Abstract
Argonaute proteins and their associated small RNAs (sRNAs) are evolutionarily conserved regulators of gene expression. Gametocyte‐specific factor 1 (Gtsf1) proteins, characterized by two tandem CHHC zinc fingers and an unstructured C‐terminal tail, are conserved in animals and have been shown to interact with Piwi clade Argonautes, thereby assisting their activity. We identified the Caenorhabditis elegans Gtsf1 homolog, named it gtsf‐1 and characterized it in the context of the sRNA pathways of C. elegans. We report that GTSF‐1 is not required for Piwi‐mediated gene silencing. Instead, gtsf‐1 mutants show a striking depletion of 26G‐RNAs, a class of endogenous sRNAs, fully phenocopying rrf‐3 mutants. We show, both in vivo and in vitro, that GTSF‐1 interacts with RRF‐3 via its CHHC zinc fingers. Furthermore, we demonstrate that GTSF‐1 is required for the assembly of a larger RRF‐3 and DCR‐1‐containing complex (ERIC), thereby allowing for 26G‐RNA generation. We propose that GTSF‐1 homologs may act to drive the assembly of larger complexes that act in sRNA production and/or in imposing sRNA‐mediated silencing activities.
Keywords: 26G‐RNA, C. elegans, GTSF, piRNA, RdRP
Subject Categories: Development & Differentiation, RNA Biology
Introduction
Endogenous small non‐coding RNAs are responsible for regulating gene expression in many organisms. These small RNAs (sRNAs) act within the context of RNA interference (RNAi) or RNAi‐like pathways. In a variety of situations, these pathways provide an RNA‐based protection against “foreign” genetic elements such as transposable elements (TEs) and viruses (Ketting, 2011; Luteijn & Ketting, 2013).
In many RNAi‐like pathways, sRNAs are generated from double‐stranded RNA (dsRNA) precursors by Dicer, a conserved RNase III‐related enzyme (Ketting, 2011). Subsequently, sRNAs associate with Argonaute family proteins and guide them to target transcripts with complete or partial sequence complementarity. Upon Argonaute binding, transcripts are usually destabilized or translationally inhibited in the cytoplasm. However, some Argonautes have nuclear localization and regulate gene expression on the transcriptional level. For instance, in Caenorhabditis elegans, NRDE‐3 and HRDE‐1 are nuclear Argonautes that silence genes on the transcriptional level in the soma and in the germline, respectively (Guang et al, 2008; Buckley et al, 2012).
Caenorhabditis elegans, like plants and yeast, has RNA‐dependent RNA Polymerases (RdRPs) dedicated to the production of sRNAs. C. elegans has four RdRP genes, RRF‐1/‐2/‐3 and EGO‐1. It is believed that these RdRPs synthesize sRNA fragments in an unprimed manner (Billi et al, 2014). Two of these RdRPs, RRF‐1 and EGO‐1, generate sRNAs after target recognition by a primary Argonaute. These secondary sRNAs (22G‐RNAs) contain a 5′‐triphosphate group, have a bias for a 5′ guanosine, and are mostly 22 nucleotides long (Billi et al, 2014). The RdRP enzyme RRF‐3 is required for the biogenesis of another endogenous sRNA population, known as 26G‐RNAs, which are mainly 26 nucleotides long, have a 5′ guanosine bias and a 5′‐monophosphate (Gent et al, 2009, 2010; Han et al, 2009; Pavelec et al, 2009; Conine et al, 2010; Vasale et al, 2010). The fourth RdRP gene, RRF‐2, has no described function in RNAi‐related pathways.
26G‐RNAs can associate with three Argonautes. During spermatogenesis, 26G‐RNAs associate with the Argonautes ALG‐3 and ALG‐4 (from here on indicated as ALG‐3/4). These Argonautes are required for normal fertility and mostly target spermatogenic transcripts, mediating post‐transcriptional gene silencing (Han et al, 2009; Conine et al, 2010, 2013). Also, ALG‐3/4 targets show a significant overlap with targets of CSR‐1, an Argonaute protein that has been suggested to potentiate gene expression, rather than gene silencing (Conine et al, 2013). During oogenesis and embryogenesis, 26G‐RNAs associate with the Argonaute ERGO‐1 (Han et al, 2009; Gent et al, 2010; Vasale et al, 2010). In contrast to the ALG‐3/4‐bound 26G‐RNAs, ERGO‐1‐bound 26G‐RNAs are 2′‐O‐methylated by HENN‐1, which increases their stability (Billi et al, 2012; Kamminga et al, 2012; Montgomery et al, 2012). The main targets of ERGO‐1 are recently duplicated paralogs and pseudogenes (Vasale et al, 2010). Upon target recognition, ERGO‐1 triggers the production of 22G‐RNAs. In turn, these 22G‐RNAs direct gene silencing and presumably associate with unknown cytoplasmic Argonautes, as well as the somatic nuclear Argonaute protein NRDE‐3 (Guang et al, 2008; Vasale et al, 2010). NRDE‐3 and other NRDE factors lead to transcriptional gene silencing of their targets, a process accompanied by H3K9 trimethylation of the target locus (Burkhart et al, 2011; Burton et al, 2011).
Mutants defective in the generation of 26G‐RNAs, in particular those associated with ERGO‐1, are hypersensitive to exogenous RNAi (exoRNAi). This enhanced RNAi (Eri) phenotype is believed to stem from the fact that 26G‐RNA pathways share common components with the exoRNAi pathway (Duchaine et al, 2006; Yigit et al, 2006; Gu et al, 2009). Interestingly, many of the identified proteins that restrict exoRNAi in wild‐type animals form a complex: the ERI complex (ERIC; Duchaine et al, 2006; Yigit et al, 2006; Thivierge et al, 2012). ERIC has a core module that has been proposed to consist of the RdRP RRF‐3 and its close interacting partners, the DExD/H box helicase DRH‐3 and the Tudor domain‐containing protein ERI‐5 (Duchaine et al, 2006; Thivierge et al, 2012). To become active, this core complex needs to interact with DCR‐1, an interaction that requires ERI‐5 (Thivierge et al, 2012). Additionally, ERI‐1 and ERI‐3 are accessory factors of the ERIC that promote 26G‐RNA biogenesis (Duchaine et al, 2006; Billi et al, 2014). Further mechanistic insights into ERIC assembly and function are severely lacking.
Besides 22G‐ and 26G‐RNAs, C. elegans produces 21U‐RNAs (Billi et al, 2014). The 21U‐RNAs interact with PRG‐1, one of the C. elegans Piwi protein homologs, and are also known as the piRNAs of C. elegans (Billi et al, 2014). In many organisms, the Piwi‐piRNA pathway provides protection against TEs (Luteijn & Ketting, 2013), and also in C. elegans, 21U‐RNAs contribute to the defense against TE activity (Bagijn et al, 2012; De Albuquerque et al, 2015; Phillips et al, 2015). Interestingly, 21U‐RNAs can initiate a nuclear, 22G‐RNA‐mediated pathway. These 22G‐RNAs bound by the nuclear Argonaute HRDE‐1 can affect histone modification patterns on targeted loci and can establish a very stably inherited form of gene silencing (named RNA‐induced epigenetic silencing or RNAe) that no longer depends on continued exposure to 21U‐RNAs (Ashe et al, 2012; Buckley et al, 2012; Lee et al, 2012; Luteijn et al, 2012; Shirayama et al, 2012).
Genome‐wide screens have uncovered many factors involved in the piRNA pathway and TE silencing in Drosophila melanogaster (Czech et al, 2013; Handler et al, 2013; Muerdter et al, 2013). Many of these factors are poorly conserved evolutionarily. Gametocyte‐specific factor 1 (Gtsf1), a double CHHC zinc finger protein, represents one of the few Piwi pathway components that displays clear evolutionary conservation. dmGtsf1 is required for fertility and associates directly with Piwi (Dönertas et al, 2013; Ohtani et al, 2013). Interestingly, in the absence of Gtsf1, Piwi is still nuclear and loaded with piRNAs, but cannot silence TEs. Hence, dmGtsf‐1 has been proposed to be required for the execution of Piwi‐mediated silencing activities following target recognition. Also in mice, Gtsf1 is required for fertility and Gtsf1‐related proteins have been shown to interact with Piwi proteins (Yoshimura et al, 2007, 2009, 2018; Takemoto et al, 2016).
The precise molecular function of GTSF1 or of its isolated domains is unknown. GTSF1 homologs have two tandem CHHC zinc finger domains and an unstructured C‐terminal tail. In silico studies showed that CHHC zinc fingers are found in three protein families (Andreeva & Tidow, 2008): (i) U11‐48K proteins, members of the alternative spliceosome; (ii) TRM13 tRNA methyltransferases; and (iii) GTSF1‐related proteins. These CHHC domains behave as independent folding units and bind stoichiometrically to zinc (Andreeva & Tidow, 2008). The CHHC zinc finger of human U11‐48K was shown to bind to the 5′ splice site of U12‐dependent introns (Tidow et al, 2009), suggesting that CHHC zinc fingers bind RNA. Interestingly, the GTSF family is the only family of proteins that has two CHHC zinc fingers in tandem (Andreeva & Tidow, 2008).
Given its strong participation in Piwi‐induced TE silencing in Drosophila and mouse, and that it is one of the few factors acting with piRNAs that displays wide conservation, we decided to characterize the function of GTSF‐1 in C. elegans. Strikingly, we find that GTSF‐1 is not involved in TE silencing and does not affect 21U‐RNA production or activity in C. elegans. Instead, GTSF‐1 associates with the RdRP RRF‐3 and is required to assemble the ERI complex. We propose that GTSF1 proteins in general may be present in smaller pre‐complexes that may promote the assembly of larger protein–RNA complexes that elicit downstream enzymatic activities, such as sRNA production or the establishment of transcriptional silencing.
Results
GTSF‐1 is enriched in the germline but not in P‐granules
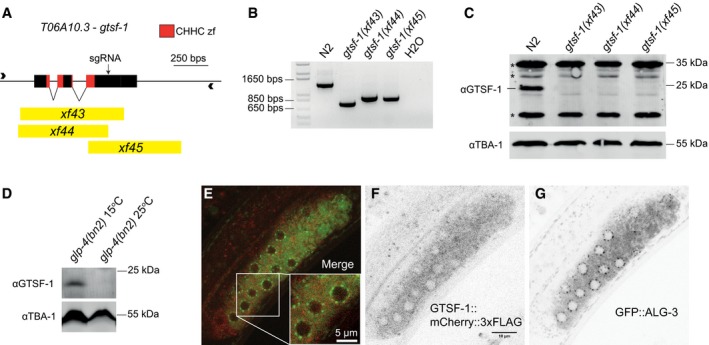
T06A10.3, the downstream partner of lsy‐13 in an operon on chromosome IV, was identified by reciprocal BLAST as the C. elegans gtsf1 homolog and was named gtsf‐1 (Fig 1A). GTSF‐1, like its mouse and fly homologs, has two predicted CHHC zinc fingers (Andreeva & Tidow, 2008). The cysteine and histidine residues of the zinc fingers, as well as several acidic residues on the C‐terminal region, are conserved from worms and flies to mouse, zebrafish, and human (Fig EV1A). We produced three independent gtsf‐1 deletion alleles using CRISPR‐Cas9 technology (Friedland et al, 2013; Figs 1A and B, and EV1B). Five times outcrossed, homozygous gtsf‐1 mutants are fertile and do not show any obvious morphological defects. No GTSF‐1 protein is detected in the mutants by Western blot, using an anti‐GTSF‐1 polyclonal antibody (Fig 1C). Expression of lsy‐13, the operon partner, does not seem to be affected in gtsf‐1(xf43) mutants (Fig EV1C).
Figure 1. T06A10.3, the Caenorhabditis elegans homolog of gtsf‐1 is expressed in the cytoplasm and is germline‐enriched.
-
AOverview of the T06A10.3 gene in chromosome IV of C. elegans. The exons are represented as black boxes, the CHHC zinc finger domains are shown in red, and the black arrow corresponds to the cut site of the sgRNA used. The deletion alleles are represented in yellow.
-
BPCR analysis of the deletion alleles using primers represented by arrowheads in (A).
-
CWestern blot analysis of mixed‐stage wild‐type and mutant worm extracts using a polyclonal anti‐GTSF‐1 antibody. TBA‐1, one of the C. elegans alpha‐tubulins, was used as a loading control. Asterisks indicate unspecific bands.
-
DWestern blot analysis of glp‐4(bn2) mutant worms grown at the non‐permissive temperature of 25°C, which precludes the development of the germline, and 15°C.
-
E–GRepresentative confocal fluorescence microscopy images showing the presence of GTSF‐1 and ALG‐3 tagged proteins in a gonad of a L4 double transgenic worm, in the alg‐3/4; gtsf‐1 triple mutant background. Scale bars correspond to 10 and 5 μm in the case of the inset.
Figure EV1. T06A10.3/CeGTSF‐1 is a conserved factor that is expressed during gametogenesis and early development (related to Fig 1).
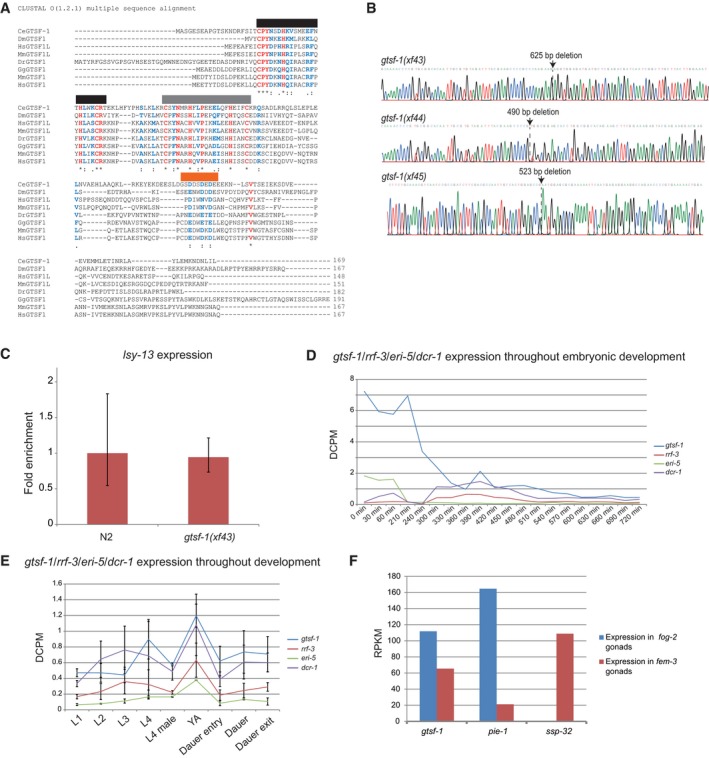
- Multiple sequence alignment, using ClustalO, of GTSF‐1 and GTSF‐1‐like proteins. An asterisk and red coloring highlights fully conserved residues. The cysteines and histidines of the CHHC zinc fingers are fully conserved. The first and second CHHC zinc fingers are highlighted with black and gray horizontal bars, respectively. A colon indicates strong conservation of the properties of the residue, while a period indicates weakly conservation of properties. Both cases are also highlighted in blue. Of note, there is a conserved acidic region on the C‐terminal tail of GTSF‐1, highlighted by an horizontal orange bar. Also, C. elegans GTSF‐1 has an extended acidic region with more glutamic and aspartic acid residues. Ce, Caenorhabditis elegans. Dm, Drosophila melanogaster. Mm, Mus musculus. Dr, Danio rerio. Gg, Gallus gallus. Hs, Homo sapiens.
- Chromatograms of Sanger sequencing of gtsf‐1 mutant alleles. Deletion sites are indicated with arrows.
- RT–qPCR of lsy‐13 in wild‐type and gtsf‐1 mutant embryos. Technical triplicates and biological duplicates were used for this experiment. Error bars represent the standard deviation of two biological replicates. pmp‐3 was used as the normalizing gene.
- mRNA expression profiles of gtsf‐1, rrf‐3, eri‐5, and dcr‐1 during embryonic development. A publicly available Poly‐A+ RNA‐seq dataset from Boeck et al (2016) was used. On the y‐axis, expression levels are shown in depth of coverage per base per million reads (DCPM).
- gtsf‐1, rrf‐3, eri‐5, and dcr‐1 expression profiles throughout larval development and dauer stage, shown in DCPM. Data points represent average between two Poly‐A+ RNA‐seq datasets from Boeck et al, 2016. Error bars represent the standard deviation of the two replicates.
- Depiction of expression levels of gtsf‐1, pie‐1 (an oogenic‐enriched gene), and ssp‐32 (a spermatogenic‐enriched gene), in both fog‐2 mutant gonads (strictly oogenic) and fem‐3 mutant gonads (strictly spermatogenic), as reported in Ortiz et al, 2014. Expression is shown in reads per kilobase million (RPKM).
To address the expression pattern of gtsf‐1 throughout development, we used publicly available RNA‐sequencing datasets (Boeck et al, 2016). During embryonic development, larval development, and adulthood, gtsf‐1 is moderately expressed (levels ranging from 0.4 to 7.2 depth of coverage per base per million reads [DCPM], Fig EV1D and E). Notably, gtsf‐1 RNA levels are highest during the first 300 min of embryonic development (2.38–7.2 DCPM), suggesting that gtsf‐1 mRNA may be maternally deposited (Fig EV1D). During larval development, gtsf‐1 mRNA reaches highest levels during the L4 and young adult stage (0.89–1.2 DCPM), correlating with germline development (Fig EV1E).
To address potential germline enrichment of GTSF‐1, we used glp‐4(bn2) worms, which lack a germline when grown at 25°C. Western blot experiments on these animals (Fig 1D) indicate that GTSF‐1 is enriched in the germline, since we could not detect GTSF‐1 in glp‐4(bn2) worms grown at 25°C. These data are supported by recent germline transcriptomes using dissected male and female gonads (Ortiz et al, 2014) that detected gtsf‐1 transcript in gonads irrespective of gender (Fig EV1F). To address subcellular localization, we produced a gtsf‐1::mCherry::3xflag single‐copy transgene controlled by the germline‐specific gld‐1 promoter (Merritt et al, 2008) and introduced it into a gtsf‐1(xf43); alg‐3(tm1155); alg‐4(ok1041) triple mutant background, also expressing a GFP::ALG‐3 fusion protein. In these animals, we observed GTSF‐1::mCherry::3xFLAG protein localized throughout the germline cytoplasm in L4 stage animals. GTSF‐1 does not appear to be concentrated in P‐granules, marked by GFP‐tagged ALG‐3 (Fig 1E–G), a known P‐granule component (Conine et al, 2010).
These data indicate that C. elegans GTSF‐1 is enriched in the germline cytoplasm, but mostly outside perinuclear granules.
GTSF‐1 is not involved in the 21U‐RNA pathway and transposon silencing in C. elegans
Next, we wanted to address whether gtsf‐1 is involved in TE silencing. To test this, we used a strain with the unc‐22(st136) allele, which has the unc‐22 gene interrupted by a Tc1 transposon (Ketting et al, 1999; Fig EV2A). Animals carrying the unc‐22(st136) allele exhibit the so‐called twitcher phenotype. When a gene that participates in TE silencing, such as mut‐7 (Ketting et al, 1999), is impaired in the unc‐22(st136) background, TEs will become mobile and phenotypical reversions to wild‐type movement can be observed. All three gtsf‐1 mutant alleles were crossed into the unc‐22(st136) background, and no reversions of the twitcher phenotype were observed after culturing the strains for several generations, in 10 biological replicates per allele (comprising a reversion frequency of < 10−5, Fig EV2B).
Figure EV2. Caenorhabditis elegans GTSF‐1 is not involved in TE silencing, the 21U‐RNA and miRNA pathways (related to Fig 2).
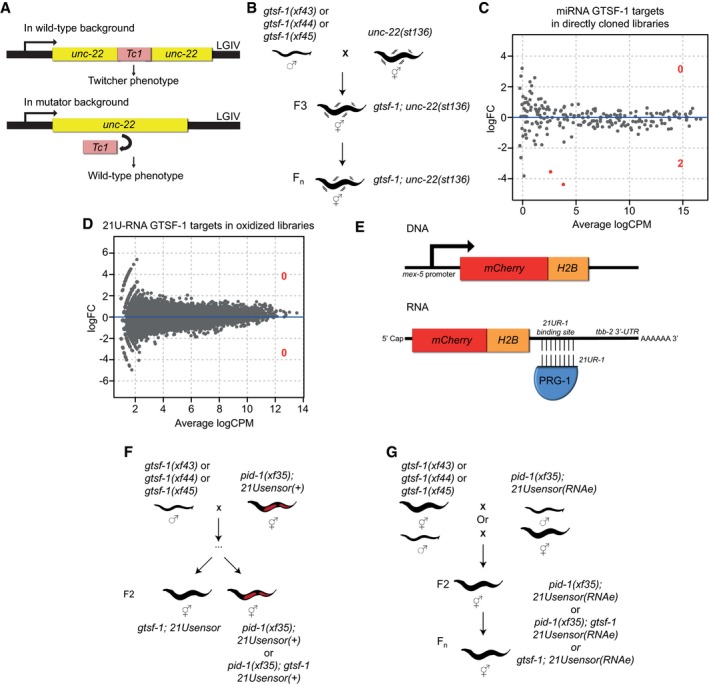
- Schematic of the unc‐22(st136) allele.
- Layout of the unc‐22(st136) x gtsf‐1 crosses to address transposon derepression. No phenotypic reversions to wild‐type were observed in ten replicate gtsf‐1;unc‐22(st136) populations, grown in parallel for several generations.
- Differential gene expression analysis of miRNAs in wild‐type versus gtsf‐1 mutant worms, to address whether miRNAs are globally deregulated in gtsf‐1. Analysis was performed in the directly cloned libraries, given the higher number of miRNA reads observed. Only two miRNAs (mir‐260, mir‐262) are significantly downregulated (1% FDR) in gtsf‐1 mutants.
- Differential gene expression analysis of 21U‐RNAs in wild‐type versus gtsf‐1 mutants. Analysis was performed in the oxidized libraries where a larger number of 21U‐RNA reads are found. No significant changes were found at 1% cutoff.
- Overview of the 21Usensor. It consists of an mCherry‐histone H2B fusion transgene with a binding site for 21U‐R1, an abundant 21U‐RNA, in the 3′UTR of the transcript.
- Testing the participation of gtsf‐1 in 21U‐RNA‐mediated silencing of C. elegans. To test this, a non‐stably silenced 21Usensor was used, and after crossing with gtsf‐1 mutant animals, the F2 of the indicated genotypes was scored for mCherry expression.
- Schematics of the crosses between gtsf‐1 mutant alleles and an RNAe 21Usensor. No derepression of the 21Usensor was observed in gtsf‐1 mutants in the F2 and in further generations.
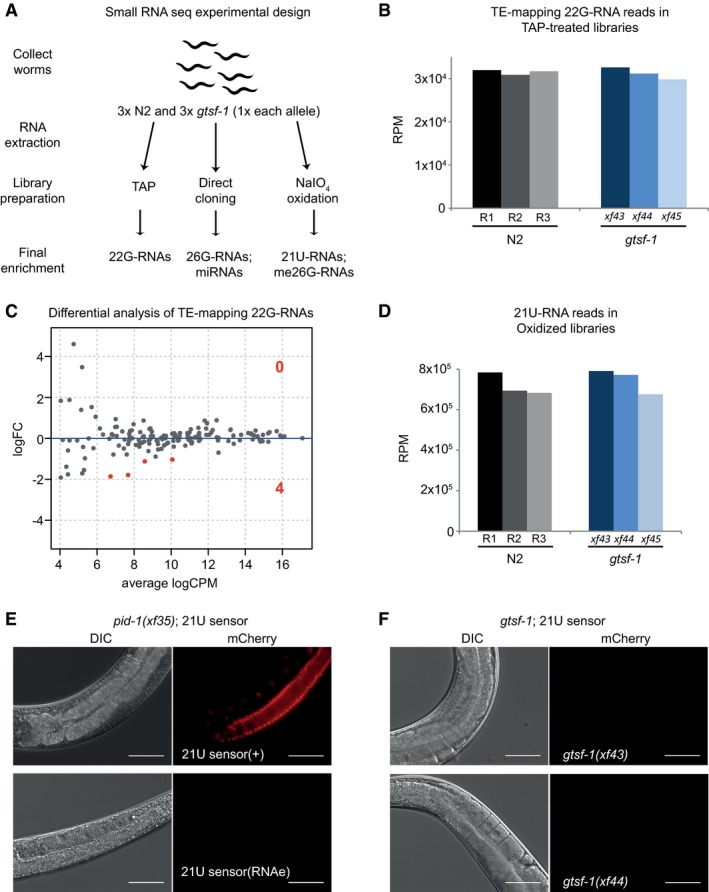
To further characterize the role of gtsf‐1 in the sRNA pathways of C. elegans, we sequenced sRNAs from wild‐type and gtsf‐1 synchronized gravid adults, in triplicates (experimental design in Fig 2A, sequencing statistics in Appendix). To enrich for different sRNA species, we employed different library preparations to each biological replicate. To increase cloning efficiency of 22G‐RNAs, which have a 5′ triphosphate, we used tobacco acid phosphatase (TAP). To enrich for sRNA species with a 2′‐O‐methyl group on their 3′ end (21U‐RNAs and ERGO‐1‐associated 26G‐RNAs), we oxidized the RNA before library preparation with NaIO4 (the 2′‐O‐methyl group protects sRNAs from oxidation). Finally, we used untreated RNA to capture a higher fraction of sRNAs carrying a 5′ monophosphate, irrespective of their 3′ end methylation status (ERGO‐1 and ALG‐3/4‐bound 26G‐RNAs and miRNAs). The latter type of libraries will be hereafter referred to as “directly cloned”. Sequences between 18 and 30 nucleotides were analyzed and read counts were normalized to the total number of mapped reads in each sample, excluding structural reads (see Materials and Methods).
Figure 2. Caenorhabditis elegans GTSF‐1 is not involved in the 21U‐RNA pathway and TE silencing.
-
AExperimental design of sRNA sequencing. Wild‐type and gtsf‐1 mutant gravid adult worms were collected in triplicates. For gtsf‐1, one sample of each allele was used as a biological replicate. Libraries were subjected to a triad of treatments to enrich for different sRNA species. TAP, tobacco acid pyrophosphatase.
-
BSimilar abundance of TE‐mapping 22G‐RNA reads in TAP‐treated libraries in wild‐type (N2) and gtsf‐1 mutants (Welch two sample t‐tests P‐value = 0.75). Normalized levels in reads per million (RPM) for each biological replicate are shown.
-
CDifferential analysis (MA‐plot) of TE‐mapping 22G‐RNAs in gtsf‐1 mutants versus wild‐type. sRNA reads from TAP‐treated libraries were used for this analysis. Only four TEs show significantly downregulated (1% FDR) sRNA levels in gtsf‐1 mutants (see Table EV1). LogFC, Log2 fold change. logCPM, log2 counts per million.
-
DSimilar abundance of 21U‐RNA reads in oxidized libraries in wild‐type (N2) and gtsf‐1 mutants (Welch two sample t‐tests P‐value = 0.62). Normalized levels in reads per million (RPM) for each biological replicate are shown.
-
E, FTesting the participation of gtsf‐1 in the 21U‐RNA pathway. For each figure, left panels are DIC, while right panels show mCherry fluorescence channel. (E) Photomicrographs of adult worms carrying a 21U‐RNA reporter in the pid‐1(xf35) background. The panels above show a strain in which the 21U‐sensor is still dependent on the 21U‐RNA pathway, because in the absence of PID‐1, mCherry can be observed in the germline. The panels below show a strain in which reporter silencing became independent of the 21U‐RNA pathway, a state known as RNAe. (F) Micrographs of 21U‐sensor;gtsf‐1 worms exhibiting the sensor repressed. This images are representative of 21U‐sensor;gtsf‐1 worms originating from the crosses with both strains shown in (E) (schematics of the crosses are shown in Fig EV2F and G). Scale bars represent 50 μm.
Consistent with the phenotypic experiments using the unc‐22(st136) Tc1‐transposition reporter, we did not observe major differences in sRNA reads mapping to TEs between wild‐type and gtsf‐1 animals (Fig 2B and C, Table EV1). Likewise, only two miRNAs were affected in gtsf‐1 mutants (Fig EV2C, Table EV1).
Also, the steady‐state 21U‐RNA levels are not significantly affected in gtsf‐1 mutants (Figs 2D and EV2D). To further test participation of gtsf‐1 in the 21U‐RNA pathway, we performed crosses combining gtsf‐1 mutant alleles with an mCherry reporter for 21U‐RNA activity (Bagijn et al, 2012; Luteijn et al, 2012; Fig EV2E). The reporter strains have a pid‐1(xf35) mutation in the background to inform on the status of the sensor (Fig 2E; de Albuquerque et al, 2014), which can be under RNAe (insensitive to the presence of PID‐1, Fig 2E, lower panels) or not (Fig 2E, upper panels). Loss of gtsf‐1 does not activate this reporter in either state, indicating it is not required for 21U‐RNA‐mediated silencing activity and RNAe (Figs 2E and F, and EV2F and G).
Overall, these data indicate that GTSF‐1 is neither involved in TE silencing, nor in the 21U‐RNA/RNAe pathway in C. elegans, in sharp contrast with the described function of Gtsf1 in mouse and fly.
gtsf‐1 mutants recapitulate phenotypes of 26G‐RNA pathway mutants
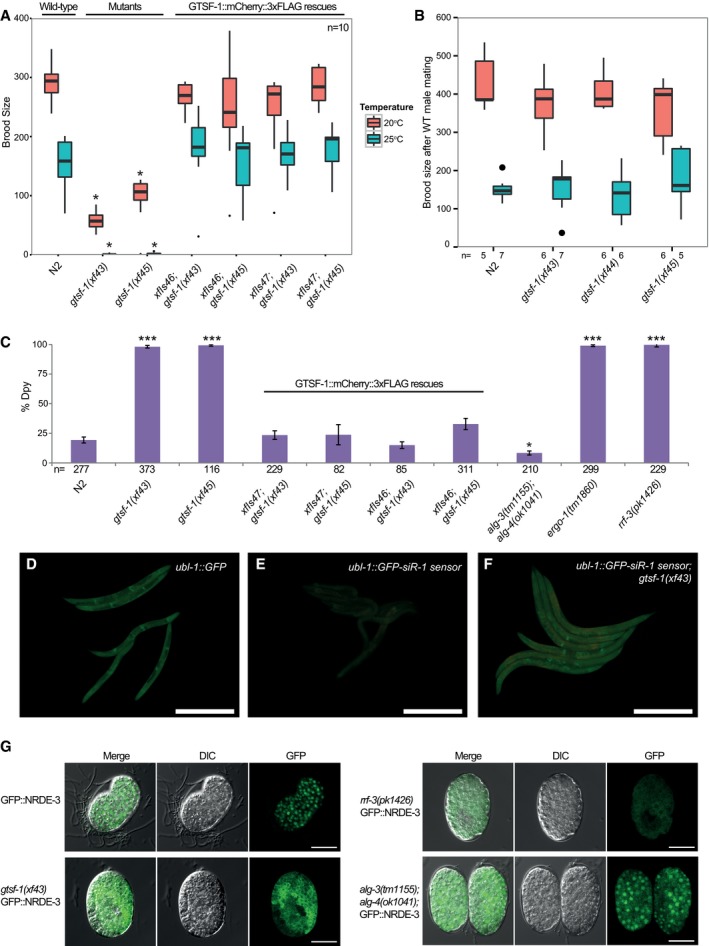
Given that gtsf‐1 is not involved in 21U‐RNA‐mediated gene silencing in C. elegans, we looked for other phenotypes that might be indicative of a role for GTSF‐1 in other endogenous sRNA pathways. We noticed that populations of gtsf‐1 mutant animals grow slower compared to wild type. This could reflect either developmental or fertility defects. When synchronized by bleaching, gtsf‐1 animals grew synchronous with wild type. In contrast, we noticed a striking reduction in brood size at 20°C, and temperature‐sensitive sterility at 25°C (Fig 3A). When grown at 25°C, gtsf‐1 mutant animals mostly produced unfertilized oocytes (Fig EV3A–C). Importantly, two independent germline‐specific gtsf‐1::mCherry::3xflag transgenes (including xfIs47, the transgene shown in Fig 1E and F) completely rescue these defects (Figs 3A and EV3C). These data clearly demonstrate that gtsf‐1 mutants display a temperature‐sensitive fertility defect.
Figure 3. gtsf‐1 animals phenocopy 26G‐RNA pathway mutants.
-
ABoxplot of brood size counts at 20 and 25°C. The progenies of 10 worms were counted for each strain and each temperature. Asterisks indicate P‐value < 0.0002 as assessed by Mann–Whitney and Wilcoxon tests comparing N2 worms with the other strains. Comparisons were done for each respective temperature. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
BHermaphrodites with the genotypes indicated on the x‐axis were mated with wild‐type males, and the progeny was counted. n for each condition is indicated in the figure below the x‐axis. Mann–Whitney and Wilcoxon tests yielded P‐values > 0.4. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
CAssaying sensitivity to somatic dpy‐13 RNAi. The rescuing transgenes shown in (A) are also assayed here. Total number of worms assayed is represented in the figure. Mann–Whitney and Wilcoxon tests were used to test whether penetrance of dpy‐13 RNAi treatment was significantly different between N2 and mutant worms. Single asterisk indicates P‐value = 0.027, while triple asterisks indicate P‐values < 2.3e‐05, P‐values calculated using Mann–Whitney and Wilcoxon tests. Error bars represent the SEM.
-
D–FGFP fluorescence images of worms carrying 22G‐siR‐1 sensor transgenes (see also Fig EV3G). Scale bars represent 0.5 mm. (D) Animals carrying the control transgene with no 22G‐siR‐1 binding site. (E) Strains carrying the 22G‐siR‐1 sensor. (F) GFP signal in the absence of GTSF‐1.
-
GMicrographs of GFP::NRDE‐3 embryos in various genetic backgrounds. Scale bars represent 10 μm.
Figure EV3. gtsf‐1 animals recapitulate the phenotypes of alg‐3/4 and ergo‐1 mutants (related to Fig 3).
-
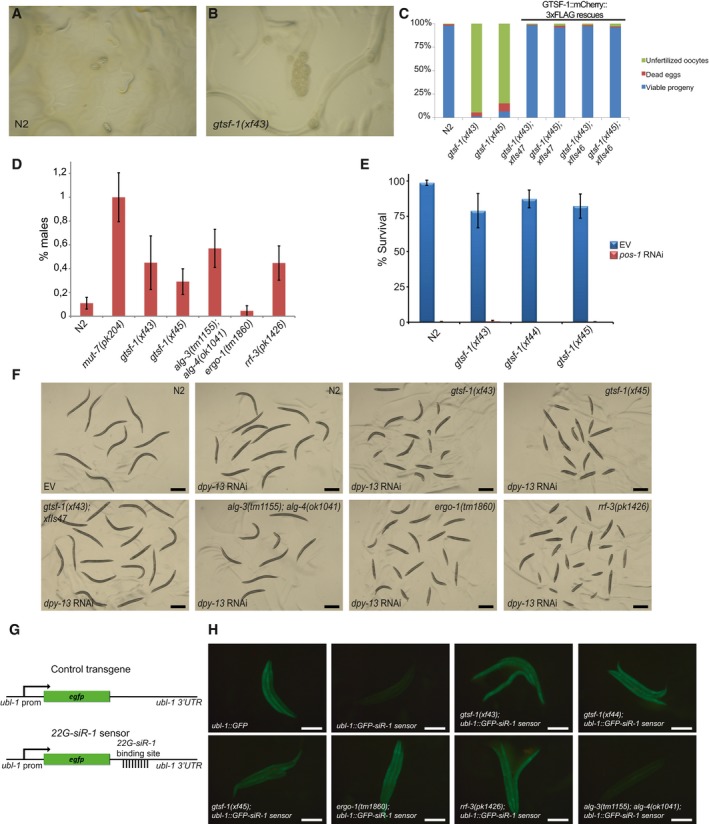
A, BRepresentative pictures of progenies from synchronized parents of wild‐type (A) and gtsf‐1 mutants (B) grown at 25°C. In (A), L1 larvae and gastrulated embryos can be observed, while in (B) only unfertilized oocytes are identified.
-
CPercentage of dead eggs and unfertilized oocytes laid by wild‐type and gtsf‐1 mutant worms when grown at 25°C. These counts were obtained from the same experiment as in Fig 3A. n = 10.
-
D% of males is plotted for the indicated strains. gtsf‐1 mutants show a percentage of males comparable to alg‐3/4 and rrf‐3 mutants, but lower than mut‐7 mutants. Spontaneous male incidence was measured in the progenies of 20 worms. For each plate, percentage of males was calculated and the percentages of the 20 plates were subsequently averaged to obtain the final male percentage value. Error bars represent SEM.
-
E% Survival of animals of the indicated genotypes on empty vector (EV) and pos‐1 RNAi. Per strain, progenies of n = 9–12 worm were scored for embryonic lethality. gtsf‐1 animals respond to pos‐1 RNAi identically to wild type. Error bars represent SD.
-
FThis panel shows a representative fraction of animals from the dpy‐13 RNAi experiment (shown in Fig 3C). Genotypes and treatment with either empty vector (EV) or dpy‐13 RNAi are indicated in the figure. Scale bars represent 0.5 mm.
-
GOverview of the 22G transgenes. The control transgene consists of GFP controlled by the ubl‐1 promoter and 3′UTR sequences, which allow ubiquitous expression. The 22G‐siR‐1 sensor is similar to the control transgene except for a binding site for 22G‐siR‐1 on the 3′UTR that reports on the activity of ERGO‐1‐dependent 22G‐RNAs.
-
HGenetic requirements for 22G‐sensor silencing. Representative fluorescence images of animals carrying 22G‐siR‐1 sensor combined with other mutations. Scale bars represent 0.5 mm.
Temperature‐sensitive sterility and embryonic lethality are recurring phenotypes of factors acting in endogenous sRNA pathways in C. elegans. For example, mutations in mutator genes, Eri genes, rrf‐3, drh‐3, and alg‐3/4, result in temperature‐sensitive sterility at 25°C (Ketting et al, 1999; Duchaine et al, 2006; Gent et al, 2009; Gu et al, 2009; Han et al, 2009; Pavelec et al, 2009; Conine et al, 2010; Billi et al, 2014). In some of those mutants, like alg‐3/4, eri‐1 and rrf‐3, these fertility defects can be rescued by wild‐type sperm, indicative of a sperm defect (Gent et al, 2009; Pavelec et al, 2009; Conine et al, 2010). Upon crossing gtsf‐1 hermaphrodites with wild‐type males, both the reduced brood size at 20°C and the temperature‐sensitive sterility at 25°C were rescued practically to wild‐type levels (Fig 3B). Furthermore, we noticed that gtsf‐1 mutants have a mild high incidence of males (him) phenotype (Fig EV3D), again, similar to alg‐3/4, many Eri and mutator mutants (Ketting et al, 1999; Gent et al, 2009; Conine et al, 2010).
One phenotype that distinguishes mutator mutants from Eri mutants is RNAi‐sensitivity. Mutators are resistant to exogenous RNAi, while Eri mutants are hypersensitive. gtsf‐1 mutants displayed normal sensitivity to RNAi against the germline gene pos‐1 (Fig EV3E, Table 1), but showed hypersensitivity to RNAi targeting somatic genes, as dpy‐13 (Figs 3C and EV3F, Table 1), lir‐1, and pop‐1 (Table 1), similarly to rrf‐3 and ergo‐1 mutant worms (Duchaine et al, 2006; Yigit et al, 2006). In contrast, alg‐3/4 double mutants did not display RNAi hypersensitivity. Two independent, germline‐specifically expressed gtsf‐1 transgenes rescued the RNAi hypersensitivity almost to wild‐type levels (Figs 3C and EV3F). We note that this rescue of a somatic phenotype with a germline‐expressed transgene likely derives from the strong maternal effect of the 26G‐RNA pathway (Zhuang & Hunter, 2011). We conclude that gtsf‐1 mutants have an Eri phenotype.
Table 1.
gtsf‐1 mutants have exogenous RNAi defects
| Germline targets | Somatic targets | |||
|---|---|---|---|---|
| pos‐1 a | dpy‐13 b | lir‐1 c | pop‐1 d | |
| N2 | + | + | − | − |
| gtsf‐1(xf43) | + | +++ | +++ | +++ |
| gtsf‐1(xf44) | + | +++ | +++ | +++ |
| gtsf‐1(xf45) | + | +++ | +++ | +++ |
“−”denotes no response to RNAi; “+” indicates response to RNAi; and “+++” indicates enhanced response to RNAi.
Scored for embryonic lethality.
Scored for strength and penetrance of Dpy phenotype.
Scored for larval arrest.
Scored for bursted and protruded vulva phenotypes.
Loss of ERGO‐1 and RRF‐3, but not ALG‐3/4, derepresses a ubiquitously expressed GFP transgene that reports on the activity of a specific 22G‐RNA (Figs 3D and E, and EV3G and H) that is produced in response to ERGO‐1 (Montgomery et al, 2012). GTSF‐1 is also required for proper silencing of this transgene, indicating that the activity of GTSF‐1 is required for ERGO‐1/RRF‐3‐driven silencing (Figs 3F and EV3H). We further tested GTSF‐1 participation in the ERGO‐1‐dependent 26G‐RNA pathway more broadly, by using a GFP::NRDE‐3 expressing transgene. GFP::NRDE‐3 in wild‐type animals displays nuclear localization, but is cytoplasmic in ergo‐1 mutants because it fails to be loaded with 22G‐RNAs (Guang et al, 2008). Nuclear localization is similarly affected by gtsf‐1(xf43) and rrf‐3(pk1426) mutation (Fig 3G). In contrast, alg‐3/4 mutations did not cause mislocalization of GFP::NRDE‐3 from the nucleus.
Overall, we conclude that gtsf‐1 mutants display phenotypes of alg‐3/4 and ergo‐1 mutants. As such, loss of GTSF‐1 perfectly phenocopies loss of the RdRP enzyme RRF‐3, suggesting that GTSF‐1 acts at a very upstream step in the 26G‐RNA pathway.
26G‐RNA levels are strongly reduced in gtsf‐1 mutants
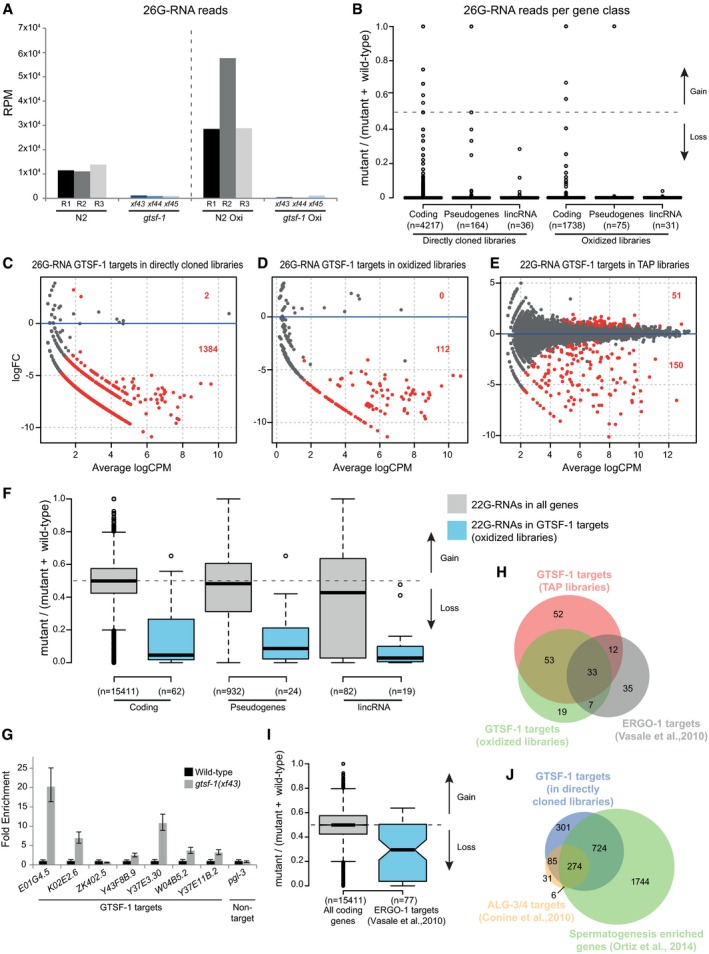
Given our phenotypic analysis, we reasoned that GTSF‐1 may affect 26G‐RNA biogenesis. Indeed, 26G‐RNA levels are severely depleted in gtsf‐1 mutants (Fig 4A). This effect is observed both in the directly cloned and in the oxidized libraries, suggesting that both classes of 26G‐RNAs, unmethylated (ALG‐3/4‐bound) and 2′‐O‐methylated (ERGO‐1‐bound), respectively, are affected by GTSF‐1 (Fig 4A). The levels of 26G‐RNAs derived from all gene classes are similarly reduced upon loss of GTSF‐1 (Fig 4B).
Figure 4. 26G‐RNAs are severely depleted in gtsf‐1 mutants.
-
AGlobal levels of 26G‐RNAs in wild‐type and gtsf‐1 mutant worms, in RPM (reads per million). Three biological replicates are shown, represented as R1–R3 for wild‐type N2 worms. The dashed line separates the levels of 26G‐RNAs in different library treatments: directly cloned libraries on the left, and oxidized libraries on the right.
-
BBoxplot showing enrichment/depletion of normalized 26G‐RNA reads per gene in gtsf‐1 mutants relative to wild type, separated by gene class. All the genes in each class that had 26G‐RNA mapped reads were used for this analysis.
-
C–EIdentification of GTSF‐1 target genes that are significantly depleted of 26G‐ or 22G‐RNA reads in the mutants in comparison with wild type. Separate MA plots are shown for the different library treatments. Statistically significant changes (1% FDR) are highlighted in red, and their number is indicated. LogFC, log2 fold change. LogCPM, log2 counts per million.
-
FBoxplot showing enrichment/depletion of 22G‐RNA reads in gtsf‐1 mutant in TAP libraries, by gene class, using all genes with mapped 22G‐RNAs (gray boxes), and only 22G‐RNAs that map to GTSF‐1 targets (blue boxes), as defined in the oxidized libraries (D). Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
GRT–qPCR of seven GTSF‐1 targets and a non‐target (pgl‐3). Error bars represent the standard deviation of two biological replicates. pmp‐3 was used as the normalizing gene.
-
HVenn diagram showing overlap of targets of the indicated libraries with previously defined ERGO‐1 targets (Vasale et al, 2010).
-
IBoxplot indicating enrichment/depletion of 22G‐RNA levels (from the TAP‐treated libraries) in all coding genes (gray box), and in ERGO‐1 targets as defined by others. We used only 77/87 ERGO‐1 RIP targets from Vasale et al, 2010, since for the remaining 10 targets, we did not have mapped reads. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile. Notches represent the 95% confidence interval for each median.
- J
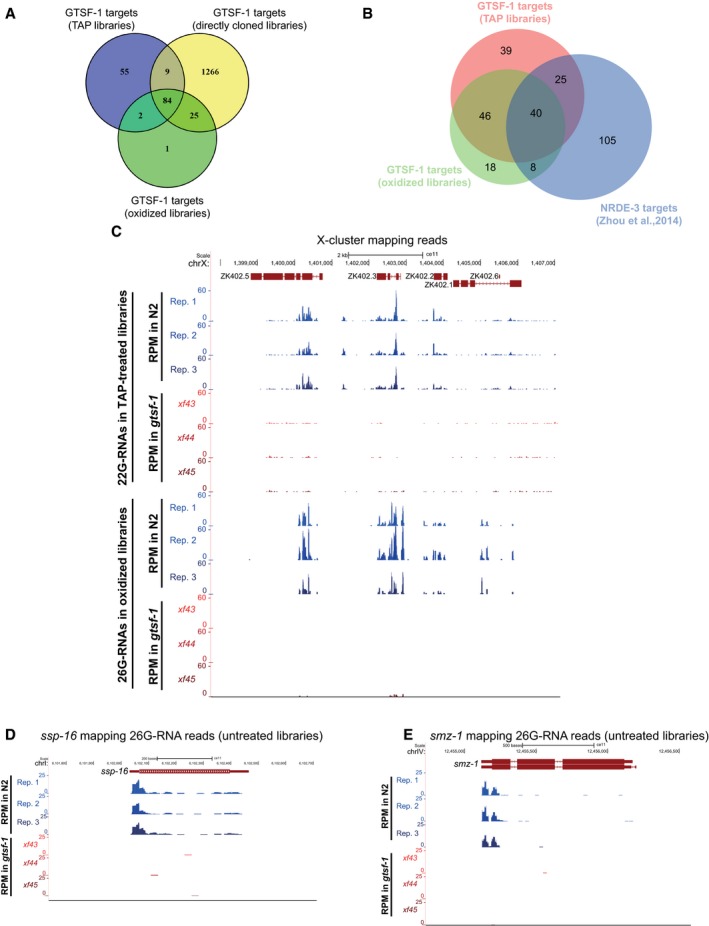
Next, we defined high‐confidence targets (at 1% FDR) of GTSF‐1‐dependent sRNAs for each library treatment (Fig 4C–E, lists of targets in Table EV1). Targets were defined as genes that have a significant depletion of sRNAs in the mutant, in comparison with wild type. The targets defined in the oxidized libraries (enriching for methylated 26G‐RNAs) significantly overlapped with the targets of the TAP‐treated libraries (enriching for 22G‐RNAs, Fig EV4A). These results suggest that genes that lose 2′‐O‐methylated 26G‐RNAs also tend to lose downstream 22G‐RNAs. This tendency is observed for all gene classes (Fig 4F). Next, we wanted to address whether there are changes in GTSF‐1 target gene expression concomitantly with loss of 26G‐/22G‐RNAs. Indeed, in the absence of GTSF‐1, its targets are upregulated as assessed by RT–qPCR (Fig 4G, in levels consistent with previously published RT–qPCR data, see Duchaine et al, 2006; Pavelec et al, 2009; Vasale et al, 2010). Furthermore, our sets of GTSF‐1 targets significantly overlap with a publicly available dataset from an ERGO‐1 RIP (Vasale et al, 2010; Fig 4H). Consistently, genes identified in the ERGO‐1 RIP are depleted of 22G‐RNAs in our TAP library dataset (Fig 4I). Of note, several of the GTSF‐1 targets that were shown to be upregulated in Fig 4G were also identified as ERGO‐1 targets (Vasale et al, 2010; namely E01G4.5, K02E2.6, W04B5.2, and Y37E11B.2). ERGO‐1 targets include paralog genes and pseudogenes (Vasale et al, 2010). Accordingly, we did not find strongly enriched gene ontology terms for the targets defined in the oxidized and TAP‐treated libraries (Table EV1). Furthermore, consistent with a role for GTSF‐1 upstream of NRDE‐3, we observed a significant overlap between GTSF‐1‐dependent sRNA targets and NRDE‐3 targets (Zhou et al, 2014; Fig EV4B).
Figure EV4. 26G‐RNAs are severely depleted in gtsf‐1 mutants (related to Fig 4).
-
AVenn diagram showing overlap between the 1% FDR GTSF‐1 targets defined in the three different NGS library preparation conditions.
-
BVenn diagram depicting the overlap between the GTSF‐1 targets defined in the oxidized and TAP‐treated libraries in this study, and NRDE‐3 targets as defined elsewhere (Zhou et al, 2014).
-
C–EExemplary UCSC genome browser tracks with RPM‐normalized sRNA coverage profiles. Three biological replicates of wild‐type N2 worms (Rep. 1–Rep. 3) and the three gtsf‐1 alleles are shown separately. (C) 22G‐RNAs (above) and 26G‐RNAs (below) mapping to the 22G‐RNA X‐cluster. 22G‐ and 26G‐RNA tracks were obtained from TAP‐treated and oxidized libraries, respectively. (D, E) 26G‐RNA reads mapping to ssp‐16 (D), a sperm‐specific family, class P protein, and to smz‐1 (E), a sperm meiosis PDZ domain‐containing protein. Directly cloned libraries were used for these tracks.
The 1,384 targets defined by the directly cloned libraries (Fig EV4A), extensively overlapped with ALG‐3/4 targets as defined by sRNA sequencing of alg‐3/4 double mutants (Conine et al, 2010; Fig 4J). Consistent with this, functional analysis for these 1,384 GTSF‐1 targets shows enrichment for sperm proteins, kinases, and phosphatases (Table EV1). As expected for ALG‐3/4 targets, these GTSF‐1‐dependent loci extensively overlapped with spermatogenesis‐specific genes as defined by others (Ortiz et al, 2014; Fig 4J, Table EV1).
To illustrate loss of sRNAs in gtsf‐1 mutants, exemplary genome tracks of GTSF‐1 targets are shown in Fig EV4C–E. Also, WormExp gene set enrichment analysis on GTSF‐1 targets retrieved ERGO‐1, ALG‐3/4, and RRF‐3 datasets, amongst many other datasets related to factors belonging to 26G‐ and 22G‐RNA pathways (Table EV1). Altogether, we conclude that both ALG‐3/4‐associated and ERGO‐1‐associated 26G‐RNA populations, as well as the 22G‐RNAs downstream of ERGO‐1, are severely impacted by the loss of GTSF‐1.
GTSF‐1 interacts with RRF‐3
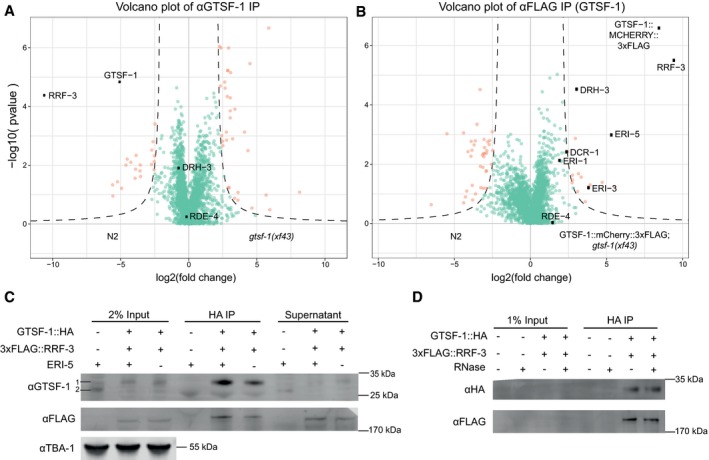
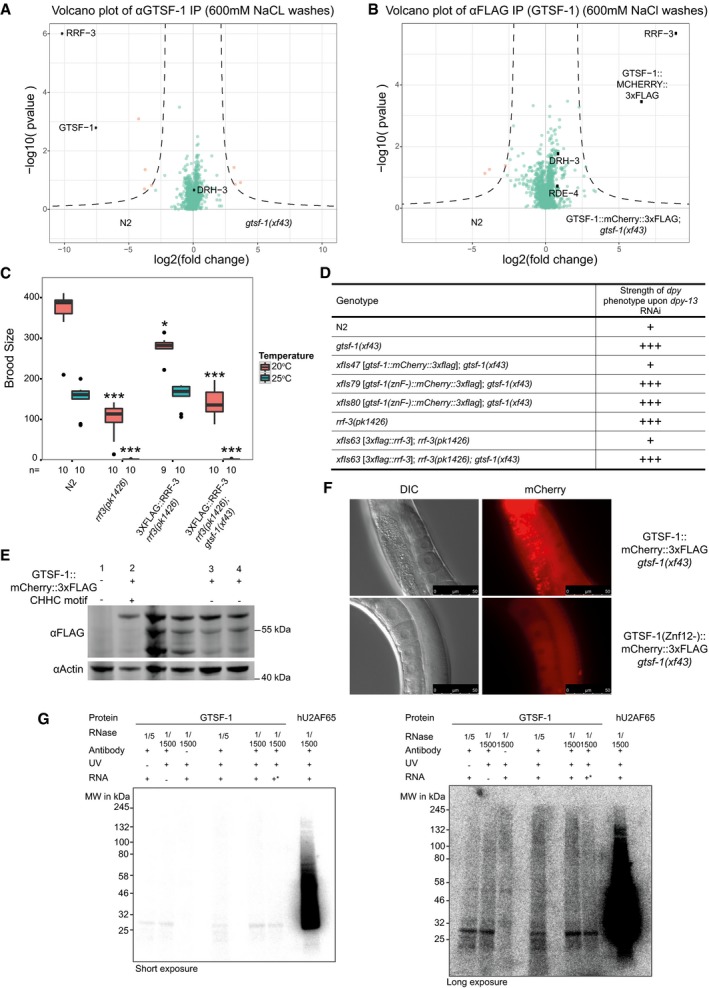
To identify interactors of GTSF‐1, we performed immunoprecipitation (IP) followed by label‐free quantitative proteomics. IPs were performed in quadruplicates, in wild‐type and gtsf‐1 mutant synchronized gravid adults using an anti‐GTSF‐1 antibody. Additionally, using an anti‐FLAG antibody, we immunoprecipitated FLAG‐tagged GTSF‐1 from a strain carrying a rescuing transgene (the same used in Figs 1E and F, and 3A and C), using wild‐type animals as a negative control. In both IP‐mass spectrometry experiments, RRF‐3 was the most enriched interactor (Fig 5A and B). Notably, in the transgene pull‐downs (potentially an overexpression setup, because of the use of the gld‐1 promoter), we also observed slight enrichment of other known cofactors of RRF‐3 in the 26G‐RNA‐producing ERIC (Fig 5B, represented by black dots). These IP experiments were also performed under more stringent wash conditions (600 mM NaCl), in which case only the RRF‐3 interaction was maintained (Fig EV5A and B). We note that previous interactomics studies on Eri factors recovered GTSF‐1 peptides, albeit with very low peptide coverage and without experiments addressing functionality (Duchaine et al, 2006; Thivierge et al, 2012). These observations support our results that GTSF‐1 is associated with RRF‐3 in the context of the ERIC.
Figure 5. GTSF‐1 interacts with RRF‐3 in the adult germline, independently of RNA .
-
A, BVolcano plots representing label‐free proteomic quantification of GTSF‐1 IPs from adult worm extracts. For each strain, IPs were performed and measured in quadruplicates. Log2 fold enrichment of individual proteins in one strain versus another is given on the x‐axis. The y‐axis indicates the Log10‐transformed probability of the observed enrichments (see Materials and Methods for details). Proteins in the background are represented as green dots, while orange dots show enriched proteins. In (A), GTSF‐1 was immunoprecipitated using our polyclonal anti‐GTSF‐1 antibody (in wild‐type and gtsf‐1 mutant worms), while in (B), an anti‐FLAG antibody was used to pull‐down GTSF‐1::mCherry::3xFLAG (in wild‐type and strains carrying the rescuing transgene).
-
CTo test interaction between GTSF‐1 and RRF‐3 in adult worms by Western blot, GTSF‐1::HA was pulled‐down via HA immunoprecipitation. Interaction was also tested in the presence/absence of ERI‐5 by introducing an eri‐5(tm2528) mutation in the background. Multi‐channel secondary antibody detection was performed with an Odyssey CLx apparatus (see Materials and Methods). For the anti‐GTSF‐1, 1 represents GTSF‐1::HA and 2 represents untagged GTSF‐1.
-
DTesting RNA dependency on the interaction between GTSF‐1 and RRF‐3 by adding RNase. Extracts from adult worms were used. Secondary antibody detection was performed with the Odyssey CLx setup.
Figure EV5. GTSF‐1 stably interacts with RRF‐3 via its tandem CHHC zinc fingers (related to Figs 5 and 6).
-
A, BVolcano plots of GTSF‐1 IP‐mass spectrometry experiments in more stringent wash conditions (600 mM NaCl). Gravid adult worms were used. IPs were performed and measured in quadruplicates. Other than the wash conditions, the setup was identical to Fig 5A and B.
-
CBrood size count of a 3xFLAG::RRF‐3 single‐copy transgene. This transgene rescues the fertility defects associated with rrf‐3 mutation almost to a complete extent. Asterisk indicates P‐value = 0.002185; triple asterisk indicates P‐value < 0.0001817. P‐values calculated with Mann–Whitney and Wilcoxon tests, using N2 brood size as a reference. n is indicated in the x‐axis. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
DOverview of dpy‐13 RNAi experiments to worms of the indicated genotypes. A “+” sign indicates that these animals show a normal response to dpy‐13 RNAi, while “+++” animals have an enhanced response to RNAi, having both a more prevalent and stronger dpy‐13 phenotype.
-
EWestern blot analysis of adult animal populations carrying GTSF‐1::mCherry::3xFLAG transgenes. Lane 1, Wild‐type, non‐transgenic worms. Lane 2, WT GTSF‐1 protein. Lanes 3 and 4 represent GTSF‐1 zinc finger mutants, wherein the zinc finger cysteines are mutated to alanines. The middle, non‐labeled lanes are of other GTSF‐1 fusion proteins not discussed in this work.
-
FWide‐field DIC and fluorescence microscopy pictures of worms expressing WT GTSF‐1::mCherry::3xFLAG (above) and zinc finger mutant GTSF‐1::mCherry::3xFLAG (below). The overall localization of mCherry is not dependent on the zinc fingers. Scale bars indicate 50 μm.
-
GIn vitro iCLIP experiment. Purified GTSF‐1 protein was incubated with Caenorhabditis elegans total RNA from wild‐type, UV cross‐linked and immunoprecipitated using the GTSF‐1 antibody used throughout this study. After the IP, the co‐purified RNA was radioactively labeled and the IPs were run on a SDS–PAGE gel followed by membrane transfer. As a positive control, human U2AF65 (hU2AF65) was used (see Sutandy et al, 2018). GTSF‐1 protein does not associate with RNA beyond background levels. Also, there are not differences between the no antibody, the no cross‐link and the no total RNA controls. Same gel is shown, on the left with short exposure and on the right with longer exposure. +* indicates that gtsf‐1(xf43) total RNA was used, not wild‐type. The rationale behind this was that in the gtsf‐1 mutant background, GTSF‐1 targets are upregulated. This was expected to increase the iCLIP signal.
Source data are available online for this figure.
To further characterize this interaction, we produced a single‐copy transgene of 3xFLAG‐tagged RRF‐3. This transgene rescues the Eri phenotype and the fertility defects associated with loss of RRF‐3 (Fig EV5C and D), indicating it recapitulates wild‐type RRF‐3 function. We then used this transgene to validate the GSTF‐1/RRF‐3 interaction via Co‐IP followed by Western blot (Fig 5C). This interaction is not abrogated by RNase A treatment, indicating it is RNA‐independent (Fig 5D).
These data clearly demonstrate that GTSF‐1 interacts robustly with the RdRP enzyme RRF‐3 and not with an Argonaute protein like its fly and mouse orthologs.
The CHHC zinc fingers of GTSF‐1 mediate the interaction with RRF‐3
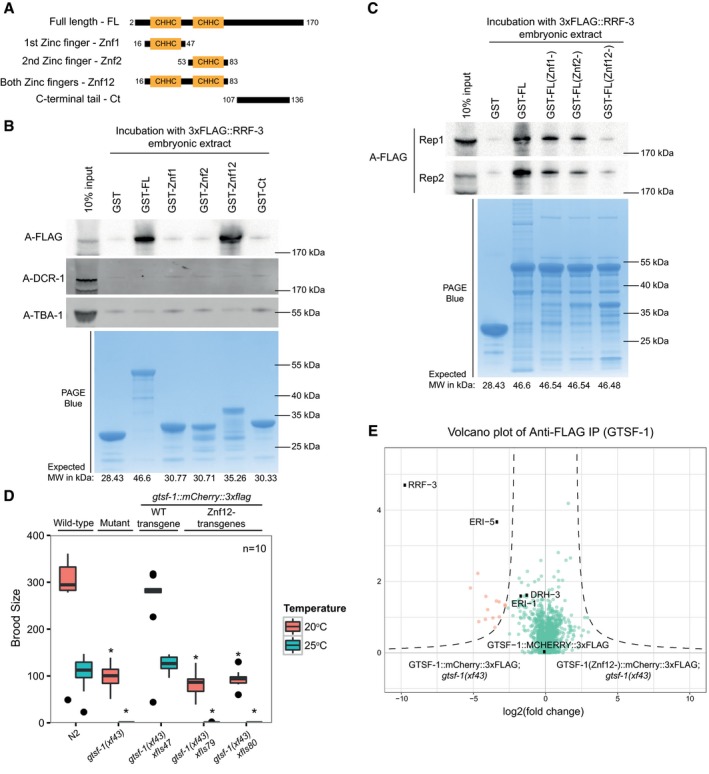
Next, we aimed to pinpoint the determinants of the GTSF‐1/RRF‐3 interaction. For this, we cloned and expressed GST‐fused constructs with different GTSF‐1 fragments (Fig 6A and B). Subsequently, we incubated these GST fusions with embryonic extracts of a 3xFLAG::RRF‐3; gtsf‐1(xf43); rrf‐3(pk1426) strain and pulled‐down GST. Full‐length (FL) GTSF‐1 pulls down 3xFLAG::RRF‐3 (Fig 6B), corroborating the results described above. The GST fusions to the individual zinc fingers and the C‐terminal tail did not pull‐down 3xFLAG::RRF‐3 over background. Interestingly, when both CHHC zinc fingers are fused to GST, 3xFLAG::RRF‐3 can be efficiently retrieved (Fig 6B). None of the fusion proteins interacted with DCR‐1 above background. We also created GST‐GTSF‐1 full‐length proteins with mutated zinc finger residues. Specifically, we mutated the cysteines of the zinc fingers to alanines (see Fig EV1A). Notably, when we mutate the cysteines of individual zinc fingers, the interaction with 3xFLAG::RRF‐3 is slightly disturbed (Fig 6C, see Znf1‐ and Znf2‐), but when all the four cysteines from both zinc fingers are simultaneously mutated, the interaction with 3xFLAG::RRF‐3 is abrogated (Fig 6C, see Znf12‐). These results demonstrate that the zinc fingers of GTSF‐1 are responsible for RRF‐3 binding and suggest that both zinc fingers may act as a unit to mediate RRF‐3 binding.
Figure 6. The tandem CHHC zinc fingers of GTSF‐1 mediate the interaction with RRF‐3.
-
ASchematic representation of GST‐fused GTSF‐1 constructs produced for this study. Amino acid residues bordering the cloned regions are indicated in the figure.
-
B, CWestern blot analysis of GST‐GTSF‐1 pull‐downs. (B) 5 μg of GST‐GTSF‐1 fusion protein (conjugated with Sepharose GSH beads, see lower PAGE Blue panel) was each incubated with approximately 1 mg of total embryonic protein extract from a 3xFLAG::RRF‐3; rrf‐3(pk1426); gtsf‐1(xf43) strain. 3xFLAG::RRF‐3 was detected using ECL, while DCR‐1 and TBA‐1 were detected using the Odyssey CLx apparatus. (C) 5 μg of various GST‐GTSF‐1 fusion proteins (conjugated with Sepharose GSH beads, see lower PAGE Blue panel), with the indicated cysteine to alanine mutations in the zinc fingers (1st Zinc finger mutated – Znf1‐; 2nd zinc finger mutated – Znf2‐; and both zinc fingers mutated ‐ Znf12‐), were each incubated with approximately 0.5 mg of total embryonic protein extract from a 3xFLAG::RRF‐3; rrf‐3(pk1426); gtsf‐1(xf43) strain. 3xFLAG::RRF‐3 pull‐down is shown for two independent biological replicates.
-
DBrood size assay at 20 and 25°C. The progenies of worms of the indicated genotype are plotted. n = 10 for every strain. Asterisks indicate P‐value < 0.0037 as assessed by Mann–Whitney and Wilcoxon tests comparing wild‐type worms with the other strains. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
EVolcano plots showing label‐free protein quantification of GTSF‐1::mCherry::3xFLAG pull‐downs. Pull‐downs were performed in quadruplicate with adult worm extract. Wild‐type GTSF‐1 fusion proteins are compared with GTSF‐1 fusion proteins with zinc finger mutations (Znf12‐). Proteins in the background are represented as green dots, while orange dots show enriched proteins.
To address the in vivo relevance of the GTSF‐1/RRF‐3 interaction, we produced single‐copy transgenes expressing GTSF‐1 with mCherry and 3xFLAG tags, in which the CHHC cysteines in GTSF‐1 were mutated to alanines [henceforth indicated as gtsf‐1(Znf12‐), see Fig EV1A]. Two independent gtsf‐1(znf12‐) transgene insertions do not rescue the Eri phenotype nor the fertility defects associated with GTSF‐1, thereby phenocopying gtsf‐1 mutants (Figs 6D and EV5D). The lack of rescue is not due to poor expression of the (Znf12‐) transgenes in the germline, although some degradation is observed (Fig EV5E). Such partial degradation might be triggered by the disruption of the structural role that the zinc fingers have in GTSF‐1. Moreover, subcellular localization of GTSF‐1 is not affected by the zinc finger mutations (Fig EV5F). FLAG pull‐down followed by quantitative proteomics revealed that the GTSF‐1(Znf12‐) protein does not stably interact with RRF‐3 (Fig 6E).
In the literature, several examples can be found of zinc fingers mediating both protein–protein and protein–nucleic acid interactions (Gamsjaeger et al, 2007). To address whether GTSF‐1 is interacting with RNA, we performed in vitro iCLIP (Sutandy et al, 2018). We sought for a holistic approach, so we used C. elegans total RNA (rRNA‐depleted) to test the binding of GTSF‐1. Surprisingly, GTSF‐1 was found not to cross‐link with RNA above background levels (Fig EV5G).
We conclude that GTSF‐1 interacts with RRF‐3 via its two tandem CHHC zinc fingers in vitro and in vivo. Since the GTSF‐1/RRF‐3 interaction is stable in presence of RNase (Fig 5D), and GTSF‐1 does not seem to interact with RNA (Fig EV5G), this suggests that the two CHHC zinc fingers in GTSF‐1 act strictly as a protein–protein interaction domain.
GTSF‐1 is both in a precursor complex that is required for ERIC assembly, and in the mature ERIC
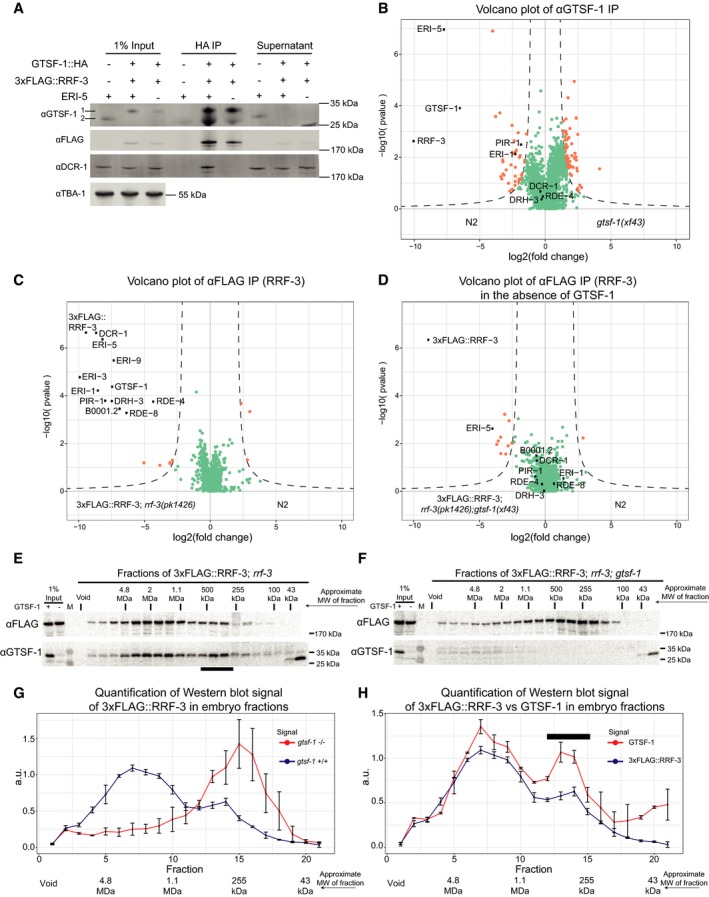
Previous studies on ERIC mostly focused on embryos (Duchaine et al, 2006; Thivierge et al, 2012). Hence, we used embryonic extracts to probe the effect of GTSF‐1 on ERIC. As in the adult germline, HA‐tagged GTSF‐1 pulls down 3xFLAG‐tagged RRF‐3 in embryos, as visualized by Western blot (Fig 7A). To circumvent potential overexpression (brought about by the transgene gld‐1 promoter), and to probe GTSF‐1 interactions more broadly, we immunoprecipitated endogenous GTSF‐1 and analyzed the precipitate with label‐free quantitative mass spectrometry (Fig 7B). In this experiment, we observed a strong enrichment for RRF‐3 and ERI‐5, while all other known components of ERIC are either only mildly enriched, or not enriched at all, contrasting with the previously published molecular niche of RRF‐3 in embryos (Duchaine et al, 2006; Thivierge et al, 2012).
Figure 7. GTSF‐1 is required for ERIC assembly.
-
AProbing the interaction between GTSF‐1 and RRF‐3 by Western blot analysis, in embryonic extracts. GTSF‐1::HA was pulled‐down via HA immunoprecipitation. Interaction was also tested in the presence/absence of ERI‐5 by introducing an eri‐5(tm2528) mutation in the background. Multi‐channel secondary antibody detection was performed with an Odyssey CLx apparatus (see Materials and Methods). For the anti‐GTSF‐1, 1 represents GTSF‐1::HA and 2 represents untagged GTSF‐1.
-
BLabel‐free quantification of GTSF‐1 IPs in embryos (comparing wild‐type and gtsf‐1 mutant worms). IPs were done in quadruplicates, and a polyclonal anti‐GTSF‐1 antibody was used.
-
C, DVolcano plots depicting quantitative proteomic analysis of RRF‐3 pull‐downs in the presence (C) and absence (D) of GTSF‐1, in embryos. IPs were performed in quadruplicates. Proteins in the background are represented as green dots, while orange dots show enriched proteins.
-
E, FSize‐exclusion chromatography of 3xFLAG::RRF‐3‐containing embryo extracts. Fractions were collected and probed for GTSF‐1 and 3xFLAG::RRF‐3. Approximate molecular weight (MW) of the fractions is indicated. The calculation of these values according to protein standards is shown in the Appendix. Fractions collected from extracts with GTSF‐1 are shown in (E), and without GTSF‐1 are shown in (F).
-
G, HComparison of size‐exclusion chromatography profiles of 3xFLAG::RRF‐3 and GTSF‐1. Relative quantification was performed with the Western blot signal using ImageJ. Error bars represent standard deviation of two biological replicates. A.u., arbitrary units. (G) Comparison of profiles of 3xFLAG::RRF‐3 in the presence of GTSF‐1 (blue line) and in the absence of GTSF‐1 (red line). (H) Comparison of profiles of 3xFLAG::RRF‐3 (blue line) and GTSF‐1 (red line).
In order to test whether we can detect ERIC in our experimental setup, we performed IP‐mass spectrometry on 3xFLAG::RRF‐3 from embryo extracts. This experiment clearly identified all known ERIC components (Fig 7C, black dots). In addition to the known ERIC components, we also found RDE‐8 to strongly co‐IP with RRF‐3 under these conditions. RDE‐8 and ERI‐9, another previously identified ERIC factor (Thivierge et al, 2012), are paralog endonucleases that have been implicated in the 26G‐RNA pathway (Gent et al, 2010; Tsai et al, 2015).
Given that the 3xFLAG::RRF‐3 IP results in the identification of ERIC in its entirety (Fig 7C), while the GTSF‐1 IP retrieves only RRF‐3 and ERI‐5, we hypothesized that GTSF‐1 binds non‐ERIC‐bound RRF‐3. Is this non‐ERIC‐bound pool of RRF‐3 perhaps a precursor complex that is required for ERIC formation? To test this, we performed a 3xFLAG::RRF‐3 IP in a gtsf‐1 mutant background and again detected RRF‐3 interactors through label‐free quantitative mass spectrometry. Strikingly, in absence of GTSF‐1, ERIC components no longer co‐IP with RRF‐3 (Fig 7D), with the sole exception of ERI‐5. We then tested whether ERI‐5 is required for interaction between GTSF‐1 and RRF‐3 and found that GTSF‐1::HA can still pull‐down 3xFLAG::RRF‐3 in eri‐5 mutants (Figs 5C and 7A). Interestingly, we noticed that in the absence of ERI‐5, both GTSF‐1 and RRF‐3 are partially destabilized in embryonic extracts (Fig EV6A), while 3xFLAG::RRF‐3 is not destabilized in the absence of GTSF‐1 (Fig EV6B). These results suggest that (i) GTSF‐1 is required to form mature ERIC from a RRF‐3/ERI‐5 precursor complex, where ERI‐5 stabilizes RRF‐3; (ii) that GTSF‐1 does not require ERI‐5 to bind to RRF‐3; and (iii) ERI‐5 does not require GTSF‐1 to bind RRF‐3.
Figure EV6. Stability of ERIC factors in different backgrounds and pre‐ERIC/ERIC profiles in young adults (related to Figs 5 and 7).
-
AWestern blot relative quantification of 3xFLAG::RRF‐3 and GTSF‐1::HA with and without ERI‐5. Absence of ERI‐5 (−) indicates the presence of eri‐5(tm2528) deletion allele. Embryonic extracts were used. Double transgenic, wild‐type ERI‐5 strain was used as the reference. Two technical replicates are shown. For detection, secondary antibodies compatible with LI‐COR Odyssey CLx were used. Quantification was done by gating the region surrounding the bands using Image Studio software (v3.1). M, molecular weight marker; TR, technical replicate. For the anti‐GTSF‐1, 1 represents GTSF‐1::HA and 2 represents untagged GTSF‐1.
-
BRelative quantification of Western blot as in (A), but to address the stability of 3xFLAG::RRF‐3 and DCR‐1 in the absence of GTSF‐1 (which indicates the presence of gtsf‐1(xf43)). Embryonic extracts were used. BR, biological replicate; TR, technical replicate. Asterisk (*) indicates unspecific signal.
-
C, DSize‐exclusion chromatography of 3xFLAG::RRF‐3‐containing young adult extracts. Fractions were collected, and Western blot was performed for GTSF‐1 and 3xFLAG::RRF‐3. Fractions collected from extracts with GTSF‐1 are shown in (C), and without GTSF‐1 are shown in (D). Approximate molecular weight (MW) of the fractions is indicated in the figure. The calculation of these values according to protein standards is shown in the Appendix.
-
E, FComparison of size‐exclusion chromatography profiles of 3xFLAG::RRF‐3 and GTSF‐1 in young adult extracts. Relative quantification was performed with the Western blot signal, using ImageJ. Error bars represent standard deviation of two biological replicates, with the exception of 3xFLAG::RRF‐3 profile in the absence of GTSF‐1 (E), red line. A.u., arbitrary units. (E) Comparison of profiles of 3xFLAG::RRF‐3 in the presence of GTSF‐1 (blue line) and in the absence of GTSF‐1 (red line). (F) Comparison of profiles of 3xFLAG::RRF‐3 (blue line) and GTSF‐1 (red line).
Source data are available online for this figure.
To further test the idea that GTSF‐1 is required to incorporate RRF‐3 into ERIC, we performed size‐exclusion chromatography with 3xFLAG::RRF‐3‐containing embryonic extracts, followed by Western blot for GTSF‐1 and FLAG. In wild‐type embryos, 3xFLAG::RRF‐3 displays a bimodal elution pattern. The main pool elutes in a broad range between 1 and 4 MDa, while a smaller fraction elutes at roughly 300–400 kDa (Fig 7E and G). In absence of GTSF‐1, 3xFLAG::RRF‐3 displays a single peak at roughly 250 kDa (Fig 7F and G), consistent with RRF‐3 (estimated molecular weight 201.4 kDa) bound to ERI‐5 (61.6 and 18.6 kDa are the predicted molecular weights for ERI‐5 isoform A and B, respectively). These data support the hypothesis that GTSF‐1 is required to incorporate an RRF‐3/ERI‐5 pre‐complex into ERIC, via an RRF‐3/ERI‐5/GTSF‐1 intermediate.
GTSF‐1 and RRF‐3 show very similar elution patterns (Fig 7E and H). This indicates that GTSF‐1 remains within ERIC, at least for some time after its assembly. Results that we obtained by size‐exclusion chromatography on young adult extracts are consistent with the embryo data: In young adults, we also find that 3xFLAG::RRF‐3 and GTSF‐1 display bimodal elution profiles (Fig EV6C–F), with GTSF‐1 again being essential to form ERIC (Fig EV6D and E). Strikingly, both 3xFLAG::RRF‐3 and GTSF‐1 show a more pronounced pre‐ERIC peak when compared to embryos (compare Fig EV6C and F with Fig 7E and H), suggesting ERIC assembly may be less active in the germline. Finally, both in embryos and in adults the ratio of pre‐ERIC:ERIC is consistently higher for GTSF‐1 than for RRF‐3 (Figs 7H and EV6F). This may indicate that GTSF‐1 can dissociate from mature ERIC to form novel pre‐ERIC complexes.
Taken together, these data show that GTSF‐1 alternates between two states: one associated with the mature ERIC and another associated with an RRF‐3‐ and ERI‐5‐containing pre‐ERIC complex. Also, and most importantly, this pre‐complex is required to form a functional ERIC, competent for driving 26G‐RNA biogenesis.
Discussion
Here, we show that GTSF‐1 does not participate in transposon silencing via the piRNA pathway in C. elegans, unlike GTSF‐1 orthologs in flies and mice. However, like its orthologs, GTSF‐1 is required for normal fertility. Surprisingly, GTSF‐1 promotes 26G‐RNA biogenesis by incorporating the 26G‐RNA generating enzyme RRF‐3 into a larger complex known as ERIC. GTSF‐1 thus provides an enticing example of a conserved protein that achieves its function in sRNA pathways via different cofactors in different species, i.e., Argonaute proteins versus RdRP enzymes. Nevertheless, we propose that the function ascribed to C. elegans GTSF‐1, of enabling the assembly of larger protein complexes from smaller subunits, may be evolutionarily conserved.
The double CHHC zinc finger as a protein–protein interaction module
Typically, zinc fingers are known to mediate interactions with nucleic acid. Nevertheless, several cases were described in which zinc fingers mediate protein–protein interactions (Gamsjaeger et al, 2007). In some of these cases, zinc fingers of one protein interact directly with the zinc fingers of another protein (e.g., like GATA‐1 and FOG; Gamsjaeger et al, 2007).
We found that GTSF‐1 interacts with RRF‐3 via its tandem CHHC zinc fingers in vitro and in vivo (Fig 6). Interestingly, the zinc fingers individually could not interact with RRF‐3 (Fig 6B). This suggests the two zinc fingers may function as one structural unit. Mutation of the cysteines of individual zinc fingers reduced but did not completely eliminate the interaction with RRF‐3 (Fig 6C, see GST‐GTSF‐1 znf1‐ and znf2‐). This could point at a certain structural robustness that allows one mutated zinc finger to fold relatively well when adjacent to a wild‐type zinc finger. Of note, GTSF‐1(znf12‐) transgenes could not rescue gtsf‐1 mutant defects (Figs 6D and EV5D), clearly showing that interaction with RRF‐3 via its zinc fingers is key for GTSF‐1 function in vivo.
These results differ from Piwi–Gtsf1 interaction data from Drosophila and mouse, in that the C‐terminal tail (also referred to as “central region”) of Gtsf1 was shown to interact with Piwi, and MIWI2 and MILI, respectively (Dönertas et al, 2013; Yoshimura et al, 2018). Also, GTSF‐1 zinc finger mutants were still found to interact with Piwi in cell culture (Ohtani et al, 2013). We note, however, that (i) the zinc fingers of DmGTSF1 were not tested directly for interaction with Piwi; (ii) the four cysteines of both zinc fingers were not simultaneously mutated, unlike our setup (Fig 6); and (iii) consistent with our observations, zinc finger mutations are required for DmGtsf1 function, as assessed by transposon derepression (Dönertas et al, 2013; Ohtani et al, 2013). Of note, the function of Gtsf1 paralogs in flies has not yet been determined. It may be that these paralog CHHC zinc finger proteins may interact with other proteins via their zinc fingers and thus have a more similar role to CeGTSF‐1 in sRNA biology.
For Gtsf1l and Gtsf2, Gtsf1 paralogs in mouse, interaction with Piwi proteins and piRNA pathway cofactors was shown to be complex (Takemoto et al, 2016). For Gtsf1l, the double CHHC zinc fingers were shown, by in vitro GST pull‐downs, to mediate interaction with MIWI and TDRD1. Interaction with MILI seems to be mediated by the “central region” encompassing the conserved acidic residues (Fig EV1A). Conversely, GTSF2 interacts with MILI and TDRD1 via its CHHC zinc fingers, while it interacts with MIWI via its “central region”. So, like CeGTSF‐1, the CHHC zinc fingers of these Gtsf1 paralogs also mediate protein–protein interactions, although the relevance of these interactions has not been demonstrated in vivo.
It seems that the CHHC zinc fingers present in GTSF proteins are not necessarily interacting with RNA, as was assumed after RNA interaction was determined for the single CHHC zinc finger of U11‐48K proteins (Andreeva & Tidow, 2008; Tidow et al, 2009). Interestingly, GTSF proteins are the only CHHC‐containing protein family that has CHHC zinc fingers in tandem. It may be that this particular feature brought about structural possibilities that facilitate specific protein–protein interactions. We hypothesize that the tandem CHHC zinc fingers of GTSF1 homologs may generally function as one structural unit, with different structural characteristics than the individual U11‐48K type CHHC zinc finger.
A parallel between GTSF‐1 in animals and Stc1 in fission yeast
In Schizosaccharomyces pombe, Stc1 is a protein that is required for sRNA‐mediated centromeric heterochromatin formation (Bayne et al, 2010). More concretely, Stc1 bridges the Ago1 RNA‐induced transcriptional silencing complex to the Clr4 methyltransferase complex. Although not phylogenetically related to GTSF‐1 homologs, Stc1 has astonishingly similar structural features. It has an N‐terminal LIM domain (which consists of two tandem zinc fingers) and a very acidic, unstructured C‐terminal domain, much like GTSF‐1 (Fig EV1A). Structure–function studies indicated that the tandem zinc fingers of Stc1 mediate a direct interaction with Ago1, while its C‐terminal tail interacts with Clr4 (He et al, 2013). These modular protein–protein interactions nicely illustrate the bridging functions of Stc1.
As discussed above, it is possible that Gtsf1 proteins, including CeGTSF‐1, may possess multiple interaction surfaces with which they may be able to bring different complexes into close contact. In a similar fashion to Stc1, C. elegans GTSF‐1 may bridge RRF‐3 and the rest of the ERIC. This would imply that the C‐terminal tail of GTSF‐1 would interact with another ERIC factor. We performed mass spectrometry of GST pull‐downs of fusion constructs containing the C‐terminal tail of GTSF‐1. However, these experiments did not enrich for any ERIC factor, nor for any other plausible candidates (M.V. Almeida and S. Dietz, unpublished observations). It may be that this interaction is too transient to be detected in our experiments. The in vitro interaction studies of Gtsf1 proteins in mouse, described above, would also lend support to such a bridging function of GTSF1 in animals, i.e., reciprocally bridging MILI and MIWI complexes undergoing the ping‐pong cycle. Also in flies, GTSF1 might function to couple Piwi to downstream effector proteins such as Panoramix (Sienski et al, 2015; Yu et al, 2015). Possibly, this would need to be tested in specific developmental stages, since Piwi activity in flies was proposed to be primarily active in embryos (Akkouche et al, 2017).
GTSF1 homologs and Stc1 are not the sole examples of tandem zinc finger proteins with roles in sRNA pathways. A family of LIM domain‐containing proteins in mammals was implicated in miRNA‐mediated gene silencing (James et al, 2010). These LIM domain proteins, LIMD1, Ajuba, and WTIP, were found to bridge Ago1/2 with other factors, like eIF4E, in the molecular surroundings of the 5′ Cap structure. This mode of action will ultimately lead to translation inhibition of Ago1/2 targets (James et al, 2010). A more recent study has determined that the LIM domains of LIMD1 are the interaction surface with TNRC6A (Bridge et al, 2017). Moreover, LIMD1 bridges AGO2 to TNRC6A/miRISC (Bridge et al, 2017).
Altogether, it seems likely that small proteins with these structural modules, tandem zinc fingers, and unstructured C‐terminal domains, have convergently evolved as versatile bridges between different protein complexes with roles in sRNA pathways.
How is the ERIC recruited to target RNA?
It is still unknown how ERIC is brought to, or assembled on target mRNA. How are the targets defined in the first place, and which ERIC component binds the RNA? To answer these questions, efforts should be made to identify the RNA‐binding protein(s) involved in the recruitment of the mRNA. This could provide nice insights into the interplay between pre‐ERIC (GTSF‐1/ERI‐5/RRF‐3), ERIC, and target mRNA.
We cannot fully exclude that the zinc fingers of GTSF‐1, either together as a unit or individually, interact to some extent with RNA. However, the interaction with RRF‐3 is not dependent on RNA (Fig 5D), and in vitro cross‐linking experiments failed to show significant GTSF‐1 association with RNA above background (Fig EV5G). Hence, we believe GTSF‐1 is unlikely to be responsible for RNA interaction during ERIC assembly.
Our FLAG::RRF‐3 pull‐downs in embryos faithfully retrieved all known ERIC factors identified previously in other proteomics studies (Duchaine et al, 2006; Thivierge et al, 2012). Interestingly, we also retrieved one new RRF‐3‐interacting factor, RDE‐8. This factor is a paralog of ERI‐9 (Pavelec et al, 2009; Gent et al, 2010; Tsai et al, 2015), which was previously shown to interact with other ERIC factors (Thivierge et al, 2012). RDE‐8 and ERI‐9 are NYN ribonucleases and have been previously shown to be involved in RNAi processes, including 26G‐RNA biogenesis (Pavelec et al, 2009; Gent et al, 2010; Tsai et al, 2015). Their roles in 26G‐RNA biogenesis seem to be independent of nucleic acid cleavage since: (i) ERI‐9 lacks the conserved catalytic residues required for nucleic acid cleavage, and (ii) RDE‐8 transgenes with mutated catalytic residues still accumulate 26G‐RNAs. Thus, it was proposed that RDE‐8 and ERI‐9 may have a structural role within the ERIC (Tsai et al, 2015). Alternatively, an attractive hypothesis is that RDE‐8 and/or ERI‐9 may be responsible for target mRNA recognition, or would play a role in stabilizing ERIC on its target RNA.
What is the exact molecular function of GTSF‐1?
We propose a model in which GTSF‐1 and ERI‐5 independently associate with RRF‐3 to form a pre‐ERIC (Fig 8). This pre‐ERIC is required to build a functional ERIC that drives 26G‐RNA biogenesis. This process seems to be developmentally regulated, since in the young adult germline there seems to be proportionally more GTSF‐1/RRF‐3 complex than in embryos (compare Fig 7E and H with Fig EV6C and F). This means that this pre‐complex may be “packaged” in the young adult germline to promptly initiate 26G‐RNA biogenesis during embryonic development. Also, within the pre‐ERIC, GTSF‐1 and ERI‐5 seem to have diverging roles. Both ERI‐5 and GTSF‐1 are required for building ERIC, but while ERI‐5 seems to be required for the stability of GTSF‐1 and RRF‐3, GTSF‐1 does not seem to be required for the stability of RRF‐3. We would like to point out that it is unclear why we do not observe all ERIC components in GTSF‐1 IP‐MS in embryos (Fig 7B), given that GTSF‐1 does co‐fractionate with mature ERIC (Fig 7E and H). There may be several reasons for this. For example, it may be that the epitope of GTSF‐1 is inaccessible within ERIC in embryos, or that GTSF‐1 more easily dissociates from mature ERIC than from pre‐ERIC. We do observe some enrichment of PIR‐1 and ERI‐1 in GTSF‐1 IP‐MS in embryos, suggesting the latter scenario may indeed apply.
Figure 8. A model for the function of GTSF‐1.
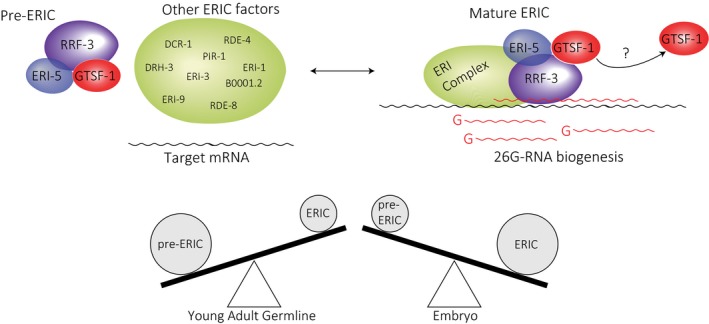
- GTSF‐1 forms a pre‐ERI complex together with RRF‐3 and ERI‐5. GTSF‐1 and ERI‐5 are both required to incorporate RRF‐3 into ERIC. Upon deposition of RRF‐3 in ERIC, GTSF‐1 may dissociate. It is unclear how stable is the association between GTSF‐1 and the mature complex. Also, this process of ERIC assembly seems to be developmentally regulated. See Discussion for details.
Then, how does GTSF‐1 exactly achieve its role? We consider a number of possibilities that are not mutually exclusive. First, GTSF‐1 may be influencing the subcellular localization of RRF‐3. Second, GTSF‐1 may be chaperoning RRF‐3 in a way that prompts conformational changes allowing RRF‐3 to interact with other proteins. Third, GTSF‐1 may allow RRF‐3 to interact with target mRNA, which in turn may trigger ERIC assembly. In order to address these issues and fully understand how RRF‐3 works, we will need to develop biochemical assays for ERIC assembly and function with purified components. Such a system would shed light on the questions stated above and other unresolved mechanistic details, for example, whether GTSF‐1 remains in the mature complex and how specific target mRNAs are selected.
Materials and Methods
Caenorhabditis elegans genetics and culture
Caenorhabditis elegans was cultured on OP50 bacteria according to standard laboratory conditions (Brenner, 1974). Unless otherwise noted, worms were grown at 20°C. The Bristol strain N2 was used as the standard wild‐type strain. The strains used and created in this study are listed in the Appendix.
Creation of gtsf‐1 mutants using CRISPR‐Cas9 technology
gtsf‐1 mutant alleles were produced as described (Friedland et al, 2013). We successfully targeted the following sequencing on the third exon of gtsf‐1: (GGAGCCGCTGGAGCTGAACG). Two other targeted sequences, cloned into p46169 in an identical fashion, did not yield any mutants, either alone or in combination (GATAACATGCCCTTACAATT and GACGTCGGAAATCGAGAAAT).
N2 worms were injected with 150 ng/μl of Cas9 construct p46168 (a gift from John Calarco, Friedland et al, 2013), 135 ng/μl of sgRNA construct pRK1134, and 15 ng/μl of co‐injection marker pCFJ104 (Pmyo‐3:mCherry:unc‐54 3′UTR, expresses mCherry in body wall muscle). F1 worms positive for mCherry expression in body wall muscle were isolated, allowed to self, and then lysed in single worm lysis buffer (5 mM KCl, 2.5 mM MgCl2, 10 mM Tris–HCl pH = 8.3, 0.45% NP‐40, 0.45% Tween‐20, and 0.01% gelatin). Subsequently, genotyping was performed with Taq Polymerase according to manufacturer's instructions (New England BioLabs, M0273X). After isolation, gtsf‐1 mutant worms were outcrossed five times.
Small RNA library preparation, sequencing, and bioinformatics analysis
Detailed procedure for RNA isolation, small RNA enrichment, library preparation, bioinformatics analysis, and sequencing statistics can be found in the Appendix.
Antibodies
Custom, affinity‐purified rabbit anti‐GTSF‐1 antibodies were ordered from SDIX. The following protein sequence, comprising the last 91 amino acid residues of GTSF‐1 (positions 79–169), was used as an antigen (KRQSADLRRQLSLEPLELNVAEHLAAQKLRKEYEKDEESLDGSDDSDEDEEEKNLSVTSEIEKSDVEEVEMMLETINRLAYLEMKNDNLIL). The antibody (animal number Q5963) was used in a 1:500 dilution on Western blots, and 2 μg was used on Immunoprecipitations. 2 μg of Anti‐FLAG antibody (M2 clone, Sigma‐Aldrich, F3165) was used on immunoprecipitations, and a 1:5,000 dilution was used for Western blot. DCR‐1 antibody was a kind gift from Thomas Duchaine, and it was used in Western blots in dilutions ranging from 1–3,000 to 1–5,000. More information on this antibody can be found elsewhere (Duchaine et al, 2006; Thivierge et al, 2012). A commercially available, mouse anti‐tubulin monoclonal antibody (clone B‐5‐1‐2, Sigma‐Aldrich, T6074) was used in Western blots in a 1:10,000 dilution to detect C. elegans TBA‐1 as a loading control. A commercially available, rabbit anti‐actin polyclonal antibody (Sigma‐Aldrich, A5060) was used in Western blots in a 1:1,000 dilution. 30 μl of suspension of EZview™ Red Anti‐HA (mouse monoclonal antibody, clone HA‐7) Affinity Gel (Sigma, E6779) was used for HA IPs. A mouse monoclonal Anti‐HA antibody (clone HA‐7, Sigma, H3663) was used in Western blots with dilutions ranging from 1–500 to 1–1,000.
Mass spectrometry
Details on worm sample collection and immunoprecipitation can be found in detail in the Appendix. Immunoprecipitates were resuspended in NuPAGE LDS Sample Buffer 1× (Life technologies, NP0007) and 0.1 M DTT and heated at 70°C for 10 min. The respective samples were separated on a 4–12% gradient Bis‐Tris gel (NuPAGE Bis‐Tris gels, 1.0 mm, 10 well, NP0321; Life Technologies) in 1× MOPS (NuPAGE 20× MOPS SDS running buffer, NP0001; Life Technologies) at 180 V for 10 min, afterward separately processed by in‐gel digest (Shevchenko et al, 2007; Kappei et al, 2013) and desalted using a C18 StageTip (Rappsilber et al, 2007).
The digested peptides were separated on a 25‐cm reverse‐phase capillary (75 μM inner diameter) packed with Reprosil C18 material (Dr. Maisch). Elution of the peptides was done along a 2 h gradient from 2 to 40% Buffer B (see Stage tip purification) with the EASY‐nLC 1,000 system (Thermo Scientific). Measurement was done on a Q Exactive Plus mass spectrometer (Thermo Scientific) operated with a Top10 data‐dependent MS/MS acquisition method per full scan (Bluhm et al, 2016). The measurements were processed with the MaxQuant software, version 1.5.2.8 (Cox & Mann, 2008) against the Uniprot C. elegans database (version of May, 2016) for quantitation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD007665.
Data availability
Sequencing data have been deposited to the NCBI Gene Expression Omnibus (GEO), and proteomics data are available at the ProteomeXchange Consortium via PRIDE. GEO: GSE103432, PRIDE: PXD007665. Source data are available at https://data.mendeley.com/datasets/xyxh6hv3mc/draft?a=03bd1ca8-dd09-44a2-a0a1-3910ad5201c9.
Author contributions
Conceptualization: MVA and RFK; Investigation: MVA, SD, SR, AH, CR; Formal Analysis: MVA, SD, SR, EK, AH; Visualization: MVA, SD, SR, EK, AH; Writing—Original Draft: MVA and RFK; Writing—Review & Editing: all authors contributed; Funding acquisition: HDU, JK, FB, RFK.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Acknowledgements
We thank all the members of the Ketting laboratory and Hans‐Peter Wollscheid for great help and discussion. We thank Yasmin el Sherif, Svenja Hellmann, and Isabel Pötzch for technical assistance. We acknowledge Chung‐Ting Han and Hanna Lukas of the IMB Genomics core facility, the IMB Microscopy core facility, and the IMB Media Lab. We are grateful to Thomas Duchaine and Ahilya Sawh for kindly providing the DCR‐1 antibody and the eri‐5(tm2528) strain. The authors thank Colin Conine and Craig C. Mello for providing the GFP::ALG‐3 strain. Also, the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), is acknowledged for providing worm strains. This work was supported by a Deutsche Forschungsgemeinschaft grant KE 1888/1‐1 (Project Funding Programme to R.F.K.) and a ERC‐AdG 323179 (to H.D.U.).
The EMBO Journal (2018) 37: e99325
References
- Akkouche A, Mugat B, Barckmann B, Varela‐Chavez C, Li B, Raffel R, Pélisson A, Chambeyron S (2017) Piwi is required during Drosophila embryogenesis to license dual‐strand piRNA clusters for transposon repression in adult ovaries. Mol Cell 66: 411–419 e4 [DOI] [PubMed] [Google Scholar]
- de Albuquerque BFM, Luteijn MJ, Rodrigues RJC, van Bergeijk P, Waaijers S, Kaaij LJT, Klein H, Boxem M, Ketting RF (2014) PID‐1 is a novel factor that operates during 21U‐RNA biogenesis in Caenorhabditis elegans . Genes Dev 28: 683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Tidow H (2008) A novel CHHC Zn‐finger domain found in spliceosomal proteins and tRNA modifying enzymes. Bioinformatics 24: 2277–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA (2012) piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans . Cell 150: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E‐M, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA (2012) Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez‐Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, Allshire RC (2010) Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK (2012) The Caenorhabditis elegans HEN1 ortholog, HENN‐1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet 8: e1002617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SEJ, Kim JK (2014) Endogenous RNAi pathways in C. elegans In The C. elegans Research Community, WormBook (ed.). WormBook. http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm A, Casas‐Vila N, Scheibe M, Butter F (2016) Reader interactome of epigenetic histone marks in birds. Proteomics 16: 427–436 [DOI] [PubMed] [Google Scholar]
- Boeck ME, Huynh C, Gevirtzman L, Thompson OA, Wang G, Kasper DM, Reinke V, Hillier LW, Waterston RH (2016) The time‐resolved transcriptome of C. elegans . Genome Res 26: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge KS, Shah KM, Li Y, Foxler DE, Wong SCK, Miller DC, Davidson KM, Foster JG, Rose R, Hodgkinson MR, Ribeiro PS, Aboobaker AA, Yashiro K, Wang X, Graves PR, Plevin MJ, Lagos D, Sharp TV (2017) Argonaute utilization for miRNA silencing is determined by phosphorylation‐dependent recruitment of LIM‐domain‐containing proteins. Cell Rep 20: 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S (2012) A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S (2011) A pre‐mRNA–associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet 7: e1002249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S (2011) Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans . Proc Natl Acad Sci USA 108: 19683–19688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC (2010) Argonautes ALG‐3 and ALG‐4 are required for spermatogenesis‐specific 26G‐RNAs and thermotolerant sperm in Caenorhabditis elegans . Proc Natl Acad Sci USA 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Yates JR, Mello CC (2013) Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans . Cell 155: 1532–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ (2013) A transcriptome‐wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell 50: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Albuquerque BFM, Placentino M, Ketting RF (2015) Maternal piRNAs are essential for germline development following de novo establishment of endo‐siRNAs in Caenorhabditis elegans . Dev Cell 34: 448–456 [DOI] [PubMed] [Google Scholar]
- Dönertas D, Sienski G, Brennecke J (2013) Drosophila Gtsf1 is an essential component of the Piwi‐mediated transcriptional silencing complex. Genes Dev 27: 1693–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR III, Mello CC (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR‐1 in multiple small‐RNA‐mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR‐Cas9 system. Nat Methods 10: 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: zinc‐fingers as protein‐recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A (2009) A Caenorhabditis elegans RNA‐directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183: 1297–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ (2010) Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR III, Mitani S, Carrington JC, Mello CC (2009) Distinct argonaute‐mediated 22G‐RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S (2008) An argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry‐Mieg D, Thierry‐Mieg J, Kim JK (2009) 26G endo‐siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans . Proc Natl Acad Sci USA 106: 18674–18679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber F, Brennecke J (2013) The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Pillai SS, Taglini F, Li F, Ruan K, Zhang J, Wu J, Shi Y, Bayne EH (2013) Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification. Proc Natl Acad Sci USA 110: E1879–E1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James V, Zhang Y, Foxler DE, de Moor CH, Kong YW, Webb TM, Self TJ, Feng Y, Lagos D, Chu CY, Rana TM, Morley SJ, Longmore GD, Bushell M, Sharp TV (2010) LIM‐domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA‐mediated gene silencing. Proc Natl Acad Sci USA 107: 12499–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJT, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF (2012) Differential impact of the HEN1 homolog HENN‐1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans . PLoS Genet 8: e1002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappei D, Butter F, Benda C, Scheibe M, Draškovič I, Stevense M, Novo CL, Basquin C, Araki M, Araki K, Krastev DB, Kittler R, Jessberger R, Londoño‐Vallejo JA, Mann M, Buchholz F (2013) HOT1 is a mammalian direct telomere repeat‐binding protein contributing to telomerase recruitment. EMBO J 32: 1681–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp THA, van Luenen HGAM, Plasterk RHA (1999) mut‐7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141 [DOI] [PubMed] [Google Scholar]
- Ketting RF (2011) The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Lee H‐C, Gu W, Shirayama M, Youngman E, Conte D Jr, Mello CC (2012) C. elegans piRNAs mediate the genome‐wide surveillance of germline transcripts. Cell 150: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, Berezikov E, Ketting RF (2012) Extremely stable Piwi‐induced gene silencing in Caenorhabditis elegans . EMBO J 31: 3422–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF (2013) PIWI‐interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet 14: 523–534 [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G (2008) 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim Y‐S, Zhang C, Dowen RH, Phillips CM, Fischer SEJ, Ruvkun G (2012) PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 Ortholog henn‐1. PLoS Genet 8: e1002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ (2013) A genome‐wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila . Mol Cell 50: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K (2013) DmGTSF1 is necessary for Piwi‐piRISC‐mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27: 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Noble D, Sorokin EP, Kimble J (2014) A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans . G3: Genes ‐ Genomes ‐ Genetics 4: 1765–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S (2009) Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans . Genetics 183: 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, Montgomery TA (2015) piRNAs and piRNA‐dependent siRNAs protect conserved and essential C. elegans genes from misrouting into the RNAi pathway. Dev Cell 34: 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M (2007) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H‐C, Gu W, Ishidate T, Conte D Jr, Mello CC (2012) piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti K‐A, Dönertas D, Tirian L, Meixner K, Brennecke J (2015) Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev 29: 2258–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutandy FXR, Ebersberger S, Huang L, Busch A, Bach M, Kang H, Fallmann J, Maticzka D, Backofen R, Stadler PF, Zarnack K, Sattler M, Legewie S, König J (2018) In vitro iCLIP‐based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res 28: 699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N, Yoshimura T, Miyazaki S, Tashiro F, Miyazaki J (2016) Gtsf1 l and Gtsf2 are specifically expressed in gonocytes and spermatids but are not essential for spermatogenesis. PLoS One 11: e0150390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, Conte D Jr, Duchaine TF (2012) Tudor domain ERI‐5 tethers an RNA‐dependent RNA polymerase to DCR‐1 to potentiate endo‐RNAi. Nat Struct Mol Biol 19: 90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidow H, Andreeva A, Rutherford TJ, Fersht AR (2009) Solution structure of the U11‐48K CHHC zinc‐finger domain that specifically binds the 5′ splice site of U12‐type introns. Structure 17: 294–302 [DOI] [PubMed] [Google Scholar]
- Tsai H‐Y, Chen C‐CG, Conte D Jr, Moresco JJ, Chaves DA, Mitani S, Yates JR III, Tsai M‐D, Mello CC (2015) A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160: 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D (2010) Sequential rounds of RNA‐dependent RNA transcription drive endogenous small‐RNA biogenesis in the ERGO‐1/Argonaute pathway. Proc Natl Acad Sci USA 107: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen C‐CG, Tolia NH, Joshua‐Tor L, Mitani S, Simard MJ, Mello CC (2006) Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Miyazaki T, Toyoda S, Miyazaki S, Tashiro F, Yamato E, Miyazaki JI (2007) Gene expression pattern of Cue110: a member of the uncharacterized UPF0224 gene family preferentially expressed in germ cells. Gene Expr Patterns 8: 27–35 [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Toyoda S, Kuramochi‐Miyagawa S, Miyazaki T, Miyazaki S, Tashiro F, Yamato E, Nakano T, Miyazaki JI (2009) Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn‐finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Dev Biol 335: 216–227 [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Watanabe T, Kuramochi‐Miyagawa S, Takemoto N, Shiromoto Y, Kudo A, Kanai‐Azuma M, Tashiro F, Miyazaki S, Katanaya A, Chuma S, Miyazaki JI (2018) Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep 19: e42054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, Hannon GJ (2015) Panoramix enforces piRNA‐dependent cotranscriptional silencing. Science 350: 339–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xu F, Mao H, Ji J, Yin M, Feng X, Guang S (2014) Nuclear RNAi contributes to the silencing of off‐target genes and repetitive sequences in Caenorhabditis elegans . Genetics 197: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Hunter CP (2011) Tissue‐specificity of Caenorhabditis elegans enhanced RNAi mutants. Genetics 111: 127209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Data Availability Statement
Sequencing data have been deposited to the NCBI Gene Expression Omnibus (GEO), and proteomics data are available at the ProteomeXchange Consortium via PRIDE. GEO: GSE103432, PRIDE: PXD007665. Source data are available at https://data.mendeley.com/datasets/xyxh6hv3mc/draft?a=03bd1ca8-dd09-44a2-a0a1-3910ad5201c9.


