Summary
Glycosylation, the covalent attachment of carbohydrate structures onto proteins, is the most abundant post-translational modification1. Over 50% of human proteins are glycosylated, which alters their activities in diverse fundamental biological processes2,3. Despite its importance in biology4, the identification and functional validation of complex glycoproteins has remained largely unexplored. Here, we developed a novel quantitative approach to identify intact glycopeptides from comparative proteomic data-sets, allowing us to not only infer complex glycan structures but also to directly map them to sites within the associated proteins at the proteome scale. We applied this method to human and murine embryonic stem cells to illuminate the stem cell glycoproteome. This analysis nearly doubles the number of experimentally confirmed glycoproteins, identifies previously unknown glycosylation sites and multiple glycosylated stemness factors, and uncovers evolutionarily conserved as well as species-specific glycoproteins in embryonic stem cells. The specificity of our method was confirmed using sister stem cells carrying repairable mutations in enzymes required for fucosylation, Fut9 and Slc35c1. Ablation of fucosylation confers resistance to the bioweapon ricin5,6, and we discovered proteins that carry a fucosylation-dependent sugar code for ricin toxicity. Mutations disrupting a subset of these proteins rendered cells ricin resistant, revealing new players that orchestrate ricin toxicity. Our novel comparative glycoproteomics platform enables genome-wide insights into protein glycosylation and glycan modifications in complex biological systems.
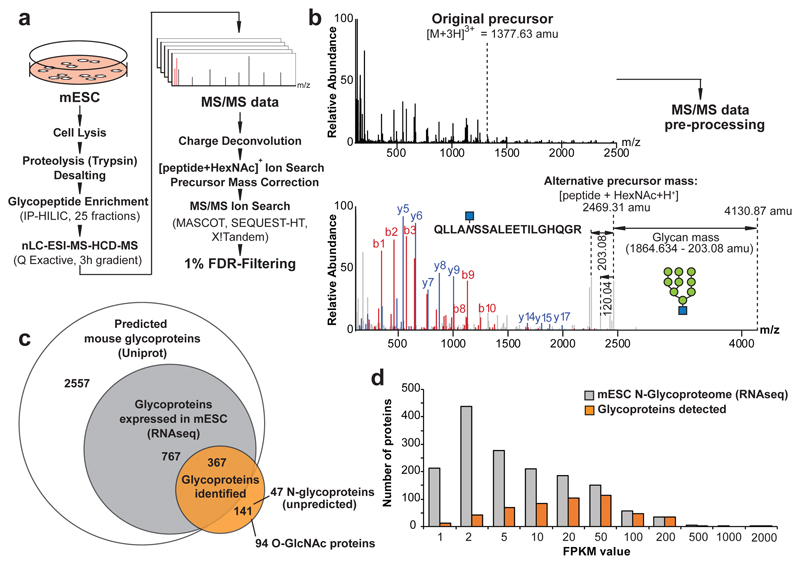
Current glycoproteomics technologies are often limited to the identification and quantification of de-glycosylated peptides7, require laborious generation of sample-specific spectral libraries8, or rely on dedicated instrumentation and data-acquisition regimes in conjunction with specialized proprietary commercial data analysis tools9. Here, we developed a glycoproteomics approach by complementing established shot-gun proteomic workflows10 with novel computational tools for the interrogation of glycopeptides. Our workflow enriches for glycopeptides after cell lysis and proteolysis using hydrophilic interaction chromatography (HILIC), followed by nano-liquid chromatography electrospray ionization tandem mass-spectrometry (nLC-ESI-MS/MS) using higher collision-energy induced dissociation (HCD) for peptide fragmentation on a Q Exactive instrument (Fig. 1a). To identify the glycan compositions and the amino-acid sequences of intact glycopeptides from MS/MS data-sets, we combined charge-deconvolution and de-isotoping algorithms11 to transform the signals of multiply charged fragment ions to the corresponding singly charged m/z values. This shifts fragment ions of the intact peptide with gradually dismembered glycans to higher m/z ranges. The less abundant fragment ions of the peptide-backbone, important for polypeptide sequence identification, remain in the lower mass-range of the MS/MS spectrum. These two regions of the MS/MS spectrum were separated by the prominent feature of the [peptide + HexNAc]+ fragment ion (Fig. 1b). Next, we identified this fragment by iteratively changing the precursor-ion mass of a given MS/MS spectrum by subtracting all glycan masses considered (Supplementary Table 1). Upon identification of a putative [peptide + HexNAc]+ fragment-ion, the MS/MS spectrum was cloned and its precursor-ion mass was set to the mass of the detected fragment-ion (Fig. 1b). To uncover the glycopeptide amino-acid sequences, we searched the processed MS/MS dataset against a protein database, using benchmarked MS/MS search engines, considering a single HexNAc moiety as a variable modification to any asparagine, serine and threonine residue (Fig. 1a). This analytical pipeline (see Methods) identifies and quantifies intact glycopeptides from complex biological samples. Of note, in all our data analyses and presentation of results, glycan structures were inferred from their monosaccharide composition and known biosynthetic rules for glycans.
Figure 1. Glycoproteomics.
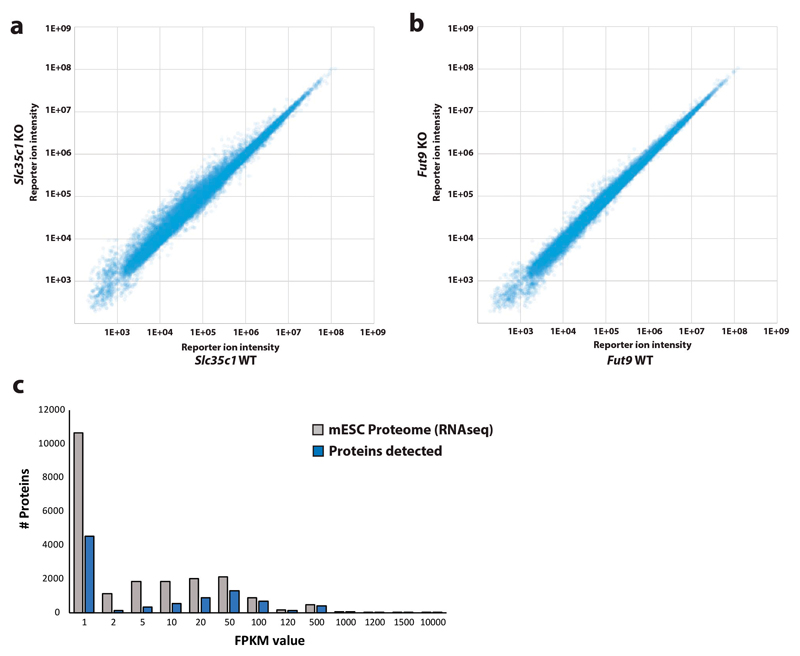
(a) Glycoproteomic workflow combining proteomics platforms with a new algorithm for the identification of intact glycopeptides from complex biological samples. (b) MS/MS raw-data pre-processing by charge deconvolution and precursor mass correction allows for the automated identification of glycopeptides, based on low-abundant peptide fragment ions in the lower mass-range of glycopeptide MS/MS spectra. (c) Coverage of experimentally identified glycoproteins (orange-filled circle; Supplementary Table 10) among all predicted mouse glycoproteins (Uniprot, white) and the glycoproteins expressed (based on RNAseq, grey) in mESC. (d) Transcript abundance of all glycoproteins expressed (RNA-seq) in our mESC clone (grey), compared to the glycoproteome experimentally detected (orange).
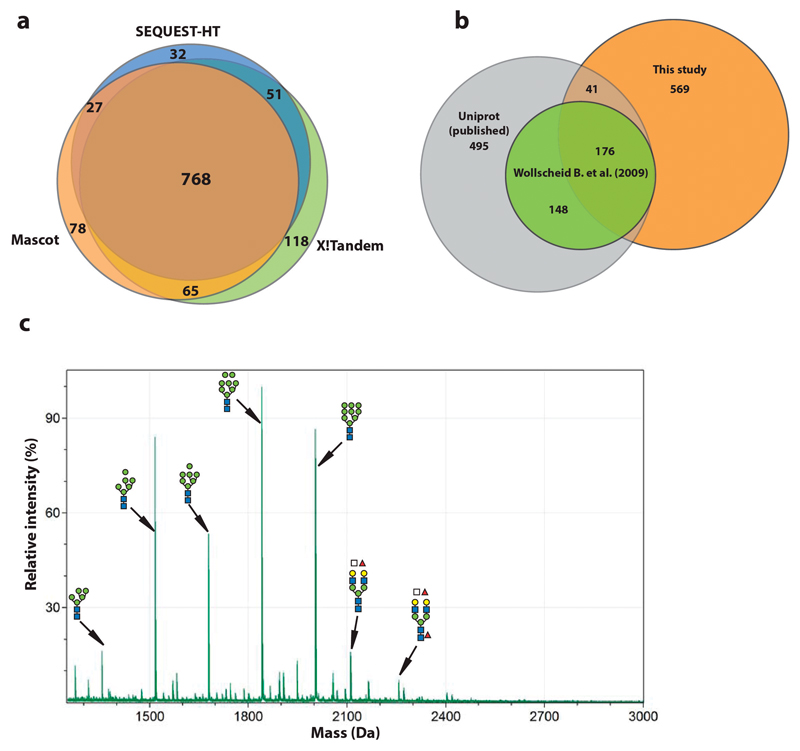
We used this approach to investigate the glycoproteome of mouse embryonic stem cells (mESC, line AN3-12, diploid state). We enriched and fractionated glycopeptides from trypsin-digested whole cell lysates of mESCs using HILIC (25 fractions) and individually analyzed them by nano-LC-ESI-MS/MS (using 3 hour gradients) (Fig. 1a). We identified 3380 unique intact glycopeptides (FDR < 1%)12, mapping to 786 glycosylation sites of 508 mESC glycoproteins (Fig. 1c), using MASCOT as the MS/MS search engine. We obtained similar results with other search engines (Extended Data Fig. 1a, Supplementary Table 2). Of note, we observed multiple HILIC fractions containing the same glycopeptides. To estimate our coverage of the mESC glycoproteome, we compared our data to the predicted murine glycoproteome, most of which have never been validated (Fig. 1c). Of the 3691 predicted mouse glycoproteins, 1134 are expressed at a FPKM > 1 in AN3-12 mESCs. Further analysis indicated approximately 50% coverage of the expressed N-glycoproteome when a FPKM > 20 was used as cut-off. Less abundant proteins were detected more sparsely (Fig. 1d). Thus, our approach uncovered nearly half of the glycoproteins predicted to be expressed in mESCs, and almost doubled the number of experimentally confirmed N-glycosylation sites of the entire murine glycoproteome (Extended Data Fig. 1b).
To control for a potential bias towards the identification of particular subsets of N-glycans, we analyzed enzymatically released N-glycans of mESC whole cell lysates by MALDI-TOF-MS. Both methods independently showed that ~70% of the mESC glycoproteome are decorated with eight neutral oligo-mannose-type structures (OMTs), whereas the group of complex type (CT) N-glycans represented about 30% of the N-glycome (Fig. 2a, Extended Data Fig. 1c). We also identified 38 mannose-6-phosphate (M6P)-containing OMT N-glycopeptides (Supplementary Table 3), mainly derived from hydrolytic proteins associated with the lysosome by Gene Ontology Term Enrichment Analysis, as expected4,13.
Figure 2. The glycoproteomes of human and mouse embryonic stem cells.
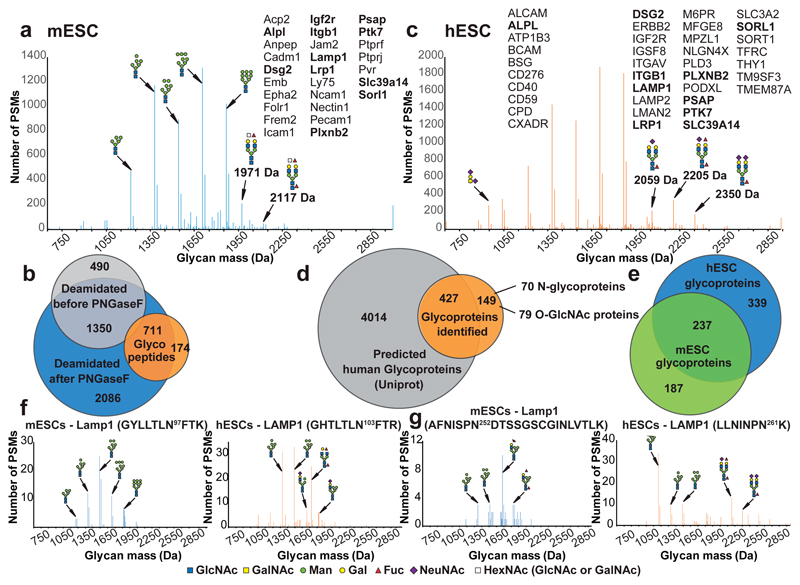
Glycan-mass histogram of all glyco-peptide spectrum matches (PSMs) identified (FDR < 1 %) from (a) murine and (c) human ESCs. Bin-width 1 Da. Indicated glycoproteins are species-specific or evolutionarily conserved (bold) and carry stemness-related glycans (labelled by their mass). (b) Deamidation of mESC derived glycopeptides upon PNGaseF treatment (blue) confirmed the identity of the N-glycopeptide sequences identified by our approach (orange). Chemical peptide deamidation was observed before PNGaseF treatment (grey). (d) Coverage of experimentally identified glycoproteins (orange) among all hypothetical (Uniprot) human glycoproteins (grey). (e) Cross-species comparison of glycoproteins identified in human (blue) and murine ESCs (green). (f and g) Comparison of glycan profiles at orthologous N-glycosylation sites of mouse Lamp1 and human LAMP1. Glycan-mass histograms of all glyco-PSMs (FDR < 1 %) specific for the N-glycosylation sites are indicated. Bin-width 1 Da. In all figures, glycan structures were inferred from their mono-saccharide compositions, based on known glycobiological rules and are represented according to the consensus pictogram symbols for different mono-saccharides30.
Further, we discovered several unanticipated glycoproteins (Fig. 1c), including nuclear envelope (e.g. Emd, Nop56, Tmpo, Rad51ap) and cytosolic proteins (e.g. Smg7, Rpl34, Map4k4, Pbxip1) modified with large N-linked OMTs (Supplementary Table 4, Supplementary Figs. 1-8). We validated these findings, warranting further investigation, by re-analyzing enzymatically de-glycosylated N-glycopeptides and their identification as deamidated non-glycopeptides7,14 (Fig. 2b, Supplementary Table 5). Furthermore, in early eluting HILIC fractions we identified mucin-type O-linked glycopeptides (e.g. Dag1, Gpc4, Fn1). These glycoproteins are usually inaccessible to conventional glycoproteomic methods7,14 and typically require specialized methodology15. In the same fractions, we identified serine and threonine residues modified with a single GlcNAc (Supplementary Table 6), an important post-translational modification of nuclear and cytosolic proteins16. Accordingly, this group of 94 O-GlcNAc proteins was significantly enriched for proteins with nucleic acid binding properties and transcription regulator activity, including the stem cell pluripotency factors Sox2 or Tet217. Thus, we uncovered multiple novel O-GlcNAc-decorated proteins, in addition to previously identified nuclear O-GlcNAc modified proteins from mESCs18.
Human embryonic stem cells (hESCs) and mESCs exhibit distinct expression profiles for carbohydrate-related stemness markers19. We identified 1106 unique glycopeptide sequences mapping to 576 glycoproteins in hESCs (Fig. 2c,d). Similar to mESCs, ~70% of the hESC glycoproteome was decorated with oligo-mannose type N-glycans, whereas complex type N-glycan structures represented ~30 % of the population (Fig. 2c). We observed M6P-containing glycan structures on 58 hESCs glycoproteins and identified several unanticipated glycoproteins (Fig. 2d and Supplementary Table 7). Among those, we detected 79 O-GlcNAc decorated proteins including the transcription factors SOX2, SP1, PHC1, or GATAD2A (Supplementary Table 8). We next compared differential protein glycosylation in mESC and hESCs. In mESCs, complex-type (CT) N-glycans were predominantly terminated with galactose and/or N-acetylhexosamine, whereas those from hESC were mainly sialylated (Fig. 2a,c). The N-glycans enriched in hESCs relative to mESCs, exemplified by the 2059, 2205 and 2350 Da glycan species in Fig. 2c, were reported as hESC-specific N-glycan structures whose abundance gradually decreases upon differentiation20,21. Similarly, the abundance of mESC-specific glycans (i.e. 1971 and 2117 Da, Fig. 2a) was reported to decrease during development20,22. We interrogated our comparative ESC glycoproteomics dataset for glycoproteins carrying exactly these species-specific stemness-related N-glycan structures and identified 27 and 37 plasma membrane proteins in mESCs and hESCs (Fig. 2a,c), respectively, most of them implicated in cell-to-cell signaling and cell interactions, as well as embryonic development.
Next, we compared the glycosylation profiles of 237 glycoproteins shared between mESCs and hESCs (Fig. 2e; Supplementary Table 9). For example, Lamp1, a major SSEA-1 reactive protein in murine neuronal stem cells23, was detected in both species (Fig. 2f). SSEA-1 (Lewis X, CD15), is a mouse-specific pluripotency marker which relates to a specific alpha-1,3-fucosylated carbohydrate epitope (Fig. 3a). Thus, we interrogated N-glycosylation sites of Lamp1 for this epitope and identified glycan compositions indicating multiply fucosylated hybrid-type N-glycans, for instance at Asn252 of mESC Lamp1 (Fig. 2g). Unexpectedly, we also observed multiply fucosylated glycan compositions at the orthologous glycosylation site of LAMP1 in hESCs, Asn261 (Fig. 2g), consistent with previous work suggesting that hESCs synthesize Lewis X CT N-glycans, despite being negative for SSEA-120. Comparative analysis of the site-specific glycan profiles revealed subtle glycan compositional differences on these proteins. Whereas Asn252 of murine Lamp1 exhibited fucosylated hybrid type N-glycans, Asn261 of human LAMP1 carried fucosylated bi-antennary N-glycan species (Fig. 2f,g). Thus, both mESCs and hESCs exhibit fucosylated glycans, even at the orthologous site on the same protein. But since the core structure of these glycans differs, the anti-SSEA-1 antibody does not recognize the Lewis X epitope on bi-antennary N-glycans.
Figure 3. Detection of specific fucosylated proteins.
(a) Slc35c1 transports GDP-fucose from the cytosol into the Golgi. Fut9 transfers fucose in α-1,3 linkage to LacNAc structures. Deletion of Slc35c1 abolishes fucosylation of secretory proteins in general. Ablation of Fut9 specifically affects synthesis of the Lewis X glyco-epitope. (b) Quantitative proteomics workflow for intact glycopeptides from Slc35c1 and Fut9 mutant (KO) and their genetically repaired (WT) mESC sister clones. (c) Quantitative glycoproteomics detects the reduction of all fucosylated glycopeptides (red) and the reciprocal increase of non-fucosylated glycopeptides (green) in Slc35c1 KO vs WT mESCs. Ablation of Fut9 affects a specific subset of fucosylated glycopeptides. Oligo-mannosidic type N-glycopeptides (blue) were not affected. Differential glycopeptide expression is shown by scatter-plotting TMT-reporter ion intensities for Slc35c1 KO (upper panel) and Fut9 KO (lower panel) versus WT cells. Data-points of Igf2r glycopeptide (ASIN1532ASYSEK) glycoforms are indicated by circles. (d) Site-specific comparison of Igf2r glycopeptides. Differential glycopeptide expression is shown as the ratio of TMT-reporter ion intensities in Slc35c1 and Fut9 KO versus WT cells. The number of PSMs (n) considered for ratio calculations and box-plot representations, are indicated.
We and others5,6 (J.T., J.S. et al., Cell Research 2017) previously established a critical sugar code whereby the presence of Lewis X glycan structures confer sensitivity to the bioweapon ricin. However, the proteins that carry this carbohydrate epitope and whether these glycoproteins are involved in ricin toxicity were unclear. In mESCs, α-1,3-fucosyltransferase Fut9 and the fucose transporter Slc35c1 are essential for the synthesis of the Lewis X carbohydrate epitope (Fig. 3a)24,25. Thus, we compared the relative glycopeptide abundances between Slc35c1 and Fut9 mutant and control mESCs sister clones, genetically repaired by the inversion of a gene trap cassette, using an in vitro stable isotope-labelling technique for peptides10,26 and analyzed by nLC-ESI-MS/MS (Fig. 3b, Supplementary Table 10). Making use of an in vitro stable isotope labeling-technique and MS/MS based quantitation also allowed for the comparative analysis of glycopeptides despite overlapping HILIC fractions.
As expected, OMT-decorated glycoproteins, M6P-containing OMTs and O-GlcNAc proteins were unaffected by loss of Slc35c1 and Fut9 (Fig. 3c). We also did not detect significant changes in overall protein expression in the mutant cells (Extended Data Fig. 2a-c). However, deletion of Slc35c1 abolished fucosylation of N- and O-glycoproteins, resulting in a reciprocal increase in the corresponding non-fucosylated glycan structures (Fig. 3c, Supplementary Table 10). These changes are exemplified by the glycosylation profiles of Igf2r (Fig. 3d). Igf2r (cation-independent mannose 6-phosphate receptor)13 has 20 potential N-glycosylation sites; only three had been experimentally confirmed previously7. We not only confirmed 7 other N-glycosylation sites, but also uncovered complex type N-glycan structures at Asn430, Asn704 and Asn1532 of Igf2r affected by inactivation of Slc35c1. Only those at Asn1532 were a target of Fut9. The glycans affected by the loss of Fut9 were a subset of those targeted by Slc35c1 (Fig. 3c-d, Supplementary Tables 11 and 12): 88 glycoproteins were sensitive to loss of Slc35c1, of which 33 were also targets of Fut9. Thus, our unbiased method detects specific changes to glycosylation profiles.
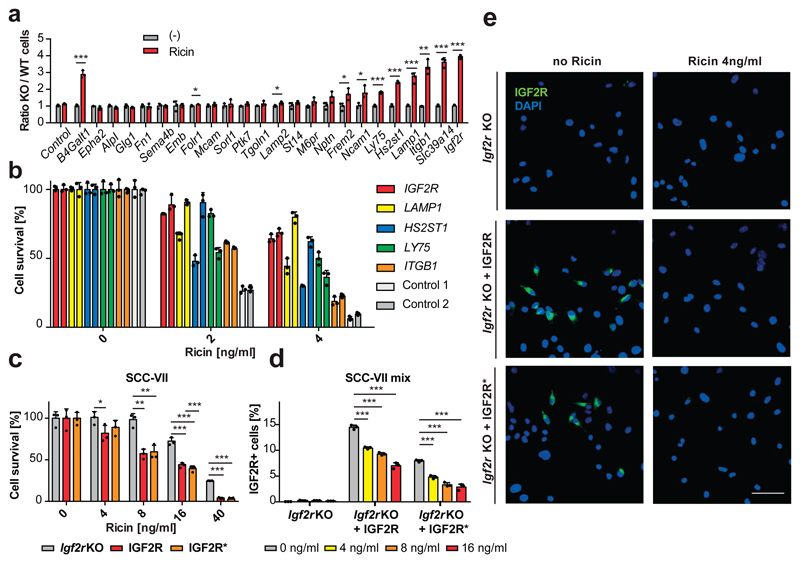
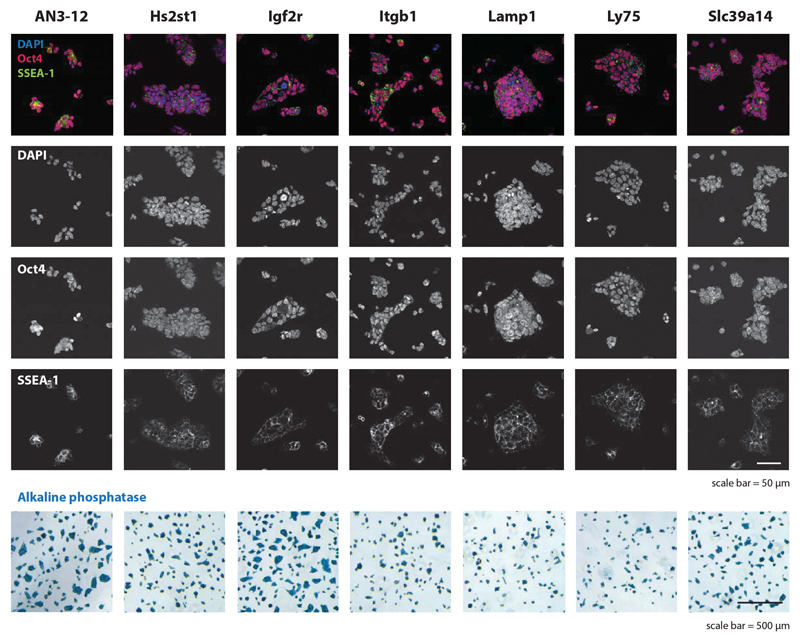
Both Fut9 and Slc35c1 knockout cell-lines were ricin resistant5,6, therefore we hypothesized that glycoproteins affected by both mutations might be required for ricin sensitivity. We generated mutant GFP-labeled clones for 24 candidate genes, and their respective repaired mCherry-labeled sister clones (Fig. 4a). Igf2r, Slc39a14, Itgb1, Lamp1, Ly75 and Hs2st1 mutant mESCs displayed improved survival in the presence of ricin compared to the sister clones (Fig. 4a, Extended Data Fig. 3). CRISPR/Cas9-mediated disruption of these genes in human HEK293 cells also increased their resistance to ricin (Fig. 4b). Further, we reconstituted an Igf2r mutant murine SCC-VII squamous cell carcinoma line (Igf2r KO)27 with either wild-type human IGF2R (IGF2R) or a mutant version inactive for M6P binding (IGF2R*)28. As ricin itself exhibits OMT N-glycan structures29, we used the IGF2R* mutant cell line to exclude possible binding of ricin to IGF2R via M6P. Parental Igf2r KO cells exhibited significant resistance to ricin treatment compared to IGF2R- and IGF2R*-expressing cells (Fig. 4c-e), indicating that human IGF2R confers sensitivity to ricin, independent of its capacity to bind M6P. Therefore, our comparative glycoproteomics approach uncovered proteins whose fucosylation is required for ricin toxicity.
Figure 4. Validation of fucosylated proteins involved in ricin toxicity.
(a) Mixed populations of mutant (KO, GFP+) and their repaired respective wild-type (WT, mCherryCre+) cells were subjected to ricin [4ng/ml] for three days or left untreated (-) and analyzed for viability. The ratios of green to red cells (KO/WT) are shown for each candidate gene. Sister clones for the known ricin sensitivity gene B4Galt131 are shown as positive control and mixtures of GFP and mCherryCre control cells as negative control. (b) HEK cells, harboring CRISPR/Cas9-induced mutations in the indicated genes (2 different sgRNAs each) and control cells (scrambed sgRNAs) were subjected to ricin or left untreated. Cell survival was monitored using Alamar Blue cell viability assay. (c) Igf2r KO murine SCC-VII cells and reconstituted IGF2R wildtype and IGF2R* M6P-binding-domain mutant SCC-VII were subjected to ricin. Cell survival was monitored as above. (d, e) Mixed populations of Igf2r KO and IGF2R/IGF2R* expressing SCC-VII cells were subjected to ricin and the relative amount of IGF2R expressing cells was determined using flow cytometry (d) and confocal microscopy (e). Scale bar 50 μm. Each image is representative of six images of three independent experiments. Data in a, b, c, d are shown as mean ± SD of triplicate cultures. Experiments were repeated three times with similar results. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student’s t-test).
In summary, we report a glycoproteomics method to identify intact glycopeptides, and directly map and quantify changes in protein glycosylation at a proteome scale. Our analysis of the hESC and mESC glycoproteomes nearly doubled the number of experimentally confirmed glycosylation sites and uncovered multiple novel glycosylated proteins, including evolutionarily conserved and species-specific glycan modifications of murine and human stemness factors. We determined the specificity of our approach by quantitatively delineating the glycoproteomes of ricin-resistant Fut9 and Slc35c1 mutants, which revealed new players orchestrating ricin toxicity. We anticipate that this freely available data-interpretation tool will be widely applied and will democratize glycoproteomics, enabling proteome-wide studies of this fundamental building block of biology in virtually all species.
Methods
Cell lines
Mouse embryonic stem cells (clone AN3-12)5 were cultured in DMEM supplemented with 10% fetal bovine serum (FCS), penicillin–streptomycin, non-essential amino acids, sodium pyruvate (1 mM), L-glutamine (2 mM), β-Mercaptoethanol (0.1 mM) and LIF (20 µg/ml). SCC-VII and HEK293 cells were cultured in DMEM supplemented with 10% FCS (fetal calf serum), penicillin–streptomycin and L-glutamine. Human embryonic stem cells (hESCs; line H9) were cultured in 20% O2 conditions on irradiated mouse embryonic fibroblast feeder layers in hESC medium (knockout-DMEM or DMEM-F12, (Gibco 121660), 15% knockout serum replacement (Gibco 10828), 1 mM glutamine (Invitrogen), 1% non-essential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), 8 ng/ml βFGF (Peprotech). H9 cultures were passaged every 5–7 days and medium changed every day. Igf2r knockout murine SCC-VII cells (parental clone) and Igf2r knockout SCC-VII cells reconstituted with human wild type (SCC-VII/IGF2R wt-3) and mutant IGF2R (SCC-VII/IGF2R Dom3/9mut-1) have been previously described28.
Glycoproteomic sample preparation and protein digestion
Murine and human ESC cultures were washed 3 times with PBS, then incubated for 5 min with PBS containing 1 mM EDTA and centrifugated at 900 x g. The supernatant was removed and the cell pellets were immediately lysed by the addition of freshly prepared 10 M urea in 120 mM triethylammonium bicarbonate buffer (TEAB, Sigma) to a final concentration of 8 M urea in 100 mM TEAB and brief ultra-sonication (ultrasonic processor UP100H, Hielscher). mESC derived samples were reduced (final concentration 5 mM Tris(2-carboxyethyl)phosphine hydrochloride, 30 min) and alkylated (final concentration 10 mM methyl-methanethiosulfonate, 30 min). Human ESC samples were reduced using a final concentration of 10 mM 1,4-dithioerythritol and alkylated using a final concentration of 20 mM 2-iodoacetamide. Protein concentrations were measured (BCA Protein Assay Kit, Pierce) and 1 mg protein per sample digested with 10 μg endoproteinase Lys-C (Wako) for 8 hours at 37°C. Subsequently, the samples were diluted with water to 6 M urea in 75 mM TEAB and incubated with 1 U Benzonase (Merck KGaA) for 1 hour at 37°C. The samples were further diluted to 4 M urea in 50 mM TEAB and incubated with 10 μg modified porcine trypsin (sequencing grade, Promega) for 12 hours at 37°C. After TMT-6plex-labelling (performed according to the supplier’s manual), the pH of the individual samples was adjusted to 2 by the addition of 10 % trifluoroacetic acid (TFA). The samples were pooled in equal amounts, desalted using reverse phase solid-phase extraction cartridges (Sep-Pak C-18, Waters) and completely dried under vacuum. Prior to glycopeptide enrichment, the samples were digested with 1 U alkaline phosphatase (from calf intestine, Roche), desalted using Sep-Pak C-18 cartridges and dried under vacuum.
N-glycan analyses
N-glycans were enzymatically released from desalted tryptic digests of ESCs using PNGaseF (from Elizabethkingia meningoseptica, Sigma), labelled with 2-aminobenzoic acid (2-AA) by reductive amination using sodium borohydride as reducing agent, essentially as described32. The 2-AA labelled N-glycans were then measured by MALDI-TOF-MS (4800 MALDI TOF/TOF Analyser, AB Sciex) operated in negative mode, using DHB as matrix component.
Glycopeptide enrichment
Glycopeptides were enriched using ion-pairing hydrophilic interaction chromatography (IP-HILIC). The dry samples were taken up in 100 μL 75% acetonitrile containing 0.1 % TFA, and subjected to chromatographic separation on a TSKgel Amide-80 column (4.6 x 250 mm, particle size 5μ) using a linear gradient from 0.1% TFA in 80% acetonitrile to 0.1% TFA in 40% acetonitrile over 35 minutes (Dionex Ultimate 3000, Thermo). The 25 collected fractions were dried in a vacuum concentrator. For validation experiments identifying de-N-glycosylated peptides, aliquots of these fractions were re-suspended in 100 mM Tris buffer, pH 8.0 and incubated with 1U PNGaseF for 12 hours at 37°C. Prior to LC-MS/MS analysis, the PNGaseF reaction was quenched by adjusting the pH of the individual samples to 2 by the addition of 10% TFA.
LC-MS/MS
The 25 IP-HILIC fractions were individually analyzed by LC-MS/MS. The samples were separated by reversed-phase chromatography (75 µm x 250 mm PepMap C18, particle size 5μ, Thermo), developing a linear gradient from 2% acetonitrile to 80% acetonitrile in 0.1% formic acid, within 3 hours (RSLC nano, Dionex – Thermo Fisher Scientific) and analyzed by MS/MS, using electrospray-ionization Quadrupole Fourier-transformed tandem mass-spectrometry (Q Exactive, Thermo) and higher collision-induced dissociation (HCD). The instrument was operated in positive mode and set to the following acquisition parameters: MS1 resolution = 70000, MS1 AGC-target = 1E6, MS1 maximum inject time = 60 ms, MS1 scan range = 350-2000 m/z, MS2 resolution = 70000, MS2 AGC-target = 1E6, maximum inject time = 256 ms, TopN = 10, isolation window = 1.2 m/z, fixed first mass = 120 m/z, normalized collision energy = 35, underfill ratio = 2.5 %, peptide match = ON, exclude isotopes = ON, dynamic exclusion = 10 s.
Data analysis
All MS/MS data were processed and analysed using Xcalibur 2.2 (version 2.2.48, Thermo) and Proteome Discoverer 1.4 (PD 1.4.0.288, Thermo).
MS/MS data processing and automated glycopeptide identification
MS/MS spectra were extracted from the .raw-file format using the generic Spectrum Exporter Node of PD 1.4 (settings: min. precursor mass = 350 Da, max. precursor mass = 10000 Da, minimum peak count = 5, S/N Threshold 1.5), charge-deconvoluted and de-isotoped (“MS2 Spectrum Processor”, in-house implementation of the algorithm described11, available as PD 1.4 Node at http://ms.imp.ac.at/?goto=kassonade), and were then scored with respect to the abundance of glycosylation-related reporter-ions (G-Score). As a general indicator for the presence of glycan-specific reporter ions within a given MS/MS spectrum, the G-Score was calculated as the reciprocal value of the negative logarithm of the summed fractions of the normalized intensities of the reporter ions (i.e. 126.05496, 138.05496, 144.06552, 163.06007, 168.06552, 186.07608, 204.08665, 274.09270, 292.10267 amu, mass-precision 10 ppm) and their respective rank in the spectrum. We empirically determined that MS/MS spectra with a G-Score of greater than 0.4 were derived from glycopeptides.
After G-scoring, the glycosylation-related reporter-ions were removed from the spectrum with a mass-precision of 10 ppm (“Reporter Ion Filter”, in-house developed and available as PD 1.4 Node at http://ms.imp.ac.at/?goto=kassonade). The glycopeptide-spectra were then analysed for the presence of potential [peptide + HexNAc]+ fragment-ions (PD 1.4 Node “Kassonade”, developed in-house). To this end, the mass of the respective precursor-ion was iteratively reduced by the masses represented in our glycan database (Supplementary Table 1) minus 203.0794 amu. For peak-matching, only fragment-ion charge-state 1 was taken into account. In cases where a corresponding potential [peptide + HexNAc]+ fragment-ion was detected (with a fragment mass-tolerance of 10 ppm) the spectrum was duplicated, with the original precursor ion-mass being set to the mass of the potential [peptide + HexNAc]+ fragment ion. For peptide sequence identification, the pre-processed MS/MS data were then searched against the Uniprot mouse reference proteome set (uniprot.org, 47435 entries; concatenated forward and reverse data-base), using MASCOT (Matrix Science Ltd., version 2.2.07), SEQUEST-HT (built-in version of PD 1.4) and X!Tandem (Sledgehammer, version 2013.09.01). The parameters for all MS/MS search engines were set to trypsin as protease, allowing for maximally two missed cleavage sites, a precursor mass tolerance of 10 ppm, a fragment mass tolerance of 25 mmu, the fixed modification of methylthiolated or carbamidomethylated cysteine, the variable modifications of oxidation (methionine), de-amidation (asparagine and glutamine) and hexosamine (asparagine, serine and threonine) and TMT-6plex (N-terminus and lysine). The resulting peptide spectrum matches (PSMs) were manually filtered (search-engine rank 1, peptide length greater than 6, and at least one HexNAc-modified residue) and filtered to 1% FDR, using the target-decoy approach12. Site-localization of N-gycans was performed using ptmRS (in-house developed33 and available as PD 1.4 Node at http://ms.imp.ac.at/?goto=kassonade), scoring the characteristic loss of 120.042259 Da from glycosylated asparagine as diagnostic ion.
Insertional mutagenesis in murine embryonic stem cells
Haploid murine ESCs were generated previously5 and used for reversible mutagenesis (i.e. derivation of genetrap-harboring knockout and genetically repaired sister cell lines). mESCs harboring a genetrap in introns of Slc35c1 and Fut9, as well as Lewis X containing candidate mutant embryonic stem cell lines were generated as described5. For the reversion of the genetrap, clones carrying the genetrap vector were infected with retroviruses carrying mCherry together with Cre Recombinase. Retroviruses carrying GFP alone were used to label knockout cells. For single cell clones, cells were transiently transfected with a plasmid encoding Cre recombinase as well as GFP, sorted and single cell clones were then analyzed using PCR. The orientation of the splice acceptor was determined using a three primer containing amplification system, consisting of a fragment binding 1st forward primer or the inverse forward primer.
GT 1st Fwd TCGACCTCGAGTACCACCACACT
GT inverse Fwd AAACGACGGGATCCGCCATGTCA
GT common Rev TATCCAGCCCTCACTCCTTCTCT
CRISPR/Cas9-mutagenesis
A lentiviral vector expressing Cas9 together with the respective sgRNA, as well as GFP, was used for mutagenesis. IGF2R, LAMP1, HS2ST1, ITGB1 and LY75 were mutated in human HEK293 cells using two different, independent CRISPR/Cas9 guide sequences, and sorted via flow cytometry for GFP+ cells. The following primers were used:
| Lamp1_1 | fwd | caccgCAACGGGACCGCGTGCATAA |
| rev | aaacTTATGCACGCGGTCCCGTTGc | |
| Lamp1_2 | fwd | caccGAAGTTGGCCATTATGCACG |
| rev | aaacCGTGCATAATGGCCAACTTC | |
| Igf2r_1 | fwd | caccgTCACTGTTGTGTTGAATTCC |
| rev | aaacGGAATTCAACACAACAGTGAc | |
| Igf2r_2 | fwd | caccgTTGAATTCCAGGAGAGATC |
| rev | aaacGATCTCTCCTGGAATTCAAc | |
| Hs2st1_1 | fwd | caccgTGTTTTCGTCTCCGTAACCC |
| rev | aaacGGGTTACGGAGACGAAAACAc | |
| Hs2st1_2 | fwd | caccgTCATAAGGGATCCTATTGAG |
| rev | aaacCTCAATAGGATCCCTTATGAc | |
| Itgb1_1 | fwd | caccgAATGTAACCAACCGTAGCAA |
| rev | aaacTTGCTACGGTTGGTTACATTc | |
| Itgb1_2 | fwd | caccgTCATCACATCGTGCAGAAGT |
| rev | aaacACTTCTGCACGATGTGATGAc | |
| Ly75_1 | fwd | caccgTCTCAGCTCATTTACCGATT |
| rev | aaacAATCGGTAAATGAGCTGAGAc | |
| Ly75_2 | fwd | caccgGCAAATGAAAGAGCCGATGC |
| rev | aaacGCATCGGCTCTTTCATTTGCc | |
| Control_ 1 | fwd | caccGGGACGCTCATCGAGTGACG |
| rev | aaacCGTCACTCGATGAGCGTCCC | |
| Control_ 2 | fwd | caccGCGGGACGTAATATTATG |
| rev | aaacCATAATATTACGTCCCGC |
Competitive growth assays
Diploid murine ESCs harboring a genetrap in introns of all candidate genes or SCC-VII IGF2R wild type and mutant cells were seeded at low density in normal growth medium and infected with two viruses, one encoding for mCherry-Cre Recombinase and one encoding GFP (together with puromycin) for 12 hours. Infected cells were selected after 24h (final concentration 1ug/ml; Puromycin, Invivogen, ant-pr-1) and expanded. Ratios of GFP to mCherry/Cre expressing cells cultured in the presence or absence of ricin were determined using high-throughput flow cytometry (BD LSRFortessa™ HTS cell analyzer).
Immunofluorescence
Paraformaldehyde (4%) fixed Igf2r negative murine SCC-VII cells and Igf2r knockout SCC-VII cells reconstituted with human wild type or mutant IGF2R were blocked and permeabilized for 1 hour at room temperature with 1xPBS, supplemented with 0.2%Triton, 1% Glycine, 5% FBS, 2% BSA. Cells were incubated with primary antibodies specific for IGF2R (anti M6PR (cation independent) antibody (MEM-238), abcam ab8093, 1:400), anti Oct-3/4 (Clone 40/Oct-3 (RUO), BD Transduction Laboratories, No.611203, 1:300) and anti-SSEA-1 (CD15, Lewis X; PE conjugated, clone: MC-480, ebioscience 12-8813-41, 1:300) o/n at 4ºC, then washed 3 times with 1x PBS, supplemented with 0.2% Triton and 1% Glycine. Binding of the primary antibodies was detected with fluorescent-labeled secondary antibodies (Streptavidin, Alexa Fluor® 633 conjugate, Invitrogen, S21375, 1:500). Images were acquired using a confocal laser scanning microscope (LSM780 Axio Observer, Carl Zeiss) and processed using Zeiss imaging software. For intracellular staining of IGF2R and subsequent flow cytometry, cells were trypsinized, fixed, blocked and stained (Foxp3 / Transcription Factor Staining Buffer Set, eBioscience 00-5523-00) according to the manufacturer’s protocols with the same anti-IGF2R antibody as above. Primary antibodies were visualized with secondary fluorescent-labeled antibodies (F(ab')2-Goat anti-mouse IgG (H+L) Alexa Fluor 488, Invitrogen A-11017, 1:500). Samples were analyzed using flow cytometry (LSR Fortessa, BD) or cell sorted using a FACS Aria III flow-cytometer (BD).
Ricin toxicity assays
Ricin extracts in PBS were generated as described34. To assess ricin toxicity in different cellular systems, cells were plated at low density (5.000-10.000 cells /96-well) in growth medium. Ricin was added to the cells at different concentrations for at least 24 hours in triplicate cultures. The cell viability at different time points was determined using the Alamar Blue Cell viability assay according to manufacturer’s protocols (Invitrogen DAL1100).
Statistics and Reproducibility
All values are given as means ± S.D., unless stated otherwise. All experiments were reproduced at least 2 independent times, with similar results. GraphPad Prism was used to generate figures and statistical analyses (GraphPad Software). An a priori sample size estimation was not performed. Data were analyzed by using the unpaired two-tailed Student's t-test, as indicated. P < 0.05 was accepted as statistically significant. Box and whisker plots depict the median as a bold line. The box ranges from the first to the third quartile. Whiskers extend to the outermost data point within 1.5-fold interquartile range outside the box. Further outliers are indicated by circles. Glycoproteomics data of mESC in Fig. 1c,d, Fig. 2a,f,g and Extended Data Figure 1a,b are representative of two independent experiments analyzing two biological replicates with similar results. Glycoproteomics data of mESC in Fig. 3 c,d and Extended Data Fig. 2a-c are representative of two technical replicates with similar results. Glycoproteomic analysis of hESCs is representative of one multiplexed experiment, analyzing two technical replicates of two biological replicates, with similar results for the individual samples.
Data Availability
The mass spectrometry proteomics data are accessible through ProteomeXchange (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the dataset identifier PXD005804. All RNAseq data are accessible through GEO accession number GSE84090 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qrgxmuaindmpjah&acc=GSE84090).
Code Availability
All specialized software tools used for the analysis of MS/MS data from glycopeptides were developed and implemented in-house as “Nodes” to the PD 1.4 software-suite. All Nodes and in silico workflows are freely available for download at http://ms.imp.ac.at/?goto=kassonade.
Extended Data
Extended Data Figure 1. Identification of N-glycopeptides and N-glycans.
(a) Glycopeptide MS/MS raw-data were pre-processed and identified using different MS/MS search engines: Mascot (orange), SEQUEST-HT (blue) and X!tandem (green). The glycopeptide sequences identified by these algorithms were highly similar, both with respect to identities and numbers. (b) The N-glycosylation sites identified in our study (orange) were mapped to published annotated N-glycosylation sites in Uniprot (grey) and those identified by Wollscheid et al.7 (green). (c) Negative-mode MALDI-TOF analysis of 2-AA labelled N-glycans from mESCs directly correlated with the N-glycan profiles identified using our glycoproteomics approach.
Extended Data Figure 2. Quantitative proteomics.
Comparative proteomics of (a) Slc35c1 knock-out (KO) and (b) Fut9 KO versus their respective genetically repaired wild-type (WT) sister murine embryonic stem cells (mESCs) did not show significant changes in the overall protein expression as determined by quantitative proteomics. Differential peptide abundances are represented as scatter-plots of the corresponding TMT-reporter ion intensities. (c) Coverage of the predicted (RNA-seq) mESCs proteome by the quantitative proteomics data set.
Extended Data Figure 3. Stemness of knock-out mESC lines.
The parental control mESC line AN3.12, as well as mutant Hs2st1, Igf2r, Itgb1, Lamp1, Ly75, Slc39a14 mESCs lines were stained for the prototypic mouse embryonic stem cell markers Oct4, SSEA-1 and alkaline phosphatase. DAPI is shown to visualize nuclei. Of note, no obvious growth defects or morphological phenotypes were observed. All mESCs were diploid as determined by Hoechst staining (not shown). Scale bars are indicated. The experiments were repeated three times. Each image is representative of five images.
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
We thank all members of our laboratories for helpful discussions, Life Science Editors for editorial support, Maria Novatchkova for RNAseq analysis, and J. Zuber for CRISPR/Cas9 vectors. K.M. is funded by the Austrian Science Fund (SFB F3402-B03, TRP308-N15 and I1469-B16). J.M.P. is supported by grants from IMBA, the Austrian Academy of Sciences, an ERC Advanced Grant, and Era of Hope Innovator award. J.S. is a Wittgenstein prize fellow.
Footnotes
Author Contributions
J.S., J.T., and J.M.P. conceived the study. J.S. designed and performed glycoproteomics experiments and conceived the bio-informatic analysis algorithm. J.T. performed in vitro cell culture experiments. D.W. provided human ESCs and U.E. murine ESCs. A.G., G.D. and F.D. programmed algorithms for glycoproteomics. L.M. provided Igf2r mutant lines. K.M. supervised glycoproteomics experiments. J.S., J.T. and J.M.P. wrote the manuscript with input from all authors.
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Bio. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. 2016;17:1244–1251. doi: 10.1038/ni.3587. [DOI] [PubMed] [Google Scholar]
- 3.Dennis JW, Lau KS, Demetriou M, Nabi IR. Adaptive Regulation at the Cell Surface by N-Glycosylation. Traffic. 2009;10:1569–1578. doi: 10.1111/j.1600-0854.2009.00981.x. [DOI] [PubMed] [Google Scholar]
- 4.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elling U, et al. Forward and Reverse Genetics through Derivation of Haploid Mouse Embryonic Stem Cells. Cell stem cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patnaik SK, Stanley P. Lectin-resistant CHO glycosylation mutants. Methods in enzymology. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 7.Wollscheid B, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eshghi ST, Shah P, Yang WM, Li XD, Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal Chem. 2015;87:5181–5188. doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin XK, et al. Glycoproteomic Analysis of the Secretome of Human Endothelial Cells. Mol Cell Proteomics. 2013;12:956–978. doi: 10.1074/mcp.M112.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauniyar N, Yates JR. Isobaric Labeling-Based Relative Quantification in Shotgun Proteomics. J Proteome Res. 2014;13:5293–5309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savitski MM, Mathieson T, Becher I, Bantscheff M. H-Score, a Mass Accuracy Driven Rescoring Approach for Improved Peptide Identification in Modification Rich Samples. J Proteome Res. 2010;9:5511–5516. doi: 10.1021/pr1006813. [DOI] [PubMed] [Google Scholar]
- 12.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: New twists in the tale. Nat Rev Mol Cell Bio. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 14.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Zhao P, Stalnaker SH, Wells L. Approaches for site mapping and quantification of O-linked glycopeptides. Methods in molecular biology. 2013;951:229–244. doi: 10.1007/978-1-62703-146-2_15. [DOI] [PubMed] [Google Scholar]
- 16.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 17.Jang H, et al. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell stem cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. P Natl Acad Sci USA. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginis I, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 20.Satomaa T, et al. The N-glycome of human embryonic stem cells. Bmc Cell Biol. 2009;10 doi: 10.1186/1471-2121-10-42. Artn 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nairn A, et al. Changes in Glycan-related Gene Transcripts Following Human Embryonic StemCell Differentiation into Cell Types Derived from Ectoderm, Mesoderm or EndodermLineages. Glycobiology. 2012;22:1590–1590. [Google Scholar]
- 22.Nairn AV, et al. Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. J Biol Chem. 2012;287:37835–37856. doi: 10.1074/jbc.M112.405233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi H, Yanagisawa M, Kato K, Yu RK. Lysosome-associated membrane protein 1 is a major SSEA-1-carrier protein in mouse neural stem cells. Glycobiology. 2010;20:976–981. doi: 10.1093/glycob/cwq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellbusch CC, et al. Golgi GDP-fucose transporter-deficient mice mimic congenital disorder of glycosylation IIc/leukocyte adhesion deficiency II. J Biol Chem. 2007;282:10762–10772. doi: 10.1074/jbc.M700314200. [DOI] [PubMed] [Google Scholar]
- 25.Lu LC, Hou XH, Shi SL, Korner C, Stanley P. Slc35c2 Promotes Notch1 Fucosylation and Is Required for Optimal Notch Signaling in Mammalian Cells. J Biol Chem. 2010;285:36245–36254. doi: 10.1074/jbc.M110.126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 27.Probst OC, et al. The mannose 6-phosphate/insulin-like growth factor II receptor restricts the tumourigenicity and invasiveness of squamous cell carcinoma cells. International journal of cancer. 2009;124:2559–2567. doi: 10.1002/ijc.24236. [DOI] [PubMed] [Google Scholar]
- 28.Probst OC, et al. The mannose 6-phosphate-binding sites of M6P/IGF2R determine its capacity to suppress matrix invasion by squamous cell carcinoma cells. Biochem J. 2013;451:91–99. doi: 10.1042/Bj20121422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foxwell BMJ, Donovan TA, Thorpe PE, Wilson G. The Removal of Carbohydrates from Ricin with Endoglycosidases-H, Endoglycosidase-F and Endoglycosidase-D and Alpha-Mannosidase. Biochim Biophys Acta. 1985;840:193–203. doi: 10.1016/0304-4165(85)90119-9. [DOI] [PubMed] [Google Scholar]
- 30.Varki A, et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Sundaram S, Shaper NL, Raju TS, Stanley P. Chinese hamster ovary (CHO) cells may express six beta 4-galactosyltransferases (beta 4GalTs) - Consequences of the loss of functional beta 4GalT-1, beta 4GalT-6, or both in CHO glycosylation mutants. J Biol Chem. 2001;276:13924–13934. doi: 10.1074/jbc.M010046200. [DOI] [PubMed] [Google Scholar]
- 32.Pabst M, et al. Comparison of fluorescent labels for oligosaccharides and introduction of a new postlabeling purification method. Anal Biochem. 2009;384:263–273. doi: 10.1016/j.ab.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 33.Taus T, et al. Universal and Confident Phosphorylation Site Localization Using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 34.Simmons BM, Russell JH. A single affinity column step method for the purification of ricin toxin from castor beans (Ricinus communis) Anal Biochem. 1985;146:206–210. doi: 10.1016/0003-2697(85)90417-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data are accessible through ProteomeXchange (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the dataset identifier PXD005804. All RNAseq data are accessible through GEO accession number GSE84090 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qrgxmuaindmpjah&acc=GSE84090).