Key Teaching Points.
-
•
Sudden infant death syndrome (SIDS) is defined as the sudden death of a healthy infant younger than 1 year of age without any obvious cause of death. Despite intensive genetic investigations, the underlying pathophysiological mechanism still remains elusive in most of the cases.
-
•
Whole-exome sequencing in a 5-month-old male infant identified a heterozygous missense variant in the β1B subunit of SCN1B. Electrophysiological recordings of Nav1.5 co-expressed with the β1B subunit variant p.R225C induced a loss of function of Nav1.5 channels.
-
•
The loss of function might have contributed to the sudden death event in this infant; however, further investigations are needed. This study demonstrates the importance of careful evaluation of likely pathogenic variants identified within next-generation sequencing approaches for an accurate interpretation of genetic results.
Introduction
Sudden infant death syndrome (SIDS) is defined as the sudden and unexpected death of an apparently healthy infant younger than 1 year of age.1 The occurrence of SIDS is described by a triple risk model involving a critical developmental period in combination with environmental and genetic risk factors; however, the pathophysiological mechanisms responsible for SIDS still remain poorly understood.2 Technical advances in high-throughput massive parallel sequencing recently enabled broad genetic analyses in large SIDS cohorts and identified likely pathogenic sequence alterations in cardiovascular disease–associated genes in up to 30% of SIDS victims.3, 4 Approximately 10%–15% of these sudden death cases are believed to be caused by cardiac channelopathies, which can cause lethal arrhythmias in absence of any structural changes in the heart.3, 4, 5
Voltage-gated sodium channels are integral membrane proteins primarily found in cardiac muscle cells, where they are involved in the generation and propagation of action potentials.6 The Na+ current (INa) is determined not only by the pore-forming α subunit (Nav1.5), but also by regulatory β subunits (β1-β4). The voltage-gated sodium channel β1B subunit gene (SCN1B) is expressed into 2 isoforms, the transmembrane β1 subunit and the soluble β1B subunit, which includes a retained intron encoding a novel C-terminus, stop codon, and polyadenylation site.7 Mutations in SCN1B have been reported in multiple inherited cardiac diseases, including congenital long QT syndrome, Brugada syndrome, cardiac conduction defect, atrial fibrillation, sick sinus syndrome, and SIDS.8 In addition, SCN1B loss-of-function mutations have been described in Dravet syndrome, a devastating pediatric epileptic encephalopathy with a high mortality during early childhood, mainly owing to sudden, unexpected death in epilepsy.9
Here, we report the electrophysiological effect of a rare SCN1B β1B subunit variant identified in the whole-exome sequencing data of a 5-month-old male SIDS case.
Case report
The 5-month old male infant (weight 6540 g; height 64 cm; European origin) was found dead in his cot early in the morning in a safe sleeping environment with no evidence of an accidental death. He was born full-term following a normal and uncomplicated pregnancy and was the third-born son of a 34-year-old healthy woman. The boy was vaccinated against diphtheria, tetanus, pertussis, and poliomyelitis, with a last vaccination 13 days prior to his death. During his months of life, no anomalous clinical events were reported except for a large head, with a head circumference greater than the 97th percentile. According to his parents, the boy slept a great deal and they often had difficulties awakening him.
A comprehensive autopsy investigation revealed normal sizes and structures of all organs and no signs of malformation, malignancy, or infections. In addition, microbiological and toxicological screening tests were all negative. As the cause of death remained unexplained, the case was assigned to SIDS category I based on the San Diego definition.1
Ethical approval for this study was provided by the local ethics committee (KEK-ZH-Nr. 2013-0086), and the study was conducted in full conformance with Swiss laws and regulations. Family members were not available for co-segregation analyses owing to the specifications in the ethical approval for this study.
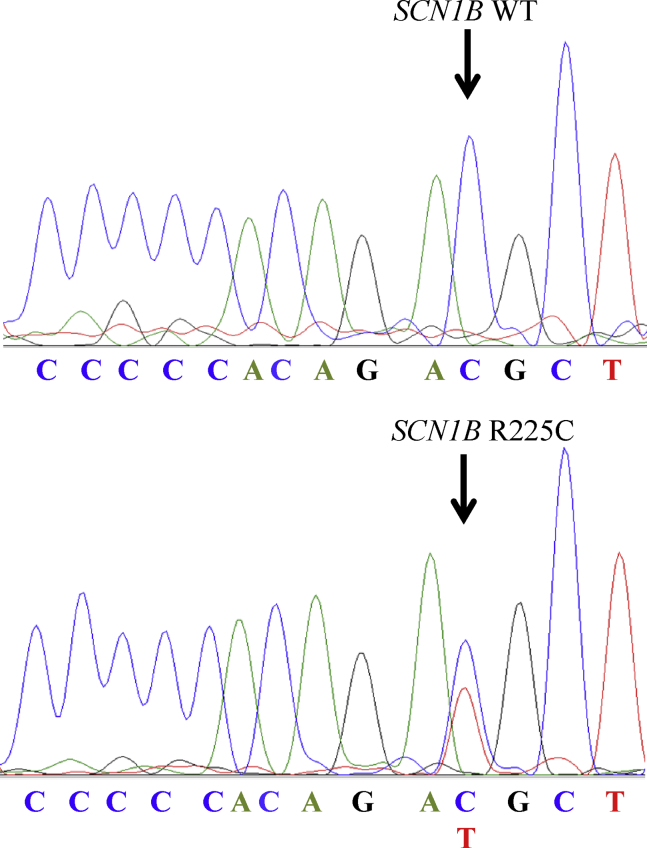
Whole-exome sequencing and variant analysis has been performed within a large genetic screening study in 161 SIDS infants.4 The 5-month-old boy carried a heterozygous missense variant (NM_199037.4, rs369588692, c.673C>T, p.R225C) in SCN1B (Figure 1). The sequence variation is located in exon 3A, which is unique to the alternative splice isoform β1B. The variant p.R225C was found to be very rare in the European (non-Finnish) population (minor allele frequency: 0.01%) in the NHLBI GO Exome Sequencing Project. The in silico protein prediction of the SCN1B variant was based on 6 different tools according to the recommendations of the American College of Medical Genetics standards and guidelines for the interpretation of sequence variants.10 Four in silico protein prediction tools categorized the variant as disease-causing (MutationTaster, SIFT, MAPP, and Grantham distance score), whereas 2 in silico protein prediction tools categorized the variant as benign (AGVGD and PolyPhen-2). In summary, the SCN1B sequence alteration was classified as a variant of uncertain significance owing to the absence of co-segregation and functional analyses.10
Figure 1.
Sanger sequencing confirmation of SCN1B c.673C>T, p.R225C.
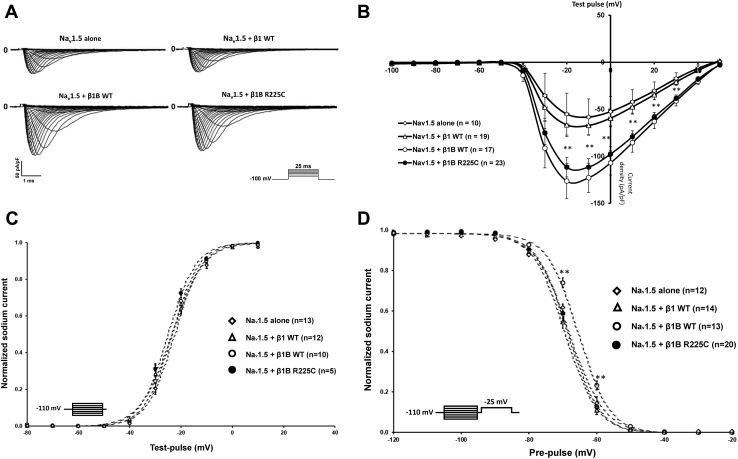
The electrophysiological effects of the 2 WT β-isoforms (β1 and β1B) and the β1B subunit variant on Nav1.5 sodium current were assessed via the whole-cell patch clamp technique using transiently transfected HEK293 cells. Parameters for Na+ current density and voltage dependence of activation and inactivation are listed in Table 1. Figure 2A shows representative current traces in cells expressing Nav1.5 alone and Nav1.5 plus β1 WT, β1B WT, or mutant β1B. Co-expression of Nav1.5 with β1B subunit WT or β1B p.R225C showed a significant increase of the sodium current density compared to Nav1.5 plus β1 WT and Nav1.5 alone (Figure 2B, Table 1). The steady-state activation was similar for all conditions (Figure 2C); however, co-expression of Nav1.5 with β1B WT induced a depolarizing shift of the steady-state inactivation curves, leading overall to a gain-of-function (Figure 2D). Interestingly, compared to β1B WT, co-expression of Nav1.5 with β1B-subunit p.R225C variant leads to a hyperpolarization of the steady-state inactivation relationship (Figure 2D). Consequently, compared to β1B subunit WT, β1B subunit p.R225C reduces the number of available channels for a given voltage, which leads to a loss of function of Nav1.5 channels.
Table 1.
Parameters for Na+-current density, activation, and inactivation
| Current density (pA/pF) |
Steady-state activation (SSA) |
Steady-state inactivation (SSI) |
||||||
|---|---|---|---|---|---|---|---|---|
| n | n | V1/2 (mV) | k | n | V1/2 (mV) | k | ||
| Nav1.5 alone | 10 | −57.94 ± 19.34 | 13 | −22.70 ± 0.77 | 5.43 ± 0.17 | 12 | −68.06 ± 0.65 | 4.90 ± 0.19 |
| Nav1.5 + β1 WT | 19 | −67.11 ± 10.04 | 12 | −23.90 ± 1.37 | 5.64 ± 0.19 | 14 | −68.94 ± 0.59 | 4.78 ± 0.11 |
| Nav1.5 + β1B WT | 17 | −106.21 ± 11.71∗ | 10 | −23.83 ± 1.29 | 5.08 ± 0.17 | 13 | −65.50 ± 0.54∗ | 4.59 ± 0.14 |
| Nav1.5 + β1B R225C | 23 | −111.41 ± 9.04∗ | 5 | −24.19 ± 0.69 | 5.32 ± 0.13 | 20 | −68.52 ± 0.52 | 4.77 ± 0.15 |
k = slope; n = number of recordings; V1/2 = voltage of half-maximal activation/inactivation.
P < .05 vs Nav1.5 alone, compared by paired 2-tailed Student t test.
Figure 2.
Electrophysiological characteristics of SCN1B. A: Representative traces of sodium current with Nav1.5 alone, β1 subunit WT, β1B subunit WT, and β1B subunit p.R225C in HEK293 cells. B: Sodium current density for Nav1.5 alone, β1 subunit WT, β1B subunit WT, and β1B subunit p.R225C. C: Voltage dependence of steady-state activation for Nav1.5 alone, β1 subunit WT, β1B subunit WT, and β1B subunit p.R225C. D: Voltage dependence of steady-state inactivation for Nav1.5 alone, β1 subunit WT, β1B subunit WT, and β1B subunit p.R225C. In B, C, and D data points represent mean ± standard error of the mean. ∗P < .05 and ∗∗P < .01, based on a paired 2-tailed Student t test.
Discussion
Here, we describe the results of whole-cell patch clamp analysis of a rare heterozygous missense variant in the β1B subunit of SCN1B found in a male SIDS infant. Sequence alterations in different β subunits (SCN1B–SCN4B) have been associated with different cardiac diseases.11 Altered Nav1.5 sodium channel function owing to β-subunit mutations may account for the molecular pathogenic mechanism in approximately 1% of SIDS cases.
In this study, the co-expression of the WT β1B subunit led to increased sodium current when compared to the WT β1 subunit or Nav1.5 alone. The effects of β1B subunit on Nav1.5 channels are controversial, as some groups have reported functional differences between the 2 isoforms, with or without affecting voltage dependences or channel kinetics, while others reported no effects.8 In addition, it is important to note that the β1B isoform is not a transmembrane protein, but a soluble peptide whose function is still poorly understood.7 The 2 β1 isoforms are expressed not only in the human heart, but also in the brain, with the highest amount of the β1B subunit during embryonic development where the soluble β1B protein functions as a ligand for β-mediated neurite migration and outgrowth.9 Genetic variants in the β1B gene were furthermore described as a risk factor for human epilepsy,7 which might be another possible terminal pathway in SIDS infants.
The p.R225C variant is located in the retained region of β1B, which is unique to this isoform. Since close located variants have been described in congenital long QT syndrome, Brugada syndrome, lone atrial fibrillation, Dravet syndrome, and idiopathic epilepsy,8, 9, 11 the identified variant in the present study represents an interesting candidate sequence alteration in the pathophysiological mechanisms contributing to the sudden death event. Therefore, the loss of function of the p.R225C variant might have contributed to the occurrence of a lethal arrhythmia or epileptic seizure in combination with other genetic or environmental risk factors. However, owing to the controversial literature reports and our own findings, further investigations are needed to clarify the role and function of the 2 β1 isoforms and the variant.
During recent years, next-generation sequencing approaches facilitated the genetic investigations of patients with complex genetic patterns or the identification of underlying genetic causes in large study populations in a time- and cost-efficient manner. However, the generation of such huge amounts of data requires very stringent variant evaluation systems in order to separate false-positive variants from truly disease-causing sequence alterations.10, 12 Several studies have demonstrated that many variants previously associated with cardiac diseases have in fact no or only minor functional effects and are therefore less likely associated with a dominant form of the disease; still, they could act as a risk modifier.13 Therefore, functional studies are required and recommended for an evidence-based classification and interpretation of the pathogenicity of variants identified in genetic SIDS studies so far.10, 14
One limitation of this study is the lack of family members for genetic testing and co-segregation analysis. This would be necessary to determine the mode of inheritance, to classify variants into the pathogenic category,15 and to identify other genetic carriers at risk for sudden cardiac death. Additional limitations are related to the functional assay. Electrophysiological recordings were conducted in a conventional heterologous expression system where the environment is different from adult rod-shaped cardiomyocytes. Therefore, the effects of many proteins known to associate with the sodium channel complex could not be investigated. In addition, only little is known about the function and physiological role of the β1B isoform, emphasizing the need to further investigate the function and expression of this isoform in native tissues.
Conclusion
Electrophysiological investigations of the β1B p.R225C variant showed significant differences compared to the WT variant, suggesting a loss of function of sodium current.
The potential effect on the cardiac or brain function of this variant needs to be further clarified in multiple populations and co-segregation studies. This study highlights the importance of a careful evaluation of likely pathogenic variants identified within next-generation sequencing data. Even if variants are categorized as disease-causing according to in silico protein predictions tools, the performance of additional functional analyses is essential for an accurate interpretation of genetic results.
Acknowledgments
Special thanks to Corinne Moser for the Sanger sequencing.
Footnotes
This project was supported by the Swiss National Science Foundation (SNF; project-Nr. 320030_149456).
References
- 1.Krous H.F., Beckwith J.B., Byard R.W., Rognum T.O., Bajanowski T., Corey T., Cutz E., Hanzlick R., Keens T.G., Mitchell E.A. Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics. 2004;114:234–238. doi: 10.1542/peds.114.1.234. [DOI] [PubMed] [Google Scholar]
- 2.Filiano J.J., Kinney H.C. A perspective on neuropathologic findings in victims of the sudden infant death syndrome: the triple-risk model. Biol Neonate. 1994;65:194–197. doi: 10.1159/000244052. [DOI] [PubMed] [Google Scholar]
- 3.Hertz C.L., Christiansen S.L., Larsen M.K. Genetic investigations of sudden unexpected deaths in infancy using next-generation sequencing of 100 genes associated with cardiac diseases. Eur J Hum Genet. 2016;24:817–822. doi: 10.1038/ejhg.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neubauer J., Lecca M.R., Russo G., Bartsch C., Medeiros-Domingo A., Berger W., Haas C. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet. 2017;25:404–409. doi: 10.1038/ejhg.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilders R. Cardiac ion channelopathies and the sudden infant death syndrome. ISRN Cardiol. 2012;2012:846171. doi: 10.5402/2012/846171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rook M.B., Evers M.M., Vos M.A., Bierhuizen M.F. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc Res. 2012;93:12–23. doi: 10.1093/cvr/cvr252. [DOI] [PubMed] [Google Scholar]
- 7.Patino G.A., Brackenbury W.J., Bao Y., Lopez-Santiago L.F., O'Malley H.A., Chen C., Calhoun J.D., Lafreniere R.G., Cossette P., Rouleau G.A., Isom L.L. Voltage-gated Na+ channel beta1B: a secreted cell adhesion molecule involved in human epilepsy. J Neurosci. 2011;31:14577–14591. doi: 10.1523/JNEUROSCI.0361-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu D., Barajas-Martinez H., Medeiros-Domingo A. A novel rare variant in SCN1Bb linked to Brugada syndrome and SIDS by combined modulation of Na(v)1.5 and K(v)4.3 channel currents. Heart Rhythm. 2012;9:760–769. doi: 10.1016/j.hrthm.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patino G.A., Claes L.R., Lopez-Santiago L.F. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan B.H., Pundi K.N., Van Norstrand D.W., Valdivia C.R., Tester D.J., Medeiros-Domingo A., Makielski J.C., Ackerman M.J. Sudden infant death syndrome-associated mutations in the sodium channel beta subunits. Heart Rhythm. 2010;7:771–778. doi: 10.1016/j.hrthm.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baruteau A.E., Tester D.J., Kapplinger J.D., Ackerman M.J., Behr E.R. Sudden infant death syndrome and inherited cardiac conditions. Nat Rev Cardiol. 2017;14:715–726. doi: 10.1038/nrcardio.2017.129. [DOI] [PubMed] [Google Scholar]
- 13.Nouhravesh N., Ahlberg G., Ghouse J., Andreasen C., Svendsen J.H., Haunso S., Bundgaard H., Weeke P.E., Olesen M.S. Analyses of more than 60,000 exomes questions the role of numerous genes previously associated with dilated cardiomyopathy. Mol Genet Genomic Med. 2016;4:617–623. doi: 10.1002/mgg3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertier G., Hetu M., Joly Y. Unsolved challenges of clinical whole-exome sequencing: a systematic literature review of end-users' views. BMC Med Genomics. 2016;9:52. doi: 10.1186/s12920-016-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Spaendonck-Zwarts K.Y., van Rijsingen I.A., van den Berg M.P. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. Eur J Heart Fail. 2013;15:628–636. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]