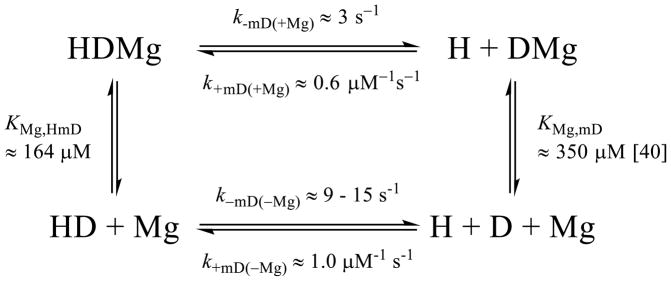Abstract
Dbp5, DDX19 in humans, is an essential DEAD-box protein involved in mRNA export, which has also been linked to other cellular processes, including rRNA export and translation. Dbp5 ATPase activity is regulated by several factors, including RNA, the nucleoporin proteins Nup159 and Gle1, and the endogenous small molecule inositol hexakisphosphate (InsP6). To better understand how these factors modulate Dbp5 activity and how this modulation relates to in vivo RNA metabolism, a detailed characterization of the Dbp5 mechanochemical cycle in the presence of those regulators individually or together is necessary. In this study, we test the hypothesis that Nup159 controls the ADP-bound state of Dbp5. In addition, the contributions of Mg2+ to the kinetics and thermodynamics of ADP binding to Dbp5 were assessed. Using a solution based in vitro approach, Mg2+ was found to slow ADP and ATP release from Dbp5 and increased the overall ADP and ATP affinities, as observed with other NTPases. Further, Nup159 did not accelerate ADP release, while Gle1 actually slowed ADP release independent of Mg2+. These findings are not consistent with Nup159 acting as a nucleotide exchange factor to promote ADP release and Dbp5 ATPase cycling. Instead, in the presence of Nup159, the interaction between Gle1 and ADP-bound Dbp5 was found to be reduced by ~18-fold, suggesting that Nup159 alters the Dbp5-Gle1 interaction to aid Gle1 release from Dbp5.
Keywords: DEAD-box protein, kinetics, Dbp5, Gle1, Nup159
Introduction
Members of the DEAD-box protein (DBP) family couple energy from cycles of ATP binding, hydrolysis, and product release to RNA and ribonucleoprotein (RNP) complex remodeling [1–3]. RNA binding activates the intrinsic ATPase activity of many DBPs [4–7] by accelerating rate limiting step(s), e.g. ATP hydrolysis and/or product release [8, 9]. Numerous regulatory proteins also tune DBP ATPase rates through modulating ATPase cycle transitions and DBP interactions, which includes promoting conformations that favor nucleotide loading, RNA binding, relieve auto-inhibition, or alter product release [10–13]. In turn, the conformations and interactions of a DBP with binding partners are often linked to the chemical states of the bound nucleotide [2, 6].
Dbp5, DDX19 in humans, is an essential DBP first described in Saccharomyces cerevisiae as being involved in mRNA export from the nucleus [14, 15]. Subsequent work has shown that Dbp5 is modulated by several regulatory factors, including RNA, the nucleoporin proteins Nup159 and Gle1, and the endogenous small molecule inositol hexakisphosphate (InsP6) [5, 16–26]. The resulting phenotypic, genetic, biochemical, and structural data provides potential models by which these regulators may control Dbp5 ATPase activity and mRNA export in vivo [27]. However, Dbp5 has also been linked to other cellular processes, including rRNA export and translation [26, 28, 29], complicating the interpretation of genetic and phenotypic data, and providing for the possibility of context-dependent regulation to facilitate multiple independent Dbp5 functions in RNA metabolism. Consequently, a detailed characterization of the Dbp5 mechanochemical cycle, and how this cycle is altered by regulators alone or in combination, is necessary to test and extend models of Dbp5 function. In other words, by determining the ATPase cycle rates and equilibrium constants in the presence of regulators, we can define the relevant events along the ATP hydrolysis pathway that represent control points for modulating Dbp5 activity in vivo and describe how regulators alter these events.
Towards this goal, our recent work showed that Dbp5 steady-state cycling (kcat) in the presence and absence of RNA is most limited by inorganic phosphate (Pi) release, and that ATP affinity for Dbp5 is approximately 10-fold weaker than ADP affinity [30]. This detailed in vitro analysis identified Pi release and nucleotide exchange as potential biochemical transitions within the Dbp5 ATPase cycle that may be modulated in vivo. In line with this, it has been proposed that Nup159 and Gle1 influence the Dbp5 nucleotide bound state, specifically by Nup159 aiding ADP release and Gle1 promoting ATP binding [22]. This model of Dbp5 regulation is consistent with high resolution structures showing that the two RecA-like domains of Dbp5 adopt an open configuration when bound by Gle1 and Nup159 [23], which may promote nucleotide exchange (i.e. ADP release and/or ATP loading).
Overall, the reported activities and structural data to date are supportive of Nup159 and Gle1 acting to alter the nucleotide state of Dbp5. One class of regulatory proteins, nucleotide exchange factors (NEFs), achieve this by accelerating the release of a protein bound nucleotide, often by disrupting coordination of the nucleotide-associated magnesium cation [31, 32]. Eviction of the nucleotide-bound cation, which mediates several key interactions between nucleotide and protein, enables rapid dissociation of the nucleotide (e.g. ADP) from the active site, subsequent binding of nucleotide (e.g. ATP), and an additional round of ATPase cycling. This mechanism of regulation has not been well described for DBPs, but Mg2+-based NEFs have been shown to regulate several GTPase proteins [33–36], actin [37], kinesin [38, 39] and myosin motor proteins [40, 41]. One exception is the S. cerevisiae NEF, Ypt1p, which accelerates nucleotide release from the TRAPPI GTPase through a Mg2+ independent pathway [42]. Consequently, it is unknown whether DBPs also employ Mg2+-based NEFs, or whether Nup159 or Gle1 act as NEFs through Mg2+ to regulate Dbp5 activity.
Here, we test the hypothesis that Nup159 and Gle1 control the ADP-bound state of Dbp5, and further assess the contributions of Mg2+ to the kinetics and thermodynamics of ADP binding to Dbp5. We report that Mg2+ slows mantADP and mantATP release from Dbp5, as observed with other NTPases [35, 39, 40], and increased the overall ADP and ATP affinities (~3-fold for mantADP, 2-fold for ADP, ~6-fold for mantATP, and 3-to-5-fold for ATP). We find that Nup159 does not accelerate mantADP or Mg2+-mantADP release, while Gle1/InsP6 slowed mantADP release ~2-fold independent of Mg2+. These findings are inconsistent with Nup159 or Gle1 acting as a NEF. Finally, binding affinity of Gle1 for the mantADP-bound Dbp5 complex was reduced ~18-fold in the presence of Nup159, suggesting that Nup159 may function in vivo to modulate the interaction between Dbp5 and Gle1.
Materials and Methods
Reagents
All reagents were of the highest purity commercially available. ATP (Sigma, A7699) and ADP (Sigma, 01879) concentrations were determined by absorbance using ε259 = 15,400 M−1 cm−1. mantATP (Jena Biosciences, NU-202 and Invitrogen, M12417) and mantADP (Jena Biosciences, NU-201 and Invitrogen, M12416) concentrations were determined by absorbance using ε255 = 23,300 M−1 cm−1. Inositol hexakisphosphate (phytic acid) was purchased from Santa Cruz Biotechnology (SC-253276). Buffers were made with either DEPC treated water (American Bio, AB021028) or Millipore MilliQ® distilled deionized water that had been filtered through a 0.2 μm filter. Experiments were performed at 25 °C in assay buffer: 30 mM Hepes (pH 7.5), 100 mM KCl, and 2 mM DTT, supplemented with the indicated [MgCl2] or [EDTA]. For all experiments, the free Mg2+ concentration ([Mg2+]free) was determined using the program WebMaxC Standard (version - 12/31/03; http://web.stanford.edu/~cpatton/webmaxcS.htm).
Protein purification
Dbp5, Gle1, and Nup159 were purified as described [43].
Transient kinetic assays
Transient kinetic measurements were performed on an Applied Photophysics SX20 stopped-flow instrument thermostatted at 25 ± 0.1 °C. mant-nucleotide binding to Dbp5 was monitored by FRET between excited tryptophans (λex = 280 nm) in Dbp5 and the bound mant-labeled nucleotide. Fluorescence intensity was measured at 90° relative to excitation light after passing through a 400 nm long-pass colored glass filter. Inner filter effects are minimal in the mant-labeled nucleotide concentration range employed [33, 44]. Time courses shown are averages of at least two traces. Fitting was performed by nonlinear least-squares regression, and uncertainties of quantities determined from fits are given as standard errors in the fits.
mantADP dissociation kinetics
Irreversible dissociation of mantADP bound to Dbp5 was achieved by mixing with a large excess of unlabeled ADP to prevent mantADP rebinding. mantADP dissociation from Dbp5 in solution was measured as a function of [Mg2+]free using two approaches. First, Dbp5-mantADP was pre-formed in the presence of saturating Mg2+ by pre-equilibrating 4 μM Dbp5 and 60 μM mantADP in assay buffer containing 2 mM MgCl2 prior to initiating mantADP dissociation by rapidly mixing with a solution of 20 mM ADP supplemented with either 40 mM (for no Mg2+ only) or 3 mM EDTA, and a range of [MgCl2] to generate 0.057–2000 μM final [Mg2+]free after mixing and accounting for EDTA, mantADP and ADP chelation. Alternatively, Dbp5-mantADP was pre-formed in the absence of Mg2+ with assay buffer containing 4 mM EDTA to ensure no free Mg2+ at the start of the reaction, before being mixed with 20 mM ADP and a range of MgCl2 to generate 0.051–2000 μM [Mg2+]free after mixing. The final concentrations after mixing are 2 μM Dbp5, 30 μM mantADP and 10 mM competing unlabeled ADP.
[Nup159]-dependent Mg2+mantADP dissociation, with saturating Mg2+ (2 mM) in solution throughout the reaction, was monitored by mixing a pre-equilibrated solution of 4 μM Dbp5 and 40 μM mantADP in assay buffer containing 2 mM MgCl2 with equal volumes of a range of [Nup159] in assay buffer supplemented with 8 mM MgCl2 and 6 mM ADP (2 mM Mg2+ in solution after ADP chelation). [Nup159]-dependent mantADP dissociation without Mg2+ was monitored by mixing pre-equilibrated solutions of 2 μM Dbp5 and 80 μM mantADP in assay buffer containing 11.05 mM EDTA with equal volumes of assay buffer containing 20 mM ADP, 11.05 mM EDTA (ca. 27 nM [Mg2+]free after mixing), and various concentrations of Nup159. [Gle1]-dependent Mg2+-mantADP (saturating Mg2+ in solution) and mantADP (no Mg2+) dissociation were measured identically to Nup159, with the modification that the assay buffer in both syringes included 15 μM InsP6. Mg2+-mantADP dissociation from Nup159-Gle1-Dbp5 complex with saturating Mg2+ in solution was performed by mixing a pre-equilibrated solution of 5 μM Nup159, 4 μM Dbp5, 40 μM mantADP, and 15 μM InsP6 with an equal volume solution of 6 mM Mg2+ADP and 15 μM InsP6 pre-equilibrated with varying concentrations of Gle1. The assay buffer in both syringes for Mg2+-mantADP dissociation from the Nup159-Gle1-Dbp5 complex included an additional 2 mM MgCl2 in excess of the nucleotide concentration.
mant-labeled nucleotide association kinetics
The kinetics of mantADP binding to Dbp5 in the absence of Mg2+ was monitored by mixing Dbp5 (final concentration after mixing is 1 μM) with various concentrations of mantADP, in assay buffer containing 10 mM EDTA. Time courses were fitted to a single exponential and the [mantADP]-dependence of the observed pseudo-first order rate constants was fitted to a linear equation to extract bimolecular binding on- and off- rate constants from the slope and y-intercept, respectively.
mantATP binding was performed identically to mantADP binding experiments, but the time courses were fitted to three exponentials. The [mantATP]-dependence of the two fastest observed rate constants were globally fitted to Equation 2 in the text [8, 30] to determine fundamental rate constants. The third observed phase for mantATP binding while in the absence of Mg2+ is very slow (0.1 s−1) compared to the two fast observed phases and may represent a downstream process or off-pathway reaction. As in our previous analysis of mantATP binding in the presence of Mg2+ [30], we have not included this phase in our reaction schemes, since it is too slow to influence the two faster processes.
Competition of mantADP and unlabeled nucleotide
Dbp5 (final concentration after mixing is 1 μM in ADP containing reactions; 2 μM in ATP containing reactions) was rapidly mixed with 40 μM mantADP (final after mixing) and varying concentrations of unlabeled nucleotide, in assay buffer containing 1.5 mM (ADP reactions) or 2.5 mM (ATP reactions) EDTA. Time courses of FRET signal change from mantADP binding to Dbp5 in the presence of varying amounts of unlabeled ATP or ADP followed double exponentials (Fig. 3A). The slow phase occurring at 0.1 – 0.6 s−1 has a small amplitude compared to the fast phases in both ATP and ADP competition cases and has no well-defined dependence on [ATP] or [ADP]. Since the processes presented by the slow phases in both cases are temporally well-separated from the initial event involving mantADP binding competition with unlabeled nucleotides, we analyzed only the fast, [ADP] or [ATP]-dependent observed rate constant without interference from the slow phase.
Results
Removal of [Mg2+] accelerates mantADP release from Dbp5
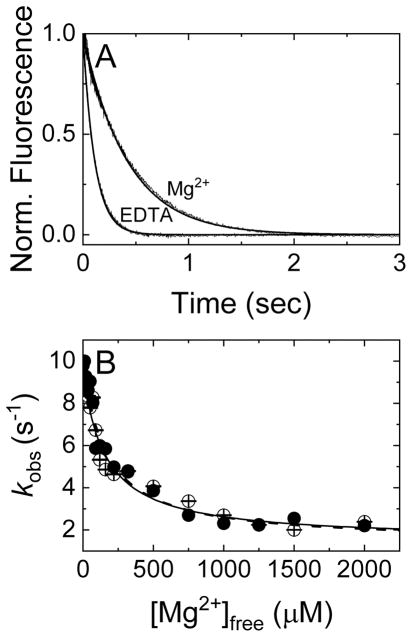
Prior to testing regulators for Mg2+-based NEF activity, it was necessary to determine how Mg2+ influences nucleotide exchange and binding. To evaluate the impact of magnesium on nucleotide-Dbp5 interactions (Scheme 1), time courses of Dbp5-mantADP dissociation in the presence of saturating Mg2+ and absence (i.e. with excess EDTA) of Mg2+ were collected (Fig. 1A). The resulting data are best fit by single exponentials where the observed rate constant depends hyperbolically on [Mg2+]free (Fig. 1B), yielding a mantADP dissociation rate constant (k−mD(+Mg)) of 2.4 ± 0.01 s−1 in the presence of Mg2+, and four-fold more rapid mantADP dissociation in the absence of Mg2+ with a rate constant (k−mD(−Mg)) of 10.0 ± 0.4 s−1 (Fig. 1A, Table 1). We interpret these and other related results with the assumption that the impact of Mg2+ on nucleotide binding arises from its direct association with the nucleotide at the Dbp5 active site, though contributions from Mg2+ binding to secondary sites on Dbp5 cannot be ruled out. Identical results are obtained when starting with a preformed Dbp5-Mg2+-mantADP complex (in the presence of excess MgCl2) or with a preformed Dbp5-mantADP complex in the absence of Mg2+ (excess EDTA present in buffer prior to mixing; Fig. 1B) before rapidly mixing with excessive competing unlabeled ADP and varying concentration of MgCl2. These results indicate that Mg2+ rapidly equilibrates between the Dbp5 active site and bulk solution on a timescale much greater than that of mantADP dissociation.
Scheme 1.
Figure 1. Mg2+ slows mantADP dissociation from Dbp5.
(A) Time courses of FRET signal changes in pre-equilibrated solutions of Dbp5 and mantADP with 2 mM MgCl2 (Mg2+ curve) or 4 mM EDTA (EDTA curve) upon rapid mixing with an equal volume of 20 mM ADP in assay buffer containing either 2 mM [MgCl2]free (Mg2+ curve) or 0 mM [MgCl2]free (EDTA curve). Continuous lines through the data represent best fits to single exponentials, yielding k−mD(−Mg) = 10.0 ± 0.4 s−1 and k−mD(+Mg) = 2.4 ± 0.009 s−1. (B) [Mg2+]free dependence of the observed rate constants of mantADP dissociation from a pre-formed Dbp5-mantADP (no Mg2+, filled circles) or Dbp5-Mg2+mantADP complex (open circles). The solid and dashed lines through the data represent the best fits of the data starting with Dbp5-mantADP (filled circles) or Dbp5-Mg2+mantADP (open circles) to Equation 1, yielding the two KMg(HmD) values (160 ± 40 – filled circles and 165 ± 26 μM – open circles) for Mg2+ binding to Dbp5-mantADP complex as well as the fundamental dissociation rate constants for mantADP (k−mD(-Mg) ≈ 10 and 9 s−1, no Mg2+) or Mg2+ mantADP (k−mD(+Mg) ≈ 2.4 and 2.2 s−1, saturating Mg2+). Uncertainty bars represent standard errors of the fits and are contained within the data points.
Table 1.
Mg2+ dependence of Dbp5-mant nucleotide interaction.
| Parameter | Value | Units | Assay |
|---|---|---|---|
| Mg2+ affinity for Dbp5-mantADP | |||
| KMg,HmD | 165 ± 40 | μM | dissociation from initial HMmD (Fig. 1) |
| 160 ± 27 | μM | dissociation from initial HmD (Fig. 1) | |
| 164 ± 25 | μM | Global fitting of HMmD and HmD data (Fig. 1) | |
| Mg2+ affinity for Dbp5-ADP | |||
| KMg,HD | ~113 | μM | Detailed balance of Scheme 1 and KMg,D ~ 676 μM [64] |
| Mg2+ affinity for Dbp5-ATP | |||
| KMg,HT | ~17 | μM | Detailed balance of Scheme 1 (modified for ATP) and KMg,T ~ 87 μM [64] |
| Unlabeled nucleotide affinity | |||
| k+D(−Mg) | ~ 2 | s−1 μM−1 | Estimated from competition with mD (Fig. 3B) |
| k−D(−Mg) | ~1600 | s−1 | Estimated from competition with mD (Fig. 3B) |
| KD(−Mg) | 0.8 ± 0.1 | mM | Kinetic competition with mD (Fig. 3B) |
| KD(+Mg) | 0.36 ± 0.05 | mM | Kinetic competition with mD [33] |
| k+T(−Mg) | > 2 | s−1 μM−1 | Estimated from competition with mD (Fig. 3C) |
| k−T(−Mg) | >30000 | s−1 | Estimated from competition with mD (Fig. 3C) |
| KT(−Mg) | 15 ± 4 | mM | Kinetic competition with mD (Fig. 3C) |
| KT(+Mg) | 3 – 6 ± 0.4 – 2 | mM | Kinetic competition with mD [33] |
| Mg2+-mantADP binding | |||
| KmD(+Mg) | 5 ± 2 | μM | k−mD(+Mg)/k+mD(+Mg) from linear fit of Mg2+mantADP binding in ref. [33]. |
| k+mD(+Mg) | 0.58 ± 0.07 | μM−1 s−1 | Linear fit of Mg2+mantADP binding in ref. [33]. |
| k−mD(+Mg) | 2.4 ± 0.009 | s−1 | MgmD dissociation from HMmD (Fig. 1) |
| 2.2 ± 0.012 | s−1 | MgmD dissociation from HmD (Fig. 1) | |
| 2.9 ± 0.9 | s−1 | Linear fit of Mg2+mantADP binding in ref. [33]. | |
| mantADP binding | |||
| k+mD(−Mg) | 1.0 ± 0.1 | μM−1 s−1 | mD binding (Fig. 2) |
| k−mD(−Mg) | 15 ± 2 | s−1 | mD binding (Fig. 2) |
| 9 ± 0.02 | s−1 | mD dissociation from HMmD (Fig. 1) | |
| 10 ± 0.4 | s−1 | mD dissociation from HmD (Fig. 1) | |
| KmD(−Mg) | 15 ± 2.5 | μM | k−mD/k+mD (Fig. 2) |
| mantATP binding | |||
| k+1mT(−Mg) | 3 ± 1 | μM−1 s−1 | mT binding global fit (Fig. 2D) |
| k−1mT(−Mg) | 125 ± 77 | s−1 | mT binding global fit (Fig. 2D) |
| K1mT(−Mg) | 42 ± 32 | μM | k−mT1/k+mT1 |
| k+2mT(Mg) | 11 ± 4 | s−1 | mT binding global fit (Fig. 2D) |
| k−2mT(−Mg) | 7 ± 3 | s−1 | mT binding global fit (Fig. 2D) |
| K2mT(−Mg) | 0.7 ± 0.2 | k−mT2/k+mT2 | |
| k−3mT,obs(−Mg) | ~0.1 | s−1 | mantATP binding (Fig. 2D) |
We fitted the [Mg2+]free-dependent observed dissociation rate constant as a weighted average of Mg2+-mantADP and mantADP dissociation according to Equation 1:
| Equation 1 |
where k−mD(+Mg) and k−mD(−Mg) are the mantADP dissociation rate constants with and without Mg2+; [HMgmD] and [HmD] are Dbp5-mantADP complex with and without Mg2+; [Mg] is [Mg2+]free, and KMg,HmD is the equilibrium dissociation constant for Mg2+ binding to Dbp5-mantADP. The simplified hyperbolic form of the HMgmD species is used since the total Mg2+ concentration in the titration range available to bind HmD complex ≫ total [HmD]. Using Equation 1, the best fit of the data for both sets of measurements in Fig. 1B yields a Mg2+ affinity for Dbp5-mantADP (KMg,HmD) of ~ 164 μM (Table 1) with a mantADP dissociation rate constant of ~ 2.4 s−1 with Mg2+ and ~ 9 s−1 without Mg2+, indicating that Mg2+ occupancy slows mantADP release.
Mg2+ modifies kinetics of mant nucleotide binding to Dbp5
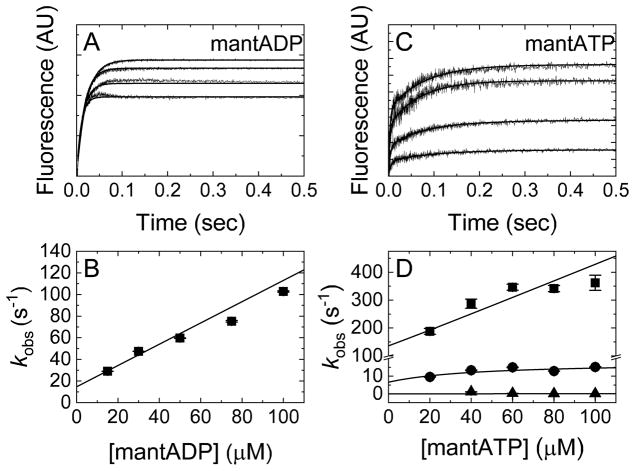
To measure the kinetics of mantADP binding in the absence of Mg2+ (Scheme 1), time courses of FRET signal change upon mantADP association were collected following rapid mixing of Dbp5 with mantADP. In the absence of Mg2+, mantADP binding traces are well-described by single exponentials with observed rate constants that depend linearly on the mantADP concentration (Fig. 2A). The association rate constant in the absence of Mg2+ (k+mD(−Mg)) determined from the slope of the best linear fit of the data is 1.0 ± 0.1 μM−1 s−1, while the dissociation rate constant (k−mD(−Mg)) estimated from the y-intercept is 15 ± 2 s−1 (Fig. 2B, Table 1), slightly faster than the value of ~9–10 s−1 determined from irreversible dissociation measurements (Fig. 1). The mantADP affinity in the absence of Mg2+ (KmD(−Mg)) calculated from the ratio of dissociation and association rate constants is 15 ± 2.5 μM.
Figure 2. mant nucleotide-Dbp5 binding kinetics in the absence of Mg2+.
(A) Time courses of FRET signal change after mixing Dbp5 with varying [mantADP] in 10 mM EDTA. Final concentrations after mixing are 1 μM Dbp5 and (lower to upper) 30, 50, 75, or 100 μM mantADP. Best fits to single exponentials are illustrated by smooth lines through the data. (B) [mantADP]-dependence of observed rate constants obtained from exponential fits in A. The line through the data represents the best linear fit, giving the fundamental association rate constant (k+mD(−Mg) = 1.0 ± 0.1 μM−1 s−1) for divalent cation-free mantADP binding to Dbp5, and fundamental dissociation rate constant, k−mD(−Mg) = 15 ± 2 s−1. (C) Time courses of FRET signal change after mixing Dbp5 with varying [mantATP], in buffer with 10 mM EDTA. Final concentrations after mixing are 1 μM Dbp5 and (lower to upper) 20, 40, 60 or 80 μM mantATP. Best fits to three exponentials are illustrated by continuous lines through the data. (D) [mantATP] dependence of observed rate constants. k1mT(−Mg),obs and k2mT(−Mg),obs were globally fit to Equation 2 [8] and the resulting kinetic rate constants are listed in Table 1. k3mT(−Mg),obs ~ 0.1 s−1 and is [mantATP]-independent over the range titrated. The [mantATP]-dependence of the fastest phase observed rate constants was not fitted to a hyperbolic function given the uncertainty associated with kobs values that approach the instrument dead time. Moreover, when these values are fitted to an unconstrained hyperbolic function, an unwarranted, large negative y-intercept value results. Therefore, we fitted to the simplest mechanism/model. Uncertainty bars represent standard errors of the fits and are contained within the data points.
We previously observed multi-step mantADP binding in the presence of Mg2+ [30], as indicated by the apparent hyperbolic [mantADP] concentration-dependence of the observed rate constant with a weak affinity of KmD0 ~100 μM for the fast binding step. In the mantADP titration range that satisfies the condition [mD] < KmD0 ~100 μM, the hyperbolic [mantADP] concentration-dependence of the observed rate constant approximates to linear [45, 46], and the overall on- and off rate constants for the combined two step binding can be approximately estimated by fitting this data [30] to a linear function, yielding Mg2+mantADP association (k+mD(+Mg)) and dissociation (k−mD(+Mg)) rate constants of 0.58 ± 0.07 μM−1 s−1 and 2.9 ± 0.9 s−1, respectively. The ratio of association and dissociation rate constants yields a Mg2+mantADP affinity of 5 ± 2 μM. Thus, Mg2+ slows mantADP dissociation and to a lesser extent mantADP association, explaining the overall weaker Dbp5-mantADP binding affinity (~ 2–3 fold) in the absence of Mg2+.
In the case of mantATP, time courses of binding to Dbp5 in the absence of Mg2+ are well-fitted by three exponentials (Fig 2C), similar to binding measured in the presence of Mg2+ [30]. We assume that the two fastest [mantATP]-dependent transitions occur in series and the first observed phase is from mantATP binding, while the second observed phase is a combined/overall step that involves downstream ATPase reactions, including ATP hydrolysis [8]. Therefore, the two observed fast phases can be globally fitted to Equation 2 to determine rate constants in the ATPase reaction scheme [8, 30].
| Equation 2 |
In Equation 2, [mT] is the total mantATP concentration, k+1 and k−1 are the fundamental forward and reverse rate constants for step 1 (mantATP binding), and k+2 and k−2 are the forward and backward rate constants for combination step 2 (including mantATP hydrolysis) [8, 30]. The mantATP association rate constant in the absence of Mg2+ determined from the fit is 3 ± 1 μM−1 s−1 (Fig. 2D, Table 1) and is comparable to the rate constant measured in the presence of Mg2+ (1.63 μM−1 s−1 from [30]). In contrast, the mantATP dissociation rate constant is an order of magnitude faster in the absence of Mg2+ (~125 s−1; Fig 2D; Table 1) as compared to the presence of Mg2+ (~11.9 ± 0.7 s−1 from [30]). As in our previous analysis of mantATP binding in the presence of Mg2+ [30], we have not included the third phase in our reaction schemes, since it is too slow to influence the two faster processes (see Materials and Methods). Together, these measurements show that Mg2+ has a strong effect on mantATP dissociation, which is ~4 fold greater than the impact of Mg2+ on mantADP dissociation.
Mg2+ tightens unlabeled ADP and ATP binding to Dbp5
To extend and validate these measurements, the affinity of Dbp5 for unlabeled nucleotides in the absence of Mg2+ was determined by kinetic competition experiments between mant-labeled and unlabeled nucleotides (Fig. 3), as previously done in the presence of Mg2+ [30, 44]. ADP or ATP competition slows the observed fast mantADP binding phase (Fig. 3B and 3C), indicating that the [ADP]- or [ATP]-dependent mantADP observed rate constants can be fitted by Equation 3 below [30, 47], derived specifically for cases in which the unlabeled competitor binds in rapid equilibrium in advance of labeled ligand binding.
Figure 3. Determination of unlabeled nucleotide affinity in the absence of Mg2+ by kinetic competition with mantADP.
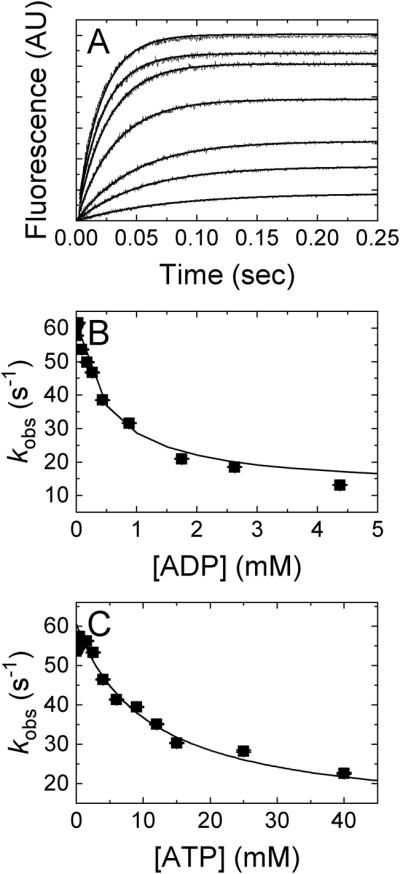
(A) Time courses of FRET signal change after mixing 1 μM Dbp5 with an equal volume solution of 40 μM mantADP and (upper to lower) 0, 0.00875, 0.0625, 0.0875, 0.175, 0.2625, 0.4375, 0.875, 1.75, 2.625 or 4.375 mM ADP in assay buffer containing excess EDTA. Concentrations are final after mixing. The lines through the data represent best fits to double exponentials. (B) [ADP]-dependence of the fast observed rate constants from the double exponential fits in Panel A. (C) [ATP]-dependence of the fast observed rate constant of two exponential fits after mixing 2 μM Dbp5 with 40 μM mantADP and excess EDTA supplemented with either 0, 0.2, 0.5, 0.9, 1.5, 2.5, 4, 6, 9, 12, 15, 25, or 40 mM ATP. Concentrations are final after mixing. The lines through the data in panels B and C represent the best fits to Equation 3 and yield apparent affinities: KD(−Mg) = 0.8 ± 0.1 mM and KT(−Mg) = 15 ± 4 mM. Uncertainty bars represent standard errors of the fits and are contained within the data points.
| Equation 3 |
In Equation 3, KD is the unlabeled ADP equilibrium binding constant. kobs,0 and kobs,inf (with unit s−1) are the observed mantADP rate constant at 0 and saturating unlabeled [ADP], i.e. kobs,0 = k+mD [mD]+k−mD and kobs,inf = k−mD. To analyze ATP competition data, replace [ADP] and KD by [ATP] and KT, respectively. Fitting the [ADP] or [ATP]-dependence of the observed mantADP binding rate constant in Fig. 3B or 3C to Equation 3 gives the apparent ATP affinity (KT(−Mg) = 15 ± 4 mM) or ADP affinity (KD(−Mg) = 0.8 ± 0.1 mM) in the absence of Mg2+ (Table 1), and a mantADP dissociation (k−mD(−Mg) = kobs,inf ~ 6 – 10 s−1) and association rate constant (k+mD(−Mg) ~ 1.2 – 1.4 μM−1 s−1) (Table 1). The equilibrium constant (15 mM) for unlabeled ATP binding to Dbp5 is ~3–5-fold weaker than the 3 – 6 mM affinity of Mg2+-ATP [30], and unlabeled ADP (0.8 mM) binding is ~2-fold weaker than that of Mg2+-ADP (Table 1) [30, 48]. These data obtained here by ADP and ATP competition are consistent with the mantADP binding and irreversible dissociation measurements made in the absence of Mg2+ in Fig. 1 and 2 (see Table 1).
A similar procedure for the derivation of Equation 3 can be performed without imposing the assumption of unlabeled competitor binding in rapid equilibrium by solving differential equations describing the competitive binding of labeled and unlabeled ligands for protein to yield Equation 3′, below:
In Equation 3′, the symbols are the same as in Equation 3 with an additional parameter, k+D, which is the unlabeled ADP bimolecular association rate constant expressed with identical concentration units as the equilibrium binding constant KD. Under conditions where , the quadratic form of Equation 3′ is simplified to a hyperbolic form given in Equation 3. Equation 3 and 3′ and the conditions and discussions above apply to ATP competition as well, while replacing [ADP] and KD by [ATP] and KT, respectively. Compared to Equation 3, Equation 3′ is more general and does not need the presumption of unlabeled competitor binding in rapid equilibrium. Moreover, Equation 3′ further permits determination of unlabeled nucleotide binding rate constants (k+D) to provide a sense of the timescale of unlabeled nucleotide binding, though it is an approximation and subject to large error due to the rapid equilibration of unlabeled competitor.
In the absence of Mg2+, unlabeled ADP and ATP competition data in Figs. 3B and C are fitted equally well to Equation 3′ and Equation 3, even though the parameter k+D (k+T) has large error. The results for the remaining parameters are essentially the same for both equations. The best fit curves generated by the two equations are indistinguishable. Fitting results by Equations 3′ estimate an ADP association rate constant (k+D(−Mg)) of ~ 2 μM−1 s−1 and an ATP association rate constant (k+T(−Mg)) that is even faster. We estimate from these association rate constants and binding affinities that ADP dissociates (k−D(−Mg)) at ~1600 s−1 and ATP dissociates (k−T(−Mg)) at > 30000 s−1 (Table 1). These extremely rapid rate constants are subject to large uncertainty and may only provide lower estimates of dissociation rate constants. However, these data highlight the fact that ADP and ATP binding to Dbp5 are in extremely fast equilibrium, similar to binding in the presence of Mg2+ [30], which suggests that a NEF would not be necessary to accelerate nucleotide exchange.
Nup159 does not accelerate mantADP release from Dbp5
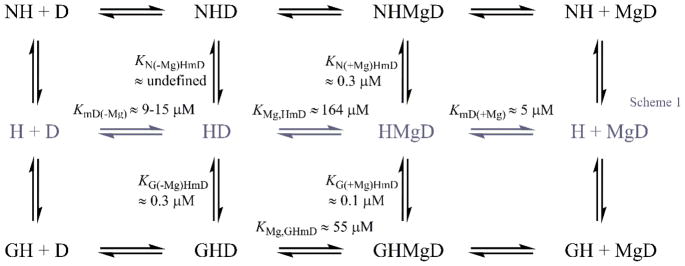
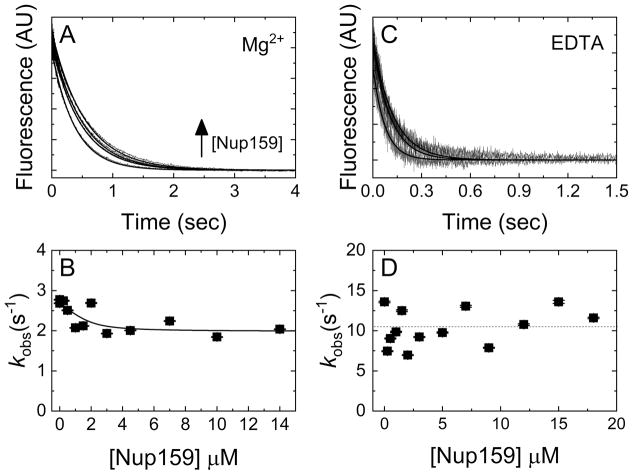
Previous work has provided evidence for Nup159 accelerating ADP release from Dbp5 [22]. Structural work has further shown that Nup159 binds Dbp5 in a manner mutually exclusive with RNA [23, 49], and may allow separation of the two RecA-like domains of Dbp5 to facilitate nucleotide exchange. The interactions of Nup159 with Dbp5-ADP and -Mg2+ADP relevant to potential Nup159 NEF activity are shown in the top half of Scheme 2, where N represents Nup159. To detail these interactions, time courses of mantADP dissociation from Dbp5 were collected in the presence of saturating Mg2+ upon rapid mixing with Nup159 and excess competing unlabeled ADP. These data followed single exponentials (Fig. 4A), with observed rate constants that slow in a weakly [Nup159]-dependent manner over the range examined (0–14 μM; Fig. 4B). The observed rate constant of Mg2+-mantADP dissociation decreased from 2.7 ± 0.1 s−1 (k−mD(+Mg)) to 2.0 ± 0.2 s−1 (k−mD(+Mg), N) in the presence of excess Nup159 (Fig. 4B). Fitting [Nup159]-dependent observed rate constants of mantADP dissociation in the presence of Mg2+ to Equation 4, a population weighted average of Nup159 (N) bound and un-bound mantADP-Dbp5 complexes:
| Equation 4 |
yields an affinity (KN(+Mg)) of ~ 0.3 ± 0.8 μM for Nup159 binding to Dbp5-Mg2+mantADP, where k−mD and k−mD,N are mantADP dissociation rate constants from Dbp5 without and with bound regulator Nup159, and KN is the equilibrium constant of Nup159 binding to Dbp5-mantADP (HmD). [HmD]tot = [HmD] + [NHmD] by mass balance. Time courses of mantADP dissociation from Dbp5 upon rapid mixing with Nup159 and excess competing unlabeled ADP in the absence of Mg2+ also followed single exponentials, with no observable [Nup159] dependence in the rate constants (~11 ± 2 s−1 Fig. 4C and 4D). Together, these measurements do not support a role for Nup159 in accelerating ADP release from Dbp5.
Scheme 2.
Figure 4. Nup159 minimally affects mantADP(±Mg2+) dissociation kinetics.
(A) Time courses of irreversible Mg2+mantADP dissociation from a pre-incubated solution of Dbp5 (2 μM) and Mg2+mantADP (20 μM) upon mixing with 3 mM Mg2+ADP and (from lower to upper) 0, 1.5, 4.5, or 10 μM Nup159 in assay buffer containing excess MgCl2. Concentrations are final after mixing. Smooth lines through the data represent best fits to single exponentials. (B) [Nup159]-dependence of the observed rate constants obtained from A. The line through the data represents the best fit to Equation 4, yielding KN(+Mg)HmD of 0.3 ± 0.8 μM, k−mD(+Mg) = 2.7 ± 0.1 s−1 and k−mD(+Mg),N = 2.0 ± 0.2 s−1. (C) Time courses of FRET signal change after mixing pre-equilibrated solutions of 1 μM Dbp5 and 40 μM mantADP with an equal volume of (lower to upper) 0, 3 or 9 μM Nup159 and 10 mM ADP in assay buffer containing excess EDTA (ca. 27 nM [Mg2+]free after mixing). The concentrations are final after mixing. Continuous lines through the data represent best fits to a single exponential. (D) [Nup159]-dependence of the mantADP dissociation observed rate constants obtained from exponential fits in C. ANOVA analysis suggests that the dependence is insignificant and the average dissociation rate constant is ~ 11 s−1 indicated by a horizontal dashed line. Uncertainty bars represent standard error of the fits and are contained within the data points.
Gle1 slows mantADP release from Dbp5
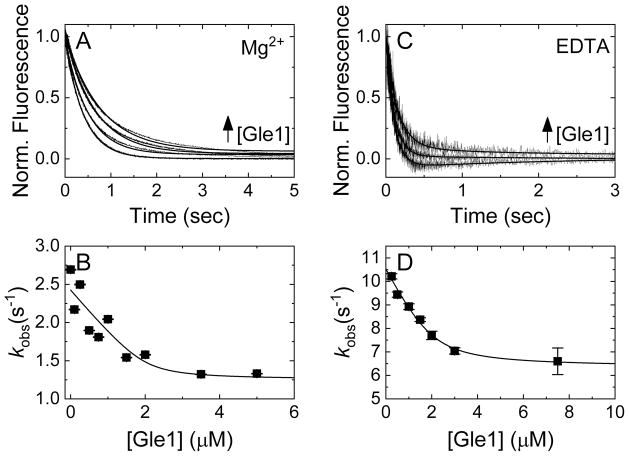
Crystal structures of the Gle1-InsP6-Dbp5-ADP complex showed that Gle1 contacts both RecA-like domains and orients them in a partially open conformation [23], and thus considered the possibility that this domain orientation may promote nucleotide exchange. The possible relationships between Gle1 and Dbp5-ADP and -Mg2+ADP interactions are detailed in the bottom half of Scheme 2. As with Nup159, time courses were collected of irreversible [Gle1]-dependent mantADP release from Dbp5 + InsP6 in the presence (Fig. 5A) and absence of Mg2+ (Fig. 5C). Data are well-fitted by single exponentials, with observed rate constants that depend hyperbolically on the [Gle1] (Fig. 5B, 5D). In the absence of Mg2+ there is a slow increase in background signal due to Gle1 itself, independent of binding, that was accounted for by an additional exponential. Under excess Gle1 conditions, mantADP release from Dbp5 slowed about two-fold from 2.4 ± 0.1 s−1 to 1.3 ± 0.3 s−1 in the presence of Mg2+ and from 10.5 ± 0.1 s−1 to 6.3 ± 0.2 s−1 in the absence of Mg2+ (Table 2). Fitting the [Gle1]-dependence of the observed rate constants to Equations 4 (substituting N with G (Gle1)) yields a Gle1 affinity for the InsP6-Dbp5-ADP complex of 0.1 ± 0.3 μM with Mg2+ (KG(+Mg), HmD) and 0.3 ± 0.1 μM without Mg2+ (KG(−Mg), HmD), suggesting the impact of Mg2+ is minimal given the measurement uncertainties. Based on these measurements, Gle1 does not accelerate ADP release from Dbp5.
Figure 5. Gle1 slows mantADP(±Mg2+) dissociation from Dbp5.
(A) Time courses of FRET signal shift upon mixing pre-equilibrated mixtures of 2 μM Dbp5 and 20 μM Mg2+mantADP with equal volumes of (lower to upper) 0, 1, 2, or 3.5 μM Gle1 and 3 mM Mg2+-ADP. Concentrations are final after mixing. The assay buffer in both syringes before mixing contains 15 μM InsP6 and an extra 2 mM MgCl2. Continuous lines through the data represent best fits to single exponential. (B) [Gle1] dependence of the observed rate constant from exponential fits in A. The continuous line through the data represents the best fit to Equation 4, yielding an affinity of Gle1 for the Dbp5-Mg2+mantADP complex of 0.1 ± 0.3 μM, k−mD(+Mg) = 2.4 ± 0.1 s−1 and k−mD(+Mg),G = 1.3 ± 0.3 s−1. (C) Time courses of FRET signal change after mixing pre-equilibrated solutions of 1 μM Dbp5 and 40 μM mantADP with an equal volume of (lower to upper) 0, 0.5 or 3 μM Gle1 and 10 mM ADP. Concentrations are final after mixing. The assay buffer in both syringes before mixing contains 15 μM InsP6 and 10.64 mM EDTA (ca. 44 nM [Mg2+]free after mixing). The time courses were fitted mainly by single exponential with an additional exponential to fit the slightly increasing part after 0.5 – 0.8 s (continuous lines through the data) due to the slow background signal increase by Gle1 alone that is not related to the binding process. (D) [Gle1] dependence of the mantADP dissociation observed rate constant obtained from exponential fits in C. The smooth line through the data represents the best fit to Equation 4, yielding an affinity (KG(−Mg)) of 0.3 ± 0.1 μM, k−mD(−Mg) = 10.5 ± 0.1 s−1 and k−mD(−Mg),G = 6.3 ± 0.2 s−1. Uncertainty bars represent standard error of the fits.
Table 2.
Regulator dependence of mantADP dissociation
| Parameter | Value | Units | Assay |
|---|---|---|---|
| Nup159-mediated Mg2+-mantADP dissociation | |||
| k−mD(+Mg) | 2.7 ± 0.1 | s−1 | mD dissociation, Nup free (Fig. 4B) |
| k−mD(+Mg),N | 2.0 ± 0.2 | s−1 | mD dissociation, Nup saturating (Fig. 4B) |
| KN(+Mg)HmD | 0.3 ± 0.8 | μM | [Nup159]-dependence of kobs(+Mg) (Fig. 4B) |
| Nup159-mediated mantADP dissociation | |||
| k−mD(−Mg),N | 11 ± 2 | s−1 | mD dissociation, all [Nup] (Fig. 4D) |
| KN(−Mg)HmD | undetermined | μM | [Nup159]-dependence of kobs(−Mg) (Fig. 4D) |
| Gle1-mediated Mg2+-mantADP dissociation | |||
| k−mD(+Mg) | 2.4 ± 0.1 | s−1 | mD dissociation from HMmD, Gle1 free (Fig. 5B) |
| k−mD(+Mg),G | 1.3 ± 0.3 | s−1 | mD dissociation from HMmD, Gle1 saturating (Fig. 5) |
| KG(+Mg)HmD | 0.1 ± 0.3 | μM | [Gle1]-dependence of kobs(+Mg) (Fig. 5B) |
| Gle1-mediated mantADP dissociation | |||
| k−mD(−Mg) | 10.5 ± 0.1 | s−1 | mD dissociation from HMmD, Gle1 free (Fig. 5D) |
| k−mD(−Mg),G | 6.3 ± 0.2 | s−1 | mD dissociation from HmD, Gle1 saturating (Fig. 5D) |
| KG(−Mg)HmD | 0.3 ± 0.1 | μM | [Gle1]-dependence of kobs(−Mg) (Fig. 5) |
| Mg2+ affinity for Gle1-Dbp5-mantADP | |||
| KMg,GHmD | 55 ± 159 | μM | Calculated from detailed balance of Scheme 2 |
| Gle1-mediated Mg2+-mantADP dissociation in the presence Nup159 | |||
| k−mD (+Mg),N | 2.2 ± 0.1 | s−1 | mD dissociation from Nup-HMmD, Gle1 free (Fig. 6B) |
| k−mD (+Mg),GN | 0.8 ± 0.9 | s−1 | mD dissociation from Nup-HMmD, Gle1 saturating (Fig. 6B) |
| KG(+Mg),HNmD | 1.8 ± 3.2 | μM | [Gle1]-dependence of kobs(−Mg)-Nup (Fig. 6B) |
Nup159 weakens Gle1 affinity for Dbp5-Mg2+mantADP complex
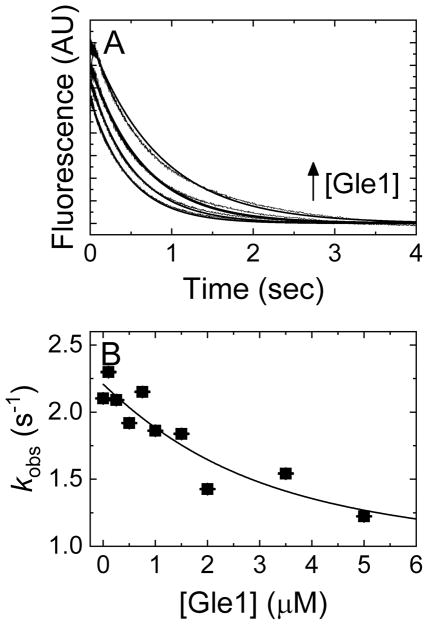
Current models propose that Dbp5 is regulated at nuclear pore complexes in a series of interactions with RNA, Gle1-InsP6 and Nup159 to facilitate mRNA export [22, 23, 27]. One component of these models is that Dbp5 sequentially interacts with Gle1 and Nup159 to spatially modulate the Dbp5 ATPase cycle. Moreover, structural work has shown that binding of Nup159 to a Gle1-InsP6-Dbp5-ADP complex induces solvent accessible separation of the two RecA-like domains of Dbp5 [23], which may favor nucleotide exchange. Consequently, to investigate the possibility that NEF activity emerges in the presence of both regulators, time courses of irreversible Mg2+-mantADP dissociation from Dbp5 in buffer containing 2.5 μM Nup159 and 15 μM InsP6 were measured upon rapid mixing with varying concentrations of Gle1 and excessive competing ADP. These data fit single exponentials (Fig. 6A), where the observed rate constant of Mg2+-mantADP dissociation slowed monotonically with [Gle1] (Fig. 6B). The best fit of the observed rate constant to Equation 4 gives the affinity of Dbp5-mantADP for Gle1 in the presence of Nup159 and InsP6 (KG(+Mg),HNmD) as 1.8 ± 3.2 μM (Fig. 6B), ~18 times weaker than that in the absence of Nup159 (0.1 ± 0.3 μM). The fit also yields a mantADP dissociation rate constants of 2.2. ± 0.1 s−1 in the presence of Nup159 (k−mD(+Mg),N) and InsP6 (no Gle1) and 0.8. ± 0.9 s−1 in the presence of Nup159, InsP6 and Gle1 (k−mD(+Mg),GN), similar to the mantADP dissociation rate constants with Gle1 and InsP6 (no Nup159; k−mD(+Mg) =2.4 ± 0.1 s−1 and k−mD(+Mg),GN = 1.3 ± 0.3 s−1; Fig. 5). These results indicate that binding of Nup159 to Dbp5 significantly alters the association between Dbp5 and Gle1, but still has minimal impact on mantADP release from Dbp5 in the presence of Gle1- InsP6.
Figure 6. Nup159 has no significant effect on Gle1-mediated Mg2+-mantADP dissociation.
(A) Time courses of FRET signal change upon mixing Dbp5 (2 μM) pre-incubated with Mg2+mantADP (30 μM), Nup159 (2.5 μM) and InsP6 (15 μM) with excess unlabeled Mg2+ADP (3 mM) and (lower to upper curves) 0, 1, 3.5, or 5 μM Gle1 in assay buffer with excess MgCl2. The concentrations are final after mixing. Smooth lines through the data represent best fits to single exponential. (B) [Gle1]-dependence of the observed rate constant of Mg2+mantADP dissociation from the exponential fit in Panel A. Continuous lines through the data represent the best fit of the observed rate constant to Equation 4, giving a KG(+Mg),HNmD of 1.8 ± 3.2 μM, k−mD(+Mg),N = 2.2 ± 0.1 s−1 and k−mD(+Mg),GN = 0.8 ± 0.9 s−1. Error bars represent standard error of the fits and are contained within the data points
Discussion
Genetics, cell biology, biochemistry and structural biology have significantly contributed to the understanding of Dbp5, including regulation of the Dbp5 ATPase cycle, leading to various models of Dbp5 mediated mRNA export [27, 50–54]. Here we present quantitative kinetic and thermodynamic data that provides evidence for an alternative model of Nup159–mediated regulation of Dbp5, achieved by mediating the interaction of Dbp5 with Gle1.
Note that the complexes and data presented here do not include RNA, which may alter Nup159 and Gle1 activities in regulating ADP release as NEFs. However, previous studies suggest that potential NEF activity from Nup159 or Gle1 binding to the Dbp5-ADP complex occurs after RNA release [22, 23]. Still, it is possible that transient co-binding of RNA, Gle1 and ADP during steady-state ATPase cycling (i.e. following hydrolysis and Pi release) may provide the opportunity for RNA to influence nucleotide and regulator interactions. For this reason, we limit our discussions and conclusions to ADP-bound complexes formed in the absence of RNA.
Mg2+-Nucleotide binding linkage in Dbp5
Although many NTPases require a Mg2+ cofactor for hydrolysis of the phosphodiester bond [55], persistence of Mg2+ in the post-hydrolysis active site can inhibit NDP dissociation and therefore recycling of the enzyme. In the Ras superfamily of small GTPases, the presence of Mg2+ contributes to extremely tight GDP binding affinities with dissociation constants that can be in the subnanomolar range [56, 57]. Consequently, by altering Mg2+ binding and accelerating off rates, NEFs are able to spatially and temporally regulate a vast number of cellular processes [58]. Like the Ras family and other NTPases, Mg2+ is also an important cofactor for nucleotide binding and hydrolysis by DBPs [2, 59]. A previous study had identified Mg2+-mediated inhibition of helicase activity for Dbp5, and another DBP Sub2, but the mechanism of inhibition was not identified and may be multifaceted given the duplex unwinding readout, which encompasses all steps of the ATPase cycle, as well as Mg2+-sensitive protein-RNA and RNA-RNA interactions [60]. Therefore, it is not known if Mg2+ is a target for regulators that function to control discrete transitions within the DBP ATPase activity.
In these studies, Mg2+ was found to bind Dbp5-mantADP with an affinity of ~164 μM. The presence of Mg2+ slowed mantADP dissociation ~4–6 fold and decreased mantADP association, which resulted in a modest tightening of mantADP binding affinity for Dbp5 (3-fold) from ~15 μM to ~5 μM (Table 1). Unlabeled ADP binding affinity was also observed to tighten 2-fold from 0.8 to 0.36 mM in the presence of Mg2+ using kinetic competition assays with mantADP (Fig. 3 and [30]). Mg2+ tightened the affinity of Dbp5 for mantATP ~6-fold (42 to 7.3 μM, K1mT or KmT comparison, Table 1 and [30]) and unlabeled ATP ~3 – 5-fold (15 to 3 – 6 mM, Table 1 and Fig. 3) as well. We assume these Mg2+ effects originate from a Mg2+ cation bound to the nucleotide in the active site of Dbp5, although Mg2+ may interact with secondary regulatory sites on Dbp5. Under this assumption, the affinity of Mg2+ for Dbp5-ADP or -ATP can be estimated to be ~113 and 17 μM, respectively (Table 1). These values are calculated using literature values for affinity of Mg2+ADP of ~676 μM and Mg2+ATP of 87 μM under our experimental conditions [61, 62] and using the subsequent detailed balance of Scheme 1, where in the case of ATP, ADP (D) is replaced by ATP. The ~7-fold tighter affinity of Mg2+ for Dbp5-ATP (17 μM) as compared to Dbp5-ADP (113 μM) might be expected from the presence of the gamma phosphate, which helps coordinates the cation in the pre-hydrolysis active site.
To our knowledge, this is the first study to investigate the role of Mg2+ in stabilizing the nucleotide-bound state of a DBP and may prove to be a more general model for DBP-nucleotide interactions given the evidence for Mg2+-linked nucleotide associations in other NTPase families [31–34, 36–42, 63], including the superfamily I helicases [64]. Notably, S. cerevisiae intracellular [Mg2+]free is estimated to be 0.1–1 mM [61], and is known to fluctuate throughout the cell cycle and in response to environmental conditions [61, 65]. As such, Dbp5-ADP is potentially responsive to these cellular fluctuations in Mg2+, raising the possibility that Dbp5 activity may be modified by environmental factors in addition to known protein regulators [5, 18, 19, 24, 26].
The impact of Gle1 on the Dbp5-ADP complex
Gle1 slowed the dissociation of mantADP(±Mg2+) from Dbp5 approximately two-fold and displayed a similar binding affinity within error for Dbp5-mantADP in the presence and absence of Mg2+ (0.1 ± 0.3 vs 0.3 ± 0.1 μM, Table 2). Although it is a trivial difference within error of the Gle1 affinity measurements, this 3-fold change produces an apparent difference in Mg2+ affinity of Gle1-InsP6-Dbp5-mantADP (55 μM) vs. Dbp5-mantADP (164 μM, Table 1) calculated from detailed balance of Scheme 2. However, the difference in Mg2+ affinity for the Dbp5-mantADP complex (±Gle1) is also not significant due to the large error propagated from the uncertainties in the Gle1 affinity measurements. Therefore, these data suggest that Gle1 has a minimal impact on interactions among Mg2+, Dbp5, and mantADP and does not act as a NEF under these solution conditions. Therefore, Gle1 stimulation of Dbp5 ATPase activity (±RNA) [5, 23] arises from a transition other than acceleration of nucleotide exchange. Given that Pi release limits both the RNA-stimulated and intrinsic Dbp5 steady-state ATPase activity [30], Gle1 presumably accelerates Pi release.
The impact of Nup159 on the Dbp5-ADP complex
It has been reported that Nup159 stimulates ADP release from Dbp5, assayed using an in vitro filter-binding assay [22]. Using rapid solution based kinetic and thermodynamic assays, we found that Nup159 did not accelerate dissociation of Mg2+mantADP or mantADP from Dbp5, nor did Nup159 alter mantADP release in the presence of Gle1 (i.e. mantADP release from Gle1-InsP6-Dbp5 is the same with and without Nup159; Table 2). We also did not observe long-lived ADP states in our mantADP release time courses through unlabeled ADP competition and Dbp5 ATPase cycling. Finally, we note that ADP release is not rate-limiting for Dbp5 ATPase activity in the presence or absence of RNA [30] and a reported slow (i.e. minutes) Nup159 stimulated ADP release [22] is unlikely to be relevant to the kinetics of mRNA export in vivo, which occurs in the sub-second time scale [66–69]. Accordingly, we conclude from these data that Nup159 and Gle1 do not function as NEFs for Dbp5.
The affinity of Dbp5 for mantADP relative to unlabeled ADP is significantly tighter due to hydrophobic interaction between the mant fluorophore and three residues of Dbp5 [30]. Thus, it is conceivable that strong interactions between Dbp5 and the mant fluorophore could potentially alter the effects of Nup159 and Gle1 in our studies. However, for the mant moiety to mask possible NEF activity of the regulators, putative NEF binding would have to enhance mant interactions with Dbp5 to an extent that the favorable interactions exceed (since mantADP release is actually slowed) the net perturbations in ADP binding. In other words, assuming the ADP dissociation activation energy is reduced to near zero following NEF perturbation (to allow for immediately release), the dissociation activation energy of the mant moiety alone must exceed the total dissociation activation energy of mantADP (both mant and ATP moieties) prior to regulator association. Consequently, we do not favor a mechanism in which the mant fluorophore masks regulator NEF activity.
Instead, our findings show that Nup159 weakens Gle1 affinity for the Dbp5-mantADP complex ~18 fold (from 0.1 to 1.8 μM, Table 2). Crystal structures of the Nup159-Gle1-Dbp5 complex demonstrate that Nup159 and Gle1 bind separate and distinct sites on Dbp5 and do not directly compete for Dbp5 binding [23]. Consequently, we expect that Nup159 likely modulates Gle1 binding to Dbp5 through an allosteric mechanism (i.e. Nup159 promotes conformational changes of Dbp5 that weaken Gle1 binding), supporting a model where Nup159 aids Gle1 release from Dbp5 to promote enzyme turnover and further rounds of ATP hydrolysis in vivo.
Highlights.
We test the hypothesis that Nup159 and Gle1 control the ADP-bound state of Dbp5.
We also assess the contributions of Mg2+ to the kinetics and thermodynamics of ADP binding to Dbp5.
Mg2+ slows mantADP and mantATP release from Dbp5 and increases the overall ADP and ATP affinities.
Nup159 does not accelerate mantADP or Mg2+-mantADP release, while Gle1/InsP6 slows mantADP release ~2-fold independent of Mg2+.
These findings are inconsistent with Nup159 or Gle1 acting as a NEF.
Acknowledgments
EVW was supported by a National Science Foundation Graduate Research Fellowship No. DGE-1122492. SG was supported by the NIH Institutional Training Grant No. 5T32GM007223-43. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM124120. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Linder P, Jankowsky E. From unwinding to clamping — the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 3.Bowers HA, Maroney PA, Fairman ME, Kastner B, Lührmann R, Nilsen TW, et al. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–12. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordin O, Tanner NK, Doère M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. The EMBO Journal. 2004;23:2478–87. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–76. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 6.Henn A, Bradley MJ, De La Cruz EM. ATP Utilization and RNA Conformational Rearrangement by DEAD-Box Proteins. Annual Review of Biophysics. 2012;41:247–67. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box Protein Required for Translation Initiation in Saccharomyces cerevisiae, Is an RNA Helicase. Journal of Biological Chemistry. 1999;274:17677–83. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 8.Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, et al. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol. 2011;409:399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2017;39:2260–70. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, et al. Structural and Biochemical Insights to the Role of the CCR4-NOT Complex and DDX6 ATPase in MicroRNA Repression. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, Le Hir H, et al. Mechanism of ATP turnover inhibition in the EJC. RNA (New York, NY) 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren L-G, et al. The DEXD/H-box RNA Helicase DDX19 Is Regulated by an α-Helical Switch. Journal of Biological Chemistry. 2009;284:10296–300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. Embo j. 1998;17:2663–76. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng SSI, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. The EMBO Journal. 1998;17:2651–62. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes & development. 2011;25:1052–64. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weirich CS, Erzberger JP, Berger JM, Weis K. The N-Terminal Domain of Nup159 Forms a β-Propeller that Functions in mRNA Export by Tethering the Helicase Dbp5 to the Nuclear Pore. Molecular Cell. 2004;16:749–60. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. Embo j. 1999;18:5778–88. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–6. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 20.Kendirgi F, Barry DM, Griffis ER, Powers MA, Wente SR. An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. The Journal of cell biology. 2003;160:1029. doi: 10.1083/jcb.200211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt C, von Kobbe C, Bachi A, Panté N, Rodrigues JP, Boscheron C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. The EMBO Journal. 1999;18:4332–47. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes & development. 2011;25:1065–77. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–42. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, et al. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 1999;18:5761–77. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K. Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proceedings of the National Academy of Sciences, USA. 2009;106:16251–6. doi: 10.1073/pnas.0902251106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alcazar-Roman AR, Bolger TA, Wente SR. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. The Journal of biological chemistry. 2010;285:16683–92. doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus (Austin, Tex) 2011;2:540–8. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, et al. The DEAD-Box RNA Helicase Dbp5 Functions in Translation Termination. Science. 2007;315:646–9. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 29.Neumann B, Wu H, Hackmann A, Krebber H. Nuclear Export of Pre-Ribosomal Subunits Requires Dbp5, but Not as an RNA-Helicase as for mRNA Export. PLOS ONE. 2016;11:e0149571. doi: 10.1371/journal.pone.0149571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong EV, Cao W, Voros J, Merchant M, Modis Y, Hackney DD, et al. P(I) Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5) J Mol Biol. 2016;428:492–508. doi: 10.1016/j.jmb.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic Analysis by Fluorescence of the Interaction between Ras and the Catalytic Domain of the Guanine Nucleotide Exchange Factor Cdc25Mm. Biochemistry. 1998;37:7420–30. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 32.Pan JY, Wessling-Resnick M. GEF-mediated GDP/GTP exchange by monomeric GTPases: A regulatory role for Mg2+? BioEssays. 1998;20:516–21. doi: 10.1002/(SICI)1521-1878(199806)20:6<516::AID-BIES11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–8. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 34.Zhang B, Zhang Y, Wang Z-x, Zheng Y. The Role of Mg2+ Cofactor in the Guanine Nucleotide Exchange and GTP Hydrolysis Reactions of Rho Family GTP-binding Proteins. Journal of Biological Chemistry. 2000;275:25299–307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg J. Structural Basis for Activation of ARF GTPase: Mechanisms of Guanine Nucleotide Exchange and GTP–Myristoyl Switching. Cell. 1998;95:237–48. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 36.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–43. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 37.Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba Profilin with Actin and Nucleotides Bound to Actin. Biochemistry. 1998;37:10871–80. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- 38.Shang Z, Zhou K, Xu C, Csencsits R, Cochran JC, Sindelar CV. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. eLife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng JQ, Jiang W, Hackney DD. Interaction of mant-adenosine nucleotides and magnesium with kinesin. Biochemistry. 1998;37:5288–95. doi: 10.1021/bi972742j. [DOI] [PubMed] [Google Scholar]
- 40.Hannemann DE, Cao W, Olivares AO, Robblee JP, De La Cruz EM. Magnesium, ADP, and Actin Binding Linkage of Myosin V: Evidence for Multiple Myosin V–ADP and Actomyosin V–ADP States. Biochemistry. 2005;44:8826–40. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 41.Rosenfeld SS, Houdusse A, Sweeney HL. Magnesium Regulates ADP Dissociation from Myosin V. Journal of Biological Chemistry. 2005;280:6072–9. doi: 10.1074/jbc.M412717200. [DOI] [PubMed] [Google Scholar]
- 42.Chin HF, Cai Y, Menon S, Ferro-Novick S, Reinisch KM, De La Cruz EM. Kinetic Analysis of the Guanine Nucleotide Exchange Activity of TRAPP, a Multimeric Ypt1p Exchange Factor. Journal of Molecular Biology. 2009;389:275–88. doi: 10.1016/j.jmb.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montpetit B, Seeliger MA, Weis K. Analysis of DEAD-box proteins in mRNA export. Methods in enzymology. 2012;511:239–54. doi: 10.1016/B978-0-12-396546-2.00011-5. [DOI] [PubMed] [Google Scholar]
- 44.Talavera MA, De La Cruz EM. Equilibrium and Kinetic Analysis of Nucleotide Binding to the DEAD-Box RNA Helicase DbpA†. Biochemistry. 2005;44:959–70. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 45.Corbett JF. Pseudo first-order kinetics. J Chem Educ. 1972;49:663. [Google Scholar]
- 46.Pollard TD, De La Cruz EM. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Molecular Biology of the Cell. 2013;24:1103–10. doi: 10.1091/mbc.E13-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods in enzymology. 2009;455:157–92. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan JS, Cheng Z, Zhang J, Noble C, Zhou Z, Song H, et al. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol. 2009;388:1–10. doi: 10.1016/j.jmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 49.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 50.Tieg B, Krebber H. Dbp5 - From nuclear export to translation. Biochim Biophys Acta. 2013;1829:791–8. doi: 10.1016/j.bbagrm.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Ledoux S, Guthrie C. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes & development. 2011;25:1109–14. doi: 10.1101/gad.2062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linder P. mRNA export: RNP remodeling by DEAD-box proteins. Current biology: CB. 2008;18:R297–9. doi: 10.1016/j.cub.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 53.Stewart M. Ratcheting mRNA out of the Nucleus. Molecular Cell. 2007;25:327–30. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Current Opinion in Cell Biology. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. JBIC Journal of Biological Inorganic Chemistry. 2008;13:1205–18. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 56.Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–72. [PubMed] [Google Scholar]
- 57.Feuerstein J, Goody RS, Wittinghofer A. Preparation and characterization of nucleotide-free and metal ion-free p21 “apoprotein”. J Biol Chem. 1987;262:8455–8. [PubMed] [Google Scholar]
- 58.Goitre L, Trapani E, Trabalzini L, Retta SF. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol. 2014;1120:1–18. doi: 10.1007/978-1-62703-791-4_1. [DOI] [PubMed] [Google Scholar]
- 59.Liu F, Putnam AA, Jankowsky E. DEAD-Box Helicases Form Nucleotide-Dependent, Long-Lived Complexes with RNA. Biochemistry. 2014;53:423–33. doi: 10.1021/bi401540q. [DOI] [PubMed] [Google Scholar]
- 60.Putnam AA, Jankowsky E. AMP Sensing by DEAD-Box RNA Helicases. Journal of Molecular Biology. 2013;425:3839–45. doi: 10.1016/j.jmb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pecoraro VL, Hermes JD, Cleland WW. Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry. 1984;23:5262–71. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 62.Martell AE, Smith RH. Critical Stability Constants. New York: Plenum Press; 1976. [Google Scholar]
- 63.Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–48. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 64.Moore KJM, Lohman TM. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 1. Use of fluorescent nucleotide analogs. Biochemistry. 1994;33:14550–64. doi: 10.1021/bi00252a023. [DOI] [PubMed] [Google Scholar]
- 65.Simon I, Zerial M, Goody RS. Kinetics of Interaction of Rab5 and Rab7 with Nucleotides and Magnesium Ions. Journal of Biological Chemistry. 1996;271:20470–8. doi: 10.1074/jbc.271.34.20470. [DOI] [PubMed] [Google Scholar]
- 66.Smith C, Lari A, Derrer CP, Ouwehand A, Rossouw A, Huisman M, et al. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. The Journal of Cell Biology. 2015;211:1121–30. doi: 10.1083/jcb.201503135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proceedings of the National Academy of Sciences. 2012;109:9426–31. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grunwald D, Singer RH. In vivo imaging of labelled endogenous [bgr]-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–7. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–52. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]