Significance
The precise location of variants in the human genome is of utmost importance. We present a unique approach, coverage-based single nucleotide variant (SNV) identification (COBASI), which uses only perfect matches between the reads of a sequence project and a reference genome to detect and accurately identify de novo SNVs. From the perfect matches, a representation of the read coverage per nucleotide along the genome, the variation landscape, is generated. SNVs are then pinpointed as significant changes in coverage and de novo SNVs can be identified with high precision. The performance of COBASI was analyzed using simulations and experimentally validated by sequencing de novo SNVs identified from a parent–offspring trio. We propose this pipeline as a useful tool for different genomic applications.
Keywords: human genome variation, genomic algorithms, de novo mutations, genomic landscape, coverage map
Abstract
The precise determination of de novo genetic variants has enormous implications across different fields of biology and medicine, particularly personalized medicine. Currently, de novo variations are identified by mapping sample reads from a parent–offspring trio to a reference genome, allowing for a certain degree of differences. While widely used, this approach often introduces false-positive (FP) results due to misaligned reads and mischaracterized sequencing errors. In a previous study, we developed an alternative approach to accurately identify single nucleotide variants (SNVs) using only perfect matches. However, this approach could be applied only to haploid regions of the genome and was computationally intensive. In this study, we present a unique approach, coverage-based single nucleotide variant identification (COBASI), which allows the exploration of the entire genome using second-generation short sequence reads without extensive computing requirements. COBASI identifies SNVs using changes in coverage of exactly matching unique substrings, and is particularly suited for pinpointing de novo SNVs. Unlike other approaches that require population frequencies across hundreds of samples to filter out any methodological biases, COBASI can be applied to detect de novo SNVs within isolated families. We demonstrate this capability through extensive simulation studies and by studying a parent–offspring trio we sequenced using short reads. Experimental validation of all 58 candidate de novo SNVs and a selection of non-de novo SNVs found in the trio confirmed zero FP calls. COBASI is available as open source at https://github.com/Laura-Gomez/COBASI for any researcher to use.
The identification of variations among genomes is the starting point for a diversity of projects to understand human health and disease. It is such an important step that several large international consortia have been established, such as the HapMap Project (1, 2) and the 1000 Genomes Project (3, 4), to catalog variations among different healthy human populations, as well as several large consortia to examine genetic variations associated with different diseases, such as the International Cancer Genome Consortium (5) and the Cancer Genome Atlas Project (6) to identify variations between normal versus cancer cells. A particularly important type of variation, de novo variants, are those variants that occur spontaneously between parents and children, and have been implicated in a variety of diseases, such as autism, intellectual disabilities, and schizophrenia (7–9).
Several bioinformatic pipelines have been developed to identify single nucleotide variants (SNVs). Most of these begin by mapping sequencing reads from the sample to the reference genome (RG), allowing some number of mismatches or indels using one of a number of short-read aligners [Burrows–Wheeler aligner (BWA), Bowtie, etc.] (10). A mapping quality score is reported to reflect the probability of the read being correctly mapped. The mapped reads are then used to make genotype assignments using computational tools, such as SAMtools (11) or Genome Analysis Toolkit (GATK) (12), which evaluate the alignment of reads at every position along the genome and assign a confidence score to indicate the probability of the existence of a variant. This is achieved using statistical inference algorithms, which are necessary because imperfect alignments create uncertainty about the position assigned to each read and sequencing errors can induce false variants (11, 12). Various correction steps, such as around-indel realignment or quality recalibration, have been proposed to correct for common artifacts. However, most of these steps require a database of known variants (13). Finally, to correctly assign each genotype, the likelihoods for each possible genotype are calculated based on the observed data, modeling both alignment accuracy and sequencing accuracy. Different scoring schemes have been used to compute the probability that the read has been correctly mapped (14) and the genotype has been correctly assigned to ultimately indicate the overall confidence in the results. Additionally, some pipelines specialized for finding de novo variants incorporate stringent filtering based on each individual genotype likelihood (15–17). These pipelines also often use population-specific samples to identify and filter out any methodological bias (15–17, 18) or they require a predetermined de novo mutation rate and population-specific allelic frequencies to calculate the probability of the called de novo variant being a false positive (FP) (19, 20).
These methods are needed to overcome an apparent paradox: when sequence reads are aligned to a reference genome, some degree of mismatch must be tolerated, since variation would not be detected by using only perfect alignments. On the other hand, because of the highly repetitive and complex structure of the human genome, the tolerance of mismatches could result in the misplacement of some reads, introducing false variants. Our group has addressed this paradox by applying a different approach to the problem of detecting SNV’s in human genomes called context-dependent individualization of nucleotides and virtual genomic hybridization (COIN-VGH) (21). It is based on perfect alignments of unique substrings of a specific size (k; kmers) of the sequencing reads to the reference genome. As a proof of concept, the COIN-VGH approach was previously used to identify SNVs in a haploid region (nonpseudoautosomal region of the chromosome X) of Craig Venter’s and James Watson’s genomes using the same Sanger or 454 sequencing data as in the original studies (22, 23). Despite the success in eliminating false-positive calls over alternative approaches, COIN-VGH has important limitations for its widespread use: (i) it can only be used in haploid regions of the genome, (ii) it requires relatively long reads, and (iii) the algorithm is time consuming and utilizes a large amount of random-access memory (RAM) and disk storage.
Addressing these issues, we have developed a unique approach, called coverage-based single nucleotide variant identification (COBASI). COBASI builds on the original COIN-VGH approach but can be used to call variants from both haploid and diploid regions of the human genome and works with 30× or greater fold coverage (it has been used in datasets with as much as 100× fold coverage) of second-generation short sequence reads. In addition to circumventing the previous limitations of COIN-VGH, the approach is particularly suited to identify de novo SNVs through the joint analysis of a parent–offspring trio sequencing data. To evaluate COBASI, we first apply it to a diverse collection of simulated sequencing data and show that its performance is similar or superior to alternative approaches. We next apply it to the whole genomes of a parent–offspring trio we sequenced using Illumina sequencing and identified de novo SNVs across the entire child genome. From this, we discover 58 de novo SNVs, and all predicted de novo SNVs were experimentally confirmed as correct (zero false positives). Furthermore, the computing time and resources required for the bioinformatics pipeline have been significantly reduced, allowing for its routine application over many human datasets or other large mammalian datasets with a high-quality reference genome. Thus, COBASI is a powerful tool to systematically scan genomes for regions of interest for a broad range of applications.
Results
Rationale of the COBASI Approach.
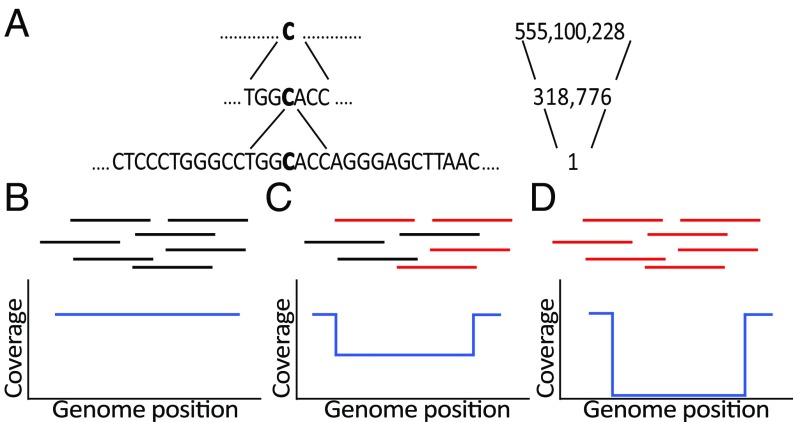
When a single specific nucleotide is searched along the genome, the position to which it belongs cannot be unambiguously determined. If two adjacent nucleotides are incorporated into the search, the set of possible locations is reduced, although it remains quite large. At some point, however, the context of the target nucleotide will contain enough information to unambiguously determine its unique origin position (Fig. 1A). In our previous research, we defined COIN-Strings (CSs) as the set of all overlapping sequences (with a one-nucleotide sliding window) from the reference genome of a specific size (k) that are uniquely localized. Thus, each nucleotide along the reference genome is contained in, at most, k CSs.
Fig. 1.
Rationale of the COBASI approach. (A) A specific nucleotide (large bold C) cannot be uniquely localized along the genome until its context is included in the search. (Left) The string to be searched; (Right) the number of positions at which such a string is found. The bottom string is a COIN-String (CS) of 30 nt. (B–D) (Upper) Schematic representation of sequence reads. (Lower) Specific regions of variation landscapes (VLs) for three scenarios. (B) No variation signal. (C) A heterozygous SNV variation signal. (D) A homozygous SNV variation signal. Black lines in B, C, and D represent reads from the genome project that contain the reference allele. Red lines represent reads from the genome project that contain the SNV allele. The sections of the VL in ref. 2 are represented by blue lines. The x axis indicates the genome position for every CS start. The y axis indicates the number of reads containing the CS sequence starting at that position.
COBASI extends this analysis of CSs to robustly find variations in the sample across the entire genome. When a SNV is present in a sample at a particular position X, it is expected that about half the reads for heterozygous SNVs, or nearly all of the reads in homozygous SNVs that overlap with X will contain the SNV. Accordingly, the CSs that include X will be present only in the reads that do not contain the alternative allele. This can be translated into specific patterns that are designated as variation signature regions (VSRs) (Figs. 1 C and D and 2A). Once candidate regions are identified, local alignments between the reads and the genome at the regions of interest will uncover the nature of the specific variants.
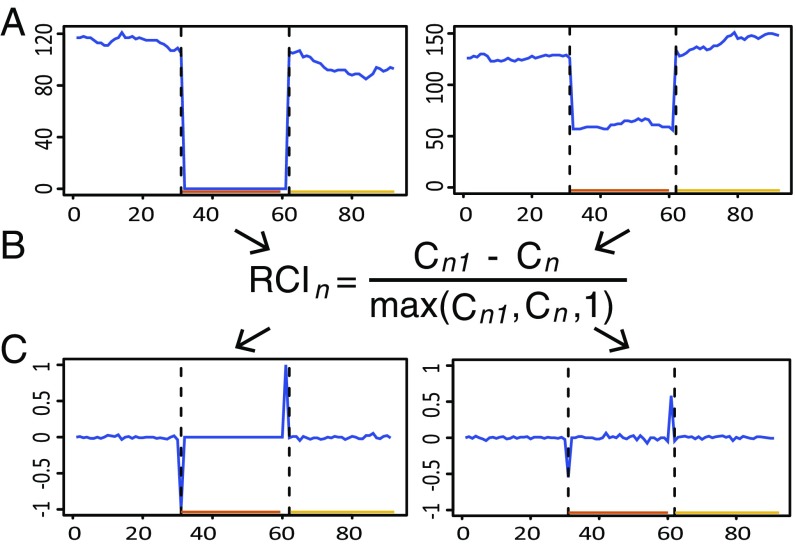
Fig. 2.
Variation landscape transformation into a relative coverage landscape. (Left) A homozygous SNV is shown. (Right) A heterozygous SNV is shown. (A) The VL for a region composed of 30 nt upstream and 30 nt downstream of each VSR is shown. The plots show the start position of each CS in that genomic region (x axis) and the coverage for each CS (y axis). (B) The VL is turned into the RVL using the RCI. RCIn refers to the relative coverage index for nucleotide n. Cn and Cn1 denote the number of reads that contain the CS starting at nucleotide n and the next downstream CS, respectively. (C) The RVL for the same regions shown in A. The plots show the start position of each CS (x axis) and RCI values associated with each CS (y axis). The VL and the RVL are represented by blue lines. The PrevCS and PostCS are shown as orange and yellow lines at the Bottom of each plot, and their start positions are highlighted with dashed black vertical lines (SI Appendix, Fig. S1).
De Novo SNV Discovery Using the COBASI Pipeline.
Based on the rationale presented, we designed and implemented a strategy to detect de novo SNVs from a parent–offspring trio. First, all of the CS positions from the reference genome are computed. We define the COBASI-accessible genome as regions at least 100 bp long for which at least 50% of the kmers starting inside the region are CSs using k = 30 bp. Even though more than 50% of the human genome is classified as repetitive sequences (24), the vast majority (around 84%) of the genome can be interrogated using COBASI (SI Appendix, Table S1).
Next, all of the SNVs from the child individual are identified by analyzing the variation landscape (VL). The VL is a representation of the number of reads that contain each CS sequence (coverage) along the whole genome (Fig. 2A). To magnify the difference in coverage between two adjacent CSs, the VL was transformed into a relative variation landscape (RVL) using a relative coverage index (RCI), measured on a scale from −1 to +1 (Fig. 2B). Under this formulation, the RCI is close to zero when there is little to no difference in coverage, and its absolute value approaches 1 when abrupt differences occur, most often because of underlying genetic variation (Fig. 2C). Since the RVL is variable in low-coverage regions, a coverage threshold was established to avoid noise in the VSR identification process (Materials and Methods).
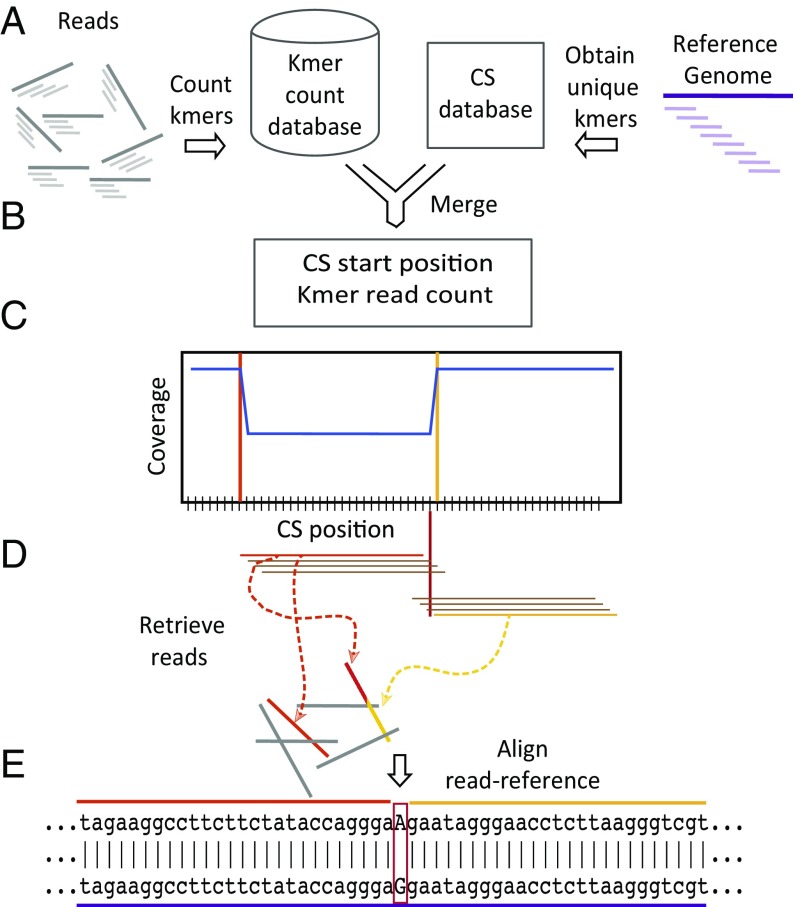
From the RVL, the VSRs can be identified spanning any candidate mutations. We define the last CS before the start of a VSR as PrevCS, and define the first CS after the end of a VSR as PostCS, and both of these CSs we call signature CSs. Next, reads containing perfect matches to the signature CSs are identified and global alignments between the corresponding region in the reads and the genome are computed. Finally, the variant nucleotides in the reads are highlighted in the local alignment to identify the specific SNV (Fig. 3). Since CSs are guaranteed to be unique in the genome, and only perfect matches are considered, no other quality filters are required.
Fig. 3.
The COBASI experimental pipeline for SNV discovery in one individual. (A, Left) Every overlapping 30-nt kmer (with a sliding window of 1 nt) along each of the reads of the sequencing project is obtained (only 3 kmers are shown per read). The counts for every kmer are stored in a database. Reads and read kmers are shown as gray and light gray lines, respectively. (A, Right) CS along the RG is obtained, and the start and end positions of all nonoverlapping unique regions is stored. RG and RG kmers are shown as purple and light purple lines. (B) The two virtual products are merged and the variation landscape (VL) is generated. (C) A region of the VL containing one heterozygous SNV is presented. The plot shows the start position of each CS along the genome (x axis) and each CS coverage (y axis). The VL is represented as a blue line. The VL is transformed into the RVL. Only the VL is depicted. The start position of the PrevCS and the PostCS are indicated by vertical orange and yellow lines, respectively. The PrevCS and PostCS are represented by horizontal orange and yellow lines, respectively. Some interCSs are shown as horizontal brown lines. The position of the SNV is shown as a red vertical line. All CSs located between the Prev- and PostCSs (interCSs) contain the SNV position. (D) The Prev- and PostCSs (signature CSs) are used as anchors to retrieve all of the reads of interest (Materials and Methods). (E) Each of the retrieved reads is then aligned with the corresponding region of the RG. An aligned read-RG region is shown. The SNV position and specific nucleotide is highlighted in a red rectangle.
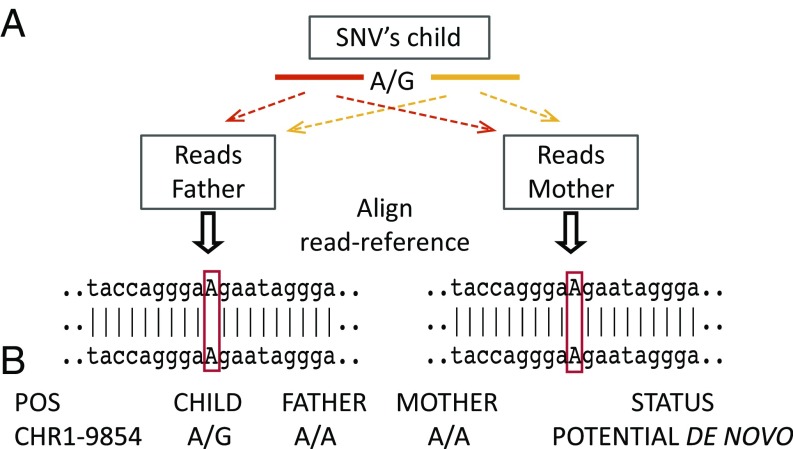
To discover the de novo SNVs, variable positions in the child are next interrogated in the parents. For each SNV in the child, its signature CSs were used as anchors to retrieve the reads of interest in the parents. Those reads from the parents are then aligned to the RG using the above procedure. A catalog containing all of the child SNVs and the alleles found in each parent for the same positions is then generated. The genotypes for each individual are assigned and compared, so that candidate de novo SNVs can be identified (Fig. 4). We considered as bona fide de novo variants those not found in either parent in more than one alignment containing both signature CSs, which are considered as high-quality alignments.
Fig. 4.
The COBASI experimental pipeline for SNV discovery in a family-based framework. (A) For each SNV in the child, its signature CSs are used as anchors to retrieve the corresponding reads in the parents. The reads are then aligned to the RG. (B) A catalog containing all child SNVs and the alleles found in each parent at the same positions is generated. The three genotypes are then compared, and the possible de novo SNVs are identified.
Performance of COBASI by Simulation Experiments.
We first evaluated COBASI relative to the most commonly used pipelines through simulation experiments considering several different sequencing depths, kmer sizes, and other internal parameters (SI Appendix, SI Materials and Methods). Mutations were introduced into one human diploid chromosome (chromosome 12), simulated reads were produced, and SNVs were called using COBASI. We quantified the performance using the widely used area under the precision-recall (AUPR) curve statistic.
The best performing parameters were derived from the simulation experiments. Over all of the tested sequencing depths, the best kmer size was 30, and the best ratio between the coverage of both signature CSs was 2.0. This maintained a low number of FPs while not significantly increasing the false negatives (FNs). Values of 0.2 or 0.3 for the RCI threshold had very similar AUPR scores. In contrast, the best value for other key parameters depended on the sequencing depth. If the sequencing depth was 35×, the minimum coverage for the signature CSs was 5, the optimal extension for alignments that contain only the PrevCS was 5 bp, and the minimum number of alignments with both CSs was 2. If the sequencing depth was 100×, the minimum coverage for the signature CSs was 10, the optimal extension for alignments that contain only the PrevCS was 5 bp or 10 bp, and the minimum number of total alignments with both CSs was 3 or 4. Once the best performing parameters were identified, the AUPR ranged from 0.94 to 0.96. To compare COBASI performance with the performance of the most commonly used variant-calling pipeline, the SNVs were also called from the simulation experiment with a sequencing depth of 100× using a combination of BWA, Picard Tools, and GATK. The AUPR was 0.99, while the AUPR obtained for COBASI was 0.96. However, the time required to obtain a list of SNVs from raw sequencing data was incredibly reduced, from more than 30 h in the case of the standard alignment-based pipeline to less than 6 h required by COBASI.
Besides, in a previous study, Hwang et al. measured the performance for any combination of three different mappers and three different callers for any of 11 datasets (10). In most cases, the AUPR for COBASI was similar to previously reported AUPRs, even though Hwang et al. used only exome data (about 2% of the genome) and COBASI was tested on the whole callable genome (about 84% of the genome) (SI Appendix, Tables S2 and S3).
We next measured the performance of de novo SNV discovery by COBASI using parent–offspring trio simulations. A trio of parent–offspring genomes was created following Mendelian inheritance along with a limited number of de novo variants (with a median of 35 de novo SNVs per simulation) (Materials and Methods), from which sequencing data were simulated. The sequencing depth was chosen to resemble our experimental sequencing data: 35× for the parents and 100× for the child. The de novo SNVs were then called using COBASI. The experiment was repeated five times, so that robust median accuracy values could be computed. The median precision obtained was 1.0 and the median recall was 0.91 with a median of 32 true positives (TPs), 3 FNs, and 0 FPs.
As with any variant detection pipeline, sufficient sequencing coverage is required to accurately detect mutations. To examine this for COBASI, we plotted the precision-recall curve ordered by the available coverage, defined as the number of alignments that contain the variant. The median AUPR across all coverage values was 0.86. However, most of the errors were found in low coverage variants, and with a reasonable coverage level (>10 reads), the median precision and recall for de novo simulations were 1.0 and 0.91, respectively. In one individual experiment, the precision and recall at the same coverage threshold were 0.9999 and 0.9613, respectively. Thus, the de novo discovery pipeline was more precise than the whole-genome pipeline at the expense of a small degree of sensitivity. Using the same simulated data, the de novo SNVs were called using the standard practices of the most commonly used alignment-based pipeline, resulting in an AUPR of 0.91. Thus, the COBASI performance can be compared with state of the art pipelines reducing the time required to complete the variant-calling process.
COBASI Application in a Family-Based Framework.
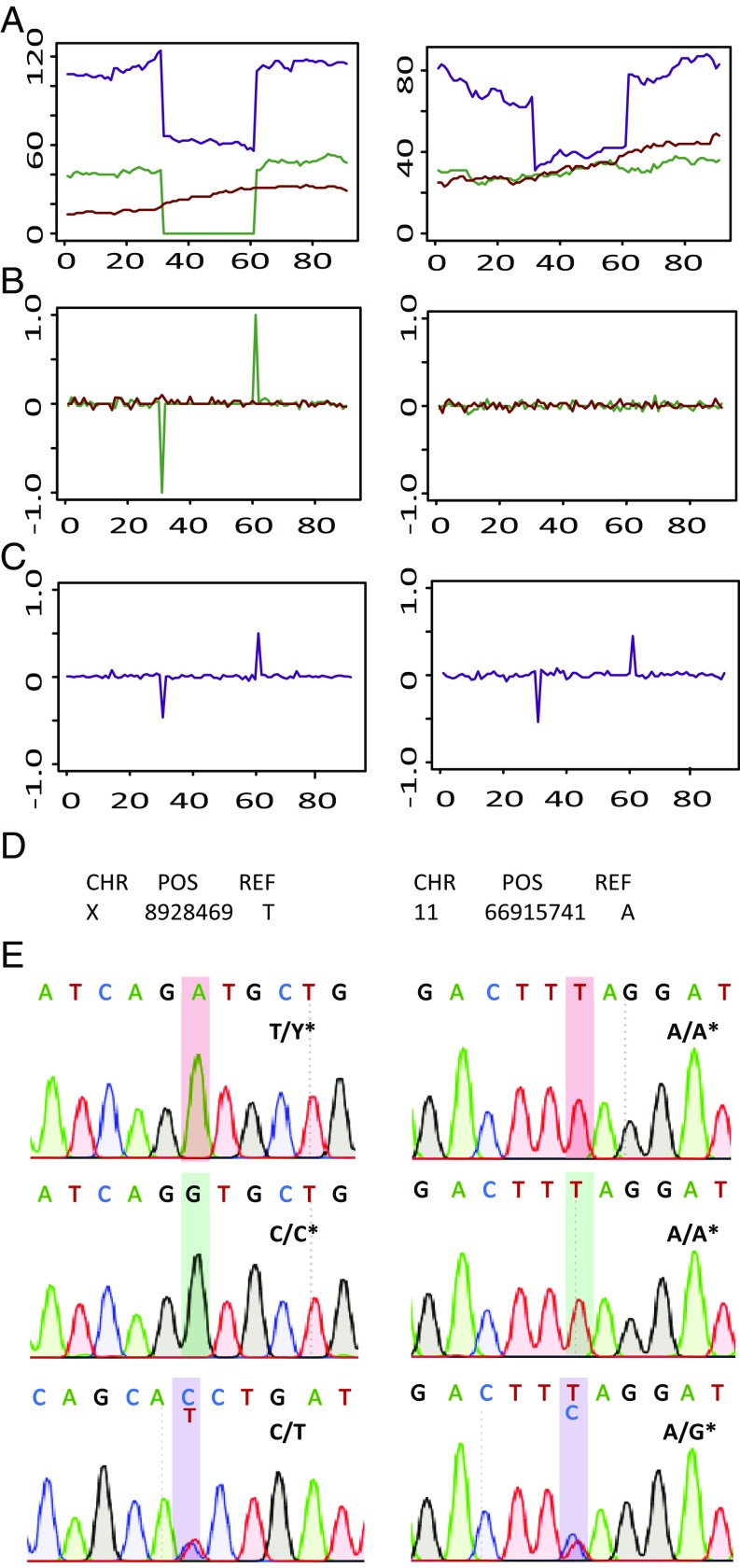
We next applied the de novo discovery COBASI pipeline to find genome-wide SNVs in a parent–offspring trio we sequenced using Illumina sequencing (Materials and Methods). Here we used the best performing parameters determined from the simulation experiments. Additionally, we considered as bona fide de novo variants those not previously reported in public databases, such as dbSNP, since the probability of two independent individuals having a de novo mutation event at the same nucleotide is very low (SI Appendix, SI Materials and Methods). Using these parameters, we found 2,912,889 SNVs in the discovery individual and 58 de novo variants (Fig. 5).
Fig. 5.
Experimental example of the COBASI strategy in the family-based framework. (Left) A Mendelian SNV is shown. Position 1 in the plots corresponds to chrX position 8928409. (Right) A de novo SNV is shown. Position 1 in the plots corresponds to chr11 position 66915681. (A) The corresponding section of the VL is shown for each parent–offspring trio individual: the red, green, and purple lines correspond to the VL for the father, mother, and child, respectively. Since the Mendelian SNV is located in the chrX, the father has around half the coverage of the mother. (B) The RVL is shown for both parents. (C) The RVL is shown for the child. (D) The nucleotide present at the RG is shown. (E) The chromatograms obtained by Sanger sequencing for these regions are shown. The genotypes obtained for each individual by the COBASI approach are shown in bold letters. An asterisk next to the individual genotype indicates that the chromatogram is in the reverse orientation. The SNV position is shadowed according to the individual color code.
The 58 de novo SNVs and a selection of two randomly chosen SNVs per chromosome (46 random variants total) identified in the child were selected for experimental validation via PCR and Sanger sequencing. In the case of the de novo variants, for five cases no PCR product could be obtained and one case could not be properly sequenced. For all 52 de novo mutations that could be sequenced, the Sanger sequencing confirmed that each predicted SNV represented a real de novo variant. SI Appendix, Table S4 presents the genomic coordinates, the genotype for each individual, and the results of the experimental validation for every de novo SNV. SI Appendix, Fig. S2 presents the experimental validation for each individual of the family trio for 10 de novo variants, chosen at random. All of the 46 Mendelian variants were successfully validated (SI Appendix, Fig. S3 and Table S5) (five examples).
Discussion
To find de novo SNVs in sequenced genomes, the COBASI approach represents a fast and precise solution to the variant-calling problem. It is based on the concept that by using only perfect matches of unique substrings to a reference genome, variation can nevertheless be found with great precision. In this study, we used unique DNA strings of 30 nucleotides, which can interrogate about 84% of all of the base pairs of the complete reference genome. Importantly, this percentage was calculated to include all repetitive sequences, such as low-complexity regions and segmental duplications of high identity. Larger strings would identify a greater percentage of the genome, although this will become more sensitive to any sequencing errors in the reads.
The VL constructed in the first stages of our approach represents a powerful tool to pinpoint regions of polymorphisms by identifying abrupt changes in local coverage. Moreover, these sharp differences were proven to be robust to noisy coverage fluctuations found in any sequencing project. The VL is generated in a fast, computationally efficient process and represents a comprehensive description of the read coverage across the genome at a single-nucleotide resolution.
The identification of de novo variants is a particularly challenging task because any false-positive calls in the child or any false-negative calls in the parents result in a variant incorrectly identified as de novo. To address this challenge, several specialized algorithms that analyze sequence data for all family individuals have been proposed. These algorithms rely on a prior probability of de novo mutations that is used to compute a posterior probability for each de novo mutation being correctly identified (11, 25). These algorithms therefore must be trained with a set of quality metrics obtained from a previously validated positive and negative set of variants (26). In addition, in previous reports, large populations are needed to remove the artifacts produced by the sequencing process, along with stringent quality filters to identify bona fide de novo variants (15–17, 27).
The strategy presented in this work is based on the most reliable types of alignments: perfect matches of unique strings of the genome followed by an analysis of the resulting alignment coverage. Other algorithms rely on less reliable alignments of imperfect matches spanning repetitive sequences and establishing probability thresholds to measure the quality of the findings. The performance of COBASI was assessed by simulation experiments, and for SNV discovery in one individual, we obtained an AUPR of 0.94 and 0.96 for a sequencing depth of 35× and 100×, respectively. In most cases, the AUPR for COBASI was similar to previously reported AUPRs (10), even though previous reports only used exome data, which represents about 2% of the genome. For de novo SNV discovery, we obtained a precision of 1.0 and a recall of 0.91 using COBASI, while a precision of 0.89 and a recall of 1 were obtained if the de novo SNV discovery was done by alignment-based approaches. COBASI achieves a good compromise between the increase of precision at the expense of a small decrease in recall. Furthermore, COBASI was tested on the whole callable genome, which constituted about 84% of the genome. It is also much faster than alignment-based approaches to achieve similar accuracy.
The precise identification of variant sites by COBASI relies on global alignments that include the variant site and two unique strings, one string located at each side of the variant site. Due to the small size of the reads, only small insertions or deletions would generate these high-quality alignments. Furthermore, in such cases, specialized aligners and detection algorithms would be required to pinpoint the variant positions. Incorporation of these specialized algorithms could be an extension of COBASI’s scope.
The computing resources and time required by COBASI enable its routine utilization. Generating a whole-genome SNV list from 35× raw sequencing data requires around 40 h on a computer server with 12 cores and 64 Gb of RAM. Moreover, the whole-genome variation landscape can be generated in only 8 h. Furthermore, if only some regions of interest are chosen to be investigated, the time required to generate a list of SNVs can be greatly reduced (SI Appendix, Table S6).
In this work, we analyzed the whole-genome sequencing of a parent–offspring trio sequenced to a genome coverage of 35× for the parents and 100× for the child. We did not assume any a priori de novo mutation rate. We applied coverage filters, but not quality filters on the reads. Regardless, we found no false positives in either our de novo SNV predictions or in the randomly selected Mendelian SNVs. Moreover, we found 58 de novo SNVs, and this number is consistent with the number of de novo SNVs expected from the previously reported germline mutation rate, 1.0–1.8 × 10−8 per nucleotide per generation, which translates into 44–82 de novo SNVs per individual (9, 28). This was accomplished because our approach combines a highly sensitive discovery in the child genome with an exhaustive validation in both parents. The number of discovered variants could be an underestimate, given that we can only interrogate 84% of the genome. However, with a world-wide sequencing capacity tending toward hundreds of thousands of genomes each year (29), our main interest is in maximizing the precision in the called variants to diminish as much as possible the extent of experimental validation that is required.
Recently, some publications have addressed the issue of calling SNVs by implementing mapping-free strategies. Known SNVs have been identified from sequencing reads if unique kmers containing the alternative allele are present in the reads (30). A Burrows–Wheeler transform of the reads was used to localize SNVs based on differences in kmer frequency (31). Changes in kmer frequency have been used to reconstruct haplotypes from genomic regions harboring long variants, this strategy focused on specific regions of the genome (32). A recently published work from our group used kmer frequency changes to identify variants along natural genomes and synthetic chromosomes of haploid yeast strains (33). However, no previous work has focused on finding de novo SNVs in human whole genomes.
COBASI could be used to identify SNVs from different organisms, since the successful application of COBASI is only limited by the ploidy of the organism and the fraction of its genome that can be represented by unique strings. Within a single genome this approach can also be used to analyze CSs from particular regions of interest, such as a cancer gene panel or other sets of genes, thus speeding up the analysis time. We propose that the general principle underlying COBASI can be used in a broad range of applications, including personalized genomics, family studies, population genetics, ancient DNA studies, and metagenomics. It could also be used for general correlations between genotype and phenotype, such as different disorders characterized by the presence of de novo mutations, such as intellectual disability, autism, and schizophrenia (7–9).
Materials and Methods
COBASI Pipeline.
The program Jellyfish (34) was used to count the number of occurrences of each kmer (k = 30) along the reads. To eliminate possible sequencing errors, all unique kmers were discarded. From the Jellyfish database, the count for every kmer along the RG was retrieved using the cov-plot script from the AMOS repository (35), and the read-based kmer counts associated with CSs were kept to generate the VL. The VL contained the start position for every CS along the genome and its number of occurrences in the reads (coverage). To identify CSs with abnormal coverage for each simulation or sequencing experiment, a coverage threshold was calculated. It corresponded to the median of the coverage [+/−]10 interquartile range (IQR), and ∼99.99% of the CSs had coverage values inside this rank. The VL was transformed into the RVL using the RCI. All CSs with an abnormal coverage were not taken into account.
In the child, the VSRs were identified from the RVL. Specifically, COBASI searches for regions with an abrupt drop in coverage followed by an abrupt rise in coverage. These partial VSRs were extended at most k nucleotides upstream and k nucleotides downstream. To characterize drastic changes in coverage, we required a minimum coverage as well as a minimum absolute value for the RCI for each of the signature CSs. Additionally, to extend the partial VSRs, a maximum ratio between the coverage of both signature CSs was established. The reference sequence for each signature CS was obtained, and all of the reads containing a signature CS were retrieved. A file containing the read identifier, the start reference position for the signature CS, and the position in the read for the match between the CS and the read and its orientation was created. Some inconsistent reads were filtered out (SI Appendix, SI Materials and Methods). For the case of the parents, the signature CSs obtained in the child were used to retrieve the reads of interest.
From reads containing both signature CSs, whole-VSR alignments were computed using a modified C++ align function from the AMOS repository. For each read, the region from the start of the PrevCS to the end of the PostCS was aligned to the corresponding RG region. These alignments were considered high-quality alignments, and only variants found in at least a certain number of these were further analyzed. For reads containing only the PrevCS, the alignment between the RG and the read was done from the start of the PrevCS to 5 nt downstream of the last variant nucleotide obtained from the high-quality alignments. In the case of the parents, if there was no variation in the whole-VSR alignments, the default extension was 5 bp. For all complete alignments, SNVs were identified.
The genotype of every SNV was assigned based on the algorithm described by Li (11), modified as described in SI Appendix, SI Materials and Methods. To identify the possible de novo SNVs, the genotypes for each of the individuals of the family trio were compared, and the potential de novo SNVs were identified. We defined criteria to establish a possible variant, such as a bona fide de novo variant (SI Appendix, SI Materials and Methods). Low-coverage sequencing experiments are prone to a higher number of both FN and FP calls. Therefore, COBASI includes additional quality requirements to avoid incorrect de novo SNV calls. Regions prone to incorrect genotype assignment were identified and excluded: (i) regions with low CS density, (ii) regions with more than one CS with a coverage higher than expected, (iii) regions with low coverage for any of the signature CSs in any individual, (iv) regions with additional significant changes in coverage inside the region corresponding to the child VSR: in the case of the child if there is any additional drop or rise it should correspond to a region with almost no coverage; in the case of the parents there should not exist any drop or rise corresponding to the child SNV position, and (v) regions with unequal coverage in both sides of the VSR for the child.
Additional Methods.
Additional methods are found in SI Appendix, SI Materials and Methods: Definition of CS Regions from the RG, Definition of Accessible Genome, Simulation Experiments, Variant Calling Using Alignment-Based Pipelines, TRIO Sequencing and COBASI Application, and Experimental Validation of de Novo SNVs.
Supplementary Material
Acknowledgments
We thank James Gurtowski and Giuseppe Narzisi (Cold Spring Harbor Laboratory) and Jair García Sotelo [Laboratorio Internacional de Investigación Sobre el Genoma Humano, Universidad Nacional Autónoma de México (UNAM)] for their technical support. L.G.-R. is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, UNAM and received Fellowship 275908 from Consejo Nacional de Ciencia y Tecnología. This work was supported, in part, by US National Science Foundation Award DBI-1350041 and US National Institutes of Health Award R01-HG006677 (to M.C.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802244115/-/DCSupplemental.
References
- 1.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson TJ, et al. International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–998, and erratum (2010) 465:966. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Cancer Genome Atlas. National Cancer Institute and National Human Genome Research Institute. Available at https://cancergenome.nih.gov/. Accessed April 22, 2018.
- 7.Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17:241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupski JR. New mutations and intellectual function. Nat Genet. 2010;42:1036–1038. doi: 10.1038/ng1210-1036. [DOI] [PubMed] [Google Scholar]
- 9.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 10.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, et al. The genome analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 17.Besenbacher S, et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat Commun. 2015;6:5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francioli LC, et al. Genome of the Netherlands consortium A framework for the detection of de novo mutations in family-based sequencing data. Eur J Hum Genet. 2017;25:227–233. doi: 10.1038/ejhg.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng G, et al. Rare variant detection using family-based sequencing analysis. Proc Natl Acad Sci USA. 2013;110:3985–3990. doi: 10.1073/pnas.1222158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyes J, et al. Context-dependent individualization of nucleotides and virtual genomic hybridization allow the precise location of human SNPs. Proc Natl Acad Sci USA. 2011;108:15294–15299. doi: 10.1073/pnas.1112567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 24.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, et al. A likelihood-based framework for variant calling and de novo mutation detection in families. PLoS Genet. 2012;8:e1002944. doi: 10.1371/journal.pgen.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaelson JJ, et al. Whole genome sequencing in autism identifies hotspots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens ZD, et al. Big Data: Astronomical or genomical? PLoS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajuste FD, et al. FastGT: An alignment-free method for calling common SNVs directly from raw sequencing reads. Sci Rep. 2017;7:2537. doi: 10.1038/s41598-017-02487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura K, Koike A. Ultrafast SNP analysis using the Burrows-Wheeler transform of short-read data. Bioinformatics. 2015;31:1577–1583. doi: 10.1093/bioinformatics/btv024. [DOI] [PubMed] [Google Scholar]
- 32.Audano PA, Ravishankar S, Vannberg FO. Mapping-free variant calling using haplotype reconstruction from k-mer frequencies. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios-Flores K, et al. A perfect match genomic landscape provides a unified framework for the precise detection of variation in natural and synthetic haploid genomes. Genetics. 2018;208:1631–1641. doi: 10.1534/genetics.117.300589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatz MC, et al. Hawkeye and AMOS: Visualizing and assessing the quality of genome assemblies. Brief Bioinform. 2013;14:213–224. doi: 10.1093/bib/bbr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.