Significance
We show that mutations in HMGA2 affect fetal resource allocation, testis descent, and the size of pigs and provides a target for gene modification that can be used to modulate size in other mammalian species. This can have implications in agriculture as well as in the development of new strains of companion animals. In addition, most xenograft pig donors have adult organs larger than those of humans. Recently, it has been shown that regulation of organ growth is donor-controlled, not host-controlled, resulting in organ overgrowth and damage after transplantation. We show here that the HMGA2 gene is a potential target for organ-size regulation in xenotransplantation.
Keywords: HMGA2, dwarfism, swine, gene editing, organ size
Abstract
Expression of HMGA2 is strongly associated with body size and growth in mice and humans. In mice, inactivation of one or both alleles of Hmga2 results in body-size reductions of 20% and 60%, respectively. In humans, microdeletions involving the HMGA2 locus result in short stature, suggesting the function of the HMGA2 protein is conserved among mammals. To test this hypothesis, we generated HMGA2-deficient pigs via gene editing and somatic cell nuclear transfer (SCNT). Examination of growth parameters revealed that HMGA2−/+ male and female pigs were on average 20% lighter and smaller than HMGA2+/+ matched controls (P < 0.05). HMGA2−/− boars showed significant size reduction ranging from 35 to 85% of controls depending on age (P < 0.05), and organ weights were also affected (P < 0.05). HMGA2−/+ gilts and boars exhibited normal reproductive development and fertility, while HMGA2−/− boars were sterile due to undescended testes (cryptorchidism). Crossbreeding HMGA2−/+ boars and gilts produced litters lacking the HMGA2−/− genotype. However, analysis of day (D) D40 and D78 pregnancies indicated that HMGA2−/− fetuses were present at the expected Mendelian ratio, but placental abnormalities were seen in the D78 HMGA2−/− concepti. Additionally, HMGA2−/− embryos generated by gene editing and SCNT produced multiple pregnancies and viable offspring, indicating that lack of HMGA2 is not lethal per se. Overall, our results show that the effect of HMGA2 with respect to growth regulation is highly conserved among mammals and opens up the possibility of regulating body and organ size in a variety of mammalian species including food and companion animals.
The high mobility group A2 (HMGA2) gene encodes a nonhistone chromosomal and architectural transcription factor that facilitates 3D changes in chromatin structure (1). HMGA2 proteins contain a binding domain peptide with three AT-hook motifs that preferentially recognize AT-rich regions in the minor groove of DNA. Of the three AT-hooks, the second motif is the most critical component for HMGA2 protein function, and the motif is highly conserved among diverse organisms (2), including pigs. Acting as an enhanceosome, chromatin-bound HMGA2 promotes the recruitment of regulatory protein complexes involved in cell proliferation and differentiation (3). During mouse embryonic development, Hmga2 expression is detected at a high level throughout the embryo except in the embryonic brain, but it is not detected in normal adult tissues except in the testes (4, 5). In humans, genome-wide association studies showed that variation in HMGA2 gene affects human height (6), and identification of an intragenic microdeletion in the HMGA2 gene in short-stature patients further supports a direct role of HMGA2 in human growth (7). HMGA2 has also been implicated in dwarfism in rabbits (8), body weight in dogs (9), and beak size in birds (10). Thus, genetic evidence supports a conserved role for HMGA2 in growth regulation.
Direct evidence for the role of Hmga2 in growth regulation comes from observations in small or “pygmy” mice with naturally occurring mutations in Hmga2 (11, 12). Hmga2 effects on body size were confirmed by inactivation of the mouse Hmga2 by insertional inactivation or homologous recombination (3, 13). Compared with wild-type controls, adult mice with inactive Hmga2, whether due to an induced or a naturally occurring mutation, were 60% smaller, and adult heterozygous mutants were 20% smaller (3, 13). Organ size was also affected (14). In contrast, overexpression of a truncated form of HMGA2 resulted in gigantism and lipomatosis in mice (15). Despite the documented role in regulation of body size in mice, the definitive function of HMGA2 in pigs remains elusive; HMGA2 has been associated with ear size in pigs (16, 17) and higher expression in fetal skeletal muscle of breeds with higher muscularity (18). Previously it has been shown that inactivation of the growth hormone receptor (GHR) in pigs results in reduced adult weight and size and increase in obesity. As of yet, however, there are no reports of the effect of HMGA2 inactivation in pigs. The goal of this project, therefore, was to understand the role of HMGA2 in body and organ-size regulation in pigs and to determine the usefulness of HMGA2 modification for the regulation of size in genetically modified pigs to be used for biomedical research including xenotransplantation. Reduced size will allow for better match between organ receipt and donor (19) as well as facilitate animal housing and management in a biomedical setting.
Results
Generation of HMGA2-Deficient Pigs.
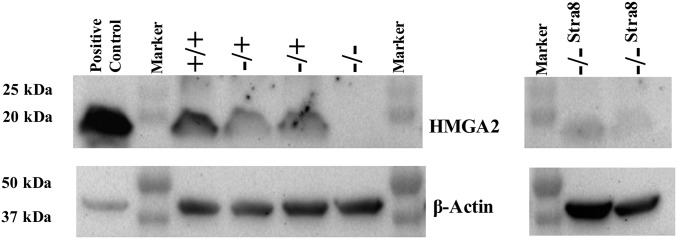
To study the effect of disruption of the HMGA2 gene on fetal and adult growth HMGA2−/+ male and female fetal fibroblasts cell lines were generated as somatic cell nuclear transfer (SCNT) donors. As HMGA2 is expressed in fetal fibroblasts, conventional homologous recombination using β-geo and a gene-trap approach was used (20) (SI Appendix, Fig. S1 and Table S1). Gene targeting events in single cell-derived colonies were confirmed as described in SI Appendix. Targeting rates were 20/34 (58.8%) for male cell lines and 4/28 (14.3%) for female cell lines. These rates are comparable to those reported in sheep somatic cells using gene trap and selection (21). HMGA2−/+ female and male pigs generated by SCNT were crossbred and litters analyzed. In addition, we generated HMGA2−/− offspring by gene editing with transcription activator-like effector nucleases (TALENs) the remaining wild-type allele in the HMGA2−/+ fetal fibroblasts. We also used TALENs to generate knock-out/knock-in lines that disrupted the endogenous HMGA2 locus while also introducing an HMGA2 coding sequence under the control of the mouse Stra8 promoter (SI Appendix, Fig. S1). This results in inactivation of the HMGA2 locus but testis-specific expression of an HMGA2 cDNA via the Stra8 promoter. This was done to potentially reduce previously reported spermatogenesis defects in Hmga2−/− mice (5), without affecting the influence of HMGA2 on overall body size. HMGA2−/−Stra8 colonies were identified by PCR screening followed by sequencing, and 8/40 (20%) cell lines analyzed had the correct modification. To confirm functional disruption of HMGA2 fibroblast cell lines derived from HMGA2−/+, HMGA2−/−Stra8, HMGA2−/−, and control fibroblast lines were analyzed by Western blot (Fig. 1). Both HMGA2−/− and HMGA2−/−Stra8 cell lines showed the absence of HMGA2, while HMGA−/+cells had reduced levels compared with wild-type controls. Densitometry analysis showed that HMGA2−/+ cells had approximately half the level of HMGA2 compared with wild-type cells (SI Appendix, Fig. S2). Using HMGA2 mutant cell lines we generated 8 HMGA2−/+ boars, 11 HMGA2−/−Stra8 boars, 5 HMGA2−/− boars, and 6 HMGA2−/+ gilts. As no statistical differences in any phenotype measured were observed between HMGA2−/− and HMGA2−/−Stra8 pigs, data from both lines were combined and referred to as HMGA2-null heretofore.
Fig. 1.
HMGA2 protein in gene-edited fetal fibroblasts. HMGA2 protein levels for HMGA2+/+, HMGA2−/+, HMGA2−/−, and HMGA2−/−Stra8 fetal fibroblasts were detected by Western blot. Proteins were extracted from primary D40 fetal fibroblasts of all three HMGA2 genotypes generated from the breeding of HMGA2−/+ founders. HMGA2−/−Stra8 samples were isolated from primary D43 fetal fibroblasts generated by SCNT. HMGA2+/+ had twice as much protein as HMGA2−/+ samples, suggesting there was no compensation from the remaining active allele (SI Appendix, Fig. S2). There was no protein detected from HMGA2−/− or HMGA2−/−Stra8 fibroblasts. β-Actin was used as internal control. HEK293 lysate was used as positive control.
Fetal Viability of HMGA2-Null Concepti.
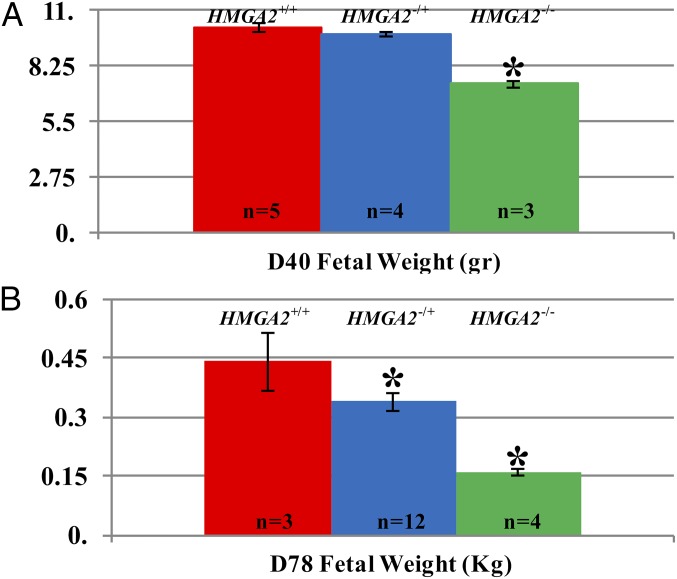
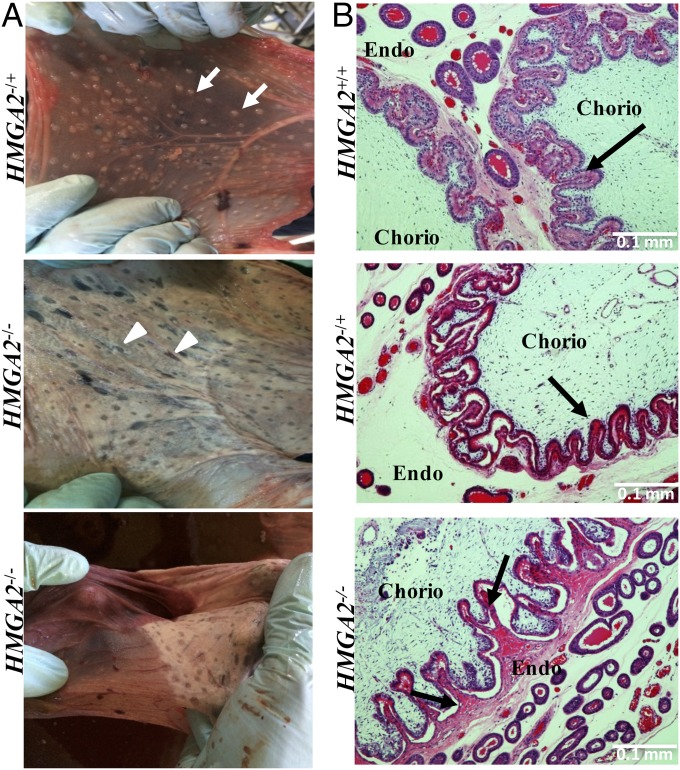
Mating of HMGA2−/+ × HMGA2−/+ founders resulted in three pregnancies to term, producing a total of 19 piglets: 6 HMGA2+/+ and 13 HMGA2−/+, no live HMGA2−/−, and 8 highly degraded dead fetuses (mummies) that could not be genotyped (SI Appendix, Fig. S3). A χ2 test to validate the goodness of fit for Mendelian ratios genotypes indicated that the live offspring data did not meet the expected ratios (P < 0.05). To determine the timing of fetal loss and to identify a cause for the loss, one pregnancy at day (D) D40 and two pregnancies at D78 were examined. At D40 and D78 HMGA2 genotyping of fetuses confirmed the presence of all genotypes, including HMGA2−/−, at the expected Mendelian frequency. Size differences were evident at fetal stages, with both male and female HMGA2-null fetuses examined at D40 and D78 of gestation being smaller and lighter than HMGA2+/+ fetuses (Fig. 2 and SI Appendix, Fig. S4). While no obvious defects beyond size reduction were detected in the HMGA2−/− D78 fetuses, abnormalities were observed in the D78 HMGA2−/− placentas at both the macroscopic and microscopic level (Fig. 3). Macroscopically, HMGA2−/− D78 placentas showed extensive degeneration and detached easily from the uterus (Fig. 3). At the histological level, HMGA2−/− placentas had abnormal rugae at the contact area between chorionic membrane and uterine endometrium. The calculated perimeter (micrometers) and area (square micrometers) in contact between fetal membranes and endometrium were significantly reduced between HMGA2+/+ and HMGA2−/− placentas by 9% and 32%, respectively (P < 0.05). To determine if there was an interaction between the HMGA2 deficiency and uterine crowding, unilateral oviduct ligation was performed on two HMGA2−/+ F1 gilts. This results in smaller litter sizes and reduced uterine crowding. The resulting pregnancies produced nine live offspring (five and four piglets per pregnancy) and one degraded mummy. Of the nine live piglets, four were HMGA2+/+ and five were HMGA2−/+. No live HMGA2−/− fetuses survived to term.
Fig. 2.
Fetal weights (D40 and D78) of HMGA2 gene-edited pigs. Pregnancies were generated by crossbreeding HMGA2−/+ gilts and boars. (A) Fetuses with all expected genotypes were detected at D40 at the expected Mendelian frequency. Weights of HMGA2−/+ fetuses were comparable to those of HMGA2+/+ fetuses, but the HMGA2−/− fetuses were significantly smaller (30% reduced) compared with the other two genotypes. Two females and one male HMGA2−/− fetuses were collected, all size-restricted. (B) At D78, HMGA2−/+ fetuses were 24% smaller and HMGA2−/− were 72% smaller than HMGA2+/+ fetuses. Both HMGA2−/+ and HMGA2−/− male and female fetuses were affected (nine females:three males for HMGA2−/+ and one female:three males for HMGA2−/−).
Fig. 3.
Placental morphology in HMGA2 concepti at D78 of gestation. (A) HMGA2−/+ placentas were healthy and well-vascularized, while HMGA2−/− placentas showed lack of absorptive structures (areolae; arrowheads compared with arrows) and vascularization. (B) Histological analysis of placenta and endometrium from HMGA2+/+, HMGA2−/+, and HMGA2−/− fetuses. Villi were severely degenerated in HMGA2−/− placenta compared with other genotypes. Furthermore, poor contact of villi between uterine endometrium (Endo) and chorionic membrane (Chorio) was detected for HMGA2−/− (arrow), suggesting the connection between uterus and placenta was deficient. (Scale bars: 100 μm.)
Effects of HMGA2 Modifications on Growth Parameters.
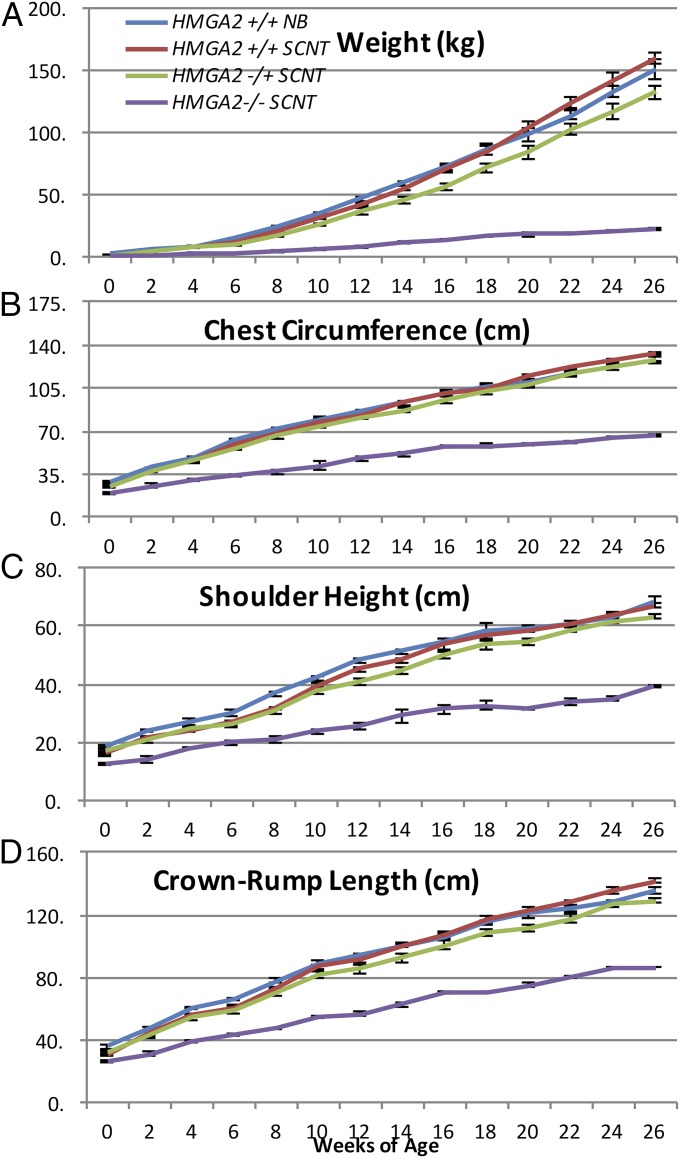
Due to the inability to generate HMGA2−/− offspring by breeding, growth comparisons were carried out using SCNT-generated animals. In boars, for the first 3 mo, length, height, circumference, and body weights of HMGA2−/+ SCNT animals were not significantly different from HMGA2+/+ SCNT controls. However, from 4 to 6 mo of age, HMGA2−/+ SCNT boars were 17% (P < 0.05) and 16% (P < 0.05) lighter compared with HMGA2+/+ SCNT boars and naturally bred boars, respectively (Fig. 4). Body length, height, and circumference measurements showed similar reductions in size (Fig. 4). HMGA2-null SCNT boars showed a dramatic reduction in all measured growth/size parameters from birth (Fig. 4). A representative photograph of all HMGA2 genotypes at 6 mo of age is shown in Fig. 5. To determine if the HMGA2 deficiency was affecting symmetry, an allometric growth analysis was performed. Each set of measurements (body length, height, circumference, and weight) was log-transformed and plotted to determine the scaling exponent, represented by the slope of the line of best fit. Standardized major axis regression analysis indicated that these slopes were not significantly different from one another (SI Appendix, Fig. S5). This suggests that none of the HMGA2 mutations affected the scaling exponent, indicating that the relationship between each measurement changes with growth is similar in both mutant and control pigs. However, significant differences in the y-intercepts of the regression lines of best fit were observed in the length vs. circumference (P < 0.05) and circumference vs. weight comparisons (P < 0.05). Pairwise comparisons revealed that at the same given length the circumference of the HMGA−/− pigs was 5% less than the SCNT and wild-type controls (P < 0.05), and that the heterozygous circumference measurements were 5% larger than the SCNT and wild-type controls (P < 0.05). At the same circumference, the weight for heterozygous mutant was lower than that of the SCNT and wild-type controls by 6% and 7%, respectively (P < 0.05), while the weight of the homozygous mutant was greater than that of the controls by 21% and 18%, respectively (P < 0.05). Essentially the HMGA2−/− mutants had a greater weight-to-size ratio (higher density/leaner) compared with a wild-type pigs of the same size. These morphological differences apply at birth and are retained throughout the animal’s life, as indicated by identical regression slopes. In gilts, disruption of one HMGA2 allele resulted in a 35% reduction in growth parameters, compared with control gilts at 30 wk of age (SI Appendix, Fig. S6). This reduction remained significant for HMGA2−/+ gilts until the end of the study, at 59 wk of age. In addition to examining allometric whole-body growth parameters, we examined whether organ weights were affected. As shown in Table 1, HMGA2−/+ boars at 9 mo of age had smaller heart and testis weights, while no statistically significant reductions were observed in other organs examined. Compared with HMGA2+/+ gilts, HMGA2−/+ gilts at 2.5 y of age also had reduced organ weights with the exception of brain and lung (Table 2). In HMGA2−/− pigs, organ sizes at 1 wk of age (Table 3) and 15 wk (SI Appendix, Fig. S7 and Table S2) were affected compared with controls and all organs showed significant reductions. Some organs, however, were proportionally more affected than others (Tables 1–3).
Fig. 4.
Growth comparison of HMGA2−/− SCNT, HMGA2−/+ SCNT, HMGA2+/+ SCNT, and HMGA2+/+ naturally bred (NB) boars. Measurements for weight (A), chest circumference (B), shoulder height (C), and crown–rump length (D) were taken every 2 wk over a 26-wk period. HMGA2+/+ NB, n = 5; HMGA2+/+ SCNT, n = 6; HMGA2−/+; n = 6; HMGA2−/−, n = 3 (one HMGA2−/− died at 22 wk so last 4 wk n = 2). Error bars indicate SEM.
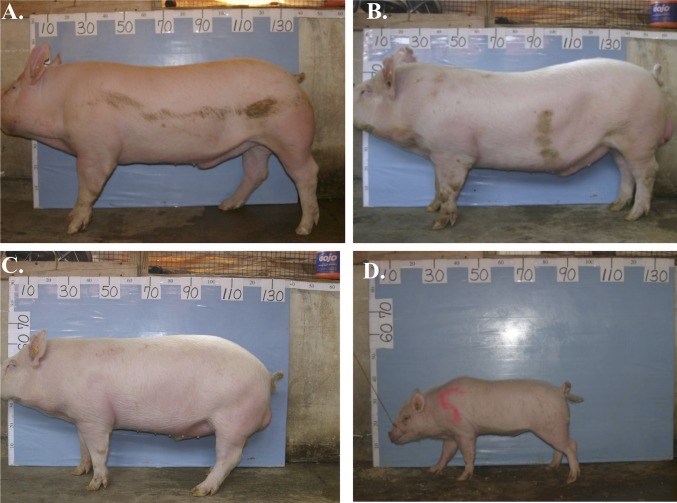
Fig. 5.
Visual comparison of representative HMGA2+/+, HMGA2−/+, and HMGA2−/− boars. At 26 wk of age boars were photographed in front of the same measuring board for to facilitate size comparisons. Board numbering is in centimeters. (A) HMGA2+/+ NB, (B) HMGA2+/+ SCNT, (C) HMGA2−/+ SCNT, (D) HMGA2−/− SCNT. Note drastic reduction in size and stocky appearance of the HMGA2−/− boar. NB, naturally bred.
Table 1.
Body and organ weight comparison of HMGA2−/+ vs. HMGA2+/+ boars at 9 mo of age
| Body and organs | HMGA2−/+ (n = 5) | HMGA2+/+ (n = 6) | Change in weight,* % |
| Body† | 165.2 ± 37.3 kg | 221.73 ± 3.6 kg | 74.5 |
| Adrenal glands† | 5.8 ± 0.2 g | 6.8 ± 0.7 g | 84.9 |
| Brain | 86.8 ± 3.2 g | 98.5 ± 2.9 g | 88.1 |
| Heart† | 504.4 ± 27.5 g | 638.5 ± 15.6 g | 79.0 |
| Kidneys | 671.6 ± 23.2 g | 656.3 ± 30.2 g | 102.3 |
| Livers | 3,056.6 ± 67.2 g | 3,145 ± 193.4 g | 97.2 |
| Lungs | 830.2 ± 85.6 g | 926.8 ± 49.0 g | 89.6 |
| Testes† | 1,081.8 ± 65.5 g | 1,282.2 ± 56.9 g | 84.4 |
Change in weight is the ratio of body or organ weights of HMGA2−/+ over HMGA2+/+.
Significant at P < 0.05.
Table 2.
Body and organ weight comparison of HMGA2−/+ vs. HMGA2+/+ nonparous gilts at 2.5 y of age
| Body and organs | HMGA2−/+ (n = 4) | HMGA2+/+ (n = 2) | Change in weight,* % |
| Body | 253.3 ± 7.5 kg | 275.5 ± 15.5 kg | 91.9 |
| Adrenal glands† | 8.0 ± 0.8 g | 11.0 ± 1.0 g | 72.7 |
| Brain | 116.0 ± 0.9 g | 118.5 ± 4.5 | 98.3 |
| Heart† | 497.3 ± 9.0 g | 853.0 ± 10.0 g | 58.3 |
| Kidneys† | 381.8 ± 18.5 g | 704.5 ± 82.5 g | 54.2 |
| Livers† | 2,372.5 ± 59.3 g | 3,224.5 ± 208.5 g | 73.6 |
| Lungs | 1,238.8 ± 136.2 g | 1,161.0 ± 59.0 g | 106.7 |
| Uterus† | 647.0 ± 67.6 g | 1,201.0 ± 25.0 g | 53.8 |
| Ovaries† | 29.7 ± 7.8 g‡ | 97.5 ± 61.5 g | 30.4 |
Change in weight is the ratio of body or organ weights of HMGA2−/+ over HMGA2+/+.
Significant at P < 0.05.
n = 3 due to cystic ovary.
Table 3.
Body and organ weight comparison of HMGA2−/− vs. HMGA2+/+ boars at 4 d of age
| Body and organs | HMGA2−/− (n = 10) | HMGA2+/+ (n = 2) | Change in weight,* % |
| Body† | 0.65 ± 0.03 kg | 1.44 ± 0.06 kg | 45.1 |
| Adrenal glands† | 0. 09 ± 0.01 g | 0. 36 ± 0.01 g | 25.0 |
| Brain† | 23.30 ± 0.38 g | 34.47 ± 0.13 g | 67.6 |
| Heart† | 4.89 ± 0.20 g | 14.07 ± 0.27 g | 34.8 |
| Kidneys† | 6.35 ± 0.39 g | 14.41 ± 0.76 g | 44.0 |
| Livers† | 26.24 ± 1.49 g | 72.76 ± 5.36 g | 36.1 |
| Lungs† | 16.22 ± 1.48 g | 30.74 ± 3.45 g | 52.8 |
| Testes† | 0.39 ± 0.03 g | 1.40 ± 0.61 g | 27.9 |
Change in weight is the ratio of body or organ weights of HMGA2−/− over HMGA2+/+.
Significant at P < 0.05.
Effect of HMGA2 on Male and Female Reproductive Characteristics.
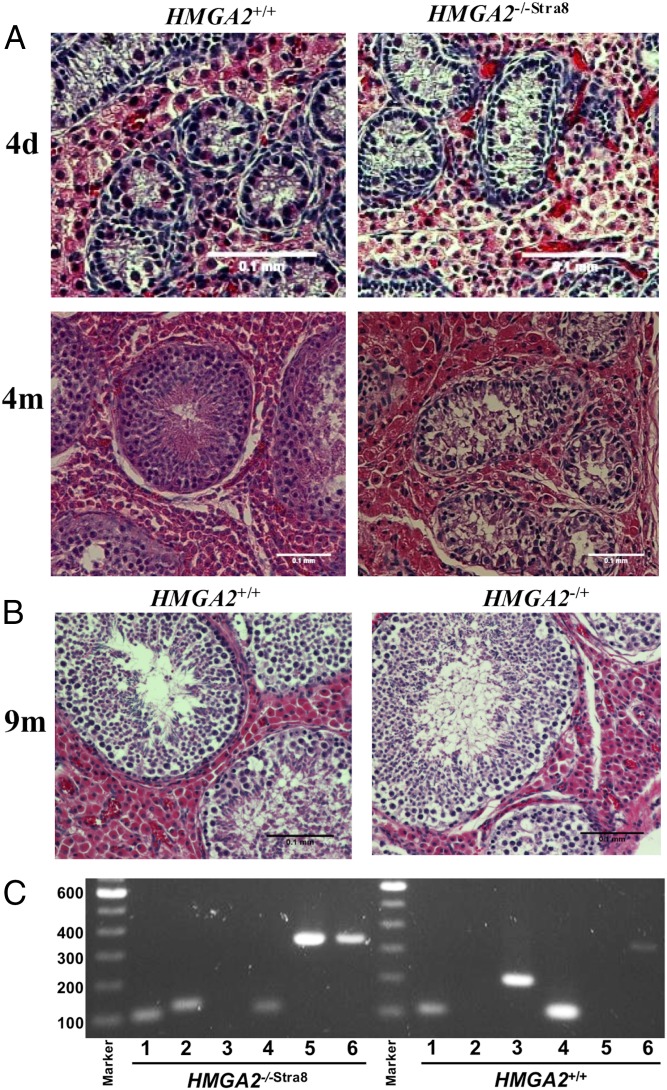
Both HMGA2−/+ boars and HMGA2−/+ F1 gilts showed normal estrus behavior and reproductive characteristics. Sperm collected from HMGA2−/+ boars had normal morphology and concentration. LacZ staining assay on embryos generated by in vitro fertilization of HMGA2+/+ oocytes with HMGA2−/+ sperm confirmed its ability to fertilize as well as germ-line transmission of the β-geo transgene (SI Appendix, Fig. S8). Gross pathological and histological examination of the testes and ovaries from the HMGA2−/+ boars and gilts revealed they were small but otherwise normal (Fig. 6 and SI Appendix, Figs. S9 and S10). In contrast, all HMGA2-null boars had undescended testes (cryptorchidism) at all time points examined, and upon dissection those testes were smaller in size than those of age-matched controls (Tables 1 and 3 and SI Appendix, Fig. S11). Comparison of neonate and 4-mo cryptorchid HMGA2−/−Stra8 boars to age-matched HMGA2+/+ testes showed that the internal structure and organization of seminiferous tubules in the HMGA2-null testes were morphologically normal in the neonates (Fig. 6). However, the seminiferous tubules in HMGA2−/−Stra8 undescended testes showed no evidence of spermatogenesis at 4 mo (Fig. 6). Analysis of expression of HMGA2 in control and HMGA2−/−Stra8 testis confirmed expression of HMGA2 from the mouse Stra8 promoter (Fig. 6C and SI Appendix, Tables S3 and S4).
Fig. 6.
Testis histology and HMGA2 expression in control and age-matched HMGA2−/−Stra8 testis. (A) Testis from control and HMGA2−/−Stra8 pigs at postnatal D4 and 4 mo of age. At D4, cell structure and arrangement of seminiferous tubules was the same as in age-matched HMGA2+/+ testis. However, at 4 mo, none of seminiferous tubules in the cryptorchid HMGA2−/−Stra8 testis showed germ-cell differentiation, while a few tubules in HMGA2+/+ testis showed evidence of spermatogenesis. (Scale bars: 100 μm.) (B) Testes from an HMGA2−/+ boar at 9 mo of age compared with the testis of an age-matched HMGA2+/+ boar, both showing evidence of spermatogenesis. (C) RT-PCR analysis of 4-mo HMGA2−/−Stra8 and age-matched HMGA2+/+ testis. Lanes: 1, GAPDH; 2, transgenic β-geo; 3, endogenous HMGA2 partial CDS (detects only endogenous HMGA2); 4, pig STRA8 CDS; 5, transgenic HMGA2 (detects only mStra8-driven HMGA2); and 6, HMGA2 full-length CDS (detects both endogenous and transgenic HMGA2). Results support inactivation of the endogenous HMGA2 and expression of the transgenic HMGA2 driven by the mStra8 promoter in the HMGA2−/−Stra8 testis.
Discussion
While the role of Hmga2 on mouse body and organ size and fertility has been examined (3, 5, 13), its role in prenatal and postnatal growth, organ size, and reproductive traits in different species is still elusive. In pigs, the effect of HMGA2 deficiency on pig body and organ size, reproduction, and fetal viability has never been reported. Our results show that adult HMGA2−/+ pigs are ∼80% size of wild-type controls, which is in agreement with reports of Hmga2+/− mice. HMGA2-null pigs were severely growth-restricted at all stages examined, with animals at 3 mo of age being one-fifth the size of controls (Fig. 4); this is a more severe phenotype than the reported 50–60% size reduction in mice (3, 13) and is more in line with the Hmga1/Hmga2-null “superpygmy” mice (22). The differences in growth reduction observed between the HMGA2−/+ lines and the HMGA2+/+ lines and the Western blot analysis (Fig. 1 and SI Appendix, Fig. S4) support HMGA2 haploinsufficiency, as reported in the mouse. Moreover, compared with HMGA2+/+ pigs, allometric analysis showed that the body-size reduction in both HMGA2−/+ and HMGA2-null pigs was proportional for length, weight, and height ratios. When the relationship of circumference to weight and length was examined, however, differences were apparent. This was most striking in the HMGA2 nulls, which were heavier than controls of similar size, suggesting a leaner phenotype. This is consistent with the reported lean phenotype of Hmga2-null mice (23). Organ sizes were also affected in the HMGA2-deficient animals (Tables 1–3). As reported previously in mice (14), some organs were proportionally more affected than others, with brain being the least affected. It should be noted that SCNT can cause epigenetic effects that affect growth/weight in mice (24). In pigs, we previously demonstrated that these weight differences are small and nonsignificant in adult SCNT pigs (25). To control for any possible donor cell line/SCNT interaction effects, however, SCNT controls were generated (Fig. 4).
Overall, these data support HMGA2’s being a potential target for reducing the size of organs for xenotransplantation into humans, if needed. This can be implemented by the use of CRISPR-Cas9 zygotic injection techniques, bypassing the need for breeding. Recent results indicate that this gene editing approach results in up to 100% of the offspring having both alleles modified (26, 27). Thus, it should be possible to conserve the complex and well-studied genetics of existing pig lines being developed for xenotransplantation and obtain reduced organ sizes by this straightforward procedure. This would bypass the need for breeding and avoid the reproductive issues we observed.
In addition to the drastic body-size reduction phenotype, we observed an unexpected loss of viability of HMGA2−/− fetuses in at-term pregnancies of HMGA2−/+ gilts bred by HMGA2−/+ boars. We observed mummified piglets at expected frequencies of the HMGA2−/− genotype in all litters from this type of mating. While we were not able to obtain DNA from the mummified fetuses for genotyping due to the high degradation of the samples it is likely that they represented the HMGA2 nulls as we were able to detect HMGA2−/− fetuses at D40 and D78 of gestation, with fetuses appearing healthy at both gestational times, but with placentas affected at D78 (Fig. 3). This suggests that gestational stage around D78 is a critical turning point for HMGA2−/− fetuses, with the affected placentas eventually leading to fetal loss and mummification (SI Appendix, Fig. S3). It has been shown that uterine capacity begins to critically affect litter size after D30 of gestation in pigs when a competition by littermates limits uterine space and resources (28, 29). Although HMGA2−/+ uteri were smaller than those in HMGA2+/+ gilts (Table 2 and SI Appendix, Fig. S6), reducing uterine crowding by unilateral tubal ligation did not rescue the HMGA2-null fetuses, suggesting that the reduced uterine size per se is not the reason. Additionally, while HMGA2−/− placental degeneration and midpregnancy fetal loss was seen in pregnancies generated by breeding the HMGA2 heterozygotes, it was not seen in pregnancies carrying only HMGA2−/− fetuses produced by TALEN-induced modification and SCNT. The main differences between the two types of pregnancies are the presence of other genotypes in the uterus in the heterozygote mating, and the fact that the uterus in the heterozygote mating was of an HMGA2−/+ genotype, while the HMGA2−/− only pregnancies were in a wild-type uterus. The evident placental degeneration at D78 in the heterozygote mating supports the notion that HMGA2−/− fetuses had difficulties surviving in the presence of other genotypes. The exact mechanism of action of this intriguing loss remains to be determined.
On the male side, HMGA2-null boars showed similar but more severe reproductive issues compared with those seen in Hmga2-null mice. MacArthur (30) described that fertility of spontaneous homozygous pygmy mice ranged from normal to sterile. However, upon separation of pygmies into two groups based on their size, a small line and a large line, testes in more than half of small-line mice did not descend into the scrotum and most of them had priapism at 6 wk of age (12, 31). In addition, some transgenic Hmga2-null mice with normally descended testes were infertile because of the lack of production of mature sperm caused by impaired spermatogenesis (5, 32). Thus, in mice, Hmga2-null males can either be fertile or sterile due to either undescended testes or abnormal spermatogenesis. In pigs, the severity of the phenotype was higher, with all HMGA2-null boars having small, undescended testes (SI Appendix, Figs. S9 and S11). As the testes are sensitive to temperature, exposure of testes to intraabdominal temperatures before adolescence affects spermatogenesis and leads to impaired fertility (33). As shown in Fig. 6, no evidence of spermatogenesis was detected in the HMGA2-null pigs despite morphologically and histologically normal structures of testes. Whether normal spermatogenesis can be rescued by surgical correction of the undescended testis (orchiopexy) (34) remains to be determined. Expression of HMGA2 in the testis only via mStra8-cHMGA2 was used in an attempt to prevent the previously reported issues with impaired meiosis of normally descended Hmga2−/− mouse testes. While this approach did result in expression of HMGA2 (Fig. 6), it did not rescue cryptorchidism, which is most likely due to a gubernaculum muscle abnormality. If breeding lines of HMGA2−/− are required this can be achieved by surgical correction of the cryptorchidic tests (orchiopexi) or by embryo complementation using embryos that can rescue the testis phenotype. This approach has been utilized to rescue nonbreeding or lethal pig lines by others (35). From a practical standpoint, gene editing through zygote injection would bypass the need to carry SCNT, allow its application to species where SCNT is not practical, bypass the reproductive issues reported above, and avoid the need to establish independent HMGA2−/− lines. As many transgenic pig lines being generated today, in particular those being used for xenotransplantation, carry multiple transgenic modifications (36), size modification by gene editing using zygotic injection would be a more practical approach than developing and maintaining independent HMGA2−/− lines.
In summary, we have been able to demonstrate that inactivation of the HMGA2 locus in pigs results in a complex phenotype with body-size reduction of over 80%, fetal competition, and cryptochidism. The magnitude of the size reduction is similar to that seen in the superpygmy mice that lack both Hmga1 and Hmga2 (22). Overall, the direct evidence from pigs and mice, and the genetic associations identified between HMGA2 and growth in dogs and humans, supports that the role of HMGA2 in body-size regulation in mammals is highly conserved. This opens up the possibility of regulating size in multiple mammalian species and the size reduction of organs to be used in xenotransplantation, thus allowing a better match between the organ size of donor and recipient and avoiding some of the issues that have been recently identified due to organ overgrowth after transplantation (19).
Materials and Methods
HMGA2 mutant pigs were generated by conventional homologous recombination or TALEN-induced gene editing of fetal fibroblasts (FF). Correctly modified FF were used for SCNT to generate animals used in this study. Growth parameters of all genotypes (HMGA2+/+, HMGA2−/+, and HMGA2−/−) were analyzed for up to 59 wk. Effects of HMGA2 mutation on reproductive parameters of males and females were examined by breeding, analysis of timed pregnancies (D40 and D78), and histological analysis of gonads. See SI Appendix for detailed information. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (37) and approved by the Institutional Animal Care and Use Committee of North Carolina State University.
Supplementary Material
Acknowledgments
We thank Dr. Shinya Yamanaka for pTV-Fbx15 and Dr. Philippe Soriano for pROSA26LAC. We also thank Kayla Howard, Charles Salmon, and Clay Byrd for help managing the animals, Dr. Luke B. Borst for advice regarding pathology, and James Cahoon for statistical analysis. This work was supported by NIH Grant R21-OD010553 (to J.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721630115/-/DCSupplemental.
References
- 1.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38:14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 2.Cattaruzzi G, et al. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007;35:1751–1760. doi: 10.1093/nar/gkl1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang X, Benson KF, Chada K. Mini-mouse: Disruption of the pygmy locus in a transgenic insertional mutant. Science. 1990;247:967–969. doi: 10.1126/science.2305264. [DOI] [PubMed] [Google Scholar]
- 4.Hirning-Folz U, Wilda M, Rippe V, Bullerdiek J, Hameister H. The expression pattern of the Hmgic gene during development. Genes Chromosomes Cancer. 1998;23:350–357. [PubMed] [Google Scholar]
- 5.Chieffi P, et al. HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene. 2002;21:3644–3650. doi: 10.1038/sj.onc.1205501. [DOI] [PubMed] [Google Scholar]
- 6.Weedon MN, et al. Diabetes Genetics Initiative; Wellcome Trust Case Control Consortium A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buysse K, et al. The 12q14 microdeletion syndrome: Additional patients and further evidence that HMGA2 is an important genetic determinant for human height. Eur J Med Genet. 2009;52:101–107. doi: 10.1016/j.ejmg.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Carneiro M, et al. Dwarfism and altered craniofacial development in rabbits is caused by a 12.1 kb deletion at the HMGA2 locus. Genetics. 2017;205:955–965. doi: 10.1534/genetics.116.196667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones P, et al. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics. 2008;179:1033–1044. doi: 10.1534/genetics.108.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamichhaney S, et al. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science. 2016;352:470–474. doi: 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- 11.MacArthur JW. Genetics of body size and related characters. I. Selecting small and large races of the laboratory mouse. Am Nat. 1944;78:142–157. [Google Scholar]
- 12.King JWB. Observations on the mutant “PYGMY” in the house mouse. J Genet. 1955;55:487–497. [Google Scholar]
- 13.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 14.Benson KF, Chada K. Mini-mouse: Phenotypic characterization of a transgenic insertional mutant allelic to pygmy. Genet Res. 1994;64:27–33. doi: 10.1017/s0016672300032511. [DOI] [PubMed] [Google Scholar]
- 15.Battista S, et al. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 16.Zhang LC, et al. 2017. mRNA and protein expression levels of four candidate genes for ear size in Erhualian and Large White pigs. Genet Mol Res, 16.
- 17.Li P, et al. Fine mapping of a QTL for ear size on porcine chromosome 5 and identification of high mobility group AT-hook 2 (HMGA2) as a positional candidate gene. Genet Sel Evol. 2012;44:6. doi: 10.1186/1297-9686-44-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muráni E, Murániová M, Ponsuksili S, Schellander K, Wimmers K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol. 2007;7:109. doi: 10.1186/1471-213X-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanabe T, et al. Role of intrinsic (graft) versus extrinsic (host) factors in the growth of transplanted organs following allogeneic and xenogeneic transplantation. Am J Transplant. 2017;17:1778–1790. doi: 10.1111/ajt.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 21.McCreath KJ, et al. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 22.Federico A, et al. Hmga1/Hmga2 double knock-out mice display a “superpygmy” phenotype. Biol Open. 2014;3:372–378. doi: 10.1242/bio.20146759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand A, Chada K. In vivo modulation of Hmgic reduces obesity. Nat Genet. 2000;24:377–380. doi: 10.1038/74207. [DOI] [PubMed] [Google Scholar]
- 24.Tamashiro KLK, et al. Cloned mice have an obese phenotype not transmitted to their offspring. Nat Med. 2002;8:262–267. doi: 10.1038/nm0302-262. [DOI] [PubMed] [Google Scholar]
- 25.Mir B, Zaunbrecher G, Archer GS, Friend TH, Piedrahita JA. Progeny of somatic cell nuclear transfer (SCNT) pig clones are phenotypically similar to non-cloned pigs. Cloning Stem Cells. 2005;7:119–125. doi: 10.1089/clo.2005.7.119. [DOI] [PubMed] [Google Scholar]
- 26.Whitworth KM, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91:78. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, et al. One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci Rep. 2016;6:20620. doi: 10.1038/srep20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenton FR, Schwartz FL, Bazer FW, Robison OW, Ulberg LC. Stage of gestation when uterine capacity limits embryo survival in gilts. J Anim Sci. 1972;35:383–388. doi: 10.2527/jas1972.352383x. [DOI] [PubMed] [Google Scholar]
- 29.Huang YT, Johnson RK, Eckardt GR. Effect of unilateral hysterectomy and ovariectomy on puberty, uterine size and embryo development in swine. J Anim Sci. 1987;65:1298–1305. doi: 10.2527/jas1987.6551298x. [DOI] [PubMed] [Google Scholar]
- 30.MacArthur JW. Genetics of body size and related characters. II. Satellite characters associated with body size in mice. Am Nat. 1944;78:224–237. [Google Scholar]
- 31.King JWB. Pygmy, a dwarfing gene in the house mouse. J Hered. 1950;41:249–252. doi: 10.1093/oxfordjournals.jhered.a106143. [DOI] [PubMed] [Google Scholar]
- 32.Di Agostino S, et al. Phosphorylation of high-mobility group protein A2 by Nek2 kinase during the first meiotic division in mouse spermatocytes. Mol Biol Cell. 2004;15:1224–1232. doi: 10.1091/mbc.E03-09-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niedzielski JK, Oszukowska E, Słowikowska-Hilczer J. Undescended testis–Current trends and guidelines: A review of the literature. Arch Med Sci. 2016;12:667–677. doi: 10.5114/aoms.2016.59940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frankenhuis MT, Wiegerinck MA, Schoorl M, Kremer J, Wensing CJ. The prevention of orchiopexy-induced testicular lesions in the pig. Eur J Pediatr. 1982;138:26–27. doi: 10.1007/BF00442323. [DOI] [PubMed] [Google Scholar]
- 35.Matsunari H, et al. Modeling lethal X-linked genetic disorders in pigs with ensured fertility. Proc Natl Acad Sci USA. 2018;115:708–713. doi: 10.1073/pnas.1715940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper DKC, Ekser B, Ramsoondar J, Phelps C, Ayares D. The role of genetically engineered pigs in xenotransplantation research. J Pathol. 2016;238:288–299. doi: 10.1002/path.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.