Significance
In this study, we demonstrate a platform for reprogramming somatic cells with high efficiency in the absence of exogenous reprogramming factors. Sustained laterally confined growth of cells on micropatterned substrates results in sequential changes to the nucleus and chromatin with each cell division, leading to the progressive erasure of lineage specific characteristics and incorporation of pluripotency. After 10 days of confined growth, the cells exhibit stemness and have multilineage differentiation potential. Our observation highlights a previously unknown role of mechanical constraints in nuclear reprogramming. Our method provides a unique approach to greatly improve stem cell technologies for developing patient specific disease models and regenerative medicine.
Keywords: lateral confined growth, nuclear reprogramming, epigenetic erasure, chromosome intermingling, cancer stemness
Abstract
Cells in tissues undergo transdifferentiation programs when stimulated by specific mechanical and biochemical signals. While seminal studies have demonstrated that exogenous biochemical factors can reprogram somatic cells into pluripotent stem cells, the critical roles played by mechanical signals in such reprogramming process have not been well documented. In this paper, we show that laterally confined growth of fibroblasts on micropatterned substrates induces nuclear reprogramming with high efficiency in the absence of any exogenous reprogramming factors. We provide compelling evidence on the induction of stem cell-like properties using alkaline phosphatase assays and expression of pluripotent markers. Early onset of reprogramming was accompanied with enhanced nuclear dynamics and changes in chromosome intermingling degrees, potentially facilitating rewiring of the genome. Time-lapse analysis of promoter occupancy by immunoprecipitation of H3K9Ac chromatin fragments revealed that epithelial, proliferative, and reprogramming gene promoters were progressively acetylated, while mesenchymal promoters were deacetylated by 10 days. Consistently, RNA sequencing analysis showed a systematic progression from mesenchymal to stem cell transcriptome, highlighting pathways involving mechanisms underlying nuclear reprogramming. We then demonstrated that these mechanically reprogrammed cells could be maintained as stem cells and can be redifferentiated into multiple lineages with high efficiency. Importantly, we also demonstrate the induction of cancer stemness properties in MCF7 cells grown in such laterally confined conditions. Collectively, our results highlight an important generic property of somatic cells that, when grown in laterally confined conditions, acquire stemness. Such mechanical reprogramming of somatic cells demonstrated here has important implications in tissue regeneration and disease models.
Nuclear reprogramming events within tissue microenvironments are critical for a number of developmental processes and tissue maintenance (1–3). In landmark experiments, biochemical factors were shown to induce nuclear reprogramming of somatic cells into induced Pluripotent Stem Cells (iPSC) in vitro (4–6). However, in vivo, cells transdifferentiate into different lineages in the absence of exogenous factors, indicating that the local mechanochemical factors could be important elements and are sufficient for inducing such transitions (7). Consistent with this, recent results have shown that culturing cells on topographic patterns combined with reprogramming factors, or on different substrate rigidity, resulted in increased efficiency of nuclear reprogramming (8–11). However, the roles played by different mechanical cues in the absence of biochemical factors in nuclear reprogramming has not been well established.
Mechanical constraints (e.g., substrate rigidity and cell morphology) are essential in controlling many cellular processes, including cellular proliferation, apoptosis, and differentiation (12–15). Biophysical forces have been shown to be very important in regulating epithelial to mesenchymal transformation (16). Matrix rigidity, cell shape, and surface topography all have been shown to direct stem cell differentiation in vitro (13, 17). In addition, in vivo experiments involving applying forces on a developing Drosophila embryo have demonstrated that altering the mechanics of the tissue can alter the differentiation programs (18, 19). Collectively, these results highlight the importance of biophysical cues in directing differentiation. However, only a few studies have looked at the potential of biophysical cues in nuclear reprogramming or transdifferentiation.
Cells of defined geometries can be obtained by culturing cells on ECM-coated micropatterned substrates. This technique has been used widely in the field to demonstrate that the cell spreading area can direct apoptosis and cell proliferation (15). Recent experiments have shown that cell geometry can modulate cytoskeletal organization, nuclear morphometrics, 3D chromosomal organization, epigenetic profiles, and, importantly, the transcription profile of the cells (20–23). Cells that are well polarized with a large spreading area have higher expression of cell–matrix and actin cytoskeletal genes compared with isotropic cells with a smaller spreading area, which express apoptotic genes at a higher level (23). Further, a recent study has shown that this cellular mechanical state is important for integrating biochemical signals such as TNF-alpha and that cells in different mechanical states have different transcriptional responses to the same signal (14). Collectively, these observations highlight the importance of cell geometry in regulating various cellular processes. Based on this, we hypothesized that culturing cells on precise geometric confinements could lead cells to obtain critical epigenetic landscapes and transcriptional profiles which could then potentially induce nuclear reprogramming.
In this paper, we report a platform to induce nuclear reprogramming through laterally confined growth of somatic cells on micropatterned substrates in the absence of any biochemical factors (Fig. 1A). Such confined growth leads to formation of spheroids from single cells and permits a precise control over the cellular geometric confinements and, to some extent, their mechanochemical properties. We show that, after 10 d of laterally confined growth, cells have reprogrammed to acquire stemness. In addition, the promoter occupancy of H3K9Ac, a transcription activation marker, and RNA sequencing (RNA-Seq) analysis reveal a monotonous decrease in the expression of mesenchymal genes, while the expression of stem cell genes shows a biphasic increase. We demonstrate the differentiation potential of the spheroids by differentiating them into endoderm and neuronal ectoderm lineages with high efficiency. Importantly, we also demonstrate, by using this platform, the induction of cancer stemness properties in MCF7 cells. These observations reveal an unknown mechanochemical mechanism of somatic cell reprogramming. While, in vivo, these transitions are highly regulated to maintain tissue homeostasis, the small probability of its occurrence, as a consequence of the heterogeneous mechanical microenvironment, could be a precursor for tissue regeneration or onset of diseases.
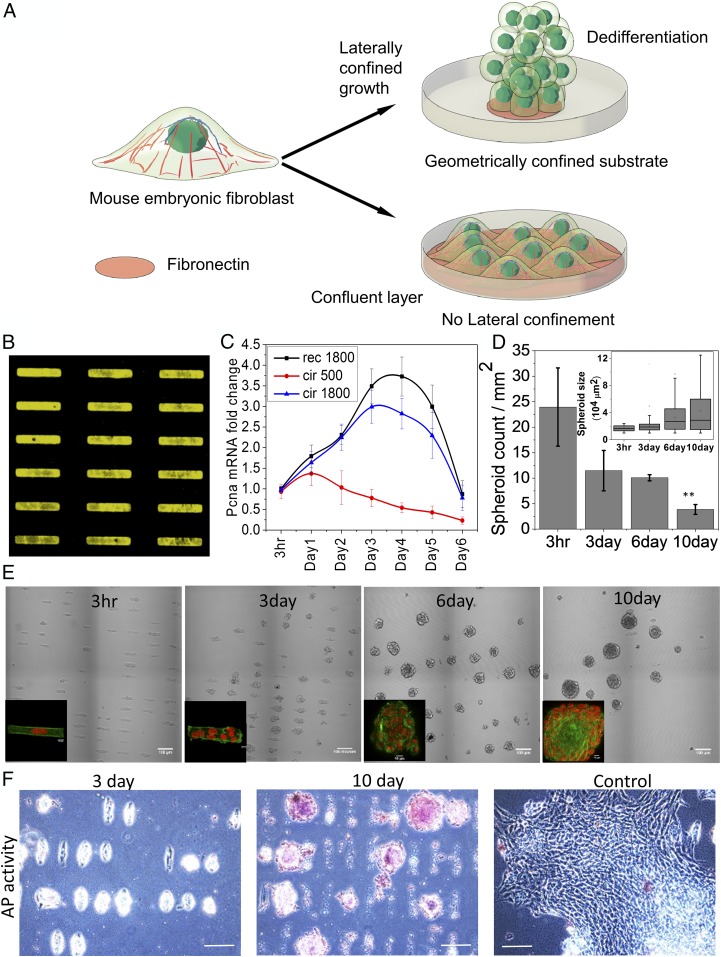
Fig. 1.
Laterally confined growth of cell facilitates induction of ES-like cells. (A) Schematic representation of the effect of geometry driven laterally confined growth of fibroblast on dedifferentiation. (B) Represents fibronectin RE micropatterns of 1,800 µm2 area. (C) Time course analysis of PCNA gene expression by qRT-PCR. (D) Plot represents number of cells or cell spheroids per unit area. (Inset) Plot represents the cells or spheroids area. Error bars represent SD. (E) The phase contrast images of NIH 3T3 mouse fibroblasts cells cultured on micropatterns up to 10 d. On tenth day, spheroids appear as ES-like colonies. Insets show fluorescent images of cells on the micropattern stained with nucleus (red) and actin (green). (Scale bar, 100 µm.) (F) These ES-like colonies on day 10 also show alkaline phosphatase (AP) activity, unlike cells cultured on RE for 3 d or culture flask for 10 d (control). (Scale bar, 100 µm.)
Results
Laterally Confined Growth of Cell Facilitates Induction of Embryonic Stem-Like Cells.
To find an optimal geometric confinement for nuclear reprogramming, mouse embryonic fibroblasts (NIH 3T3 cell line) were cultured on fibronectin-coated micropatterns with different geometries: 1,800 μm2 rectangle with aspect ratio 1:5(RE), 1,800 μm2 circle (BC) and 500 μm2 circle (SC) (Fig. 1B and SI Appendix, Fig. S1A) and were allowed to grow with such lateral confinement for 10 d (Fig. 1A). Measuring proliferating cell nuclear antigen (PCNA, a cellular proliferation marker) levels in cells grown under these laterally confined growth conditions suggested that RE was a more suitable geometry for nuclear reprogramming (24, 25) with spheroid forming efficiency of ∼20% after 10 d (Fig. 1 C–E and SI Appendix, Fig. S1A). With increased proliferation on RE, the nuclei became more circular, and their orientations were aligned along the growth (z) axis (SI Appendix, Figs. S1 and S2). In addition, growth of spheroids was also accompanied by the down-regulation of Lamin A, suggesting that these cells were poised for acquiring stemness (SI Appendix, Fig. S3). Consistent with these observations, the projected nuclear area fluctuations increased with spheroid formation (SI Appendix, Fig. S4), indicating that the onset of reprogramming is accompanied by the softening of the nucleus. This also correlates with low levels of Lamin A, increased projected nuclear area fluctuations, and chromatin plasticity observed in embryonic stem cells (ESCs) (26, 27). To test whether these spheroids indeed possessed stem cell-like properties, we stained for alkaline phosphatase activity. Interestingly, we found that a sizeable number (∼60%) of spheroids cultured for 10 d originating from single RE cells were positive for alkaline phosphatase activity (Fig. 1F and SI Appendix, Fig. S5). In contrast, unpatterned surface cells grown for 10 d with no lateral confinement showed negative staining (Fig. 1F).
Time Course Transitions in Epigenetic Landscape Facilitating Nuclear Reprogramming.
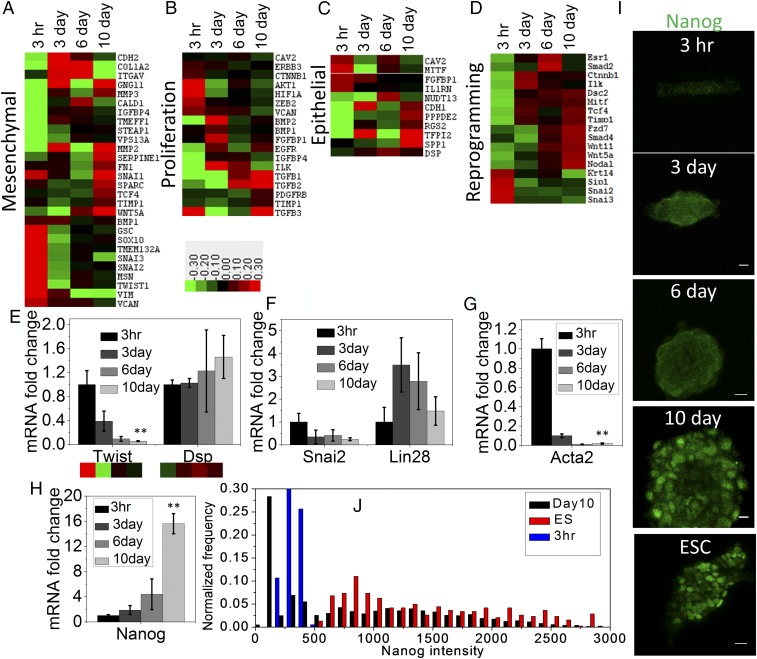
To analyze the time course transitions in epigenetic landscape of promoters accompanied with nuclear reprogramming, we carried out Epitect ChIP qPCR assay on immunoprecipitated chromatin fragments using antibodies to H3K9Ac, a marker of transcription activation (28, 29). A schematic of this assay is shown in SI Appendix, Fig. S6A. We screened promoters of 84 candidate genes which characterize mesenchymal, proliferation, epithelial, and reprogramming properties (SI Appendix, Fig. S6B). As shown in the heat maps in Fig. 2 A–D, there was a gradual reduction in the promoter occupancy of some of the mesenchymal genes (Snai2/3, Twist1, Sox10, Vim, and Vcan), while there was a substantial increase in the promoter occupancy of proliferative (Egfr, Ilk, Pdgfrb, and Tgfb3), epithelial (Cdh1, Dsp, Spp1, and Rgs2), and reprogramming genes (Esr1, Nodal, Timp1, Wnt5a, Wnt11, and Tcf4) (30–32). Further, we have quantified the global change of H3K9Ac by immunostaining. Interestingly, the average global H3K9ac level was gradually increasing from 3 d to 10 d (SI Appendix, Fig. S7 A and C). From the box plot, it is evident that, within this time course, an increasing fraction of cell populations possess the elevated global H3K9ac marks (SI Appendix, Fig. S7C). However, the average level was maximum in 3 h compared with other three conditions. The most common antagonistic epigenetic regulators found together on bivalent chromatin domains are methylation marks on H3K4me3 and H3K27me3 (33). The H3K27me3 mark silences the gene, while the H3K4me3 mark allows the gene to not be permanently silenced, and activated when needed. During reprogramming, these bivalent histone marks play a vital role in silencing the differentiation genes and simultaneously activating the stemness-related genes. Interestingly, with time, we observed two opposite shifts of the global level of these two histone marks. Particularly, H3K4me3 levels were elevated from 3 h onward, whereas H3K27me3 levels were reduced for the same period (SI Appendix, Fig. S7 B and D). This result suggests that, during such lateral confined growth of fibroblast, substantial resetting of histone marks may influence differential gene regulation. Consistent with this, the early induction of nuclear reprogramming revealed specific alterations in the chromosome intermingling degrees (22), suggesting that there were large-scale changes to the genome architecture (SI Appendix, Fig. S8). To test whether these changes in promoter occupancy mapped using H3K9Ac were linked to gene expression, we measured mRNA levels, using qRT-PCR of relevant transcription factors associated with nuclear reprogramming (34). We showed that the mRNA expression levels of Twist1, Snail, and Acta-2 (mesenchymal transcription factors) decreased, while that of the epithelial signature gene Dsp, the early reprogramming factor Lin28, and ESC marker Nanog increased (Fig. 2 E–H). These results were further supported by the increased Nanog protein expression levels in these spheroids, which were similar to that of ESCs using immunofluorescence (Fig. 2 I and J). The inherent heterogeneity in the expression levels of Nanog and Oct4 stem cell marker genes within the spheroids are shown in SI Appendix, Fig. S9 A–C. Interestingly, we found that cells at the first layer of spheroid from the bottom surface are more elongated and polarized compared with higher layers of cells, which are mostly circular and isotropic. We then quantified the actin level at different heights of the spheroids. Importantly, the first layer of cells has a higher actin level, while the Nanog levels are lower in the first layer of the cells and increase with height of the spheroid, as shown in SI Appendix, Fig. S9 D–G.
Fig. 2.
Time course transitions in epigenetic landscape facilitating nuclear reprogramming. Epitect ChIP-qPCR array for differential histone acetylation represented as heat maps shows relative changes of H3K9Ac occupancy at the promoter regions of genes characterizing (A) mesenchymal, (B) proliferation, (C) epithelium-like, and (D) reprogramming properties. Mean-normalized data were analyzed by Cluster 3.0. Ordering method is complete linkage. The heat map with clustering was acquired using Java Tree View. Positive, red; negative, green; zero, black. (E) Twist and Dsp mRNA in these four conditions obtained by qRT-PCR shows similar trend of their corresponding promoter occupancy of H3K9Ac. Error bars represent SD; **P < 0.01; Student’s t test. (F) The mRNA level of two transcription factors Snai2 and Lin28 obtained by qRT-PCR associated with negative and positive regulation, respectively, in nuclear reprogramming. (G and H) The mRNA level of mesenchymal and ESC characteristic genes Acta2 (αSMA) and Nanog, respectively, normalized with respect to NIH 3T3 grown on RE for 3 h cells (n = 3 samples). Error bars represent SD; **P < 0.01; Student’s t test. (I) Representative Nanog immunofluorescence micrographs of NIH 3T3 cells grown under laterally confined conditions for 10 d on RE and mouse ESC. (Scale bars, 10 µm.) (J) Normalized frequency distribution plot of nuclear Nanog fluorescence intensity in 3 h rectangle cells, 10 d spheroids, and ESCs.
Temporal Changes in the Transcriptome During the Nuclear Reprogramming.
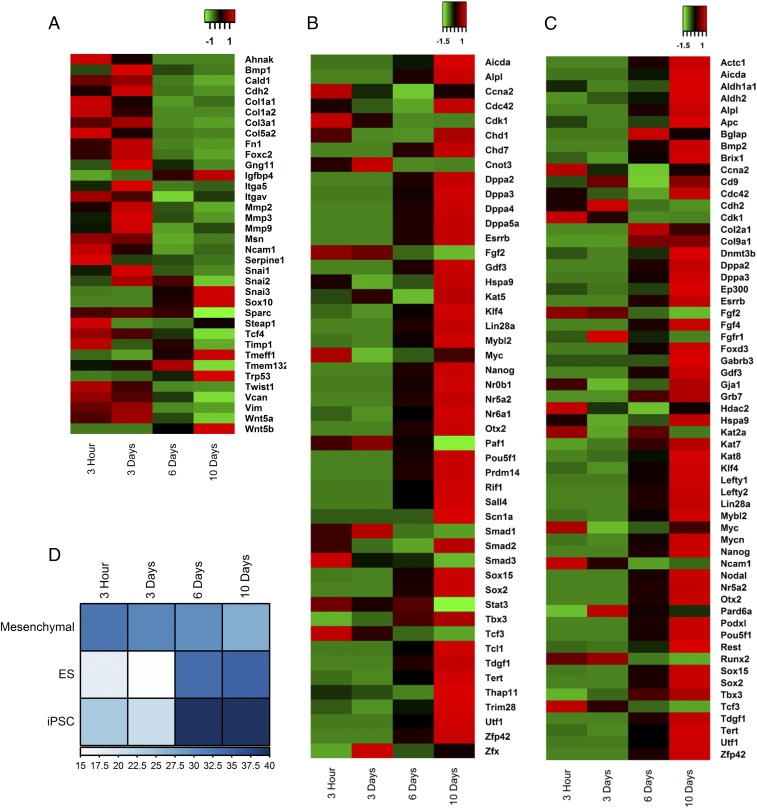
We next went on to quantitatively characterize the transcription profile during reprogramming by RNA-Seq at the four different time points. Heat maps in Fig. 3 A–C depict the changes in the expression of characteristic mesenchymal, ESC, and iPSC genes. Consistent with the promoter occupancy and qRT-PCR measurements, the relative expression of the characteristic mesenchymal genes was reduced, while the expressions of characteristic ESC and iPSC genes were increased in cells grown for 3 h to 10 d. Mesenchymal genes were prominently repressed from 6 d onward, whereas ESC and iPSC genes were maximally expressed on day 10, suggesting a temporal order in gene expression during the reprogramming process. These expression levels are a result of nuclear reprogramming events and not due to changes in the chromosomal copy numbers, which were maintained during the induction process (SI Appendix, Fig. S9). The expression of ESC and iPSC genes was found to increase by a median fold change of 8 (SI Appendix, Fig. S11C) during the process. The time course expression levels of candidate genes are shown in SI Appendix, Fig. S11 D and E. To establish the link between promoter occupancy and expression levels, we plotted the heat map of the expression levels of the genes used in the Epitect assay (SI Appendix, Fig. S12). Interestingly, a number of candidate mesenchymal and reprogramming genes showed a significant correlation, highlighting the coupling between the alterations in the epigenetic landscape and gene expression. Additionally, the fraction of the stem cell marker transcripts among the highly expressed transcripts (top 10%) sharply increased after 6 d, while there was a monotonic decrease observed for mesenchymal marker transcripts (Fig. 3D and SI Appendix, Fig. S11 A and B). These results suggest a biphasic transcription profile during the induction of pluripotent states by laterally confined growth of cells.
Fig. 3.
Temporal changes in the transcriptome during the nuclear reprogramming. Heat maps showing the relative gene expression patterns of (A) mesenchymal, (B) ESC (ES), and (C) iPSC genes. (D) A heat map representing the number of transcripts of candidate genes in the highly expressed transcripts (top 10%) in each sample.
Transcriptome Analysis Reveals Altered Biological Pathways Leading to Nuclear Reprogrammming.
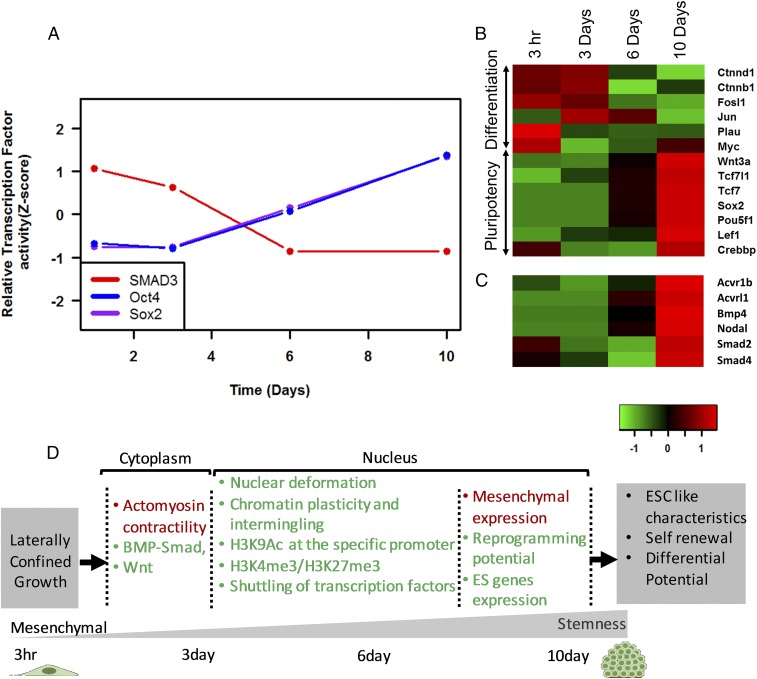
In addition to the global shift in transcriptional profile, we used RNA-Seq data to obtain insights into some of the underlying signaling involved in nuclear reprogramming induced by laterally confined growth of fibroblasts. As expected, the expression of the genes involved in Actin filament organization and contractility decreased from day 1 to day 6 (SI Appendix, Fig. S13). In addition, there were temporal changes to histone methyl and acetyl transferases which reflected the changes in the epigenetic histone modifications mentioned in Fig. 2 A–E and SI Appendix, Figs. S7 and S14. To identify which pathways were getting activated during this process, we summed up the expression of the participating genes in each pathway, and the top 30 pathways which showed maximal activation from day 1 to day 10 were plotted in SI Appendix, Fig. S15 (see Materials and Methods). Similar analysis was carried out to identify which transcription factors are activated (SI Appendix, Fig. S16). We find that the transcriptional activity of SMAD3, a repressor for hiPSC, decreased, while that of OCT4 and SOX2 increased (Fig. 4A). Importantly, we report the transcriptional activation Wnt and pluripotency pathway and BMP−SMAD pathway (Fig. 4 B and C and SI Appendix, Fig. S17). Wnt pathway has been established to be intimately connected to the core circuitry of pluripotency, with Wnt being one of the naturally occurring soluble signals that promote reprogramming and maintain pluripotency (35). Laterally confined growth of fibroblasts leads to an increase in the expression levels of several genes that are involved in the Wnt-regulated pluripotency pathway, such as Wnt3a and TCF3, and down-regulation of the genes that promote differentiation, such as Jun and Myc (Fig. 4B). Tcf3 is a key transcriptional regulator, downstream of the Wnt pathway, which cooccupies almost all promoter regions occupied by ESC-specific transcription factors, including Oct4 and Nanog (35). It has been previously reported that Wnt3a could promote the reprogramming of fibroblasts toward pluripotency in absence of c-Myc (35). Further, the expression of Acvr1, BMP4, Smad4, and Nodal, which are important regulators of BMP−SMAD signaling, is also up-regulated during the process. BMP−SMAD signal has been shown to have important roles in the induction and maintenance of pluripotency and self-renewal of mouse ESCs (36). Therefore, based on the transcriptional stimulation of these pathways through the geometric confined growth of cells, we hypothesize that BMP−SMAD and Wnt signaling pathways could play an important role in inducing and maintaining pluripotency. A schematic summary of the nuclear reprogramming process induced by laterally confined growth of RE cells is shown in Fig. 4D.
Fig. 4.
Transcriptome analysis reveals altered biological pathways leading to nuclear reprogramming. (A) The transcriptional activity of the target genes of the respective transcription factors’ gene expression changes that indicate transcriptional activation of (B) Wnt pathway and (C) BMP−SMAD pathway. (D) Schematic summary of the induction of reprogramming by laterally confined growth of cells. Red and green colors represent the events that decrease and increase, respectively.
Maintenance of Stem-Cell Like State and Differentiation Potential of the Reprogrammed Spheroids.
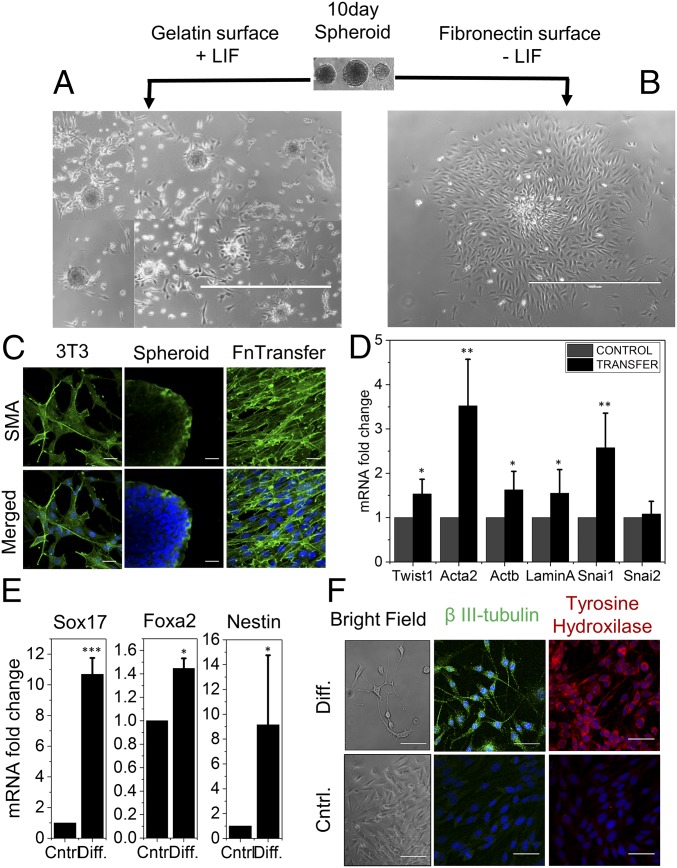
Collectively, our results showed that the induction of nuclear reprogramming by mechanical constraints is a highly efficient process. The next step, therefore, was to test whether these spheroids could be maintained as stem cells and whether they could be differentiated into specific lineages. As can be seen clearly in Fig. 5A, resuspending the 10-d spheroids on gelatin-coated dishes in the presence of leukemia inhibitory factor (LIF), a stem cell growth factor (37), maintained the stemness for a few days. However, when these spheroids were resuspended on fibronectin-coated dishes in the absence of LIF, they spread and displayed fibroblast-like morphologies (Fig. 5B). Next, we wanted to test the transfer efficiency of these spheroids to another substrate. We show that the 10-d spheroids were robust enough for efficient transfer to another substrate for further processing (SI Appendix, Fig. S18). Moreover, the fibroblast characteristic genes, including Twist1, Acta2, and Snai1, showed higher expression in the redifferentiated fibroblast (fibronectin-transfer) compared with parental NIH 3T3 cells (Fig. 5D). In agreement with the mRNA level, the immunofluorescence micrograph of alpha-SMA showed higher levels in the redifferentiated fibroblast (fibronectin-transfer) (Fig. 5C). These redifferentiated fibroblasts also showed elevated levels of Lamin A and Actb (Fig. 5D). These results suggest that reprogrammed cells can be maintained as spheroids and undergo redifferentiation when cultured under differentiation medium. Next, we demonstrated standard in vitro lineage differentiation potency of these 10-d spheroids toward endoderm and neuroectoderm lineages. As shown in Fig. 5F, there was an up-regulation of definitive endoderm marker Sox17 (11-fold) in the matured differentiation condition relative to their respective expression in control (38). However, the other endoderm marker, Foxa2, displayed only a marginal increase in expression (1.5-fold). Production of neuroectoderm marker Nestin was increased noticeably after the ectodermal lineage differentiation (Fig. 5E). Additionally, we next showed that these 10-d spheroids, when subjected to neuronal differentiation culture conditions, exhibited long axonal growth (Fig. 5F and SI Appendix, Fig. S20). We used β-III-tubulin and tyrosine hydroxylase staining [dopaminergic neuronal markers (39)] and confirmed that these cells were indeed neuron-like (Fig. 5F and SI Appendix, Fig. S21). As a negative control, culturing these spheroids in the absence of neuronal differentiation factors did not result in their differentiation to neuronal-like cells.
Fig. 5.
Maintenance of stem cell-like state and differentiation potential of the reprogrammed spheroids. (A) Isolated 10-d-old spheroids maintain their cellular confinement memory or ES-like colony state for a few days upon culture on gelatin-coated dish in LIF media, as represented in the composite image obtained from different field of view of the same culture dish; (B) appearance of fibroblast morphology upon culture on fibronectin-coated dishes without LIF media. (Scale bar, 1 mm.) (C) Elevated level of αSMA in the fibronectin (Fn)-transferred reverse fibroblasts compared with parental NIH 3T3 and gelatin+LIF transferred spheroids. (Scale bar, 20 µm.) (D) Relative expression profile of gene by qPCR assay in fibronectin-transferred reverse fibroblasts and NIH 3T3. Error bars represent SD; *P < 0.05; **P < 0.01; Student’s t test. (E) Relative expression profile of endodermal lineage marker gene (Sox17 and Foxa2) and neuroectodermal marker gene (Nestin) upon differentiation of the 10-d spheroids in corresponding differentiation culture condition. Error bars represent SD; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test. (F) Representative micrograph of dopaminergic neuronal markers β-III-tubulin and tyrosine hydroxylase in transformed spheroids taken through differentiation culture condition and without differentiation factors. (Bright Field: scale bar, 1 mm.) (IF: scale bar, 50 µm.)
Laterally Confined Growth of Human Breast Cancer Cell Facilitates Induction of Cancer Stemness.
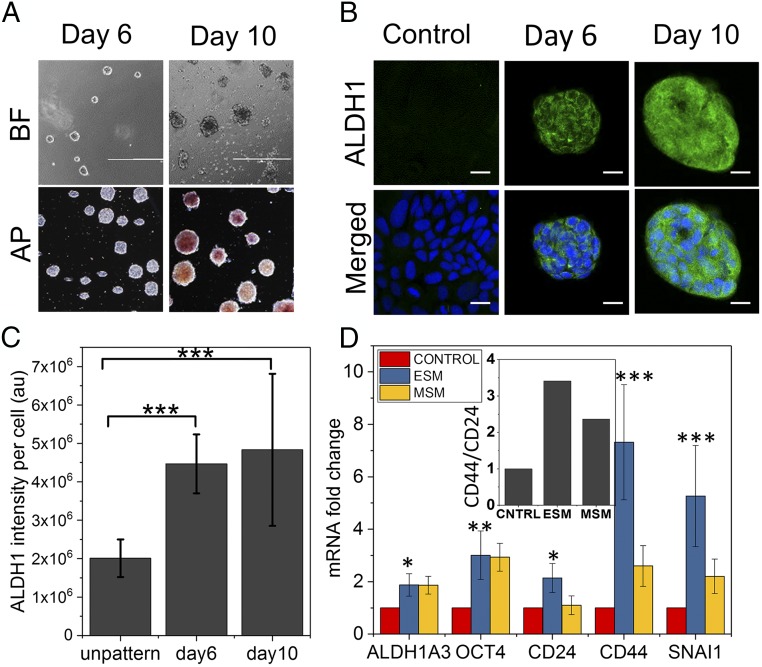
To test the effect of laterally confined growth on another cell line, we used breast cancer cell line MCF-7. Studies have shown that culturing MCF-7 cells on nonadherent and defined medium condition promotes mammosphere formation, which is found to be enriched with cancer stem cells (CSCs) (40). CSCs are defined as the transformed type of cancer cells that drive tumorigenesis with properties of self-renewal and stemness. Similar to NIH 3T3 cells, MCF-7 cells were cultured on fibronectin-coated RE micropatterns and were allowed to grow with such lateral confinement for 10 d. In agreement with previous observations, MCF-7 cells grown under this condition were able to form spheroids with positive alkaline phosphatase activity after 10 d (Fig. 6A). In contrast, comparatively smaller spheroids grown for 6 d showed negative staining. A complementary strategy for identifying cells with a stem/progenitor phenotype involves measurement of ALDH1 activity (41). ALDH1, an aldehyde dehydrogenase enzyme, which has been shown be highly active in stem/progenitor cells, was positively stained in spheroids grown for 6 d onward, which is concurrent with the RT-qPCR results (Fig. 6 B–D). For the parental cells (MCF7) cultured on fibronectin-coated unpatterned surfaces, the vast majority of cells showed negative staining for ALDH1. Further, to define whether the spheroids acquire stem cell-like characteristics, spheroids were characterized based on the expression of several breast CSC markers. We next compared mRNA levels of candidate CSC marker genes in the spheroids grown for 10 d on RE ESC medium (ESM)], and the MCF7 mammospheres (MSM), which were obtained via a serum-free culture which has been reported to be an efficient way to enrich CSCs with respect to the mRNA levels in parental MCF7 cultured on fibronectin-coated unpatterned surfaces (Fig. 6D). The expression of ALDH1A3 and OCT4 was augmented in both ESM and MSM conditions, whereas the expression of CD24, CD44, and SNAI was higher in ESM than in MSM. In addition, the CSC characteristic surface markers ratio (CD44+/CD24low) was higher in ESM than in MSM. These results suggest that lateral confined growth condition can also augment the cancer stemness phenotype in MCF-7 cells, similar to mammosphere culture condition.
Fig. 6.
Laterally confined growth of human breast cancer cell facilitates induction of cancer stemness. (A) The bright-field images (BF) of MCF7 cells cultured on RE micropatterns up to 10 d. These MCF7 spheroids on day 10 also show alkaline phosphatase (AP) activity, unlike cells cultured on RE for 6 d. (Scale bar, 1 mm.) (B) Representative confocal micrographs of MCF7 cells grown for 6 and 10 d under lateral confinement and in control cell grown on unpatterned culture plate for 10 d. Cells were immunostained for cancer stemness marker aldehyde dehydrogenase (ALDH1) (green). (Scale bar, 20 µm.) (C) Normalized ALDH1 fluorescence intensity plot on day 6 and day 10 compared with cell grown on unpatterned culture plate for 10 d. Error bars represent SD; ***P < 0.001; Student’s t test. (D) Cancer stemness-related gene expression profile in MCF7 cells grown in RE micropatterns up to 10 d (ESM) and in MCF7 mammosphere culture (MSM) compared with cell grown on unpatterned culture plate for 10 d (CONTROL). Error bars represent SD; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test. (Inset) The ratio of expression between CD44 and CD24.
Discussion
Critical processes such as tissue maintenance and regeneration involve reprogramming of cells. Seminal studies have induced reprogramming by the introduction of external transcription factors (4–6). In our study, we present a strategy to reprogram cells by subjecting cells to latterly confined growth conditions for a prolonged period of 10 d. Our study highlights a previously unknown window of cellular reprogramming that could be important for understanding transdifferentiation in tissue context. Fibroblasts are connective tissue cells which are highly attached to the matrix. Local changes in the tissue mechanical homeostasis due to a wound or physiological defects could lead to these cells getting locally confined in tissue interstitial spaces where they can experience atypical mechanical confinement. While, in vivo, cell reprogramming in such abnormal conditions is highly regulated, our results indicate that presence of such reprogramming events could potentially be exploited either for local tissue regeneration or for the onset of diseases, if genomic integrity is compromised.
Mechanical induction of reprogramming, reported in this paper, leads to cells undergoing a series of changes which ultimately lead to cells acquiring pluripotency or stemness. One of the key events in nuclear reprogramming is the increase in nuclear plasticity and chromatin reorganization (26, 27). As cells divide on defined micropatterned substrates which promote efficient spheroid formation, they undergo cytoskeletal reorganization, which changes nuclear shape and facilitates nuclear orientation along the growth axis. These spheroids are stable and can be maintained for a few days. With time, the Lamin A levels reduce, resulting in higher nuclear area fluctuations and increase in nuclear plasticity. Such changes to the nuclear mechanics also lead to the rewiring of the 3D chromatin organization, as indicated by changes to the chromosome intermingling partners (42). In addition, the epigenetic landscape of the chromatin, particularly the levels of H3K9Ac, H3K4Me3, and H3K27Me3, changes with time. The increase in nuclear plasticity along with the reorganization of epigenetic and chromosome packing within the nucleus with time leads to the rewiring of the nuclear architecture in a manner that primes the nucleus for reprogramming.
Another hallmark of nuclear reprogramming is the loss of lineage-specific transcripts and gain of reprogramming and stem cell-specific transcripts. In line with this, we see a temporal reduction of H3K9Ac levels (a transcription activation marker) in mesenchymal gene promoters, while that of the proliferative and reprogramming gene promoters increases. As a result, there is monotonous decrease in the expression of mesenchymal genes, while there is an increase in stem cell and iPSC genes. Hence, the sequential rewiring of the nuclear architecture with each cell division leads to the progressive erasure of lineage-specific chromatin characteristics, a critical step for the crossing of transient barriers toward pluripotency (30). Interestingly, the stem cell-specific genes show a biphasic expression trend with a sudden increase at day 6, suggesting that cells need to be maintained in the confined growth conditions for a long time in order for cells to transition. Therefore, the sustained growth of cells in a confined environment for prolonged periods is pivotal for inducing nuclear reprogramming.
Gain of stemness or pluripotent properties is characteristic of mature reprogramming process. The transcriptional data along with the positive staining with alkaline phosphatase activity and stem cell transcription factors like Nanog and Oct4 provide compelling evidence that supports the stemness of the day 10 spheroids. In addition to these measures, the defining stemness or pluripotent property is the ability to differentiate into different lineages. Indeed, day 10 spheroids, when subjected to the specific differentiation conditions, differentiated into endoderm and neuronal ectoderm or redifferentiated into fibroblasts with high efficiency thereby, demonstrating their multilineage differentiation potential. In agreement with the aforementioned observations, we see an increase in the expression of chromatin remodeling genes and a reduction in the expression of genes regulating actin contractility from day 1 to day 6. Finally, we report the transcriptional activation of Wnt and pluripotency and BMP−SMAD pathway from day 1 to day 10, and we hypothesize that these pathways are part of the underlying mechanisms involved in the mechanical induction of nuclear reprogramming.
Fibroblasts are the most frequently utilized somatic cell type for the production of iPSCs because of their high availability and ease of cultivation and propagation. The potency to reprogram several adult somatic cell types requires different experimental methods that are unique to the various cell types (43). Motivated by a number of experiments in the literature related to mammosphere and CSCs, we tested whether cancer cells (MCF7) can be reprogrammed to the stem cell state in spheroids obtained by their laterally confined growth. We demonstrate the mechanical induction of nuclear reprogramming in metastatic breast cancer cells (MCF7 cell line) using the same platform as that for fibroblasts. In both fibroblast and MCF7 cells, the reprogramming process takes a few days. The day 10 MCF7 spheroids mimic a mammosphere, which is indicative of the cancer stemness (40). Consistent with this, day 10 MCF7 spheroids and mammospheres highly express CSC characteristics marker genes. This observation indicates that 3D confinement effects of local tumor microenvironments can lead to cancer stemness. However, whether mammary cancer cells are at any time subjected to transient geometric confinement in vivo is still unexplored. Additionally, it shows that the mechanical induction of nuclear reprogramming using our platform is not restricted to fibroblasts.
We demonstrate a previously unknown, fundamental role for mechanical constraints imposed on cells, namely, laterally confined growth of cells, in the induction of nuclear reprogramming with high efficiency. We believe these results have important implications in physiological contexts, namely, that there exists a finite probability for cells to reprogram within the tissues. The model system used in this study could help in understanding biophysical factors involved in cell fate regulation and transdifferentiation in disease conditions. This observation also opens up new directions for innovations in tissue engineering. The high efficiency of mechanical reprogramming in our platform can be used to reprogram somatic cells to develop patient-specific disease models and tissue grafting. In addition, the highly efficient induction of reprogramming in our platform paves the way for the exciting prospect of shifting from genetic to microenvironmental mechanical control of somatic cell fate, which would greatly improve the design and effectiveness of next-generation stem cell technologies and tissue engineering applications for regenerative medicine.
Materials and Methods
Microcontact Printing.
Fibronectin micropatterning was performed as described previously (23). Briefly, 1,800 μm2 rectangles (aspect ratio 1:5) (RE), 1,800 μm2 circles (BC), and 500 μm2 circles fibronectin (Sigma F1141-2MG) micropatterns (area = 1,800 μm2 and aspect ratio = 1:5) were made on uncoated Ibidi dishes (81151). These micropatterned dishes were then passivated with 0.2% pluronic acid (Sigma P2443) for 10 min and washed with PBS.
Culturing NIH 3T3 Cells to Form Spheroids.
Wild-type NIH 3T3 fibroblasts were grown in mouse embryonic fibroblast (MEF) media composed of high-glucose DMEM (Gibco; Life Technologies) supplemented with 10% (vol/vol) FBS (Gibco; Thermo Fisher Scientific) and 1% penicillin−streptomycin (Gibco; Thermo Fisher Scientific) and maintained at 37 °C in a humidified atmosphere with 5% CO2. Cells between passages 10 and 20 were trypsinized and seeded on fibronectin-micropatterned µ-dish at a concentration of ∼8,000 cells/mL. Cells on the micropatterns were grown in previously described culture media for up to 6 d at 37 °C and 5% CO2. The media was replaced with fresh culture media on every alternate day unless otherwise stated. Within 6 d of culture on micropatterns, a majority of cells formed tightly packed multicellular spherical bodies (spheroids). On day 6, the MEF media was replaced with complete ESM (ready to use medium; Merck-Millipore) formulated with 15% FBS and mouse LIF. Spheroids were cultured in ESM from day 6 to day 10, and media was replaced on every other day unless otherwise stated. Similarly, NIH 3T3 fibroblasts stably expressing H2B-EGFP were grown on fibronectin-micropatterned µ-dish for studying their nuclear area fluctuations. Similarly, for cancer stemness assay, MCF-7 cells were grown on RE platform for 6 d in MEF media, and, on day 6, the MEF media was replaced with ESM.
Transfer Assay.
NIH 3T3 cells were grown on fibronectin-micropatterned µ-dish for 6 d or 10 d in MEF media as mentioned in Culturing NIH 3T3 Cells to Form Spheroids. Spheroids were transferred from the micropatterned dish by placing an uncoated coverslip on top of the spheroids on day 6 or 10. Further, to investigate the nuclear dynamics only of those colonies forming cells from 6-d transfer and to evade the impact of geometry on dynamics, only cells from those colonies were collected and seeded on 1,800 μm2 BC micropattern.
Subculture and Differentiation Assay of Transformed Spheroids.
To explore the ability to retain the stemness properties, the 10-d-old spheroids were isolated from the RE micropattern by mild trypsinization (30 s to 1 min), followed by collection of the detached but intact spheroids by allowing the suspension to stand for ∼5 min in a 15-mL tube. The collected spheroids were cultured in ESC media (composition described in ES cell culture section), SI Appendix, SI Materials and Methods with and without mouse LIF on 1% gelatin and fibronectin-coated culture plate (ThermoFisher), respectively. For differentiation assay, 10-d-old spheroids were isolated using the aforementioned protocol and cultured for another 20 d in endoderm and dopaminergic neuronal (neuroectoderm) differentiation condition according to manufacturer’s protocol (R&D System) (SI Appendix, SI Materials and Methods and Fig. S19).
Quantitative Real-Time PCR (qRT-PCR).
The qRT-PCR was performed to quantify the level of expression of multiple genes. Total mRNA was isolated using RNeasy Mini kit (Qiagen) according to manufacturer’s protocol, followed by cDNA synthesis using iScript cDNA Synthesis kit (Bio-Rad). The qRT-PCR was performed using SsoFast qPCR kit (Bio-Rad) for 40 cycles in a Bio-Rad CFX96. To quantify relative fold change in the level of genes, the qRT-PCR data were analyzed using the ∆∆Ct methods with respect to GAPDH levels. The primer sequences used are listed in SI Appendix, Table S1.
Immunostaining.
Cells were fixed in 4% Paraformaldehyde (Sigma) in PBS buffer (pH 7.4) for 20 min, followed by washing with PBS (15 min × 3). Cells were permeabilized using 0.5% Triton (Sigma-Aldrich) in PBS for 10 min. After incubating with blocking solution [3% BSA (A3059; Sigma-Aldrich) in PBS with 0.1% Triton] overnight at 4 °C, cells were incubated overnight at 4 °C with the following primary antibodies: Lamin A (1:400; Abcam), Nanog (1:100; Abcam), Oct4 (1:200; Abcam), αSMA (1:200), and ALDH1(1:100), diluted in blocking solution. Cells were washed with PBS (15 min × 4) and incubated with corresponding fluorescent-labeled secondary antibodies diluted in blocking solution for 1 h at room temperature, followed by washing with PBS (15 min × 4). The nucleus was stained with NucBlue Live ReadyProbes (Molecular Probes; Thermo Fisher Scientific) in PBS for 10 min at room temperature, and filamentous actin was labeled using phalloidin Alexa Fluor 488 or 568 (1:100; Molecular Probes; Thermo Fisher Scientific) for 45 min.
Alkaline Phosphatase Staining.
Cells and compact colonies were fixed with 4% Paraformaldehyde (Sigma) at room temperature for 30 min, followed by washing with PBS and Tris buffered saline (100 mM Tris and 5 mM MgCl2 in deionized water). The cells were incubated with the alkaline phosphatase substrate 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) (Sigma-Aldrich) at room temperature under slowly rocking conditions. The pinkish blue-colored colonies were developed with time. To avoid oversaturation, plates were monitored under bright-field microscopy, and the reaction was stopped, as required, by aspirating the substrate solution and rinsing with PBS. The number of colonies that were positive for alkaline phosphatase activity was quantified from the bright-field images.
Chromosome Fluorescence in Situ Hybridization.
Chromosome fluorescence in situ hybridization in cells under various conditions was done as described previously (22) (SI Appendix, SI Materials and Methods).
Imaging.
Wide-field images of a large field of view were acquired using EVOS FL Cell Imaging System (Thermo Fisher Scientific). Confocal and epifluorescence images were acquired using Nikon A1R using either 63× with 1.25 NA oil objective or 100× with 1.4 NA oil objective and EVOS using 10× objectives. Confocal images (512 × 512 pixels with pinhole size 1 airy unit) were captured in z depth with a step size of 0.5 mm to 1 mm. Time lapse imaging was done in confocal mode with 60-s or 90-s time intervals for up to 60 min in each condition.
Image Analysis.
Colonies were determined to be positive for Nanog, Oct4, and alkaline phosphatase expression on the basis of positive (reprogrammed) and negative (nonreprogrammed mouse fibroblasts) thresholds. The total fluorescence intensity was measured for each protein in its respective channel using custom-written code in MATLAB and IMARIS8. Details of the specific analysis are given in SI Appendix, SI Materials and Methods.
Epitect (ChIP-qPCR) Assay.
NIH 3T3 cells were grown on rectangular patterns for 3 h, 3 d, 6 d, and 10 d and subjected to Epitect (ChIP-qPCR) assay. The protocol used for this assay is divided into three steps: (i) fixing and preparation for immunostaining, (ii) coupling with beads, and (iii) reverse cross-linking, as described in SI Appendix, SI Materials and Methods.
RNA-Seq Sample Preparation and Analysis.
Cells were grown on RE pattern for 10 d, and mRNA was isolated using RNeasy Plus Micro Kit (Qiagen). The library preparation and sequencing on HiSeq 2000 platform was carried out at the Genome Institute Singapore. In summary, we had four conditions, 3 h, 3 d, 6 d, and 10 d, and each condition had three biological replicates which in turn had four technical replicates (run on four different lanes). The sequence quality was accessed by FASTQC (44). All sample reads were of high quality, with a Phred score of above +27 and a length of 70 bases. For the assembly of RNA-Seq reads into the genome, TopHat (45) was used, and the number of aligned sequences to a transcript was quantified using high-throughput sequencing (46) based on the Ensemble (v7.9) for Mus musculus. These steps were carried out in Galaxy (47). Other statistical analysis was carried out in R (48). The read counts across technical replicates were highly similar and not significantly different from one another and were summed up for each sample. The read counts were normalized by calculating the Transcript per kilobasepair per million (TPKM). RPK = (read count/1,000). TPKM = RPK/(Σ(RPK)/106). The normalized read counts across biological replicates were then averaged. The mesenchymal, ESC, and iPSC gene lists were obtained from Qiagen website (49–51). Pathway and transcription factor target genes were obtained from Gene Set data for pathway analysis in mouse (gksb; BioConductor). For each pathway or transcription factor, the sum of all participating genes in each sample was used to plot heat maps or line graphs. The ratio of this sum between day 10 and day 1 samples was ordered to get the top 30 repressed/activated transcription factors and pathways.
Statistical Analysis.
All data are expressed as mean ±SD or ±SEM as noted. Each experiment was repeated at least three times, and >5 cells were imaged and analyzed under each condition. We evaluated statistical significance with the Student’s unpaired two-tailed t test was performed between sample of interest and corresponding control. *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Information.
Any additional materials and methods, display items, table and source data and references are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Diego Pitta de Araugo for the illustrations. We thank the Ministry of Education Tier-3 program (2012-T3-1-001), Institute of Molecular Oncology, Italian Foundation for Cancer Research, and Mechanobiology Institute Joint-Research Laboratory for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714770115/-/DCSupplemental.
References
- 1.Halley-Stott RP, Pasque V, Gurdon JB. Nuclear reprogramming. Development. 2013;140:2468–2471. doi: 10.1242/dev.092049. [DOI] [PubMed] [Google Scholar]
- 2.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 3.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 7.De Matteis R, et al. In vivo physiological transdifferentiation of adult adipose cells. Stem Cells. 2009;27:2761–2768. doi: 10.1002/stem.197. [DOI] [PubMed] [Google Scholar]
- 8.Downing TL, et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caiazzo M, et al. Defined three-dimensional microenvironments boost induction of pluripotency. Nat Mater. 2016;15:344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 10.Guo J, Wang Y, Sachs F, Meng F. Actin stress in cell reprogramming. Proc Natl Acad Sci USA. 2014;111:E5252–E5261. doi: 10.1073/pnas.1411683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su G, et al. The effect of forced growth of cells into 3D spheres using low attachment surfaces on the acquisition of stemness properties. Biomaterials. 2013;34:3215–3222. doi: 10.1016/j.biomaterials.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 12.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci USA. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Mitra A, et al. Cell geometry dictates TNFα-induced genome response. Proc Natl Acad Sci USA. 2017;114:E3882–E3891. doi: 10.1073/pnas.1618007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 16.McGrail DJ, Mezencev R, Kieu QMN, McDonald JF, Dawson MR. SNAIL-induced epithelial-to-mesenchymal transition produces concerted biophysical changes from altered cytoskeletal gene expression. FASEB J. 2015;29:1280–1289. doi: 10.1096/fj.14-257345. [DOI] [PubMed] [Google Scholar]
- 17.Kshitiz, et al. Control of stem cell fate and function by engineering physical microenvironments. Integr Biol. 2012;4:1008–1018. doi: 10.1039/c2ib20080e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Shivashankar GV. Mechanical force alters morphogenetic movements and segmental gene expression patterns during Drosophila embryogenesis. PLoS One. 2012;7:e33089. doi: 10.1371/journal.pone.0033089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desprat N, Supatto W, Pouille PA, Beaurepaire E, Farge E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev Cell. 2008;15:470–477. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Kumar A, Makhija E, Shivashankar GV. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 2014;35:961–969. doi: 10.1016/j.biomaterials.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Makhija E, Jokhun DS, Shivashankar GV. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci USA. 2016;113:E32–E40. doi: 10.1073/pnas.1513189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Nagarajan M, Uhler C, Shivashankar GV. Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression. Mol Biol Cell. 2017;28:1997–2009. doi: 10.1091/mbc.E16-12-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci USA. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz S, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martí M, et al. Characterization of pluripotent stem cells. Nat Protoc. 2013;8:223–253. doi: 10.1038/nprot.2012.154. [DOI] [PubMed] [Google Scholar]
- 26.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talwar S, Kumar A, Rao M, Menon GI, Shivashankar GV. Correlated spatio-temporal fluctuations in chromatin compaction states characterize stem cells. Biophys J. 2013;104:553–564. doi: 10.1016/j.bpj.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Ratna P, Shivashankar GV. Superresolution imaging of nanoscale chromosome contacts. Sci Rep. 2017;7:42422. doi: 10.1038/srep42422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 30.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2012;76:211–220. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marson A, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi Y, et al. BMP-SMAD-ID promotes reprogramming to pluripotency by inhibiting p16/INK4A-dependent senescence. Proc Natl Acad Sci USA. 2016;113:13057–13062. doi: 10.1073/pnas.1603668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray P, Edgar D. The regulation of embryonic stem cell differentiation by leukaemia inhibitory factor (LIF) Differentiation. 2001;68:227–234. doi: 10.1046/j.1432-0436.2001.680410.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim PTW, et al. Differentiation of mouse embryonic stem cells into endoderm without embryoid body formation. PLoS One. 2010;5:e14146. doi: 10.1371/journal.pone.0014146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng X, et al. Dopaminergic differentiation of human embryonic stem cells. Stem Cells. 2004;22:925–940. doi: 10.1634/stemcells.22-6-925. [DOI] [PubMed] [Google Scholar]
- 40.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 41.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhler C, Shivashankar GV. Chromosome intermingling: Mechanical hotspots for genome regulation. Trends Cell Biol. 2017;27:810–819. doi: 10.1016/j.tcb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews S. 2010 FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed May 1, 2018.
- 45.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Development Core Team 2008. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
- 49.SABiosciences(Qiagen) 2017 SABiosciences Corporation Privacy Policy. Available at www.sabiosciences.com/dna_methylation_product/HTML/EAMM-901Z.html. Accessed March 15, 2017.
- 50.SABiosciences(Qiagen) 2017 SABiosciences Corporation Privacy Policy. Available at www.sabiosciences.com/rt_pcr_product/HTML/PAMM-092Z.html. Accessed April 8, 2017.
- 51.SABiosciences(Qiagen) 2017 SABiosciences Corporation Privacy Policy. Available at www.sabiosciences.com/rt_pcr_product/HTML/PAMM-405A.html. Accessed April 8, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.