Significance
Night shift work is associated with adverse health effects, including diabetes, cardiovascular disease, and cancer. Understanding the molecular mechanisms that underlie this association is instrumental in advancing the diagnosis, prevention, and treatment of shift work-related health concerns. We characterized the effect on genome-wide gene expression levels of a 4-day protocol simulating night shifts in healthy human subjects under highly controlled laboratory conditions. We demonstrate that this night shift protocol leads to a dampening of gene expression rhythms and a desynchrony between rhythmic transcripts and the shifted sleep/wake cycle. Moreover, we uncovered key biological processes and regulatory molecules that are altered during this night shift protocol and that may contribute to the development of health problems on the long term.
Keywords: chronobiology, circadian rhythms, transcriptomics, night shift work
Abstract
Misalignment of the endogenous circadian timing system leads to disruption of physiological rhythms and may contribute to the development of the deleterious health effects associated with night shift work. However, the molecular underpinnings remain to be elucidated. Here, we investigated the effect of a 4-day simulated night shift work protocol on the circadian regulation of the human transcriptome. Repeated blood samples were collected over two 24-hour measurement periods from eight healthy subjects under highly controlled laboratory conditions before and 4 days after a 10-hour delay of their habitual sleep period. RNA was extracted from peripheral blood mononuclear cells to obtain transcriptomic data. Cosinor analysis revealed a marked reduction of significantly rhythmic transcripts in the night shift condition compared with baseline at group and individual levels. Subsequent analysis using a mixed-effects model selection approach indicated that this decrease is mainly due to dampened rhythms rather than to a complete loss of rhythmicity: 73% of transcripts rhythmically expressed at baseline remained rhythmic during the night shift condition with a similar phase relative to habitual bedtimes, but with lower amplitudes. Functional analysis revealed that key biological processes are affected by the night shift protocol, most notably the natural killer cell-mediated immune response and Jun/AP1 and STAT pathways. These results show that 4 days of simulated night shifts leads to a loss in temporal coordination between the human circadian transcriptome and the external environment and impacts biological processes related to the adverse health effects associated to night shift work.
On the long term, night shift and rotating shift work are associated with an increased prevalence of various medical disorders, such as diabetes, cardiovascular disease, and cancer (1). With 20–30% of the workforce in North America and Europe involved in shift work (2, 3), it is crucial to gain a better understanding of the physiological mechanisms contributing to these adverse health effects. Maladaptation of the endogenous circadian timing system to the altered sleep/wake schedule is thought to be an important contributor (4); however, the underlying molecular processes are unknown.
Through the coordination of 24-h rhythms of many physiological and behavioral processes, the circadian timing system allows organisms to anticipate daily variations in light, temperature, and food availability (5). At the molecular level, these rhythms are generated via transcriptional/translational feedback loops involving the periodic expression of clock genes (6). By regulating the expression of many output genes, clock genes and their protein products have a profound effect on physiology. This is exemplified by the recent observation that the expression of 6–9% of the human blood transcriptome shows a circadian rhythm (7, 8).
Night shift work has been associated with alterations in a wide range of physiological parameters, including elevated postprandial glucose, insulin, and triacylglycerol levels as well as increased body mass index and waist–hip ratio (9–11). Furthermore, long-term exposure to night shift work is associated with higher levels of total white blood cells, neutrophils, lymphocytes, and monocytes, indicating dysregulation of the immune system (12).
The study of the human transcriptome may yield important clues as to how physiology is affected by night shift work and how this may contribute to the development of health problems on the long term. Recently, it has been shown that a forced desynchrony protocol, in which subjects are exposed to 28-h days so that the endogenous circadian timing system dissociates from the sleep/wake cycle, or sleep restriction leads to a reduction in the number and amplitude of cycling transcripts (7, 8, 13). Various biological processes are affected under these conditions, many of which have been implicated in the adverse health effects associated with night shift work (14). However, it is still unknown to what extent night shift work affects the circadian regulation of the human transcriptome. Here, we study the effect of simulated night shift work, in which the sleep period is acutely delayed by 10 h for 4 d, on genome-wide gene expression levels in peripheral blood mononuclear cells (PBMCs) under highly controlled conditions to better understand the molecular underpinnings of the negative health effects associated with night shift work.
Results
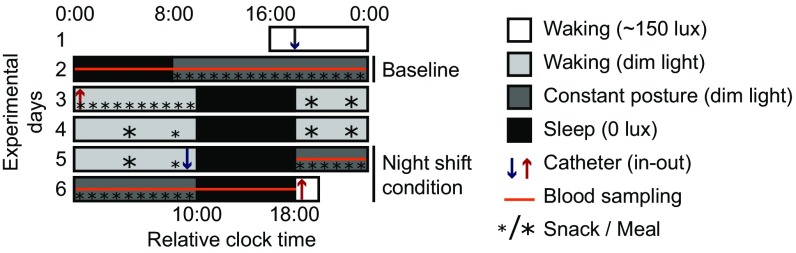
Eight healthy human subjects were exposed to a 4-d simulated night shift work protocol in which their habitual sleep period was delayed by 10 h (Fig. 1; see SI Methods for details). Blood samples for microarray analysis were taken every 4 h during two 24-h measurement periods before and on the fourth day of the simulated night shift protocol under highly controlled laboratory conditions. A main effect of study night on actigraphy-based sleep duration was found (P = 0.013; mixed-effects model with subject as random effect), with significantly less sleep during the last sleep opportunity at the end of the second measurement period compared with the first and second sleep periods (Fig. S1). On average, subjects slept 38 ± 10 min less in the 8-h sleep opportunities during the night shift protocol than at baseline.
Fig. 1.
Study protocol. Subjects entered the time isolation laboratory on day 1. The first 24-h measurement period started at the beginning of day 2. Subjects slept according to their habitual sleep/wake schedule, followed by a 16-h constant posture procedure, during which they remained in a semirecumbent position and received hourly isocaloric snacks. On day 3, the sleep episode was delayed by 10 h relative to their habitual sleep schedule, followed by an 8-h sleep period. This night shift schedule was maintained for the subsequent nights. Following the third night on this schedule, subjects underwent another 24-h measurement period. Blood samples (10 mL) for microarray analysis were collected every 4 h during both measurement periods.
Circadian Rhythms in the Human Transcriptome at Baseline and During the Night Shift Condition.
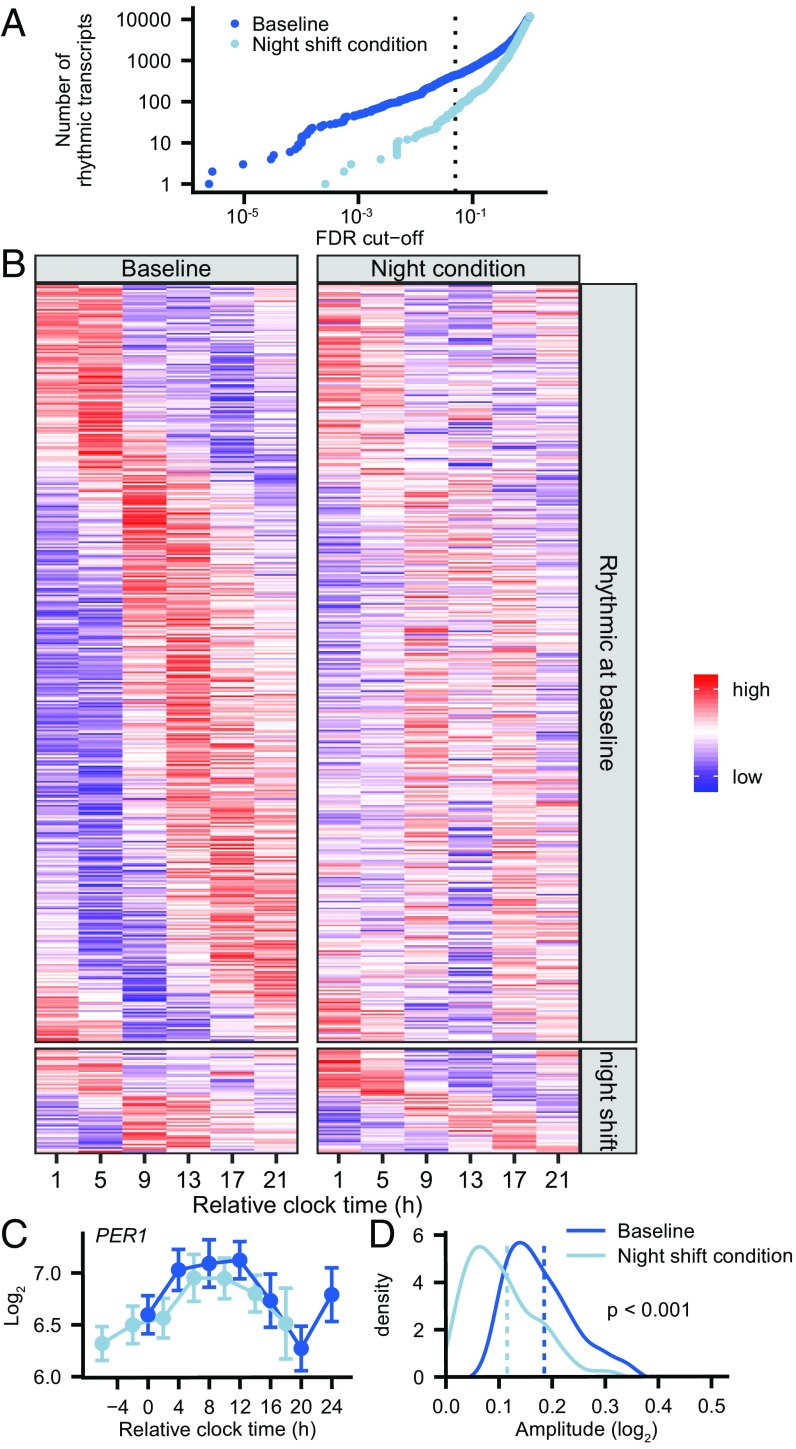
Mixed-effects cosinor analysis on genome-wide gene expression levels at baseline and during the night shift condition separately revealed that the number of probe sets classified as rhythmic was consistently lower during the night shift condition compared with baseline across a continuum of false-discovery rates (FDRs) (Fig. 2A). For example, at an FDR of 0.05, the number of rhythmic probe sets decreased from 3.8% [n = 444 probe sets, targeting 442 unique transcripts (4.0%)] at baseline to 0.5% in the night shift condition [n = 62 probe sets, targeting 62 unique transcripts (0.6%)] (Fig. 2A). At this FDR cutoff, 35 probe sets were significantly rhythmic in both conditions, including transcripts related to the immune system (FCGR3A, FCGR3B, GNLY, and SLC11A1), regulation of transcription (FOXP1, TSC22D3, PAX5, and PRDM1), and metabolism (MPI, PDK4, and MARC1) (Dataset S1). Additionally, cosinor analysis based on individual expression profiles, which takes into account the possibility that transcripts may be cycling in different subjects with a different phase, revealed that the number of rhythmic probe sets decreased in seven out of eight subjects in the night shift condition compared with baseline (Fig. S2A). Variability was observed regarding the probe sets that cycle in different subjects, with more heterogeneity in the night shift condition compared with baseline (Fig. S2B). Combining the individual P values using Fisher’s method, we found that, across all subjects, the number of rhythmic transcripts dropped from 11.8% at baseline to 6.5% in the night shift condition (Fig. S2C and Dataset S1). Expression profiles of rhythmic circadian clock-related transcripts at baseline in our study show similar phase relationships as previously reported (Fig. S3 and SI Discussion).
Fig. 2.
Group-level cosinor analysis. (A) Number of rhythmic transcripts identified at baseline and during the night shift condition across the continuum of FDR cutoffs. The dotted vertical line represents an FDR cutoff of 0.05. (B) Heatmap showing the expression profiles during baseline (Left) and the night shift condition (Right) of probe sets that are rhythmic at baseline (Top) and during the night shift condition (Bottom). Rows are ordered by phase of the peak expression, as determined by cosinor analysis. (C) Example of the expression profile (mean ± SEM) of a transcript (PER1) that is identified as significantly rhythmic at baseline but not during the night shift condition. (D) Comparison of amplitude of all 471 probe sets that are rhythmic either at baseline and during the night shift condition as identified by group-level cosinor analysis on the two conditions separately. Amplitudes in the night shift condition [0.115 (0.077); mean (SD)] were significantly lower than those at baseline [0.185 (0.076); P < 0.0001, Mann–Whitney test]. Dotted lines show the mean amplitudes.
These findings indicate a large decrease of significantly rhythmic transcripts in the simulated night shift work schedule. Interestingly, close inspection of a heatmap depicting all transcripts that were identified as significantly rhythmic by the group-level cosinor analysis (Fig. 2B) suggested that the apparent decrease in the proportion of rhythms transcripts might be due, at least partly, to a reduction in the amplitude of many transcripts. For example, the clock gene PER1 was classified as significantly rhythmic at baseline but not during the night shift condition. However, its expression profile seems to vary over the 24-h period in the night shift condition as well, albeit with a reduced amplitude (amplitude at baseline, 0.404, vs. night shift condition, 0.313; Fig. 2C). In addition, comparing the amplitudes of all 471 probe sets that are significantly rhythmic at baseline, during the night shift condition, or both, it was found that the amplitudes in the night shift condition were significantly lower than those at baseline (Fig. 2D). Therefore, the lower number of rhythmic transcripts in the night shift condition may indicate dampened rhythms rather than a complete loss of rhythmicity per se. These observations prompted us to use an approach that allows for the direct comparison of rhythms between the two conditions.
Circadian Rhythms in the Transcriptome and the Effect of Night Shift Protocol.
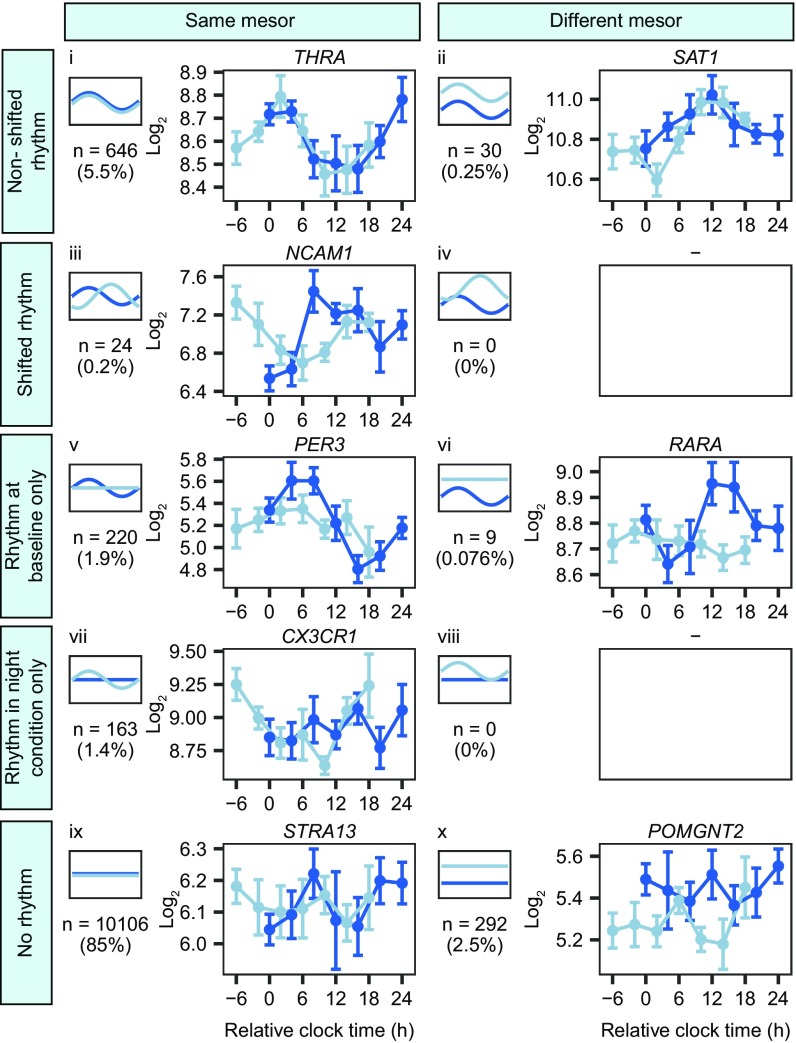
To assess the expression profiles at baseline and during the night shift condition simultaneously, a model selection approach based on the Bayesian information criterion (BIC) was used to group gene expression profiles into 10 different possible categories based on changes in the 24-h rhythmic pattern and/or changes in mesor in the night shift condition compared with baseline (Fig. 3). Using a cutoff of 0.4 on the BIC weights, 11,490 probe sets (97.1% of probe sets expressed above background on the microarray) were assigned to a category (Dataset S2). Two of the possible categories were not assigned any transcripts, indicating that none of the transcripts provided an optimal fit to these two categories.
Fig. 3.
Summary of rhythmic gene expression and the effect of the simulated night shift protocol. Dark blue lines, baseline; light blue lines, night shift condition. Different panels represent different model categories. Small plots on the Left of each panel provide an example of each model category. Note that a change in mesor or a shift in rhythmicity can occur in either direction. The number represents the number of probe sets assigned to each category; the percentage represents the percentage of probe sets assigned to each category relative to all probe sets detected as expressed on the microarray. Plots on the Right of each panel show examples of the expression levels of a transcript in each category (mean ± SEM).
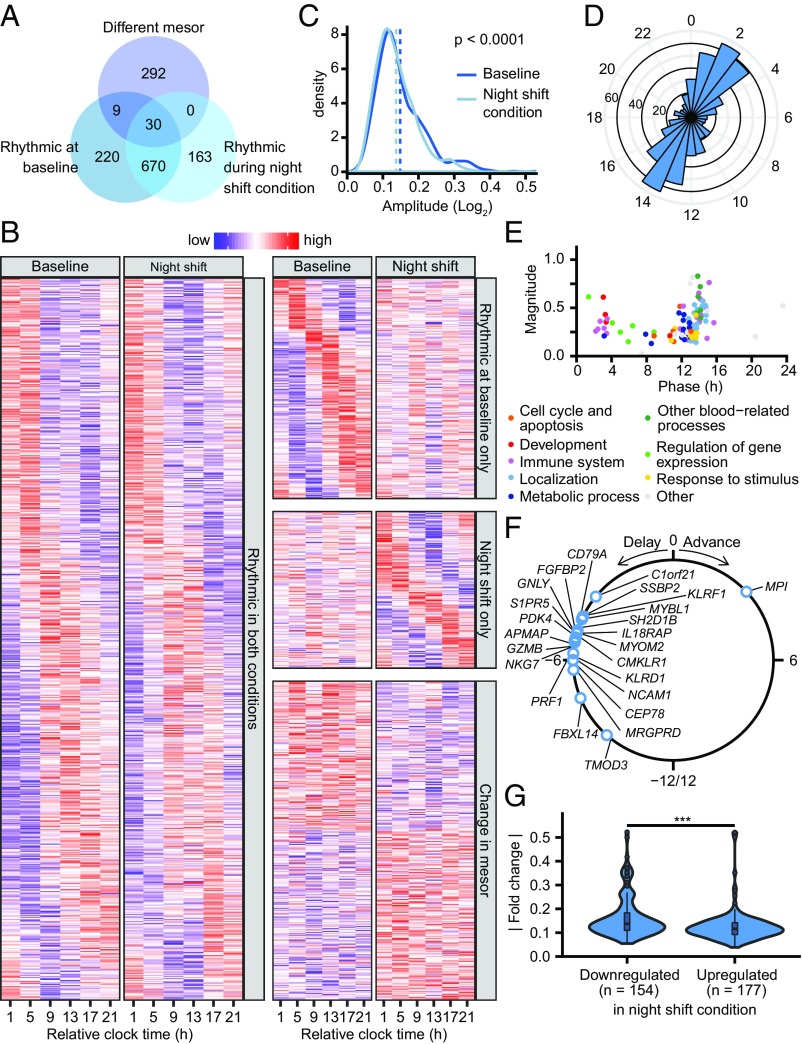
Twenty-four-hour rhythms were found in the expression levels of 1,092 probe sets targeting 1,078 transcripts (9.7% of the transcripts detected as expressed on the microarray) (Fig. 4A). Among these, 700 probe sets were rhythmic in both conditions (among which 24 displayed a shifted rhythm) (categories i–iv in Fig. 3), 229 were rhythmic at baseline only (categories v and vi in Fig. 3), and 163 were only rhythmic during the night shift condition (categories vii and viii in Fig. 3). Interestingly, this approach revealed that 73% of the probe sets (676 out of 929 probe sets) that were rhythmic at baseline remained cycling with a similar rhythmic pattern during the night shift condition. This indicates that the majority of rhythmic probe sets at baseline were misaligned with respect to the sleep period during the night shift condition.
Fig. 4.
Effect of the simulated night shift protocol on rhythmic gene expression. (A) Venn diagram showing the number of probe sets rhythmic at baseline and during the night shift condition identified by the model selection approach, as well as probe sets that show a change in mesor (Fig. 3). (B) Heatmap of probe sets identified as having a rhythm in both conditions (Left; categories i, ii, iii, and iv in Fig. 3), a rhythm only at baseline (Top Right; categories v and vi), a rhythm only during the night shift condition (Middle Right; categories vii and viii), or an overall change in baseline levels (Bottom Right; categories ii, iv, vi, viii, and x). Rows are ordered by phase or the extent of up- or down-regulation. Note that the rows in the different panels are not mutually exclusive as some probe sets that are identified as rhythmic also show a change in mesor. (C) Density plot of the amplitude of transcripts that are significantly rhythmic both at baseline and during the night shift condition as estimated by cosinor analysis performed on both conditions separately. The mean amplitude (dotted vertical lines) is significantly reduced from 0.149 at baseline to 0.137 during the night shift condition (P < 0.0001, paired Mann–Whitney test). (D) Phase distribution of probe sets that are rhythmic in both conditions. (E) Magnitude and phase of enriched biological processes (q < 0.05 vs. uniform phase distribution) identified using phase set enrichment analysis, among transcripts that are rhythmic in both conditions, colored by functional category. (F) Change in phase of transcripts that show altered rhythmicity during the night shift condition relative to baseline (categories iii and iv in Fig. 3). (G) Distribution of absolute up- and down-regulation in the night shift condition compared with baseline. (***P < 0.001, Mann–Whitney test.)
A heatmap confirms the large degree of overlap in rhythmic genes in the two conditions that was not detected by the previous analysis (Fig. 4B). Various circadian clock-related genes showed oscillations in both conditions, such as PER1, DBP, ARNTL, NR1D1, NR1D2, SIN3A, and NFIL3, while others showed only a rhythm at baseline (e.g., RAI1 and PER3) or were arrhythmic in both conditions (e.g., RORA, RORC, and CLOCK) (Fig. S4A). In general, amplitudes of probe sets that were rhythmic in both conditions were significantly reduced in the night shift condition compared with baseline (Fig. 4C).
The phase distribution of the 676 probe sets (669 unique transcripts) that were rhythmic in both conditions with a similar amplitude and phase (categories i and ii in Fig. 3) revealed a bimodal pattern (Fig. 4D). Phase set enrichment analysis (15) revealed temporal segregation of significantly enriched biological processes (Fig. 4E and Dataset S3), including chromatin modification (clustering ∼1 h after habitual bedtimes), transcriptional regulation (∼4–9 h after habitual bed times), metabolic processes (∼12–13 h after habitual bedtimes), and blood-specific processes (∼14 h after habitual bedtimes). Processes related to the immune system were clustered in the early night (∼2 h after the habitual bedtimes), including leukocyte activation and differentiation, and in the midafternoon (∼14 h after the habitual bedtimes), including the innate immune system, cytokine production, and migration of leukocytes.
Twenty-four rhythmic probe sets (targeting 24 unique transcripts) showed a phase shift and/or a change in amplitude in the night shift condition compared with baseline (category iii and iv in Fig. 3). Among these, 23 out of 24 transcripts showed a phase delay [average: 5.4 (1.2) h, circular mean (SD)]. The exception was MPI, which was phase advanced by 3 h (Fig. 4F). Of note, 7 out of the 23 phase-delayed transcripts were related to natural-killer (NK) cell-mediated immune response (SH2D1B, GNLY, PRF1, KLRF1, KLRD1, GZMB, and NKG7), which oscillated in a highly synchronous manner (Fig. S4B).
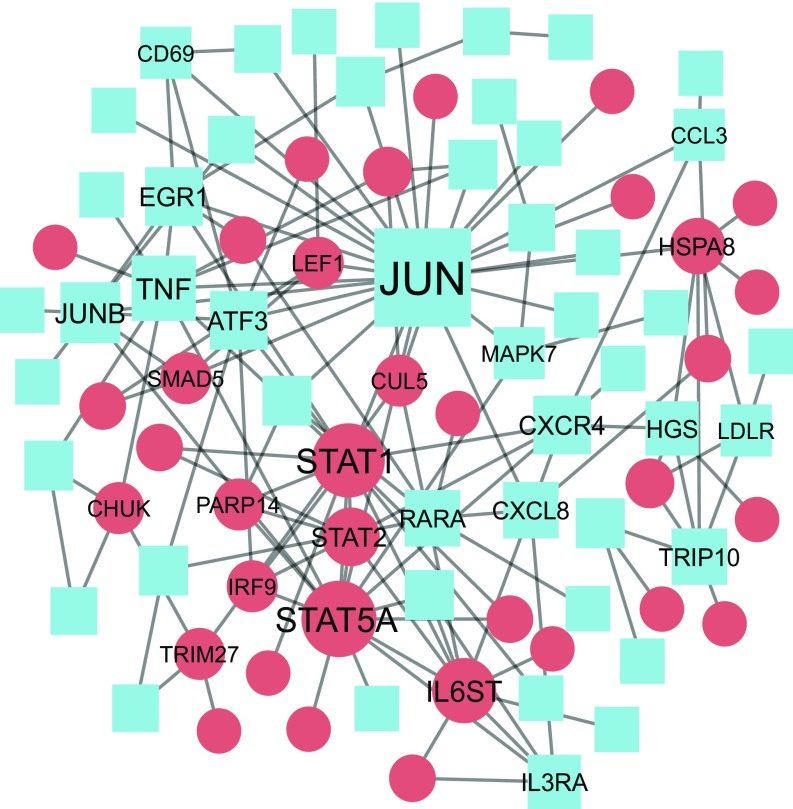
An effect of the simulated shift work protocol on overall expression levels was found in 331 probe sets targeting 330 unique transcripts (3.0% of the transcripts detected as expressed on the microarray; categories ii, iv, vi, vii, and x in Fig. 3), of which 154 probe sets were down-regulated and 177 probe sets were up-regulated during the night shift condition. It was found that the fold change of down-regulated probe sets [0.164 (0.087); mean (SD)] was significantly larger than that of the up-regulated probe sets [0.126 (0.067)] (P < 0.0001; Mann–Whitney test; Fig. 4G). A functional interaction network revealed the transcription factors JUN, STAT5A, and STAT1 as highly interconnected nodes among the transcripts that were differentially regulated during night shift condition (Fig. 5).
Fig. 5.
Functional interaction network of transcripts that were up-regulated and down-regulated during the night shift condition. Node size represents the number of connections within the network. Lines represent the functional interactions between gene products as identified in the Reactome database (version 2016). Red circles, up-regulated genes; blue squares, down-regulated genes. Transcript names are shown for transcripts with four or more connections.
Discussion
It is well known that rotating and permanent night shift work is associated with an increased prevalence of various health problems, but a detailed understanding of the underlying molecular mechanisms is lacking. The objective of this study was to assess the effect of simulated night shift work on the circadian regulation of the human PBMC transcriptome. Our initial analyses based on a binary rhythmic/arrhythmic classification showed that the number of rhythmic transcripts dropped during the night shift work condition compared with baseline on both group and individual levels. This is in line with previous research in humans and animals showing a large reduction in the number of rhythmic transcripts in various tissues when sleep occurs out of phase with the endogenous circadian clock (7, 16, 17) (see SI Discussion for a detailed comparison between our study and previous human circadian transcriptomic studies). However, our subsequent analysis using a model selection approach to assess 24-h rhythms in both conditions simultaneously revealed that 73% of the transcripts that were rhythmic at baseline remained oscillating during the night shift condition with a similar phase relative to the habitual bedtimes, albeit with reduced amplitudes. This creates a state of misalignment between the rhythmic transcripts and the shifted sleep/wake and feeding cycles and demonstrates that temporal coordination is disrupted between the transcriptome and the external environment after 4 d of simulated night shift work.
Cosinor analysis, or other statistical methods to detect rhythmic time series [such as MetaCycle (18) or RAIN (19)], are typically used to compare the proportion of rhythmic transcripts between two conditions based on a classification of rhythmic vs. arrhythmic at specific statistical cutoff values (see SI Discussion for more details on the group-level and individual-level cosinor analysis). Moving beyond this binary classification, as recently recommended in a guidelines paper endorsed by many chronobiologists (20), we employed a model selection approach to directly compare the rhythmic parameters of gene expression profiles between the two conditions, as previously done in the context of differential rhythmicity analysis among pre-mRNA, mRNA, and ribosome footprints (21). This allowed us to infer that the large reduction in significantly rhythmic transcripts is not due to a loss of rhythmicity per se, but rather to overall dampening of rhythmicity.
Peak times of transcripts that were identified as rhythmic during both baseline and the night shift condition showed a bimodal distribution, as observed previously (7, 8, 13). The majority of transcripts peaked around 2 and 14 h after the habitual bedtimes. Phase set enrichment analysis (15) revealed profound temporal clustering of significantly enriched biological processes, including lipid metabolism, platelet activation, response to stress, and regulation of cell proliferation. These observations show that many biological processes remain aligned to baseline during 4 d of simulated night shift work, resulting in a state of desynchrony between the internal circadian timing system and the altered sleep/wake cycle. Interestingly, the temporal clustering of biological processes is similar to previous publications on the human circadian transcriptome, with processes related to the regulation of gene expression peaking during the biological night and processes related to response to stimuli peaking during the biological day (7, 8).
We identified 24 transcripts whose expression profile had shifted during the 4 d of simulated shift work. The expression of these transcripts is most likely influenced by the shifted rest/activity cycle or feeding schedules rather than by the central circadian clock. Strikingly, among these 24 transcripts, 7 (including PRF1 and GZMB that encode cytolytic factors perforin 1 and granzyme B) were related to NK cells, which are key components of the innate immune system and play a crucial role in the killing of tumors and virally infected cells (22). This is reminiscent of previous findings showing that circadian disruption alters perforin 1 and granzyme B levels and suppresses NK cell-mediated killing of tumor cells in mice (23, 24).
Furthermore, the model selection approach showed that 3.0% of the human transcriptome is either up-regulated or down-regulated during the night shift condition. A functional interaction network revealed that key regulatory transcripts were affected by the night shift work protocol. The most connected node in the network, JUN, was down-regulated during the night shift condition. JUN encodes a basic leucine zipper protein that dimerizes with FOS to form the AP1 transcription factor, which regulates various cellular processes, such as apoptosis, cell proliferation, and differentiation (25). Disruption of circadian clock function in mice has been linked to AP1-controlled oncogenic activation of Myc, uncontrolled cell proliferation, and tumor growth (26), which was suggested to explain the increased risk of cancer associated to circadian disruption (27). The observation that JUN is down-regulated in the night shift condition provides additional evidence for altered AP1 signaling in circadian disruption. Furthermore, several members of the signal transducer and activator of transcription (STAT) protein family (STAT1, STAT2, STAT5A) were up-regulated in the night shift condition and appeared as highly interconnected nodes in the gene interaction network. STAT family members regulate the expression of hundreds of genes involved in defense mechanisms against viruses and tumors (28). Their up-regulation during the simulated shift work protocol may alter the expression of many of their targets and thereby affect these defense mechanisms.
Our protocol was designed to study the acute effect of a shifted sleep period and feeding behavior, as experienced on a regular basis by millions of shift workers around the world. The central circadian clock, as measured by plasma melatonin levels, did not adapt to the shifted sleep/wake cycle in our study subjects (29) as well as in a similar protocol (30). Although some of the entrainment cues that may be present in actual night shift workers, such as physical exercise and exposure to brighter light, are not mimicked by our protocol, it should be noted that in the majority of actual shift workers, the central circadian clock does not adjust to the night shift schedule (31, 32). Furthermore, our protocol may have caused partial sleep deprivation, as subjects were kept awake during the first night of the shift work protocol and sleep duration was significantly reduced during the final sleep period. Although sleep restriction was shown to affect the human blood transcriptome (8), we expect the effect to be relatively small as the degree of sleep deprivation was minimal. Since sleep restriction is commonly experienced by night shift workers (1), our protocol reflects a situation that is experienced by a large proportion of shift workers not only in terms of circadian misalignment but also in terms of sleep restriction.
We determined transcriptional changes in PBMCs, a subset of white blood cells that, besides their prominent role in the immune system, have been successfully used to evaluate genome-wide responses to a variety of interventions (33–35). Even though PBMCs are not representative of all physiological processes that can be measured in blood, our results show that PBMCs are suitable to evaluate genome-wide changes in rhythmic gene expression and provide insight on a wide range of biological processes related to metabolism, transcriptional regulation, cell proliferation, and stress response.
Several limitations of our study should be considered. First, the sample size was relatively small. Since the baseline and night shift condition were completed by the same individuals, we can be confident that any effect of the night shift condition is due to the intervention rather than to the small sample size. Furthermore, we detected significant 24-h rhythms in the expression of known clock-related genes at baseline, demonstrating the ability to identify rhythmicity in known cyclic genes. The effect of the night shift condition on the expression profile of clock genes shown in this study is similar to our previous report based on quantitative PCR expression analysis in the same subjects (36). Second, the majority of our study population was male. Interestingly, the only subject that showed an increase in the number of rhythmic transcripts during the night shift condition was female. Further research is required to investigate the effect of sex on the circadian regulation of the human transcriptome.
In this study, we show that 4 d of simulated night shift work leads to a disruption of temporal coordination in the circadian regulation of the human transcriptome, which is characterized by reduced amplitudes of rhythmic transcripts and overall misalignment of rhythmic transcripts with the shifted sleep/wake cycle. Various key biological processes are affected by the shift work protocol that may have implications for long-term health effects. Further research is warranted to extend these findings to actual shift workers and other populations at risk for circadian misalignment.
Methods
Study Protocol.
For microarray analysis, a total of 103 samples were available from eight healthy subjects [age, 22.5 (18–29) y, mean (range); seven men/one woman] that participated in the simulated shift work protocol (Fig. 1). All subjects provided written informed consent before the study. The study was approved by the Douglas Institute Ethics Board and was conducted according to the Declaration of Helsinki. Details on the experimental protocol were previously published (29) and are available in SI Methods. A total of 103 RNA samples (n = 12–14 samples per subject) were analyzed using microarray (Fig. S5). Details on the isolation of PBMCs, RNA extraction, microarray hybridization, and data preprocessing are available in SI Methods. Microarray data have been deposited in the Gene Expression Omnibus database (accession no. GSE107537). All analyses are performed on log2-transformed expression values.
Cosinor Analysis.
A linear mixed-effects model was used to identify 24-h rhythms of gene expression profiles on a group level at baseline and during the night shift condition separately. A cosinor model with a period of 24 h was fit to the expression values of each probe set using a linear mixed-effects model (R package lme4, version 1.1-14):
| [1] |
In this model, yijk is the log2 expression of gene k in individual i at time point j, ak is the fitted average log2 expression (mesor), bk and ck are the cosinor coefficients, tij is the time after lights off in individual i at time point j (in hours), ηik represents the interindividual variability associated to subject i, and εijk is the residual variability. The likelihood ratio test was used to compare the fit of this model to the null model in which bk = 0 and ck = 0. The resulting P values were corrected for multiple testing using the Benjamini–Hochberg method (FDR < 5%). Amplitude and phase (time of peak) were calculated from the cosinor coefficients b and c (37). For details regarding the individual-level cosinor analysis, see SI Methods.
Differential Rhythmicity Analysis.
To study rhythmicity at baseline and during the night shift condition simultaneously, a model selection approach was used (21). This approach allowed us to directly assess rhythmic patterns in gene expression profiles and the effect of the simulated night shift condition on these patterns. Different models were applied to the data in which the mean a and/or the cosinor coefficients b and c in Eq. 1 were either shared or different between the two conditions and in which the coefficients b and c could either be zero (no rhythmicity) or nonzero (rhythmicity). The fit of the different models was compared using the BIC, using a threshold of 0.4 on the BIC weights. For additional details, see SI Methods. To quantify and compare the amplitude of transcripts identified as rhythmic in both conditions, group-level mixed-effects cosinor analysis was applied on data from the conditions separately, and the cosinor coefficients were used to calculate the amplitude as described above. For details on phase set enrichment analysis (15) and the functional interaction network (38), see SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. S. Rhéaume as well as A. Azzoug and M. R. Guertin (research nurses) for medical supervision. We acknowledge the contribution of the McGill University and Génome Québec Innovation Centre for performing the microarray hybridization and related services. This research was supported by operating grants from the Canadian Institutes of Health Research [MOP-102724 (to D.B.B. and N.C.)]. L.K. and M.C. received postdoctoral fellowships from the Fonds de Recherche du Québec–Santé.
Footnotes
Conflict of interest statement: D.B.B. provides conferences and legal expert advice on various shift work-related cases.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107537).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720719115/-/DCSupplemental.
References
- 1.Kecklund G, Axelsson J. Health consequences of shift work and insufficient sleep. BMJ. 2016;355:i5210. doi: 10.1136/bmj.i5210. [DOI] [PubMed] [Google Scholar]
- 2.Alterman T, Luckhaupt SE, Dahlhamer JM, Ward BW, Calvert GM. Prevalence rates of work organization characteristics among workers in the U.S.: Data from the 2010 National Health Interview Survey. Am J Ind Med. 2013;56:647–659. doi: 10.1002/ajim.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eurofound 2015. Sixth European Working Conditions Survey 2015 (Eurofound, Dublin)
- 4.Boivin DB, Boudreau P. Impacts of shift work on sleep and circadian rhythms. Pathol Biol (Paris) 2014;62:292–301. doi: 10.1016/j.patbio.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Schibler U, et al. Clock-Talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb Symp Quant Biol. 2015;80:223–232. doi: 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2016;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer SN, et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci USA. 2014;111:E682–E691. doi: 10.1073/pnas.1316335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110:E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S, et al. Effects of rotating shift work on biomarkers of metabolic syndrome and inflammation. J Intern Med. 2007;261:285–292. doi: 10.1111/j.1365-2796.2007.01766.x. [DOI] [PubMed] [Google Scholar]
- 10.Lund J, Arendt J, Hampton SM, English J, Morgan LM. Postprandial hormone and metabolic responses amongst shift workers in Antarctica. J Endocrinol. 2001;171:557–564. doi: 10.1677/joe.0.1710557. [DOI] [PubMed] [Google Scholar]
- 11.Morris CJ, Purvis TE, Mistretta J, Scheer FA. Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J Clin Endocrinol Metab. 2016;101:1066–1074. doi: 10.1210/jc.2015-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirth MD, et al. Association of shiftwork and immune cells among police officers from the Buffalo Cardio-Metabolic Occupational Police Stress study. Chronobiol Int. 2017;34:721–731. doi: 10.1080/07420528.2017.1316732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnardottir ES, et al. Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. Sleep. 2014;37:1589–1600. doi: 10.5665/sleep.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laing EE, et al. Exploiting human and mouse transcriptomic data: Identification of circadian genes and pathways influencing health. BioEssays. 2015;37:544–556. doi: 10.1002/bies.201400193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Podtelezhnikov AA, Hogenesch JB, Anafi RC. Discovering biology in periodic data through phase set enrichment analysis (PSEA) J Biol Rhythms. 2016;31:244–257. doi: 10.1177/0748730416631895. [DOI] [PubMed] [Google Scholar]
- 16.Husse J, et al. Tissue-specific dissociation of diurnal transcriptome rhythms during sleep restriction in mice. Sleep. 2017;40 doi: 10.1093/sleep/zsx068. [DOI] [PubMed] [Google Scholar]
- 17.Barclay JL, et al. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7:e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes ME, et al. Guidelines for genome-scale analysis of biological rhythms. J Biol Rhythms. 2017;32:380–393. doi: 10.1177/0748730417728663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atger F, et al. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci USA. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16:112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 23.Logan RW, et al. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. J Immunol. 2012;188:2583–2591. doi: 10.4049/jimmunol.1102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469–476. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Fu L, Lee CC. The circadian clock: Pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kettner NM, Katchy CA, Fu L. Circadian gene variants in cancer. Ann Med. 2014;46:208–220. doi: 10.3109/07853890.2014.914808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuesta M, Boudreau P, Dubeau-Laramée G, Cermakian N, Boivin DB. Simulated night shift disrupts circadian rhythms of immune functions in humans. J Immunol. 2016;196:2466–2475. doi: 10.4049/jimmunol.1502422. [DOI] [PubMed] [Google Scholar]
- 30.McHill AW, et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci USA. 2014;111:17302–17307. doi: 10.1073/pnas.1412021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folkard S. Do permanent night workers show circadian adjustment? A review based on the endogenous melatonin rhythm. Chronobiol Int. 2008;25:215–224. doi: 10.1080/07420520802106835. [DOI] [PubMed] [Google Scholar]
- 32.Boudreau P, Dumont GA, Boivin DB. Circadian adaptation to night shift work influences sleep, performance, mood and the autonomic modulation of the heart. PLoS One. 2013;8:e70813. doi: 10.1371/journal.pone.0070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burczynski ME, Dorner AJ. Transcriptional profiling of peripheral blood cells in clinical pharmacogenomic studies. Pharmacogenomics. 2006;7:187–202. doi: 10.2217/14622416.7.2.187. [DOI] [PubMed] [Google Scholar]
- 34.Carlson LA, et al. Changes in transcriptional output of human peripheral blood mononuclear cells following resistance exercise. Eur J Appl Physiol. 2011;111:2919–2929. doi: 10.1007/s00421-011-1923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Mello VD, Kolehmanien M, Schwab U, Pulkkinen L, Uusitupa M. Gene expression of peripheral blood mononuclear cells as a tool in dietary intervention studies: What do we know so far? Mol Nutr Food Res. 2012;56:1160–1172. doi: 10.1002/mnfr.201100685. [DOI] [PubMed] [Google Scholar]
- 36.Cuesta M, Boudreau P, Cermakian N, Boivin DB. Rapid resetting of human peripheral clocks by phototherapy during simulated night shift work. Sci Rep. 2017;7:16310. doi: 10.1038/s41598-017-16429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornelissen G. Cosinor-based rhythmometry. Theor Biol Med Model. 2014;11:16. doi: 10.1186/1742-4682-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: A Cytoscape app for pathway and network-based data analysis. F1000 Res. 2014;3:146. doi: 10.12688/f1000research.4431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.