRF3. In contrast, RNF43 is not required for testis determination. Loss of ZNRF3 results in ectopic canonical WNT signaling in XY gonads at the sex-determining stage of 11.5 days post coitum (dpc) and a consequent reduction in Sox9 expression. We also report variants in human ZNRF3 associated with 46, XY DSD and show that two missense substitutions identified can disrupt ZNRF3’s anti-WNT activity in a cell line and zebrafish embryos. Our data reveal an antagonistic molecular interaction, between RSPO1 and ZNRF3, at the heart of the sex-determining mechanism.
Results
We examined Znrf3 expression in somatic (Sf1(Nr5a1)-positive) cells of the developing XY and XX gonads between 10.5 and 16.5 dpc using single-cell RNA sequencing (sc-RNAseq) (SI Appendix, Fig. S1). This revealed early Znrf3 expression in progenitor cells and supporting cells in both sexes, but with no significant sexual dimorphism before 13.5 dpc, by which time primary sex determination is complete. In contrast, Sox9 expression was enhanced in XY supporting cells and Rspo1 expression was enhanced in XX supporting cells from around 11.5 dpc (SI Appendix, Fig. S1 B and C), as previously described (12). Quantitative RT-PCR (qRT-PCR) analyses of Znrf3 expression in XY and XX Sf1-positive cells and whole gonads at 11.5 dpc also revealed no sexual dimorphism (SI Appendix, Fig. S1 D and E). These data indicate that ZNRF3 activity in developing gonadal somatic cells may be regulated in a sexually dimorphic fashion primarily due to the enhanced expression of Rspo1 in XX supporting cells: ZNRF3 activity is predicted to be higher in XY cells than in XX cells due to RSPO1-mediated membrane clearance in XX cells.
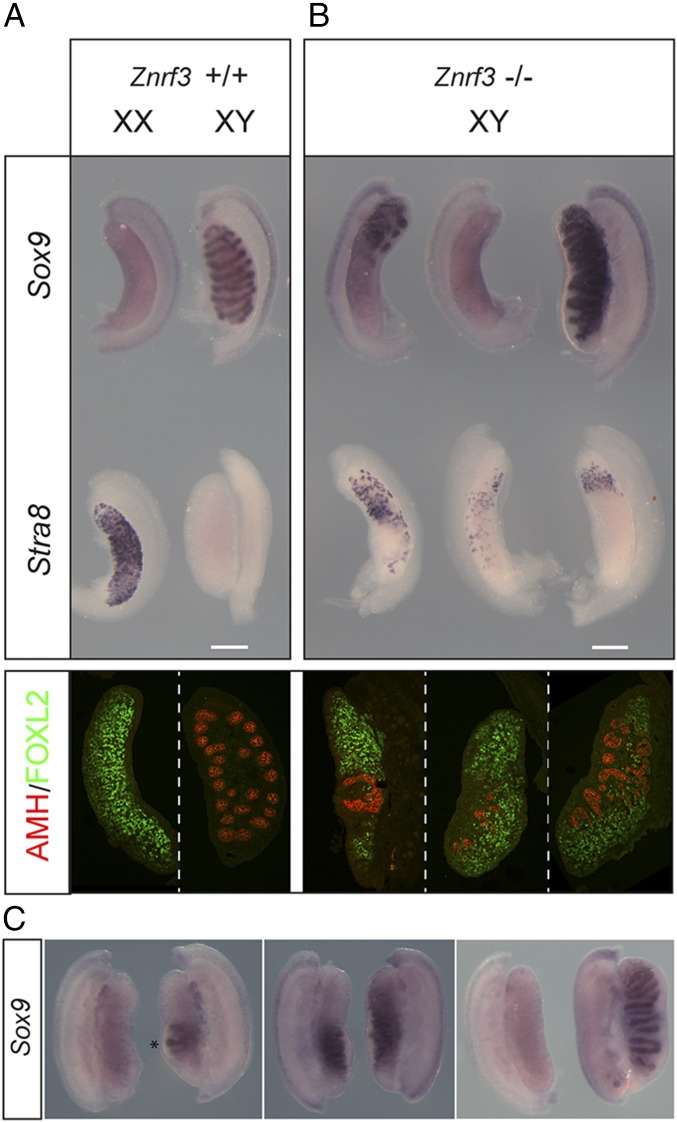
We then tested directly whether loss of ZNRF3 disrupted testis determination in the mouse. XY fetal gonads homozygous for a Znrf3 null allele (16) on the C57BL/6J (B6) background exhibited pronounced defects in testis determination with variable severity at 14.5 dpc (Fig. 1). The most severely affected appeared sex-reversed, with an ovarian morphology characterized by the absence of Sox9 expression and patchy Stra8 expression throughout (Fig. 1 A and B). Stra8 expression is indicative of germ cell meiotic entry, normally only detectable in XX gonads at this stage. Other mutants developed ovotestes, with only particular regions of the gonad expressing significant Stra8 or Sox9. The least affected class exhibited disruption to morphology with irregular testis cords and restricted areas containing Stra8-positive cells. XY mutant gonads had significant numbers of FOXL2-positive cells, depending on the degree of sex reversal, indicating ovarian granulosa cell differentiation (Fig. 1 A and B). Significantly, phenotypic variability in the degree of sex reversal was observed between gonads of an individual mutant fetus at 14.5 dpc (Fig. 1C), indicating innate phenotypic stochasticity in the absence of ZNRF3, in addition to any contributions made by residual genetic background differences.
Fig. 1.
Testis determination defects in XY gonads lacking ZNRF3. (A) Wild-type controls. (B) XY gonads (14.5 dpc) lacking ZNRF3 exhibit severe morphological abnormalities in comparison with wild-type controls. WMISH with Sox9 (Top, upper row) reveals mutant gonads with a sex-reversed (ovarian) morphology (left and center) and either a major reduction in levels of Sox9 or no detectable expression. Less affected gonads have higher Sox9 levels but abnormal testis cords (right). WMISH with Stra8 (Top, lower row) reveals a significant number of meiotic germ cells in ZNRF3-deficient gonads. Gonadal sex reversal in XY gonads lacking ZNRF3 is confirmed by immunostaining of 14.5 dpc gonadal sections for AMH (red), a Sertoli cell marker, and FOXL2 (green), a granulosa cell marker (Bottom). (C) Pairs of gonads dissected from individual Znrf3 fetuses (14.5 dpc) exhibit asymmetry. Note rudimentary testis cord formation in one gonad (asterisk), but not the other (Left). Center shows distinct Sox9 expression profiles in two fetal gonads. Right shows, remarkably, an ovary on one side and a well-formed testis on the other. (Scale bar, 250 μm.)
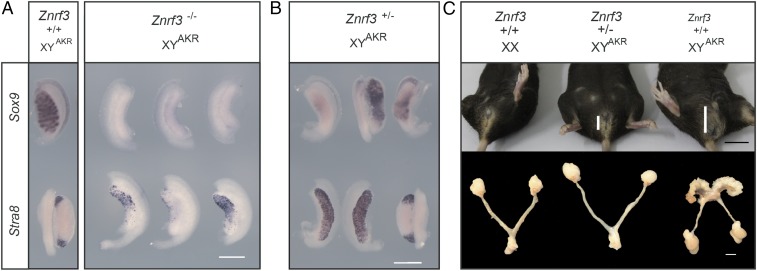
We also examined the consequences of loss of ZNRF3 for XY gonad development on the B6.YAKR genetic background, which is highly sensitized to disruptions to testis determination due to the presence of the Mus domesticus AKR Y chromosome (22, 23). Embryos lacking ZNRF3 on B6.YAKR consistently exhibited complete gonadal sex reversal, having an ovarian morphology and lacking Sox9 expression and often containing large numbers of Stra8-positive cells (Fig. 2A). Heterozygous embryos lacking just a single copy of Znrf3 also had severe gonadal defects, but with variable severity (Fig. 2B). Some developed ovotestes, with central gonadal Sox9 expression, some exhibited Sox9 expression but lacked testis cords, and others had an ovarian morphology with no detectable Sox9 expression. Adult B6.YAKR heterozygous mice are viable, unlike homozygotes, which die at around birth (15). B6.YAKR adult heterozygotes exhibited a range of gonadal abnormalities; remarkably, a minority (2/5) developed as phenotypic XY females with internal genitalia and anogenital distances grossly similar to XX controls (Fig. 2C and SI Appendix, Fig. S2). Others either contained an ovary-like gonad on one side and a very small contralateral testis or contained two small testes (SI Appendix, Fig. S3). These data establish ZNRF3 as testis-determining in the mouse. In contrast, B6 XY embryos lacking the functionally related paralogue RNF43 (16, 18, 24) develop testes as normal (SI Appendix, Fig. S4).
Fig. 2.
Znrf3 mutants exhibit gonadal sex reversal phenotypes on B6.YAKR. (A) B6.YAKR embryonic gonads lacking ZNRF3 at 13.5 (Right, upper row) and 14.5 dpc (Right, lower row) have an ovarian morphology. They lack Sox9 expression and have variable numbers of Stra8-positive germ cells. Stra8 is also expressed in a more restricted and transient fashion at the poles of the B6.YAKR wild-type controls (Left, lower row). Sox9 expression is expressed in testis cords of controls (Left, upper row). (B) Fetal XY gonads lacking a single functional copy of Znrf3 exhibit variable degrees of sex reversal, revealed by Sox9 WMISH (upper row) and Stra8 WMISH (lower row). (C) Female phenotypic sex in an adult B6.YAKR mouse lacking one copy of Znrf3 (Top, center). Sex-reversed animals have an anogenital distance (vertical white bar) usually found in XX controls. Mammary gland pigmentation is also visible in the XY female mouse (center). Examination of internal genitalia in XY heterozygotes reveals uterine horns and bilateral ovaries (Bottom, center). [Scale bar, 500 μm (A and B); 1 cm (C, Top, in black); 0.5 cm (C, Bottom, in white).]
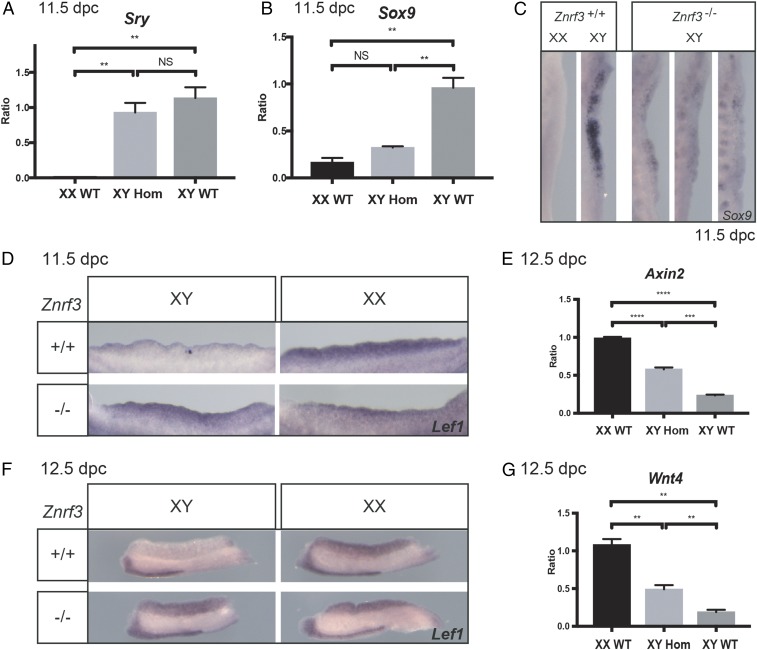
To determine the basis of the observed testis determination defects at 14.5 dpc, we performed molecular analyses around the time of sex determination, from 11.5 dpc to 12.5 dpc, in XY Znrf3 mutant gonads and controls. No overt disruption to Sry expression was detected by qRT-PCR (Fig. 3A). In contrast, Sox9 expression was either reduced or absent compared with XY controls (Fig. 3 B and C). These data suggest that disruption to Sox9, rather than Sry, accounts for the failure to undergo normal testis determination in homozygous XY embryos. Gonadal somatic cell proliferation was also reduced in Znrf3 mutants at 11.5 dpc, consistent with their feminization (SI Appendix, Fig. S5). We then tested whether disruption to Sox9 expression is caused by elevated canonical WNT signaling in XY mutant gonads due to the absence of ZNRF3-mediated clearance of Frizzled receptor. Lef1, which is positively regulated by WNT/β-catenin signals, was detected at higher levels in XY mutant homozygotes than XY controls at both 11.5 and 12.5 dpc (Fig. 3 D and F). Similarly, Axin2 levels were also elevated in XY gonads lacking ZNRF3 (Fig. 3E). By contrast, expression of the somatic cell marker Sf1 and the primordial germ cell marker Oct4 were not affected at 12.5 dpc (SI Appendix, Fig. S6). Interestingly, proovarian Wnt4 expression was enhanced by loss of ZNRF3, both at 11.5 and 12.5 dpc, with noticeable variability in expression levels between distinct mutant XY gonads (Fig. 3G and SI Appendix, Fig. S6). These data suggest that testis determination defects in XY gonads lacking ZNRF3 are caused by ectopic canonical WNT signaling resulting in significant reduction in levels of Sox9 in supporting cell precursors at 11.5 dpc. The inhibitory effect of β-catenin on gonadal Sox9 expression has been previously reported and explained by disruption to SF-1−mediated activation of its gonad-specific enhancer, testis-specific enhancer core (TESCO) (25), although disruption to other Sox9 enhancers is conceivable (26). SRY-dependent promotion of SOX9 transcription may sometimes tilt the balance toward the testicular fate in XY mutants, generating an inherently unstable sex-determining network, accounting for some of the phenotypic variability observed.
Fig. 3.
Ectopic activation of canonical WNT signaling and disruption to Sox9 expression during sex determination in XY Znrf3 homozygotes. (A) The qRT-PCR shows that Sry is expressed in Znrf3 homozygous mutant embryos at 11.5 dpc with no significant difference to controls. (B) The qRT-PCR showing Sox9 expression is significantly reduced in mutants. (C) Reduced Sox9 expression is observed in some mutant gonads (left-hand mutant) following WMISH, while expression is negligible in others. Sox9 expression in XY control gonads is strong. (D) WMISH shows Lef1 expression is elevated in XY homozygous mutant gonads at 11.5 dpc. (E) Expression of Axin2 is also elevated in XY homozygous mutant gonads at 12.5 dpc, although not to XX control levels. (F) Lef1 expression is also clearly detectable in XY homozygotes at 12.5 dpc, in contrast to controls. (G) Expression of Wnt4 is also elevated in XY homozygotes at 12.5 dpc, although, again, not to XX control levels. **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
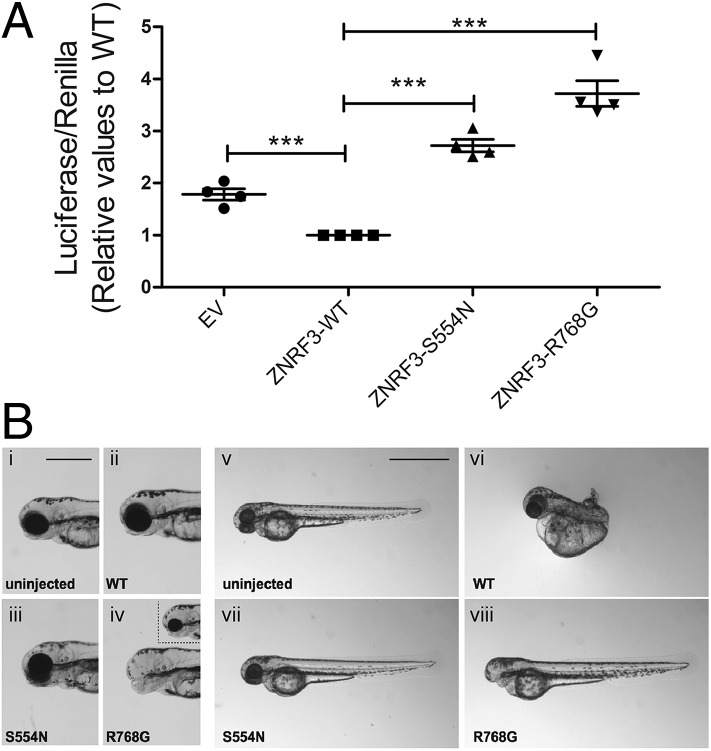
Finally, we tested whether ZNRF3 mutations might disrupt human sexual development, by performing next-generation sequencing of whole exomes from a cohort of patients with very rare 46,XY DSD (Table 1 and SI Appendix, Supplementary Methods). We identified two novel ZNRF3 variants and two known variants in five individuals, including four with 46,XY female presentation (Table 1). No other DSD-associated variants were present in these patients. We then assessed the functional consequences of two of the missense variants. Human cell line transfection assays (Fig. 4A) and zebrafish embryo assays (Fig. 4B) indicate that the Ser554Asn amino acid substitution (patient 2, 46,XY complete gonadal dysgenesis) disrupts the ability of ZNRF3 to inhibit canonical WNT signals compared with wild-type ZNRF3. Both assays also demonstrated that the Arg768Gly substitution (patients 3 and 4, 46,XY DSD of unknown gonadal phenotype) acts to enhance WNT/β-catenin signaling (Fig. 4). Thus, both human variants tested can disrupt ZNRF3’s role in inhibiting canonical WNT signals in both a cellular and organismal context.
Table 1.
ZNRF3 variants detected by exome sequencing of DSD patients
| Variable | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 |
| Ancestry | Tunisian | North African | Indian | Indian | North African |
| Karyotype | 46,XY | 46,XY | 46,XY | 46,XY | 46,XY |
| Age at presentation | 17 y | 16 y | 19 y | 17 y | Birth |
| External genitalia | Female | Female | Female | Female | Perineal hypospadias |
| Internal genitalia | Hypoplastic uterus | Uterus present | Absent uterus | Absent uterus | Male |
| Gonads | Not seen by US | “streak” gonad | Not seen by US | Not seen by US | Intrascrotal testis |
| Endocrine data (reference values) | T, 0.13 ng/mL (2.7 to 9); FSH, 132 U/L (ND to 13.5); LH, 35.9 U/L (2.4 to 13) | T, 0.2 ng/mL (2.7 to 9); FSH, 37 U/L (ND to 13.5); LH, 42 U/L (2.4 to 13) | NA | NA | 3 mo: T, 0.96 ng/mL (0.75 to 4.0); FSH < 0.4 U/L (ND to 13.5); AMH AMH 1,305 pmol/l (100–3,300) |
| ZNRF3 mutation | Splice site exon8:c.2767+5G > A | c.1661G > A p.Ser554Asn | c.2302A > G p.Arg768Gly | c.2302A > G p.Arg768Gly | c.1861T > G p.Arg621Ser |
| Allele Freq (ExAC) and Population | Novel | Novel | 0.00487 South Asian | 0.00487 South Asian | 0.00003258 European |
| Diagnosis | Suspected 46,XY gonadal dysgenesis | 46,XY Complete gonadal dysgenesis | 46,XY DSD | 46,XY DSD | 46,XY DSD |
Normal range refers to the range of basal levels in control subjects matched according to age and chromosomal sex with the case subjects. AMH, anti-Müllerian hormone; FSH, follicle stimulating hormone; LH, luteinizing hormone; NA, not available; ND, not detectable; T, testosterone; US, ultrasound.
Fig. 4.
Human ZNRF3 variants disrupt ZNRF3 activity. (A) Effects of overexpressing wild-type and variant ZNRF3 on WNT/β-catenin signaling was tested by SuperTOPFlash luciferase reporter assay in KGN (ovarian granulosa cell tumor) cells for 48 h. The reporter constructs were transfected into KGN cells with either the empty vector (EV), wild-type (WT), or variant ZNRF3 expression vectors. The values are expressed relative to ZNRF3-WT. Each data point represents the mean of the value of each experiment ± SEM (n = four independent experiments; each experimental measurement was performed in quadruplicate). Pairwise comparison between the effect of ZNRF3-WT and all of the other conditions was performed using a two-tailed Student t test. ***P ≤ 0.0001. (B) Lateral views of 58-h postfertilization uninjected control (i and v) or injected (12.5 pg, ii–iv; 50 pg, vi–viii) zebrafish embryos. Embryos injected with 12.5 pg of ZNRF3-WT or ZNRF3-S554N show no defects, but ZNRF3-R768G−injected embryos display reduction of the forebrain and either loss (iv) or reduction (Inset in iv) of the eyes. At the 50-pg dose, ZNRF3-WT embryos displayed dorsalization (vi), while ZNRF3-S554N−injected embryos showed no axis patterning defects (vii). At 50 pg, ZNRF3-R768G−injected embryos exhibited posteriorization of anterior structures (viii).
Discussion
Here we report detailed analysis of a developmental role for ZNRF3, a known tumor suppressor (27, 28). The testis determination defects described here in ZNRF3-deficient fetuses are associated with a clear elevation of canonical WNT signaling in the developing XY gonad, consistent with the known role of ZNRF3 in increasing membrane turnover of Frizzled receptor and thereby inhibiting WNT signaling (15, 16). The basis of the sex reversal phenotypes observed at 14.5 dpc is the reduction of Sox9 seen at 11.5 dpc, when commitment to either the testicular or ovarian fate is decided. The simplest explanation of this loss of Sox9 expression is the elevation in canonical WNT signals: In other genetic contexts, forcing stabilization of β-catenin can disrupt testis determination (21). Phenotypic variability in the severity of the XY gonadal defects in mutants indicates a degree of instability in the sex-determining mechanism in the absence of ZNRF3; however, while the Znrf3 mutant line was maintained on C57BL/6J for several generations, we cannot exclude residual genetic background differences as a contributory factor. It is worth noting here too that large-scale phenotypic studies of mouse knockouts reveal that incomplete penetrance and variable expressivity are common even on pure genetic backgrounds (29). This phenomenon, like pronounced phenotypic variability between the gonads of a single fetus (Fig. 3C), cannot clearly be explained by genetic mechanisms.
The role of ZNRF3 in testis determination and that of RSPO1 in ovary development (12–14) indicates an antagonistic molecular interaction at the heart of mammalian gonad development (SI Appendix, Fig. S7). Our studies suggest that Znrf3 is expressed at equivalent levels in supporting cells of the XY and XX gonads at the time of sex determination. This suggests that Znrf3 is not an obvious target for transcriptional up-regulation by testis-determining factors such as SRY, SOX9, or FGF9, despite the known role of FGF signals in opposing WNT signals in the mouse (7, 20). We do not, of course, rule out posttranscriptional mechanisms of activation. RSPO1 promotes WNT signaling by inducing membrane clearance of ZNRF3, and the related molecule RNF43 (17), and given the elevated levels of Rspo1 in the XX gonad at the sex-determining stage, we conclude that ZNRF3 activity is reduced in XX supporting cells and therefore elevated in XY cells. In XY cells, ZNRF3 acts to inhibit canonical WNT signaling by increasing membrane turnover of Frizzled receptor, and the consequences of removing that inhibition in the XY gonad are the testis determination defects described here, mediated by suppression of Sox9 expression. Thus, during gonad development, RSPO1 acts to inhibit an inhibitor of canonical WNT signaling, a double-negative operation that is familiar in models of sex determination (30). While loss of RSPO1 results in masculinization of the XX supporting cell lineage only in the postnatal period, this may be due to a number of factors, including the availability of proovarian FOXL2 and/or other pro-WNT signaling factors, the complexity of the ovarian somatic cell lineages, or the contribution of germ cells to ovary development (13, 31, 32). It will be interesting to determine the consequences of removal of both ZNRF3 and RSPO1 on XX and XY gonad development. Likewise, it will be interesting to determine the consequences of Znrf3 overexpression on XX gonad development.
We report that two human ZNRF3 variants, associated with 46,XY DSD and causing amino acid substitutions, can disrupt the role of ZNRF3 in inhibiting canonical WNT signaling. All variants detected map to the long intracellular domain of ZNRF3, the function of which is unclear (24). Affected individuals are heterozygous carriers of each variant identified, and not all are novel. However, in this context, it is very important to reconsider the role played by genetic background in the penetrance and expressivity of sex determination phenotypes. Sex reversal in some B6.YAKR mice lacking just one copy of Znrf3 suggests that heterozygosity for a disruptive ZNRF3 mutation may be sufficient to cause 46,XY DSD, including complete gonadal dysgenesis, in the relevant human genetic background. Taken together, our data establish a testis-determining function for ZNRF3. ZNRF3 inhibits canonical WNT signaling in XY gonads during sex determination, permitting the establishment of robust Sox9 expression. Our data reveal a molecular conflict between ZNRF3 and RSPO1 that governs mammalian sex determination.
Methods
Mouse Strains and Ethical Approval.
Mice used were bred with licensed approval from the UK Home Office (PPL 30/2877 and 70/8988). Mice were housed in individually ventilated cages in a specific pathogen-free environment. Mice harboring Znrf3 and Rnf43 floxed alleles have been previously described (16). Sf1-eGFP reporter mice (33) were a kind gift from J. Bowles and P. Koopman (University of Queensland, Brisbane, QLD, Australia). All lines were maintained on C57BL/6J (B6) for at least six generations.
Generation of Embryos and Whole-Mount in Situ Hybridization.
Noon on the day of the copulatory plug was counted as 0.5 dpc. Embryos collected at 11.5 dpc were staged accurately based on the number of tail somites. Whole-mount in situ hybridization (WMISH) analysis of embryonic tissues and probes for Sox9, Wnt4, and Stra8 have been previously described (34). The Lef1 probe (13) was a kind gift from Marie-Christine Chaboissier, Université Côte d’Azur, CNRS, INSERM, iBV, Nice, France. At least three independent biological samples from a given group were analyzed with a particular marker.
qRT-PCR.
Total RNA was extracted using RNeasy plus micro kit (Qiagen), and reverse transcription (RT) was carried out with 250 ng of total RNA using the High capacity cDNA RT kit (Applied Biosystems). The qRT-PCR was performed with Fast SYBR Green Master Mix (Life Technologies) on a 7500 Fast Real-Time PCR system (Applied Biosystems). RNA expression levels were normalized to those of Hrpt1 (endogenous control) using the ΔΔCt method. At least three samples for each genotype were analyzed. See SI Appendix for primer sequences.
Antibodies and Immunohistochemistry.
Antibodies used in this study were FOXL2 (1:100; a kind gift from Dagmar Wilhelm, The University of Melbourne, Parkville, Australia), AMH (1:100; Santa Cruz sc28912), and GATA4 (1:50; Santa Cruz 25310). At least two independent biological replicates for each sex and genotype were analyzed in immunohistochemistry experiments.
Cell Proliferation Assays.
Dams were injected with 9 mg/kg 5-ethynyl-2′-deoxyuridine (C10339; Thermofisher) and killed 1 h later. Gonads/mesonephroi were dissected from embryos and fixed in 4% paraformaldehyde for 4 h at 4 °C or 2 h at room temperature. Gonads underwent Click-iT chemistry according to manufacturer’s instructions. GATA4 immunostaining was then performed before imaging using a Zeiss 710 multiphoton microscope.
TOPFlash Assays.
The wild-type ZNRF3 construct (ZNRF3-HA) has been previously described (15). Point mutations were generated using site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit; Agilent) according to manufacturer’s instructions. T-cell factor/lymphoid enhancer factor (TCF/LEF) binding-site/mutated binding-site (TOPFlash/FOPFlash) reporter plasmids (M50 Super 8x TOPFlash and M51 Super 8x FOPFlash mutant; Addgene) were used for the detection of β-catenin−driven WNT transcriptional activity. See SI Appendix for further details.
Zebrafish Embryo Assays.
To determine the effect of human variants on ZNRF3 function, we used an overexpression assay in zebrafish that assesses the impact of low (12.5 pg) and high (50 pg) doses of injected ZNRF3 messenger RNA on early embryogenesis. Plasmids containing ZNRF3-HA were mutagenized using the QuikChange II Site-Directed Mutagenesis kit (Agilent) to incorporate the S554N and R768G point mutations. See SI Appendix for details.
Subjects, Samples, and Exome Sequencing.
All patients with 46,XY DSD met the revised criteria of the Pediatric Endocrine Society European Society for Pediatric Endocrinology. This study was approved by the local French ethical committee (2014/18NICB; registration number IRB00003835), and consent to genetic testing was obtained from adult probands or from the parents when the patient was under 18 y. Exon enrichment was performed with Agilent SureSelect Human All Exon V4. Paired-end sequencing was performed on the Illumina HiSeq2000 platform with TruSeq v3 chemistry. See SI Appendix for details.
Single-Cell RNAseq.
The Fluidigm C1 Autoprep system was used for cell capture and processing. Methodological details can be found in Stévant et al. (35) and SI Appendix.
Supplementary Material
Acknowledgments
We thank the husbandry team in Ward 5 of the Mary Lyon Centre at Harwell and the Frozen Embryo and Sperm Archive (FESA) and histology teams. We thank Dagmar Wilhelm for the kind gift of anti-FOXL2 antibody. We thank Phil Johnson for zebrafish husbandry. We acknowledge European Cooperation in Science & Technology (COST) Action BM1303 (DSDnet). This work was supported by the Medical Research Council by core funding Grant MC_U142684167 (to A.G.) at the Harwell Institute, and the Agence Nationale de la Recherche Grant ANR-10-LABX-73 (to K.M.). S.N. acknowledges support from Swiss National Science Foundation Grant 31003A_173070. M.S. was a visiting scientist supported by the Strategic International Research Exchange Program between Princeton University and National Institutes of Natural Sciences, Japan. D.T.G. was supported by National Institute of Arthritis and Mucoskeletal and Skin Diseases Pathway to Independence Award 1K99AR070905. Work in the R.D.B. laboratory is supported by the National Institute of Child Health and Development Grant 2R01HD048584.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801223115/-/DCSupplemental.
References
- 1.Cool J, DeFalco T, Capel B. Testis formation in the fetal mouse: Dynamic and complex de novo tubulogenesis. Wiley Interdiscip Rev Dev Biol. 2012;1:847–859. doi: 10.1002/wdev.62. [DOI] [PubMed] [Google Scholar]
- 2.Warr N, Greenfield A. The molecular and cellular basis of gonadal sex reversal in mice and humans. Wiley Interdiscip Rev Dev Biol. 2012;1:559–577. doi: 10.1002/wdev.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 4.Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 5.Barrionuevo F, et al. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- 6.Hiramatsu R, et al. A critical time window of Sry action in gonadal sex determination in mice. Development. 2009;136:129–138. doi: 10.1242/dev.029587. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, et al. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 10.Vainio S, Heikkilä M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 11.Biason-Lauber A, Konrad D, Navratil F, Schoenle EJ. A WNT4 mutation associated with Müllerian-duct regression and virilization in a 46,XX woman. N Engl J Med. 2004;351:792–798. doi: 10.1056/NEJMoa040533. [DOI] [PubMed] [Google Scholar]
- 12.Parma P, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 13.Chassot AA, et al. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet. 2008;17:1264–1277. doi: 10.1093/hmg/ddn016. [DOI] [PubMed] [Google Scholar]
- 14.Tomizuka K, et al. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet. 2008;17:1278–1291. doi: 10.1093/hmg/ddn036. [DOI] [PubMed] [Google Scholar]
- 15.Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 16.Koo BK, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, et al. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 2013;14:1120–1126. doi: 10.1038/embor.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zebisch M, et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4:2787. doi: 10.1038/ncomms3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen PH, Chen X, Lin Z, Fang D, He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27:1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jameson SA, Lin YT, Capel B. Testis development requires the repression of Wnt4 by Fgf signaling. Dev Biol. 2012;370:24–32. doi: 10.1016/j.ydbio.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maatouk DM, et al. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet. 2008;17:2949–2955. doi: 10.1093/hmg/ddn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouma GJ, Washburn LL, Albrecht KH, Eicher EM. Correct dosage of Fog2 and Gata4 transcription factors is critical for fetal testis development in mice. Proc Natl Acad Sci USA. 2007;104:14994–14999. doi: 10.1073/pnas.0701677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warr N, et al. Transgenic expression of Map3k4 rescues T-associated sex reversal (Tas) in mice. Hum Mol Genet. 2014;23:3035–3044. doi: 10.1093/hmg/ddu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard P, et al. Wnt signaling in ovarian development inhibits Sf1 activation of Sox9 via the Tesco enhancer. Endocrinology. 2012;153:901–912. doi: 10.1210/en.2011-1347. [DOI] [PubMed] [Google Scholar]
- 26.Gonen N, Quinn A, O’Neill HC, Koopman P, Lovell-Badge R. Normal levels of Sox9 expression in the developing mouse testis depend on the TES/TESCO enhancer, but this does not act alone. PLoS Genet. 2017;13:e1006520. doi: 10.1371/journal.pgen.1006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, et al. ZNRF3 acts as a tumour suppressor by the Wnt signalling pathway in human gastric adenocarcinoma. J Mol Histol. 2013;44:555–563. doi: 10.1007/s10735-013-9504-9. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, et al. ZNRF3 contributes to the growth of lung carcinoma via inhibiting Wnt/β-catenin pathway and is regulated by miR-93. Tumour Biol. 2016;37:3051–3057. doi: 10.1007/s13277-015-3949-2. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson ME, et al. International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McElreavey K, Vilain E, Abbas N, Herskowitz I, Fellous M. A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc Natl Acad Sci USA. 1993;90:3368–3372. doi: 10.1073/pnas.90.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maatouk DM, Mork L, Chassot AA, Chaboissier MC, Capel B. Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev Biol. 2013;383:295–306. doi: 10.1016/j.ydbio.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rastetter RH, et al. Marker genes identify three somatic cell types in the fetal mouse ovary. Dev Biol. 2014;394:242–252. doi: 10.1016/j.ydbio.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet. 2006;15:417–431. doi: 10.1093/hmg/ddi463. [DOI] [PubMed] [Google Scholar]
- 34.Bogani D, et al. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 2009;7:e1000196. doi: 10.1371/journal.pbio.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stévant I, et al. Deciphering cell lineage specification during male sex determination with single-cell RNA sequencing. Cell Rep. 2018;22:1589–1599. doi: 10.1016/j.celrep.2018.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.