Significance
This study expands our knowledge of protein hydration, which is highly related to the macromolecular antifreeze property of proteins. We examined a polypentagonal network formation of waters for a series of artificial variants of a 65-residue ice-binding protein. The polypentagonal waters were created solely on the surface of an activity-improved variant, which appeared to contain two sets of water clusters exhibiting a perfect position match to the waters constructing the first prism and pyramidal ice planes. These data suggest that a minute structural change in a protein organizes the surface waters into a polypentagonal arrangement, which merges with the intrinsically disordered ice surface and freezes to specific ice crystal planes.
Keywords: hydration shell, ice-binding protein, crystal structure, water cluster, polypentagonal waters
Abstract
Polypentagonal water networks were recently observed in a protein capable of binding to ice crystals, or ice-binding protein (IBP). To examine such water networks and clarify their role in ice-binding, we determined X-ray crystal structures of a 65-residue defective isoform of a Zoarcidae-derived IBP (wild type, WT) and its five single mutants (A20L, A20G, A20T, A20V, and A20I). Polypentagonal water networks composed of ∼50 semiclathrate waters were observed solely on the strongest A20I mutant, which appeared to include a tetrahedral water cluster exhibiting a perfect position match to the first prism plane of a single ice crystal. Inclusion of another symmetrical water cluster in the polypentagonal network showed a perfect complementarity to the waters constructing the pyramidal ice plane. The order of ice-binding strength was A20L < A20G < WT < A20T < A20V < A20I, where the top three mutants capable of binding to the first prism and the pyramidal ice planes commonly contained a bifurcated γ-CH3 group. These results suggest that a fine-tuning of the surface of Zoarcidae-derived IBP assisted by a side-chain group regulates the holding property of its polypentagonal water network, the function of which is to freeze the host protein to specific ice planes.
Proteins are generally surrounded by a hydration shell composed of a number of water molecules in the vicinity of their surface. This has been known to play a critical role in the structural construction and ligand interactions of proteins (1, 2). Sun et al. (3) reported that a 33-kDa ice-binding protein (IBP) named “Maxi” locates an extremely unique shell composed of more than 400 waters forming a polypentagonal network. Maxi is folded into a four-helix bundle with this network, and a part of it extending outward was thought to work for interaction between the protein and an ice crystal surface. Water arrangement in a single ice crystal is hexagonal but not pentagonal, so that Maxi’s polypentagonal water network was thought to merge with, and freeze to, an intrinsically disordered water layer creating the ice surface (3, 4). In the insect IBPs, contiguous troughs uniquely created on their molecular surface are known to trap the waters and rank them at regular intervals, which exhibited a perfect position match with the hexagonal waters (5–7). Such organized waters on the protein were named anchored-clathrate waters (ACWs), which have been assumed to combine the host protein with specific ice-crystal planes (8). Less is known, however, about how a polypentagonal water network emerges on a protein and works for protein–ice interactions.
Supercooled water freezes in two steps. First, a nucleation of single ice crystals occurs, and then these crystals grow to form ice blocks (9, 10). IBPs are capable of binding to each single ice crystal to inhibit the ice block formation (11). The mechanism of ice-binding is not fully understood. A single ice crystal consists of water molecules forming a hexagonal unit defined by a1–a3 and c axes, where a set of the water molecules forming an ice plane is represented by Miller–Bravais indices (12). For example, indices (0001), , and represent a basal plane normal to the c axis, a primary prism plane parallel to the c-axis, and a pyramidal plane defined for a sloped slice inclined by 14.9° to the c-axis, respectively. IBPs are unique macromolecules that are capable of binding to one or more ice planes (5) and creating a convex ice front on the plane between the bound IBPs through a Gibbs–Thomson effect (13). Such fronts are energetically unfavorable for further adsorption of bulk water molecules, leading to termination of growth of the ice crystal (14). When the growth of prism and/or pyramidal planes into facets is terminated, the ice crystal is changed into a hexagonal bipyramid or hexagonal trapezohedron (15). The ice-binding ability of IBP also depresses the nonequilibrium freezing point (Tf) and slightly elevates the nonequilibrium melting point (Tm) (16). The resultant difference between Tf and Tm, termed “thermal hysteresis” (TH), determines the ability of ice-growth–inhibition of IBP (13–16). IBPs exhibiting TH values of 3 °C–6 °C and 1 °C–2 °C are termed “hyperactive” and “moderately active” species, respectively (6). For IBPs that cannot perfectly arrest the ice crystal growth, ice-growth speed (μm/min) is used as an alternative measure of ice-growth inhibition (17).
IBPs have been isolated from cold-adapted fishes, insects, plants, and microorganisms, and they display a remarkable diversity in their amino acid sequences and structures (5). A species of Zoarcidae, Macrozoarces americanus (ocean pout), contains a total of 12 isoforms of IBPs, which contain 65–66 amino acid residues and are monomeric globular proteins (molecular weight: 6.5–7.0 kDa) (18). These IBP isoforms are also known as type-III antifreeze proteins. The X-ray crystal structure of two isoforms denoted HPLC3 and HPLC12 has been determined (19–21). Both isoforms comprise many twisted loops folded into triple-strand β-sheets, the N- and C-terminal parts of which are related with a local twofold axis of symmetry. A compound ice-binding site (IBS) was postulated on one side of this structural motif, composed of two adjacent planar surfaces forming an angle of 150° (22, 23). One of the surfaces contributed by Q9, P12, N14, T15, A16, and N44 is thought to adsorb to the pyramidal plane, and the other containing T18, L19, and V20, to the first prism plane. They were named pyramidal-plane–binding IBS (denoted “pyramidal IBS” here) and prism-plane–binding IBS (“prism IBS”), respectively. The residues on the pyramidal IBS are well conserved in this protein family, while the residues on the prism IBS are frequently replaced (18). Computer simulations have suggested the existence of ordered water molecules around this compound IBS, which was thought to work for ice binding (24, 25). The insect IBPs composed of repetitive amino acid sequences form 2D arrays of ice-binding residues to create the contiguous troughs (5). The ice-binding residues of Zoarcidae-derived IBPs, however, are not aligned on the protein surface.
A midlatitude fish, Zoarces elongatus Kner (Notched-fin eelpout, NFE), is known to contain at least 13 isoforms of this type of IBP (26). As a model protein to observe the polypentagonal ice-like water networks on its surface, we selected a 65-residue defective isoform of this IBP denoted NFE6, which only slows but cannot perfectly terminate the ice crystal growth (17). The ice-binding ability of NFE6 was improved dramatically by the replacement of P19 and A20 with Leu and Val, respectively (denoted P19L/A20V-mutant) (22). The ice-binding ability of another defective isoform (NFE11) also improved with the V9Q/V19L/G20V triple mutation (23). Similar engineering results of this type of IBP allowed us to speculate that amino acid replacement of the 20th residue of NFE6 drastically changed its ice-binding property. We hence prepared a wild type (WT) of NFE6 and its five single mutants by replacing the 20th residue with Leu, Gly, Thr, Val, and Ile (A20L, A20G, A20T, A20V, and A20I, respectively) and examined their fluorescence-based ice plane affinity (FIPA) (15), strength of ice-growth inhibition (17), and X-ray crystal structure. The mechanism of ice-binding with regard to the formation of a polypentagonal semiclathrate water network was observed solely in the A20I mutant that exhibited the strongest ice-binding ability.
Results and Discussion
Mutants Exhibited Different Ice-Binding Abilities.
Recombinant proteins of WT and five single mutants of NFE6 were prepared using the standard overlap extension PCR methodologies (27). The amino acid sequence and primer construction of each sample are described in SI Appendix, Material S1. We cultured Escherichia coli BL21 (DE3), which contained the expression vector, with Luria–Bertani medium to produce the recombinant samples. The products were then purified via cation-exchange chromatography (Econo-Pac High S cartridge, Bio-Rad) with a linear NaCl gradient (0–0.5 M) with 50 mM citric acid buffer (pH 2.9). The eluted samples were dialyzed against Milli-Q water overnight to be lyophilized. Purity of the final products was verified with SDS/PAGE and silver staining. The protein yields per 1 L culture were 300 (WT), 50 (A20V), 37 (A20T), 58 (A20I), 39 (A20L), and 105 mg (A20G), respectively.
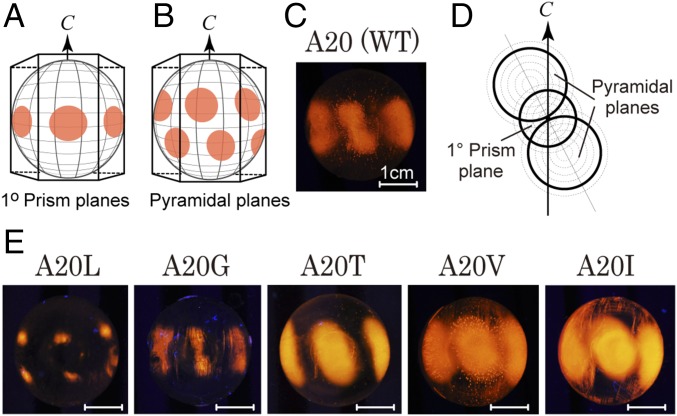
The ice-binding property of all recombinant proteins was evaluated on the basis of their patterns of FIPA (15). We prepared a golf-ball–sized single-ice-crystal hemisphere (ϕ = 3 cm) and attached it to a frozen probe in order to make its c axis perpendicular to the probe and then soaked it in a 0.02 mg/mL solution of each protein labeled with the fluorescent detergent tetra-methyl-rhodamine [5 (6)-TAMRA-X, SE; Thermo Fisher Scientific]. A FIPA pattern, like the pattern observed in Fig. 1A, implies that the IBP sample binds to the first prism plane of an ice hemisphere. Similarly, a pattern like Fig. 1B indicates IBP binding to the pyramidal plane. The dumbbell-like pattern in WT (Fig. 1C) may be interpreted as a superimposition of Fig. 1 A and B, where two pyramidal-plane–binding areas are larger than one first prism-plane–binding area, as illustrated by the thick lines in Fig. 1D. It could be speculated that in Fig. 1E (A20L–I) this dumbbell pattern is modulated by the alteration of the first prism- and pyramidal-plane–binding area. For example, both areas decreased in A20G, whereas A20V consists of one prism- and two pyramidal-plane patches with almost the same size. A20L showed the smallest patterns at higher latitudes of the ice hemisphere, indicating its poor ability to bind to fewer sets of waters in the pyramidal plane. In contrast, the widest ellipse observed for A20I is interpreted as this protein binding to a substantial area of both the first prism and the pyramidal ice planes. The slits observed between the fluorescent ellipses of A20T are explained by the area of its target ice planes being a little narrower than those of A20V and A20I.
Fig. 1.
Definition of ice planes and FIPA patterns observed on a single ice crystal hemisphere. (A) First prism planes corresponding to the facets of a hexagonal ice unit. The arrow indicates the crystallographic c axis. (B) Pyramidal planes of the ice hemisphere, the orientation of which is the same as in A. (C) FIPA pattern of a WT Zoarcidae-derived IBP containing A20. (D) Simulation of FIPA patterns by superimpositions of A and B of different sizes. The size is modulated according to the sample’s affinity to an ice crystal plane. (E) Actual FIPA patterns observed for A20L, A20G, A20T, A20V, and A20I mutants. (Scale bars, 1 cm.)
A20I Is the Strongest Mutant.
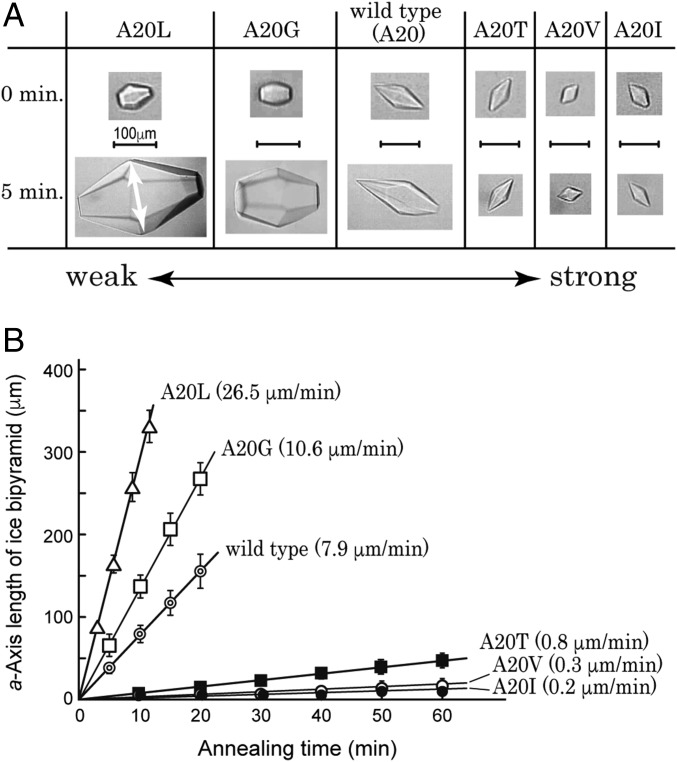
All of the samples were capable of modifying a single ice crystal into a bipyramid (Fig. 2A), indicating their ice-binding ability (28). The crystal images captured before and after 5 min of annealing time below 0 °C showed that the ice bipyramid undergoes crystal growth in A20L, A20G, and WT solutions, while the growth is inhibited in A20T, A20V, and A20I solutions. Their detailed time-dependent data (Fig. 2B), measured at the Tm − 0.05 °C with 0.2 °C/min of cooling, showed that each ice bipyramid grows in proportion with the annealing time. The ice-growth rates for A20L, A20G, WT, A20T, A20V, and A20I were evaluated at 26.5, 10.6, 7.9, 0.8, 0.3, and 0.2 μm/min, respectively. Larger growth rates imply weaker inhibition of ice growth, so that the strength order of ice-growth inhibition is A20L < A20G < WT < A20T < A20V ≤ A20I. Maximal thermal hysteresis activity for A20I was evaluated at ∼1.2 °C. FIPA analysis (Fig. 1) showed that the weakest A20L binds to an ice hemisphere only partly, whereas the strongest A20I binds to a substantial area of both pyramidal and first-prism planes. The size of the IBP-bound area on an ice hemisphere, therefore, is well correlated with the strength of ice-growth inhibition. It is significant that only one amino acid replacement at the 20th residue causes such a systematic change of the inhibitory action of ice crystal growth.
Fig. 2.
Comparison of the strength of ice-growth inhibition between WT and mutant proteins. (A) Images of bipyramidal ice crystals captured for A20L, A20G, WT, A20T, A20V, and A20I mutants before and after 5 min of annealing time. The arrow indicates the length of the a axis. (B) Dependence of the length of the a axis on the annealing time measured at the Tm − 0.05 °C with 0.2 °C/min of cooling. The larger growth rate (μm/min) implies a weaker ability for the ice to inhibit growth.
Poor ice-growth inhibition was reported previously for WT (17). This is verified by both the FIPA pattern and the fast ice-growth rate in our study (Figs. 1 and 2). The ice-growth inhibition ability of WT was decreased by the A20G mutation, in which the side-chain group of the 20th residue became smaller (CH3 → H). The ice-growth inhibition ability of A20G was further decreased by the A20L mutation, in which the side-chain group became larger [H → -CH2-CH-(CH3)2]. The ice-binding ability is therefore not correlated with the size of the side-chain group. Indeed, A20I containing the side-chain CH(CH3)-CH2-CH3 group, the molecular weight of which is exactly the same as that of A20L, behaved in an opposite fashion to A20L (Fig. 1). Both A20I and A20V exhibited superior ice-binding ability and contain bifurcated β-carbon located in the γ-CH3 group. Another mutant A20T containing a γ-CH3 group exhibited similar ability to A20I and A20V. It is therefore speculated that the location of the γ-CH3 group at the 20th residue plays a crucial role in the ice-binding ability of this IBP to both the first prism and the pyramidal ice planes. The γ-CH3 group of V20 is contained in the HPLC12 isoform that also exhibits superior ice-binding ability, which might be further evidence of the significance of this side-chain group location in this series of IBP.
Structures of the Mutants Are Highly Identical.
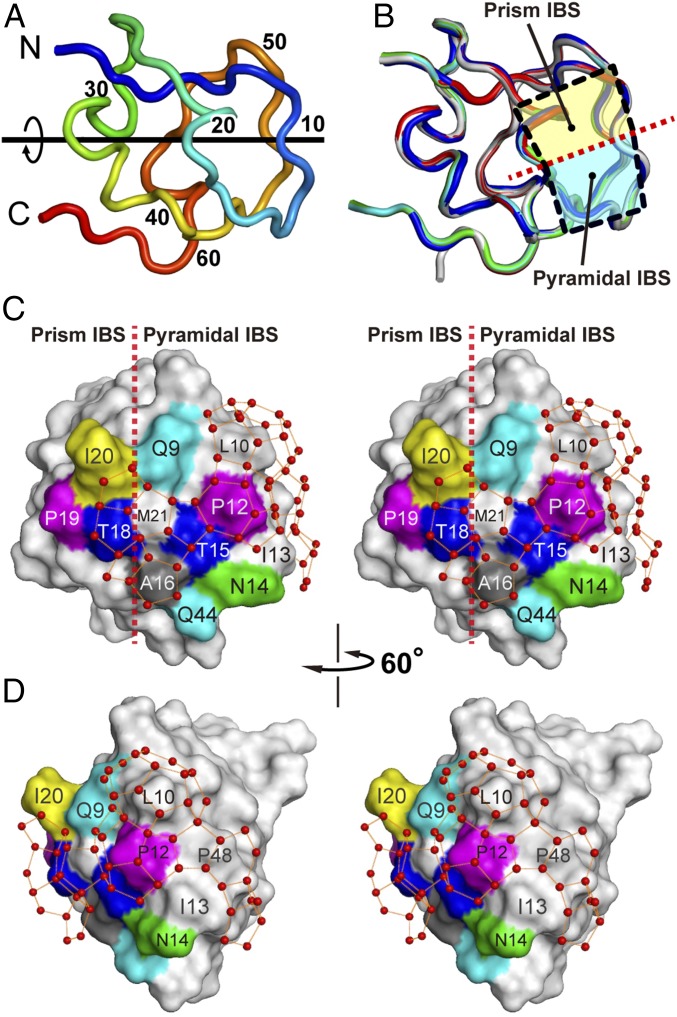
We prepared 75–200 mg/mL of the protein solutions and mixed 1- to 4-μL samples of these solutions with an equal amount of the reservoir solution (0.1 M sodium citrate, pH 3.5, with 2.0–2.5 M ammonium sulfate) (29), which generated the protein crystals of A20V, A20I, A20L, and A20T. The WT sample was crystallized into two different forms belonging to the space groups C2221 and P212121, denoted WT1 and WT2, respectively. Structures of WT1, WT2, A20V, A20I, A20L, and A20T were determined by the molecular replacement method utilizing the coordinates of HPLC3 [Protein Data Bank (PDB) ID 1OPS] and/or HPLC12 (PDB ID 1HG7). The structural refinement of these molecules was successively performed using 33,361–167,949 reflections to achieve 0.97–1.19 Å of high resolution with 0.11–0.13 of fine crystallographic R-factor (SI Appendix, Table S1). Every structure was characterized by an internal-dyad motif with a local twofold axis of symmetry contributed by many short twisted loops (Fig. 3A), which is the well-known structural property of Zoarcidae-derived IBPs (19–21). Averaged values of interatomic distances between HPLC3 and the determined structures were evaluated, with root-mean-square deviation (rmsd) of only 0.35 Å (Fig. 3B). This indicates that the backbone conformations of all samples are highly identical to each other. All of the mutants had a 10–20th loop segment on their surface, and a region encircled by this loop was assigned to a compound IBS consisting of the prism and pyramidal IBSs (Fig. 3B) (22). The former is contributed by the putative ice-binding residues of T18, P19, and A20, and the latter by those of Q9, P12, N14, T15, A16, and Q44 (Fig. 3C). Overall, alteration of the ice-binding property originated solely from a limited structural change near the 20th residue by the present mutations.
Fig. 3.
Construction of polypentagonal water networks on the A20I mutant. (A) Loop representation of the backbone structure of WT1 (PDB ID 5XQN) characterized with an internal-dyad motif with a local twofold axis of symmetry typical for Zoarcidae-derived IBPs. (B) Overlay of the backbone structures of WT1, WT2, A20L, A20T, A20V, A20I, and HPLC3 (PDB ID 1OPS). Two squares separated by a broken line indicate the location of two IBSs: one is the first prism plane (prism IBS, yellow) and another the pyramidal plane (pyramidal IBS, cyan). (C) Stereoview of the A20I structure with ∼50 semiclathrate waters constructing the polypentagonal network. A broken line indicates the boundary of the two IBSs. (D) A 60°-rotated image of the A20I structure showing the side of the polypentagonal networks that surround edge of the pyramidal IBS.
In general, IBPs tend to crystallize so as to locate their ice-binding surface face-to-face in an asymmetric unit because of their relatively flat and hydrophobic nature (30). Such a preferred protein–protein contact has been thought to displace the surface waters from their original positions. For example, no ordering of water molecules was detected in the crystal structure of an insect IBP, whereas ranks emerge if the waters on the multiple structures in an asymmetric unit are merged onto one structure (7). The distance between the merged waters in the rank showed an excellent match to that of waters constructing an ice plane, so they are assumed to anchor the IBP–ice complex formation. To forcibly expose an ice-binding surface to solvent and to intentionally observe ice-like waters, Sun et al. (31) crystallized the HPLC12 isoform as a fusion molecule with a maltose-binding protein. They detected no robust network of clathrate waters on this synthesized molecule, but revealed several waters located on the pyramidal IBS. Here, it appeared that one molecule of A20T, A20L, and A20I, two molecules of A20V and WT1, and four molecules of WT2 were crystallized in an asymmetric unit of each protein crystal (SI Appendix, Fig. S1). The compound ice-binding site of all proteins was exposed to the solvent regardless of their crystal packing, and formation of ice-like waters was comparable between the molecules.
Waters Construct Polypentagonal Networks on A20I.
Computer simulations have shown that an interfacial region between bulk waters and bulk ice consists of a 10- to 15-Å thick, intrinsically disordered quasi-liquid layer, which is more ordered than bulk waters, but less ordered than that of an hexagonal ice lattice (32, 33). The pentagonally arranged waters located outside of a four-helix bundle structure of Maxi were thought to merge with those in the disordered layer in conjunction with the ice growth (3, 4). The A20I mutant (PDB ID 5XQU) similarly located ∼50 semiclathrate waters consecutively jointed to form 18 polypentagonal networks (Fig. 3C and SI Appendix, Fig. S2). For the other mutants, pentagonal waters exist only dispersively and do not construct any robust network. These ice-like waters of A20I are mostly situated along the side-chain atoms of the ice-binding residues, which are roughly divided into two groups according to their locations. The first group, composed of seven pentagons (1–7 in SI Appendix, Fig. S2) with 19 semiclathrate waters, is located near the boundary of the two ice-binding sites contributed by Q9, P12, T15, A16, T18, P19, I20, M21, and Q44 (Fig. 3C). Five waters of 19 are hydrogen-bonded to either the ε-amino group (denoted NE2) of Q9, the γ-hydroxyl group (OG1) of T15 and T18, or the backbone N and O atoms of A16, which strengthen this network formation. The second group consisting of 10 pentagons (8–18) with ∼25 waters propagates from a region containing P12 to the end of the pyramidal-plane–binding IBS contributed by L10, I13, T47, and P48 (SI Appendix, Fig. S2). Hydrogen bondings between 7/25 waters and L10 (N, O), I13 (N, O), T47 (OG1), and K51 (N) strengthen the network formation of this second group. Before this study, five pentagonal ice-like water clusters were observed in crystals of crambin (PDB ID 1CRN), a 46-residue seed storage protein, as a result of intermolecular packing (34). Water polygons (pentagons or hexagons) were also created on hydrophobic regions formed between the helices of tetrameric malate dehydrogenase from the bacterium Chloroflexus aurantiacus (Ca MalDH) (PDB ID 4CL3) (35). Since its ortholog from a halophilic bacterium Salinibacter ruber (PDB ID 3NEP) contains no organized waters, acidic residue substitutions are thought to disrupt the pentagonal water formation. It was speculated from these results that surface waters tend to organize themselves as polygons with their nearest stable waters on a hydrophobic surface, not always related to the ice-binding ability.
Polypentagonal Network Contains Two Water Clusters.
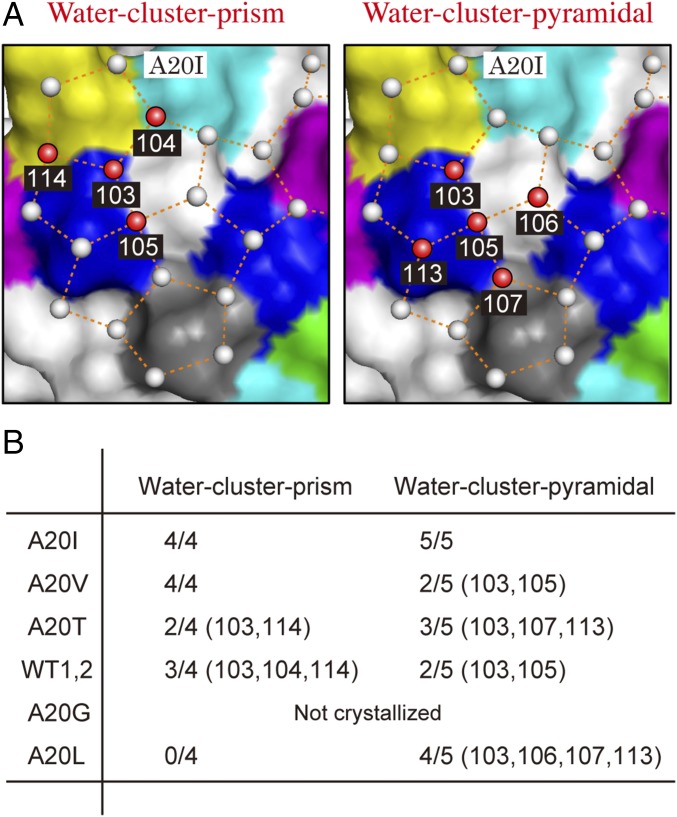
A neutron diffraction study has revealed the location of a tetrahedral water cluster in the first hydration layer of the HPLC12 isoform (PDB ID 3QF6) (36), the position of which matched those of the waters constructing the first prism plane. This water cluster is constructed from four surface waters and one γ-hydroxyl group of T18, where the average distance between the central water and the four vertices is 2.77 ± 0.02 Å, and the average H–O–H angle is 114.5 ± 1.3°. Here, we prepared a water coordinate file of the first prism plane with a size of 86 × 70 × 2 Å by employing a software VESTA (jp-minerals.org/vesta/jp) (37), and manually docked it onto the A20I structure (PDB ID 5XQU) to overlap a selected oxygen atom in the prism plane onto that in the polypentagonal waters. The coordinate file consists of only first and second layers of the prism plane, which avoided congestion of waters during the docking process and facilitated our position-match evaluations. Since a water cluster should be symmetrically located on the protein, we manually rotated this prism plane coordinate around the selected water to x, y, and z axes to achieve multiple superpositions as possible. The best-fit result was verified with evaluation of rmsd between the distances of all of the superimposed waters. As a consequence, we identified a water cluster prism (Fig. 4A, Left) in the first group of the polypentagonal network on the A20I mutant, the geometry of which is highly similar to that of the tetrahedral waters on the HPLC12 isoform. The identified waters were labeled 103–105 and 114, in which 103 and 104 are hydrogen-bonded to T18-OG1 and Q9-NE2, respectively. The rmsd between the oxygen atoms of the water cluster prism and those of the first prism plane was only 0.14 Å, indicating their perfect position match. Significantly, this water cluster prism is located over the first prism plane IBS and the pyramidal plane IBSs; the water 103 is situated on the boundary, 114 on the former IBS, while 104 and 105 are on the latter IBS. The second best mutant A20V locates this water cluster prism in the same position, although it has no polypentagonal network. In WT1, WT2, A20T, and A20L mutants, however, at least one of the waters appeared to be displaced from the tetrahedral geometry described above, as shown in Fig. 4B. In A20L, for example, four corresponding waters were displaced toward the 20th residue position without keeping the tetrahedral formation. This is probably due to a lack of a γ-CH3 group that creates a space to pull these waters inside, disrupting the water arrangement necessary to bind to the ice prism plane.
Fig. 4.
Two water clusters exhibiting a space-match to ice crystal planes. (A) A water cluster prism and a water cluster pyramidal involved in the polypentagonal network on the A20I mutant. The spatial arrangements of each cluster exhibit perfect position matches to those constructing the first prism and the pyramidal planes, respectively. (B) Search results for the water cluster prism and water cluster pyramidal for each mutant. The 2/4 (103, 104), for example, indicates that A20T contains only the waters corresponding to 103 and 104 in the water cluster prism.
To search for additional water clusters, we manually docked a water coordinate of the (1000) basal and the pyramidal planes in the size of 86 × 70 × 2 Å onto the polypentagonal waters of the A20I mutant to superpose a selected oxygen atom of each plane onto another selected atom from the network waters. By employing the procedure utilized to find the water-cluster prism, we manually rotated each ice plane around the selected water to x, y, and z axes to achieve multiple superpositions of the other water molecules (SI Appendix, Fig. S3). The final docking model was evaluated by calculation of an average value of rmsd for all superimposed waters. Although no clustering waters corresponded to the (1000) basal plane, another water cluster consisting of five water molecules labeled 103, 105, 106, 107, and 113 (Fig. 4A, Right) was detected. These exhibited a perfect position match to the waters constructing the pyramidal plane, with only 0.28 Å rmsd, and were named “water cluster pyramidal” (SI Appendix, Fig. S3). Among them, four water molecules, 103, 106, 107, and 113, are symmetrically located around the central water (105) to form four identical isosceles triangles, where the average distance between the four vertices and the center is 2.76 ± 0.03 Å. The waters 103 and 105 are also the constituents of the water cluster prism, and 103 and 107 are hydrogen-bonded with T18 (OG1) and A16 (O), respectively. This water cluster pyramid is constructed over the prism and pyramidal IBSs (Fig. 3), in which only 106 and 107 are located on the latter. For the A20V, A20T, WT1&2, and A20L mutants, at least one of the waters is shifted from the ideal symmetrical geometry, as shown in Fig. 4B. The difference between the ice-binding area (Fig. 1) and the strength (Fig. 2) of the variants is therefore ascribable to the degree of perfection of the water cluster prism and water cluster pyramidal constructed in each molecule, but not to the construction of the polypentagonal water network.
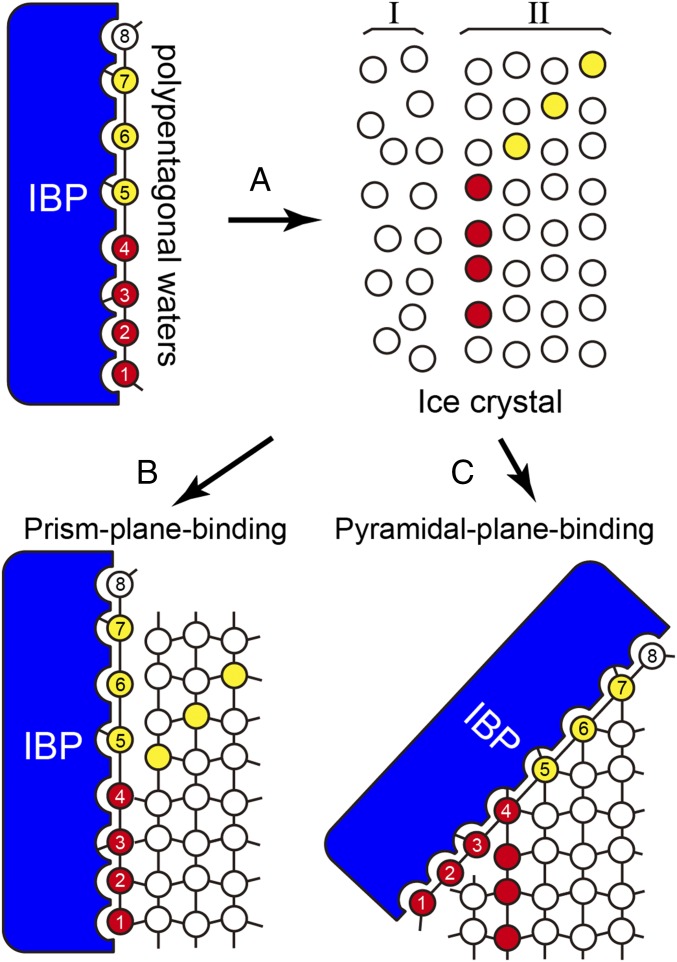
Polypentagonal Networks Merge with the Quasi-Liquid Layer and Freeze to Ice Lattice.
Molecular dynamics calculation of the growth kinetics of the ice-water interface suggests that IBPs initially present an intermediate state in the quasi-liquid layer (Fig. 5) to dynamically associate with both mobile and restricted waters on the ice surface (38). To recognize such a disordered water layer, the hydration waters are thought to form a preconfigured structure (39). Our detection of a polypentagonal water network on an activity-improved variant of IBP is consistent with these previous indications; these organized waters in the hydration shell assist the A20I mutant in immersing into the quasi-liquid layer, which leads to both ice-binding sites being located in close proximity to the waters constructing the solid ice lattice (Fig. 5A). This facilitates successive binding of this activity-improved variant to both the prism and the pyramidal ice planes through the water cluster prism and water cluster pyramid composing the polypentagonal network (Fig. 5 B and C). The involvement of both ice-like semiclathrate waters and perfect lattice-match waters in the network waters must be effective for these two ice-binding steps to occur. Known examples of such space-match waters are seven linearly aligned waters on a hyperactive IBP from the beetle Tenebrio molitor (40) and the waters constructing extensive arrays on another hyperactive IBP from the bacterium Marinomonas primoryensis (41), which are thought to connect these proteins to an ice crystal plane through the ACW mechanism. The present mutants are also capable of binding to the ice planes through this mechanism with their constituents of the water cluster prism and water cluster pyramidal. Inclusion of an additional water cluster corresponding to the basal plane in the polypentagonal network may combine the host protein with the multiple ice planes of an ice crystal.
Fig. 5.
Function of polypentagonal waters. (A) The 50 semiclathrate waters are initiatively merged with an intrinsically disordered quasi-liquid layer (region I) constructed on the ice crystal lattice (region II). (B) The water cluster prism (e.g., the waters labeled 1−4) successively shares the positions of the waters constructing the first prism plane through the ACW mechanism. (C) The water cluster pyramidal (e.g., the waters labeled 5−7) also shares the positions of those constructing the pyramidal ice plane.
Materials and Methods
Measurement of Ice-Growth Rate and TH Activity.
We used a Leica DMLB100 photomicroscope system equipped with a Linkam THMS 600 temperature controller to examine the morphology of a single ice crystal and its growth rate. A 0.7-μL droplet of the IBP solution was placed near the center of a capillary tube (ø = 0.92 mm, l = 10 mm), the termini of which were sealed with mineral oil. This capillary tube was inserted into a disk-shaped holder (ø = 17 mm, thickness = 2.4 mm), which was placed on the cooling stage of the photomicroscope system (28). The temperature of the protein solution in the capillary tube was controlled with an accuracy of ±0.2 °C. The tubes were frozen entirely at once by lowering the temperature to −25 °C to form a multicrystalline state and melted back to near 0 °C to carefully leave one single ice crystal with a size of 10–20 μm, which enabled the melting point (Tm) to be determined. The single ice crystal in the protein solution was modified into a bipyramid by lowering the temperature at a constant cooling rate (0.1 °C/min), which started off bursting ice crystal growth at a certain temperature. This observation allowed us to determine both Tburst and the ice-growth speed (μm/min). The values of Tm, Tburst, and the ice-growth speed were measured at least three times, and their averaged value was used for comparison.
X-Ray Crystallography.
All proteins were crystallized at 277 K by the hanging-drop vapor-diffusion method using EasyXtal 15-Well Tool (Qiagen). Before crystallization, the solutions of all mutants were dialyzed against 20 mM Tris⋅HCl buffer (pH 8.0) and concentrated by ultrafiltration using Amicon ultra 3000NMWL (Merck). The crystallization droplets were equilibrated to 500 µL of the reservoir solution. The X-ray diffraction data were obtained at 100 K in Photon Factory (BL5A, 6A, and 17A) and processed with the program HKL2000 (42). It should be noted that both the crystal and hydration structures of HPLC12 determined at 110 K (PDB ID 1HG7) (21) and 277 K (PDB ID 1MSI) (19) were not significantly different; their backbone structures exhibited only 0.32 Å of rmsd. The crystallographic refinements of the protein structures were performed with the program PHENIX (43). When the structure was refined by replacing one of the polypentagonal waters with an Na+ ion, a negative density of the difference Fourier map at the replaced position was observed. The replacements also increased the B-factor of that position, implying that the Na+ ion is not positioned in the water network. The deposited PDB codes (https://www.rcsb.org/) of each sample are 5XQN (WT1), 5XQP (WT2), 5XQR (A20V), 5XQU (A20I), 5XQV (A20L), and 5XR0 (A20T). Statistics of obtained diffraction data and crystallographic parameters are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This study was performed with Dr. Ryota Kuroki (deceased August 8, 2015) (Quantum Beam Science Center, Japan Atomic Energy Agency). We thank Dr. Yuichi Hanada and Maya Tamura for their help in preparing the recombinant proteins. This study was supported by a Grant-in-Aid (15K13760) for scientific research from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes: 5XQN, 5XQP, 5XQR, 5XQU, 5XQV, and 5XR0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800635115/-/DCSupplemental.
References
- 1.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 2.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 3.Sun T, Lin F-H, Campbell RL, Allingham JS, Davies PL. An antifreeze protein folds with an interior network of more than 400 semi-clathrate waters. Science. 2014;343:795–798. doi: 10.1126/science.1247407. [DOI] [PubMed] [Google Scholar]
- 4.Sharp KA. The remarkable hydration of the antifreeze protein Maxi: A computational study. J Chem Phys. 2014;141:22D510. doi: 10.1063/1.4896693. [DOI] [PubMed] [Google Scholar]
- 5.Davies PL. Ice-binding proteins: A remarkable diversity of structures for stopping and starting ice growth. Trends Biochem Sci. 2014;39:548–555. doi: 10.1016/j.tibs.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Scotter AJ, et al. The basis for hyperactivity of antifreeze proteins. Cryobiology. 2006;53:229–239. doi: 10.1016/j.cryobiol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Graether SP, Sykes BD. Cold survival in freeze-intolerant insects: The structure and function of beta-helical antifreeze proteins. Eur J Biochem. 2004;271:3285–3296. doi: 10.1111/j.1432-1033.2004.04256.x. [DOI] [PubMed] [Google Scholar]
- 8.Garnham CP, Campbell RL, Davies PL. Anchored clathrate waters bind antifreeze proteins to ice. Proc Natl Acad Sci USA. 2011;108:7363–7367. doi: 10.1073/pnas.1100429108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiani H, Sun D-W. Water crystallization and its importance to freezing of foods: A review. Trends Food Sci Technol. 2011;22:407–426. [Google Scholar]
- 10.Capicciotti CJ, Doshi M, Ben RN. Recent Developments in the Study of Recrystallization. INTECH Open Access; Rijeka, Croatia: 2013. Ice recrystallization inhibitors: From biological antifreezes to small molecules. [Google Scholar]
- 11.Doherty RD, et al. Current issues in recrystallization: A review. Mater Sci Eng A. 1997;238:219–274. [Google Scholar]
- 12.Hobbs PV. Ice Physics. Oxford Univ Press; London: 1974. pp. 461–523. [Google Scholar]
- 13.Raymond JA, DeVries AL. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc Natl Acad Sci USA. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett J. Thermal hysteresis proteins. Int J Biochem Cell Biol. 2001;33:105–117. doi: 10.1016/s1357-2725(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 15.Basu K, et al. Determining the ice-binding planes of antifreeze proteins by fluorescence-based ice plane affinity. J Vis Exp. 2014;83:e51185. doi: 10.3791/51185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celik Y, et al. Superheating of ice crystals in antifreeze protein solutions. Proc Natl Acad Sci USA. 2010;107:5423–5428. doi: 10.1073/pnas.0909456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamichi M, Nishimiya Y, Miura A, Tsuda S. Fully active QAE isoform confers thermal hysteresis activity on a defective SP isoform of type III antifreeze protein. FEBS J. 2009;276:1471–1479. doi: 10.1111/j.1742-4658.2009.06887.x. [DOI] [PubMed] [Google Scholar]
- 18.Hew CL, Slaughter D, Joshi SB, Fletcher GL, Ananthanarayanan VS. Antifreeze polypeptides from the Newfoundland ocean pout, Macrozoarces americ- anus: Presence of multiple and compositionally diverse components. J Comp Physiol B. 1984;155:81–88. [Google Scholar]
- 19.Jia Z, DeLuca CI, Chao H, Davies PL. Structural basis for the binding of a globular antifreeze protein to ice. Nature. 1996;384:285–288. doi: 10.1038/384285a0. [DOI] [PubMed] [Google Scholar]
- 20.Yang DSC, et al. Identification of the ice-binding surface on a type III antifreeze protein with a “flatness function” algorithm. Biophys J. 1998;74:2142–2151. doi: 10.1016/S0006-3495(98)77923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antson AA, et al. Understanding the mechanism of ice binding by type III antifreeze proteins. J Mol Biol. 2001;305:875–889. doi: 10.1006/jmbi.2000.4336. [DOI] [PubMed] [Google Scholar]
- 22.Garnham CP, et al. Compound ice-binding site of an antifreeze protein revealed by mutagenesis and fluorescent tagging. Biochemistry. 2010;49:9063–9071. doi: 10.1021/bi100516e. [DOI] [PubMed] [Google Scholar]
- 23.Garnham CP, Nishimiya Y, Tsuda S, Davies PL. Engineering a naturally inactive isoform of type III antifreeze protein into one that can stop the growth of ice. FEBS Lett. 2012;586:3876–3881. doi: 10.1016/j.febslet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Gallagher KR, Sharp KA. Analysis of thermal hysteresis protein hydration using the random network model. Biophys Chem. 2003;105:195–209. doi: 10.1016/s0301-4622(03)00087-5. [DOI] [PubMed] [Google Scholar]
- 25.Yang C, Sharp KA. The mechanism of the type III antifreeze protein action: A computational study. Biophys Chem. 2004;109:137–148. doi: 10.1016/j.bpc.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Nishimiya Y, Sato R, Takamichi M, Miura A, Tsuda S. Co-operative effect of the isoforms of type III antifreeze protein expressed in Notched-fin eelpout, Zoarces elongatus Kner. FEBS J. 2005;272:482–492. doi: 10.1111/j.1742-4658.2004.04490.x. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamichi M, Nishimiya Y, Miura A, Tsuda S. Effect of annealing time of an ice crystal on the activity of type III antifreeze protein. FEBS J. 2007;274:6469–6476. doi: 10.1111/j.1742-4658.2007.06164.x. [DOI] [PubMed] [Google Scholar]
- 29.McPherson A. Current approaches to macromolecular crystallization. Eur J Biochem. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- 30.Leinala EK, Davies PL, Jia Z. Crystal structure of beta-helical antifreeze protein points to a general ice binding model. Structure. 2002;10:619–627. doi: 10.1016/s0969-2126(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 31.Sun T, Gauthier SY, Campbell RL, Davies PL. Revealing surface waters on an antifreeze protein by fusion protein crystallography combined with molecular dynamic simulations. J Phys Chem B. 2015;119:12808–12815. doi: 10.1021/acs.jpcb.5b06474. [DOI] [PubMed] [Google Scholar]
- 32.Mantz YA, Geiger FM, Molina LT, Molina MJ, Trout BL. First-principles molecular-dynamics study of surface disordering of the (0001) face of hexagonal ice. J Chem Phys. 2000;113:10733–10743. [Google Scholar]
- 33.Hayward JA, Haymet ADJ. The ice/water interface: Molecular dynamics simulations of the basal, prism, {2021}, and {2110} Interfaces of Ice Ih. J Chem Phys. 2001;114:3713–3726. [Google Scholar]
- 34.Teeter MM. Water structure of a hydrophobic protein at atomic resolution: Pentagon rings of water molecules in crystals of crambin. Proc Natl Acad Sci USA. 1984;81:6014–6018. doi: 10.1073/pnas.81.19.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talon R, Coquelle N, Madern D, Girard E. An experimental point of view on hydration/solvation in halophilic proteins. Front Microbiol. 2014;5:66. doi: 10.3389/fmicb.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard EI, et al. Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface. J Mol Recognit. 2011;24:724–732. doi: 10.1002/jmr.1130. [DOI] [PubMed] [Google Scholar]
- 37.Momma K, Izumi F. VESTA3 for three-dimensional visualization of crystal, volumetric and morphology data. J Appl Cryst. 2011;44:1272–1276. [Google Scholar]
- 38.Nada H, Furukawa Y. Antifreeze proteins: Computer simulation studies on the mechanism of ice growth inhibition. Polym J. 2012;44:690–698. [Google Scholar]
- 39.Smolin N, Daggett V. Formation of ice-like water structure on the surface of an antifreeze protein. J Phys Chem B. 2008;112:6193–6202. doi: 10.1021/jp710546e. [DOI] [PubMed] [Google Scholar]
- 40.Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a β-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- 41.Garnham CP, et al. A Ca2+-dependent bacterial antifreeze protein domain has a novel beta-helical ice-binding fold. Biochem J. 2008;411:171–180. doi: 10.1042/BJ20071372. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.