Abstract
Cytoplasmic lipid droplets are important organelles in nearly every eukaryotic and some prokaryotic cells. Storing and providing energy is their main function, but they do not work in isolation. They respond to stimuli initiated either on the cell surface or in the cytoplasm as conditions change. Cellular stresses such as starvation and invasion are internal insults that evoke changes in droplet metabolism and dynamics. This review will first outline lipid droplet assembly and then discuss how droplets respond to stress and in particular nutrient starvation. Finally, the role of droplets in viral and microbial invasion will be presented, where an unresolved issue is whether changes in droplet abundance promote the invader, defend the host, to try to do both. The challenges of stress and infection are often accompanied by changes in physical contacts between droplets and other organelles. How these changes may result in improving cellular physiology, an ongoing focus in the field, is discussed.
Keywords: cellular stresses, lipid droplet assembly, metabolism, microbial invasion, starvation
Subject Categories: Metabolism; Microbiology, Virology & Host Pathogen Interaction
Introduction
Cytoplasmic lipid droplets are components of virtually all eukaryotic and some prokaryotic cells (Waltermann et al, 2005; Fujimoto & Ohsaki, 2006). They are efficient (high calories per gram ratio) depots of energy that can be released as required, either for the hosting cell or the organism at large. The neutral lipids stored in droplets can also contain precursors of more complex molecules such as sphingolipids, signaling molecules (e.g., eicosanoids), and hormones (retinoic acid and steroids). While the control of storage and release of fatty acids from triacylglycerols have been well studied, the corresponding importance of the stored component of signaling molecules and lipophilic hormones is largely unknown.
Droplet neutral lipids are enveloped by a phospholipid monolayer into which is embedded proteins. Many of these are unique to the droplet compartment (Yang et al, 2012). The perilipins in animals and fungi, and the oleosins in plants, provide physical stability to the droplet and a barrier to adventitious lipolysis (Schmidt & Herman, 2008; Gao et al, 2017; Sztalryd & Brasaemle, 2017). Enzymes of neutral lipid synthesis and hydrolysis also reside on droplets and can be modulated by perilipins (Sztalryd & Brasaemle, 2017). Organelle trafficking molecules, particularly Rab proteins, are also droplet components, the functions of which at the droplet surface have begun to be understood (Li et al, 2017). Recent theoretical and physical studies indicate that the droplet surface may be unique in providing a more hydrophobic environment than that of a normal phospholipid bilayer (Thiam et al, 2013; Kory et al, 2016). This may explain the dependence of viral assembly [for hepatitis C, dengue, and rotaviruses, among others (Herker & Ott, 2012), and see below] and the storage of particular proteins [such as insect histones (Welte & Gould, 2017)] at this site.
Much has been learned about the function of droplets as energy storage organelles, their most important function. However, droplets are emerging as intermediaries in, or platforms for, more complex cell biological functions. There are several excellent recent reviews that discuss the multiplicity of known droplet functions (Barbosa & Siniossoglou, 2017; Gluchowski et al, 2017; Roingeard & Melo, 2017; Welte & Gould, 2017). Here, we focus on the role of droplets in cell stress and related challenges, including starvation, infection, and the immune response. Their roles in cancer and liver steatosis have been recently covered elsewhere in comprehensive reviews (Gluchowski et al, 2017; Tirinato et al, 2017). The lipid droplet can be an indicator, a vehicle, and a facilitator for stress responses. Droplets are attuned to and respond to starvation, the unfolded protein response, and oxidative stress. They are the vehicle of choice for viral assembly, but, in an ironic twist, they also signal the cell to mount immune responses. Before we cover these behaviors in more detail, we present an outline of droplet assembly.
Steps in droplet assembly
The state of knowledge of droplet assembly is addressed in a comprehensive recent review (Walther et al, 2017). This following section outlines the process, adding a few hypothetical details regarding the nature of the ER–droplet interface and the notion of droplet release. The steps outlined below correspond to the diagram illustrated as in Fig 1.
Figure 1. Steps in lipid droplet assembly.
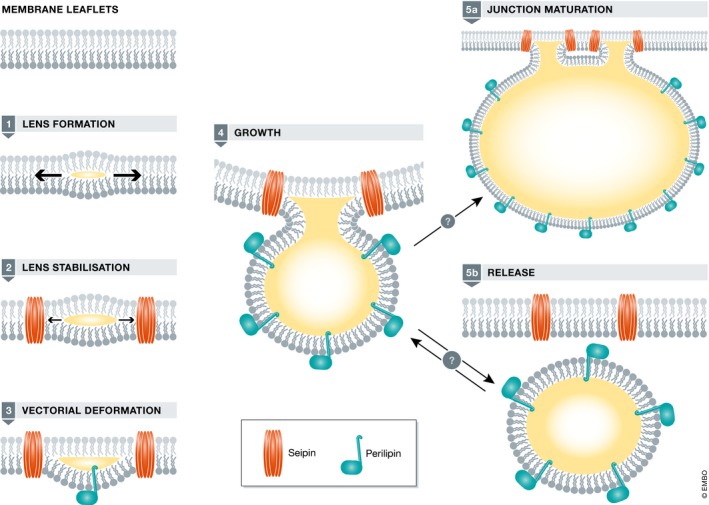
See text for details. Both seipin, involved in nascent droplet stability, and perilipin(s), which can deform membranes, have been shown to bind early in the pathway, and they may not bind in the order indicated. Maturation of junctions is considered irreversible, while both droplet release and rebinding have been postulated.
1) Lens formation – Terminal enzymes in neutral lipid synthesis reside largely in the ER. Newly generated neutral lipid molecules are presumed to be deposited among the acyl chains of the lipid bilayer, sequestered from the polar head groups of phospholipids. As their concentration increases, molecules of neutral lipid, laterally diffusing, coalesce to form a lens, the energy cost for interaction with each other is less than their interaction with phospholipid acyl chains (Thiam & Foret, 2016). Such lenses, hypothetical for many years, have been observed recently in yeast by electron microscopy following a brief induction of droplet formation (Choudhary et al, 2015). At some point in the growth of the lens, fluorescent probes of neutral lipid such as BODIPY 493/503 or Nile Red can visualize these neutral lipid accumulations as diffraction‐limited punctae, although their sensitivity—the number of triacylglycerol molecules at their detection limits—has not been determined. However, before puncta representing nascent droplets are detectable by fluorescent dyes, they can be revealed with specific fluorescently conjugated proteins or peptides. In animal cells, these include a fluorescently tagged‐amphipathic helix derived from GPAT4, termed LiveDrop (Wang et al, 2016) and a fusion product, termed HPos, consisting of the hydrophobic domain of associated with lipid droplet protein 1 (ALDI) and the droplet targeting signal of caveolin‐1 (Kassan et al, 2013). Similarly, the perilipin Pln1p (formerly Pet10p) can often be visualized at punctae before they stain with BODIPY (Gao et al, 2017).
2) Nascent droplet stabilization – In wild‐type Drosophila S2 cells subjected to oleic acid, new LiveDrop punctae quickly appeared and within a few minutes became visible with BODIPY, indicating growth (Wang et al, 2016). These BODIPY‐stained particles were relatively immobile. Without seipin, which is required for adipogenesis and normal droplet morphology (Cartwright & Goodman, 2012), however, the small LiveDrop particles usually failed to stain with BODIPY and were mobile (Wang et al, 2016). This elegant experiment suggests that seipin stabilizes the nascent droplets and permits them to mature. How seipin performs this activity early in the droplet life cycle is still uncertain. It is known to regulate protein and lipid trafficking into the droplet (see below). We propose that seipin also destabilizes the ER lipid bilayer to allow a growing lens to deform it.
3) Vectorial membrane deformation – Lipid droplets bud toward the cytosol. The control of this vectorial budding is not fully understood although recent progress has been made. Both phospholipids and proteins are involved. The shape and packing of phospholipids at the ER outer leaflet–bud interface likely play a role. Outward wedge‐shaped lipids such as diacylglycerol (DAG) or phosphatidic acid (PA) would be permissive for this distortion. There is mounting evidence that both of these lipids are indeed involved in droplet budding (Skinner et al, 2009; Adeyo et al, 2011; Grippa et al, 2015; Han et al, 2015; Wolinski et al, 2015; Gaspar et al, 2017). Relatedly, slight differences in surface tension between the ER leaflets are sufficient to induce vectorial budding in model systems (Chorlay & Thiam, 2018).
Proteins likely play key roles in vectorial budding, with recent work focusing on two protein families in particular. One of them, the ER protein FIT2, binds directly to diacylglycerol (DAG) and triacylglycerol (TAG; Gross et al, 2011), and deletion of the two yeast FIT2 orthologs Yft2p and Scs3p results in a failure of droplets to emerge from the ER (Choudhary et al, 2015). FIT2 has weak PA activity that is tied to its function (Hayes et al, 2017). Perilipins, especially mammalian Plin3 and yeast Pln1p, bind early to nascent droplets from the cytosol, and their abundance makes them good candidates for the membrane deformation that must accompany budding (Skinner et al, 2009; Gao et al, 2017). This idea is supported by the ability of purified Plin3 to deform bilayers (Bulankina et al, 2009).
Proteins that control ER shape play an important role in droplet budding. The reticulons, REEP proteins, and atlastins, which promote tubular ER morphology and connections (Voeltz et al, 2006; Hu et al, 2009; Shibata et al, 2010), are important for normal droplet morphology. Loss of function of atlastin led to smaller lipid droplets and less TAG, while overexpression results in large droplets (Klemm et al, 2013). Relatedly, a REEP1 null mouse displays a partial lipodystrophy reminiscent of the absence of seipin (Renvoise et al, 2016). This work also showed co‐precipitation of REEP1 and seipin, reinforcing the idea that manipulation of ER dynamics may be essential to appropriate droplet budding. However, knockdown of reticulon‐1C resulted in accumulation of lipid droplets; this may be secondary to disturbed communication between ER and mitochondria (Reali et al, 2015).
4) Droplet growth – As the nascent droplet grows, the core of neutral lipid becomes largely surrounded by phospholipids from the ER outer leaflet. The surface area of the ER outer leaflet thus expands, and it is unclear how the cell accommodates this increase. Relevant questions include the following two: Is there an increase in total ER phospholipids? Is there an increase in flippase activity between the leaflets, and is there local synthesis of new phospholipids at the developing ER/droplet junction? To our knowledge, there have been no studies of the localization or recruitment of phospholipid‐terminal enzymes on the ER during active droplet formation. However, in insect cells the rate‐limiting initial enzyme in PC synthesis, CTP: phosphocholine cytidylyltransferase (CTT), associates with growing droplets from the nucleus (Krahmer et al, 2011). This association may promote PC formation at the droplet bud site not by recruitment of the terminal enzyme but by providing a pipeline of substrate to the enzyme localized nearby. Interestingly, the enzyme targets instead to the ER in mammalian cells, perhaps reflecting a different ratio of phospholipids in the droplet monolayers of the two cell types attracting the enzyme (Aitchison et al, 2015).
The extrusion of neutral lipid from the ER into the bud does not typically result in total sequestration of neutral lipid away from the ER bilayer. If newly synthesized droplet proteins are diverted to the ER lumen instead of the cytosol, they still are able to target to droplets (Mishra et al, 2016). The simplest interpretation of this result is that the luminal ER surface apposed to the droplet is still bathed in neutral lipid that is continuous with the corresponding droplet and as such is recognized by droplet proteins. Other possible topologies, such as extended phospholipid bilayer necks connecting droplets to the outer ER leaflet, would not be compatible with these data. The result may indicate that much of the droplet surface normally faces the lumen. While maintaining a close relationship with the ER has been a hallmark of droplets since early studies (Blanchette‐Mackie et al, 1995), the extent to which the surface of an attached droplet is exposed normally to the ER lumen is not known; it may be different depending upon metabolic state and cell type.
Growth of nascent lipid droplets in yeast is accompanied by recruitment of the neutral lipid terminal enzymes Dga1p to the droplet surface from the ER (Jacquier et al, 2011). However, in fly and mammalian cells, a subset of droplets expand (“eLDs”) while others stay small (“iLDs”; Wilfling et al, 2013). The reason for maintaining a pool of small droplets is still unclear, but may reflect functional differences, as small droplets in flies accumulate the perilipin LSD2 and are enriched near the cell periphery in the fat body, the adipose tissue of insects (Bi et al, 2012). As in yeast, these eLDs contain enzymes of neutral lipid synthesis, notably GPAT4 and DGAT2. The factors or forces that direct these enzymes to the eLDs and not the iLDs are not clear, but hydrophobic hairpins in their structures are targeting motifs that likely are retained by the unique environment of the droplet phospholipid monolayer (Kory et al, 2016).
Droplets are often seen to be connected at the end of ER tubules, termed ER/droplet bridges (Wilfling et al, 2014). The transfer of GPAT4 from ER to droplets has been seen to occur through such structures (Wilfling et al, 2013). The origin of bridges is not clear. They may form by elongation of the region between ER and the initial droplet bud (perhaps such droplets would not be accessible to luminally expressed perilipins), or simply represent a new droplet that forms at the tip of an ER tubule, the tip becoming the “bridge”. Bridges have also been hypothesized to represent re‐attachment of a droplet to the ER following an increase in droplet surface tension as a result of nano‐droplet release by COPI from an existing isolated droplet (Wilfling et al, 2014).
5a) Formation of an extended ER/droplet junction – Lipid droplets are typically seen closely associated with the ER over a significant fraction of their surface (Blanchette‐Mackie et al, 1995). Junctions of considerable length (150 nm in human A431 cells, ~1 μm in Drosophila S2 cells) and electron density have been observed, suggesting a robust communication link between the two organelles (Wilfling et al, 2013; Salo et al, 2016). Several proteins have been identified at ER/droplet junctions, including seipin (Szymanski et al, 2007; Fei et al, 2008; Salo et al, 2016), the acyl‐CoA synthetase FATP1 (Xu et al, 2012), and Rab18 and the associated NRZ tethering complex (Xu et al, 2018). While the junction can give the impression of an intact droplet lying on the surface of the ER, we hypothesize multiple fenestrations in the ER, through which neutral lipids, phospholipids, and proteins can flow from the ER to the droplet, as shown in Fig 1 panel 5A. This notion is consistent with seipin, an ER protein which transports substrates in this direction, existing at many points at the junction (Fei et al, 2008; Salo et al, 2016).
5b) Droplet release? In most models of lipid droplet maturation, droplets are shown to separate from the ER. Evidence for droplet release is strongest in mammary tissue, where cytoplasmic lipid droplets are released from cells as milk fat globules, enveloped by apical plasma membrane of the mammary epithelial cells (McManaman, 2014). Although important proteins for the interaction of plasma membrane and droplet have been identified—Plin2, butyrophilin, and xanthine dehydrogenase (McManaman, 2009)—how these bring about globule secretion is still unclear. The stage at which the ER is separated from the droplet in mammary epithelia, whether early in secretion or as it is enveloped by plasma membrane, is unknown. In the developing Drosophila oocyte, lipid droplets move from nurse cells into the oocyte by cytoplasmic streaming, and later, in the early embryo, they undergo bidirectional migrations between yolk and apical surface, catalyzed by motors on microtubules (Welte, 2015). These processes may occur independent of the ER, but this has not been formally shown. In Drosophila S2 cells, depletion of a COPI constituent, βCOP, resulted in lack of trafficking of GPAT4 to droplets (Wilfling et al, 2014). Addition back of this protein subsequent to fusion of the cell with one containing intact COPI resulted in repopulating droplets with GPAT4 through the bridges discussed above. The authors concluded that COPI (and ARF) were necessary to reattach droplets to the ER upon depleting phospholipids from dis‐attached droplets (Wilfling et al, 2014). Although there was much supporting evidence for this hypothesis from elegant model systems, the absence of COPI could prevent trafficking of ER proteins to droplets by more indirect means, such as depletion of critical trafficking factors. We have failed to observe this process in the yeast system (Cartwright et al, 2015 and unpublished data); 85% of droplets in COS‐7 cells were clearly localized with the ER, suggesting that a small number of droplets were indeed untethered from its organelle of origin (Valm et al, 2017). Nevertheless, the field still lacks convincing evidence that droplets regularly separate from the ER in metazoan cells. The alternative is that droplets remain connected to membranes, sometimes marking a highly specialized region of the ER, or, under some conditions discussed next, ER junctions with other organelles.
Lipid droplets in cellular stress responses
Cells constantly sense and respond to external stresses. Cellular adaptations to stress are often manifested in metabolic remodeling, and one of the most universally observed adaptations is the increased production of lipid droplets. Recent studies are revealing why: Droplets serve a multi‐faceted role as nutrient reservoirs, as cytoplasmic chaperones for toxic proteins and lipids, and as signaling platforms for immune response pathways. Not surprisingly, cancer cells have recently been observed to take advantage of the many benefits droplets provide, implicating droplet homeostasis in cancer onset and progression. In this section, we dissect the many ways that lipid droplets contribute to adaptive stress responses and cell survival.
Lipid droplets as nutrient reservoirs
As a major organelle for energy storage, droplets are closely linked to the metabolic adaptations that cells initiate when responding to changes in nutrient availability. In fact, their role as nutrient reservoirs may represent the most ancient function of droplets, as even simple eukaryotes such as budding yeast use droplets to survive in nutrient‐poor environments. This is exemplified by the observation that yeast grown in nutrient‐rich media exhibit high phospholipid production and rapid cell growth, and correspondingly low levels of neutral lipids and droplets (Fig 2A; Radulovic et al, 2013; Markgraf et al, 2014). As nutrients such as amino acids and sugars become exhausted, yeast sense this imminent starvation and decrease their phospholipid production, shunting precursor lipids such as sterols and DAG into the synthesis of TAG and sterol esters at the ER, which are ultimately packaged into droplets that bud from this organelle. This transition phase is defined as the diauxic shift, the phase of growth where yeast shift from fermentation to respiration, and corresponds to a burst of LD biogenesis that will promote cell survival in the ongoing nutrient‐poor environment (Wang et al, 2014; Barbosa et al, 2015). Consistent with this, droplet‐deficient yeast are less capable of long‐term growth in nutrient‐deficient media (Seo et al, 2017).
Figure 2. Lipid droplet behaviors.
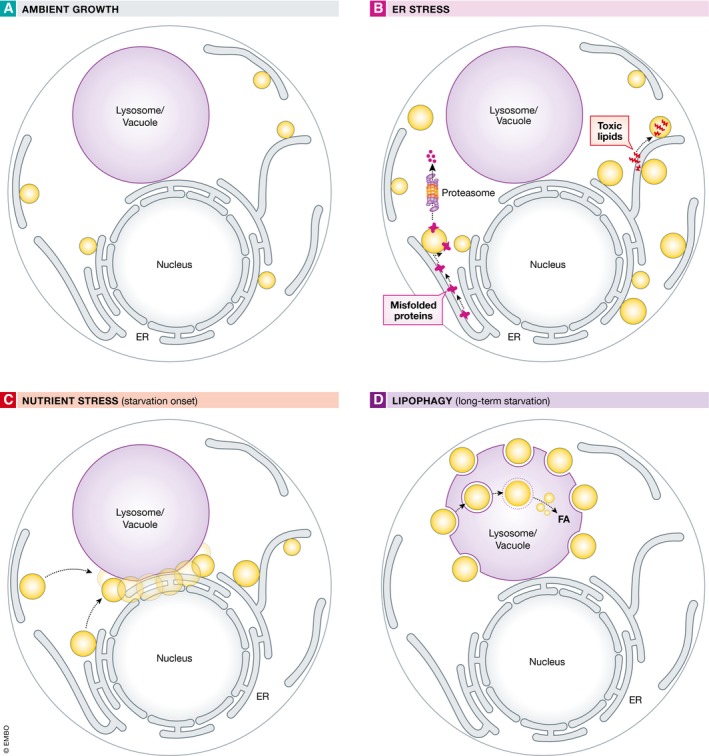
Schematic cartoons of droplets in yeast in ambient growth (A) and under various stresses including ER stress induced by protein mis‐folding or toxic lipid accumulation (B), and nutrient stress induced by the onset of glucose starvation (C). Long‐term nutrient deprivation can also promote vacuolar micro‐autophagy (lipophagy) of lipid droplets (D).
Lipid droplets and neutral lipids in “sensing” and responding to starvation
How is nutritional stress sensed, and the decision to create droplets made by cells? Emerging evidence suggests that droplets and the precursor lipids used to make TAG play key roles in both sensing and responding to nutritional stresses. In fact, both DAG and its direct precursor PA function as signaling lipids, the levels of which quickly change in response to nutritional demands. In yeast, this is regulated through Opi1p, a PA‐binding transcription factor and master regulator of inositol metabolism. In rapidly growing yeast, phosphoinositide metabolism and phospholipid production are highly active. However, this sustained growth is dependent upon the availability of exogenous inositol, which is required for the production of phosphatidyl inositides from the precursor PA. If cells are starved of inositol, the PA concentration in the ER rises, with a fraction converted into DAG and subsequently to TAG, which is then stored in lipid droplets. The ER‐localized PA, along with the ER protein Scs2p, sequesters Opi1p at the membrane. This prohibits Opi1p from translocating into the nucleus and silencing genes required for de novo inositol synthesis (Carman & Zeimetz, 1996; Loewen et al, 2004). In a remarkable switch, when exogenous inositol is added, PA is rapidly consumed from the ER surface to make phospholipids, Opi1p is released from the ER, and it enters the nucleus to repress key inositol synthesis genes. Thus, the new flood of exogenous inositol drives membrane biosynthesis, and DAG is shunted away from TAG and droplet production to phospholipid and membrane biogenesis via the CDP‐DAG pathway (Han et al, 2008). Neutral lipids and PA in particular therefore serve as important signaling factors modulating cell growth, with droplets serving as lipid storage organelles when general biomembrane production is low.
Beyond its role in transcriptional regulation, PA itself also serves as a physico‐chemical “pH sensor” that signals starvation stress to various stress–response pathways. Indeed, glucose‐starved yeast exhibit a significant drop in cytoplasmic pH from ~7 to ~6 (Martinez‐Munoz & Kane, 2008). At this lower pH, the electrostatic charge of the monoester PA headgroup favors −1 rather than −2, thereby reducing its affinity for PA‐effector proteins, allowing them to relocalize from the ER to other cellular sub‐regions to drive stress responses (Young et al, 2010; Barbosa et al, 2015). Indeed, droplet production is up‐regulated as part of this general metabolic remodeling. Consistent with this, experimentally lowering the cytoplasmic pH is sufficient to drive droplet biogenesis (Gubern et al, 2009).
Lipid droplets in starvation stress response in metazoa
Following their production, droplets serve important pro‐survival roles in fasting and long‐term starvation in both insects and mammals. As a consequence, droplet dynamics are tightly regulated by the metabolic status of the organism. Amino acid starvation, or treatment with the mTOR inhibitor rapamycin, suppresses TORC signaling and stimulates TAG synthesis and droplet production across eukaryotes, being observed in yeast as well as in both Drosophila and cultured mammalian cells (Dubots et al, 2014). Indeed, the increased TAG observed from treating Drosophila flies with rapamycin can be subsequently mobilized if these flies are starved, and blocking this initial droplet biogenesis causes extreme starvation sensitivity in Drosophila (Bjedov et al, 2010). Similarly, mammalian cells exhibit a burst of droplet biogenesis in the hours following the removal of amino acids from their culture media (Nguyen et al, 2017). Thus, droplet production following the initial phases of starvation represents a highly conserved metabolic program to survive the unknowns of a nutrient‐poor future.
From where do the lipids used to make these starvation‐induced droplets come? At least one source appears to be macro‐autophagy, which is stimulated in response to the reduced TORC signaling following nutrient deprivation. Indeed, autophagy represents a nearly universal cellular stress response, and the raw lipid metabolites that are generated in autolysosomes appear to directly contribute to droplet production (Rambold et al, 2015; Nguyen et al, 2017).
Spatial organization of lipid droplet sub‐populations in the starvation response
Although an increase in droplet biogenesis following starvation‐induced autophagy in mammalian cells has been observed, the emerging picture in yeast is less clear. In yeast, starvation‐induced droplet production appears to precede large‐scale autophagy (van Zutphen et al, 2014; Seo et al, 2017). Indeed, yeast cells exhibit a blooming of droplets shortly following initial starvation. Surprisingly, this starvation‐induced droplet production is highly spatially organized such that droplets are quickly observed clustered around the yeast ER–vacuole inter‐organelle contact site, called the nuclear ER–vacuole junction (NVJ; Barbosa et al, 2015; Hariri et al, 2018; Fig 2C). The NVJ‐tethering protein Mdm1p appears to play a critical role in this droplet accumulation, as its overexpression is sufficient to drive NVJ‐associated droplet clustering even under exponential growth conditions (Henne et al, 2015; Hariri et al, 2018). Indeed, Mdm1p was sufficient to cluster droplets at ER–vacuole contacts even in yeast devoid of other NVJ tethers, indicating that Mdm1p functions in droplet dynamics at the NVJ. Following their production, these NVJ‐associated droplets will gradually be absorbed into the yeast vacuole in a specialized form of micro‐autophagy known as lipophagy (Fig 2D). This process is required for long‐term survival during prolonged starvation, and it utilizes sterol‐enriched liquid‐ordered domains on the vacuolar surface as sites for these micro‐autophagy events (Toulmay & Prinz, 2013; Wang et al, 2014; Fig 2D). These observations in yeast suggest that droplets serve as a major nutrient reservoir for cells facing long‐term subsistence in a nutrient‐poor environment.
Several studies indicate that specific droplet sub‐populations with unique protein compositions exist within a single cell, suggesting they play specific roles in cellular metabolism. The NVJ‐associated droplet sub‐population is decorated with Pdr16p, a lipid transfer protein and Sec14p homolog, implying a role for these droplets in lipid trafficking. Recent studies also define proteins required for targeting Pdr16p to these droplets. Ldo45p and Ldo16p are two protein products of a unique intergenic splicing event and encode ancillary subunits of the seipin protein complex (Eisenberg‐Bord et al, 2018; Teixeira et al, 2018). Intriguingly, Ldo45p is required for targeting Pdr16p to droplets. Similar to the NVJ tether Mdm1p, overexpression of Ldo45p also is sufficient to cluster droplets at the NVJ, indicating that it controls aspects of both droplet composition and spatial distribution (Fig 3). Similarly, loss of Ldo16p reduces the number of droplets clustered at the NVJ in stationary phase. Consistent with this, both Ldo45p and Ldo16p are highly expressed in yeast undergoing exponential growth, but only Ldo16p expression is maintained in stationary phase, implying a role for the latter in stationary‐phase droplet dynamics and vacuolar turnover (Teixeira et al, 2018). Collectively, these studies suggest the spatial distribution of droplets is highly dependent on the metabolic status of the cell, as yeast entering stationary phase exhibit a significant accumulation of NVJ‐associated droplets that associate with the NVJ tether Mdm1p (Hariri et al, 2018).
Figure 3. Lipid droplet interactome with other organelles.
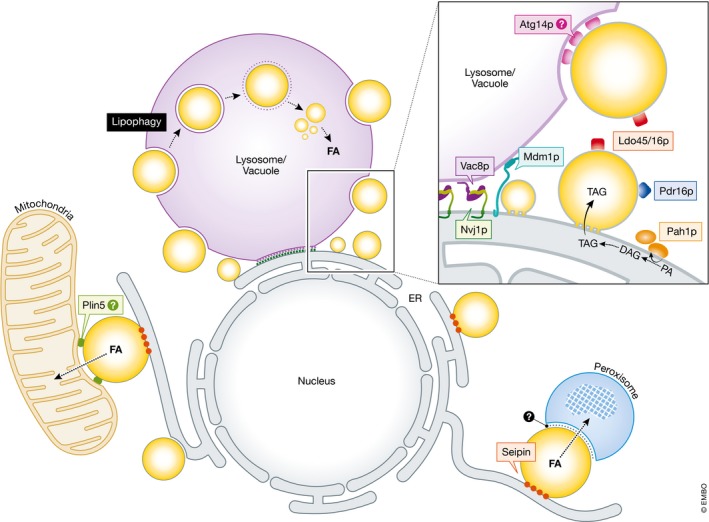
Droplets form contacts with numerous other organelles, including the endoplasmic reticulum (ER) via the protein seipin (red). Mitochondria–droplet contacts promote the transfer of fatty acids (FA) for β‐oxidation in mitochondria, and these contacts may be stabilized by perilipin 5 (Plin5, green). Similarly, in yeast peroxisome–droplet contacts are sites of FA transfer. Also in yeast, nuclear ER–vacuole junctions (NVJ) are sites of droplet budding. NVJ contact sites are maintained by the tethering proteins Nvj1p (green) and Vac8p (purple). NVJ‐associated droplets co‐localize with the NVJ tether Mdm1 (black), and the droplets are decorated with specific proteins including Prd16p (pink) and Ldo45p/16p (aqua). The PA phosphatase Pah1p (orange), which generates DAG from PA, is enriched at the NVJ periphery during the diauxic shift. NVJ‐derived droplets may be eventually digested via lipophagy at sterol‐rich microdomains at the vacuole surface in an ATG14‐dependent (gray) manner.
Further studies are needed to elucidate the specific spatial and temporal events that govern droplet accumulation at the NVJ. Are all NVJ‐associated droplets made at the NVJ, or do some migrate there from elsewhere in the ER network where they are produced? Furthermore, what are the roles of these droplets beyond their eventual digestion in vacuolar lipophagy, and why are they enriched with lipid transfer proteins like Pdr16p? One intriguing possibility is that they may donate lipids such as sterol to the vacuole that aid in the metabolic remodeling the vacuole must undergo in stationary phase. Finally, do droplets also cluster at ER–lysosome contacts in metazoan cells as they do in yeast? Several NVJ tethers including Nvj1p are not conserved in metazoans, but Mdm1p exhibits clear homologs in mammals. Consistent with this, Mdm1 homolog SNX14, which is associated with pediatric cerebellar ataxia, was recently shown to associate with droplets formed in oleate‐treated mammalian cells, suggesting a conserved role for this protein in droplet dynamics (Bryant et al, 2018).
Lipid droplets in membrane and protein quality control
In addition to serving as nutrient reservoirs during starvation, droplets also function as storage sites for otherwise‐toxic lipids and proteins that accumulate during metabolic stresses. Mammalian cells undergoing active macro‐autophagy accumulate mitochondrially derived toxic lipids such as acylcarnitine, which droplets sequester to prevent lipotoxicity and subsequent apoptosis (Nguyen et al, 2017). Indeed, both yeast and mammalian cells with defects in droplet biogenesis are sensitive to fatty acid (FA)‐induced lipotoxicity, indicating that the conversion of FAs to TAG in droplets is generally protective (Fig 2B; Listenberger et al, 2003; Petschnigg et al, 2009). Similarly, yeast with defects in pathways of phospholipid synthesis up‐regulate droplet biogenesis as a mechanism to control their lipid synthesis at the ER and thus better maintain cellular lipid homeostasis (Vevea et al, 2015).
A second way droplets function in protein quality control is by regulating another major protein quality control pathway: autophagy. Specifically droplets can serve as direct lipid donors in the formation of autophagosomes and have been suggested to directly contribute lipids for the formation of Atg8p‐positive autophagic compartments (Shpilka et al, 2015). Consistent with this idea, yeast unable to produce droplets are viable, but display significant defects in macro‐autophagy and accumulate aberrant Atg8p foci in their cytoplasm (Velazquez et al, 2016).
More recently, droplets have been proposed to function in protein quality control a third way: as cytoplasmic chaperones that prevent toxicity associated with protein mis‐folding. In fact, mis‐folded proteins have also been directly observed to accumulate in the droplets of yeast cells exposed to proteotoxic heat stress, a major driver of protein mis‐folding (Fig 2B; Moldavski et al, 2015). Here, droplets are thought to function similar to inclusion bodies of prokaryotes, and these are ultimately turned over. In mammalian cells, proteins such as apolipoprotein B (apoB) are targeted to droplets for proteasomal degradation. In this case, they are thought to serve as a reservoir for hydrophobic proteins destined for proteasomal breakdown, which may also form toxic aggregates in the ER before they can be digested (Ohsaki et al, 2006).
Lipid droplets in ER homeostasis
As droplets bud from the ER, they represent a convenient sequestration for otherwise‐toxic lipids and proteins within the ER network. Not surprisingly, the unfolded protein response (UPR), a major stress response pathway, is intimately connected with droplet production. In fact, cells with active UPR response such as hepatocytes manifest active droplet budding when UPR is active (Chen et al, 2017). As mentioned above, droplets likely serve as cellular chaperones for mis‐folded proteins produced in the ER, providing a convenient organelle for storing otherwise‐toxic proteins and lipids. Consistent with this, mice up‐regulate UPR signaling and droplet production in their liver and adipose tissues during nutritional stresses when ER homeostasis is challenged.
Several secreted hormones have also been identified that aid in mammalian metabolic adaptations to stress. In fasting mice, the liver senses a decrease in blood circulating nutrients and secretes hormones such as fibroblast growth factor‐21 (Fgf21) to promote droplet mobilization in neighboring adipose tissues (Potthoff et al, 2009). Fgf21 production is regulated by the XBP1 branch of the UPR pathway and thus directly links cellular stress response at the ER to organismal‐level adaptations for nutritional stress (Jiang et al, 2014).
Lipid droplets in inter‐organelle crosstalk
In addition to their roles as storage reservoirs for lipids and proteins, droplets also directly contact and communicate with other organelles. Their network of organelle interactions places droplets at the crossroads of numerous important metabolic pathways that drive stress response. The most obvious droplet contact is with its mother organelle, the ER. ER–droplet junctions are maintained by the protein seipin (Sei1p in yeast) and control both droplet lipid and protein composition (Fei et al, 2011; Grippa et al, 2015; Salo et al, 2016). During fasting, mitochondria require fatty acids as an energy source and tap into fatty acid‐rich lipids such as TAG to maintain their homeostasis and ATP production via β‐oxidation. Numerous studies indicate that starvation‐induced droplets are a major source of these energy‐rich lipids. Using fluorescently tagged FAs, Rambold et al (2015) detected the net movement of FAs from droplets to mitochondria over the course of sustained starvation. Indeed, perilipin 5 (Plin5) appears to play a key role in creating droplet–mitochondrial junctions and in the exchange of fatty acids (Wang et al, 2011). While yeast perform β‐oxidation only in peroxisomes, the situation appears to parallel metazoan mitochondrial β‐oxidation, as clear droplet–peroxisome contact sites have been observed in yeast undergoing stationary‐phase growth (Binns et al, 2006). To date, no droplet–peroxisomal tethers have been identified. Finally, as mentioned above, droplets accumulate at ER–vacuole/lysosome interfaces, forming a tri‐organelle junction with proteins such as Mdm1p that coordinate droplet biogenesis with eventual turnover in lipophagy (Hariri et al, 2018).
Lipid droplets and infectious disease
Hijacking of lipid droplets by intracellular pathogens
Intracellular pathogens are polyauxotrophs and thus dependent on a number of host nutrient resources for growth and replication. It is therefore perhaps unsurprising that droplets, as concentrated sources of lipids and cholesterol, can be hijacked by intracellular pathogens. In fact, it appears that the majority of intracellular infectious agents, be they viral, bacterial, or protozoan, rely on host droplets for some portion of their life cycle (Table 1). For instance, both the bacteria Chlamydia (Cocchiaro et al, 2008) and the protozoan parasite Toxoplasma (Hu et al, 2017; Nolan et al, 2017) appear to salvage lipids by trafficking host cell droplets to the vacuolar spaces in which the pathogens replicate. Furthermore, Toxoplasma actively induces an increase in DGAT catalytic activity, ensuring a large supply of TAG droplets (Hu et al, 2017). In addition to providing essential lipid metabolites, these droplets also shuttle other essential biomaterials to intracellular pathogens. For example, Mycobacterium tuberculosis induces trafficking of host droplets to the phagosomes containing replicating bacteria in infected macrophages, and these droplets function to concentrate and deliver cellular iron via lipophilic siderophores that are secreted by the bacteria (Luo et al, 2005).
Table 1.
Pathogens that are known interactors with lipid droplets
| Pathogen | Interaction with droplets | Effectors | References |
|---|---|---|---|
| Bacteria | |||
| Chlamydia | Droplets traffic to bacterial inclusion | Unknown | Cocchiaro et al (2008) |
| M. tuberculosis | Droplets traffic to infected phagosome; iron acquisition via siderophores | Mycobactins | Luo et al (2005) |
| Orientia tsutsugamushi | Up‐regulates TAG synthesis | Unknown | Ogawa et al (2014) |
| Viruses | |||
| Hepatitis C | Reduces ATGL localization on droplets; Viral assembly on droplet surface | HCV core protein | Harris et al (2011) |
| Dengue | Induces lipophagy; C protein interacts with droplets | Unknown; Dengue C protein | Jordan & Randall (2017), Samsa et al (2009) |
| Polio | Increases fatty acid import and synthesis, competes with TAG synthesis | Unknown | Nchoutmboube et al (2013) |
| Rotavirus | Virus replicates at droplet contacts | Unknown | Cheung et al (2010) |
| Protozoa | |||
| Toxoplasma | Droplets traffic to vacuole; infection up‐regulates TAG synthesis | Unknown | Hu et al (2017), Nolan et al (2017) |
| Leishmania | Transcriptional changes in genes associated with neutral lipid metabolism | Unknown | Lecoeur et al (2013) |
| Trypanosoma brucei | Adipose tissue forms extracellular parasite reservoir | Unknown | Trindade et al (2016) |
| Trypanosoma cruzi | Infects adipocytes, forming parasite reservoir | Unknown | Shoemaker et al (1970), Ferreira et al (2011) |
Enveloped viruses use host membranes as platforms for their replication and structural organization, as well as an ample of source lipids. For example, dengue (Samsa et al, 2009), hepatitis C (Romero‐Brey et al, 2012), poliovirus (Suhy et al, 2000), and rotaviruses (Crawford & Desselberger, 2016) all replicate at ER sites in close proximity to droplets. Unsurprisingly, these interactions alter droplet homeostasis, though the mechanisms and cellular outcomes can vary widely, even among closely related viruses. For instance, hepatitis C virus blocks ATGL localization to droplets, thus inhibiting lipolysis and increasing droplet surface area to ensure its replication (Harris et al, 2011). On the other hand, dengue infection induces lipophagy to facilitate robust replication (Jordan & Randall, 2017). Poliovirus appears to interact with droplets in a more indirect manner. Polioviral membranes are derived from autophagic remodeling of the host ER (Suhy et al, 2000), which, as noted above, are likely regulated by interaction with droplets (Hamasaki et al, 2013; Shpilka et al, 2015; Velázquez & Graef, 2016). In addition, poliovirus infection increases long‐chain fatty acid synthesis and import into the host, but hijacks the increased fatty acids for creation of its own membranes rather than deposition in host droplets (Nchoutmboube et al, 2013).
Although there are now clear connections between droplet homeostasis and pathogen replication, the mechanisms by which host droplets interact with the specialized structures that form pathogens’ replicative niches remain largely unknown. Pathogen survival requires an interface with several host organelles, and these interactions are driven by pathogen‐derived proteins that have evolved to manipulate fundamental host cellular processes. Elucidating the molecular mechanisms of these organellar interactions has the potential to shed light on droplet functions in both pathogenic and normal contexts.
Lipid droplets as signaling platforms for regulating host responses to infection
Given the diverse interactions between pathogens and droplets, and the evolutionary pressure that infection represents to the host cell, it is perhaps unsurprising that droplets are emerging as critical mediators of host immune response. Strikingly, Drosophila droplets were found to sequester histones via the receptor Jabba (Li et al, 2012) and to release them in response to bacterial infection (Anand et al, 2012). Droplets thereby mediate a direct antimicrobial response that restricts bacterial infection in vivo. It is unclear whether this function is conserved in vertebrates, but this finding certainly raises the possibility of additional roles for droplets in fighting pathogen infection.
There is additional evidence linking droplet homeostasis to host immune antimicrobial pathways. The immune‐related GTPases (IRGs) are critical components of the non‐primate vertebrate cell autonomous immune responses (Hunn et al, 2011). Intriguingly, in cells lacking the negative regulator of this system (a protein called IRGM), activated IRGs preferentially mistarget to droplets, resulting in drastically increased lipophagy (Haldar et al, 2013). In fact, human IRGM is an important regulator of autophagy and lipophagy (Grégoire et al, 2012; Lin et al, 2016). It is thus tantalizing to hypothesize that the vertebrate effector IRGs evolved from a role in autophagic regulation to target organisms known to hijack host droplets, such as Chlamydia and Toxoplasma (Lilue et al, 2013), precisely because droplets are so important to these pathogens. As we begin to better understand the interplay between pathogens and host lipid metabolism, it is likely that such evolutionary competition will unveil new modes in which host immunity directly intersects droplet function.
Lipid droplets and immune activation
The adipose tissue that surrounds lymph nodes is specialized to provide both fuel and immuno‐modulatory molecules to immune cells, and is thus an important collaborator in tuning immune activation (Knight, 2008). In addition, immune cell differentiation and activation require switching between specific metabolic programs (Odegaard & Chawla, 2011; Buck et al, 2015). Through their intimate connections with other organelles, droplets play an essential role in modulating cellular metabolism and thus in innate immune cell function and activation. For instance, macrophages lacking ATGL are unable to efficiently use fatty acids as fuel for ATP generation and have a pronounced reduction in phagocytosis of bacteria (Chandak et al, 2010). In fact, the inflammatory state of a macrophage is integrally linked to its metabolism. Pro‐inflammatory “M1‐polarized” macrophages generate the majority of their ATP through glycolysis rather than through oxidative phosphorylation (Rodríguez‐Prados et al, 2010). In anti‐inflammatory, “M2‐polarized” macrophages, however, mitochondria have up‐regulated oxidative phosphorylation and drive the β‐oxidation of fatty acids (and thus to TAG turnover from droplets) as a fuel (Martinez et al, 2006). In fact, β‐oxidation is not only required for alternative activation of macrophages, but can also divert an otherwise pro‐inflammatory response toward M2‐polarization (Vats et al, 2006). CD4+ T cells exhibit similarly polarized metabolic programs as they differentiate into effector T cells, fueled by glucose, or into regulatory T‐cells, fueled by fatty acid β‐oxidation (Michalek et al, 2011). Finally, a study in mast cells demonstrated that droplets can also serve as the main storage site for arachidonic acid, which is the precursor to eicosanoid hormones such as the prostaglandins and leukotrienes that regulate inflammation and vasodilation (Dichlberger et al, 2014).
Lipid droplets as stress buffers and non‐infectious disease
Through both their role in immune modulation and their diverse functions in other cell types, droplets have also been implicated in the pathogenesis of a number of non‐infectious diseases. The role of immune activation in metabolic diseases such as obesity and diabetes has been reviewed elsewhere (Odegaard & Chawla, 2011), though there is growing evidence for a specific role of droplets in modulating these diseases. In particular, droplets are associated with protection from reactive oxygen species (ROS) and sequestration of otherwise‐toxic lipophilic species, and there is evidence that tissues can be protected from stress by the accumulation of droplets in specific cellular sub‐populations (Herms et al, 2013). In fact, glial droplets ensure normal Drosophila neuronal development by protecting neural stem cells from oxidative stress (Bailey et al, 2015). Over time, the insults due to oxidative stress and increased droplets can themselves cause disease. For instance, oxidative stress due to mitochondrial dysfunction leads first to increased glial droplets and finally to neurodegeneration in both Drosophila and mouse models (Liu et al, 2015). Similarly, cancer cells often accumulate droplets compared to normal cells, and droplets are associated with cancer resistance to ROS (Bensaad et al, 2014). Furthermore, droplet accumulation has been suggested to be tumorigenic as the fatty acid‐binding proteins that assist droplet function by shuttling fatty acids between membranes have been implicated as oncogenes (Levi et al, 2013; Powell et al, 2015; Kawaguchi et al, 2016).
Given their close association with oxidative health of a cell, disrupting the level of cellular droplets can be devastating to the health of oxidative tissues. For instance, accumulation of droplets in cardiomyocytes is associated with reduced cardiac function and the development of severe heart disease (Goldberg et al, 2012). Plin5 is highly expressed in oxidative tissues such as heart, muscle, and liver, and it regulates β‐oxidation through modulating TAG storage and release (Fig 3; Wolins et al, 2006). Mice lacking Plin5 have fewer TAG droplets in cardiac tissue, leading to increased oxidative stress, which, in turn, causes age‐related heart defects (Kuramoto et al, 2012). Strikingly, the same lack of cardiac droplets in PLIN5‐knockout mice protect them from diabetes‐induced heart disease (Kuramoto et al, 2014). Thus, droplet homeostasis can be a critical factor in tipping the balance between cellular health and pathogenesis.
The involvement of droplets in a variety of infectious diseases gives them the potential to be central (albeit presently understudied) players in host–pathogen interaction. In particular, pathogenic manipulation of droplet homeostasis may underpin comorbidity of a number of infectious and non‐infectious diseases. The molecular mechanisms of the interactions between lipid droplets and the diverse pathogeneses described above remain largely a mystery. Given the breadth of the alterations to lipid droplet functions driven by pathogenesis, uncovering the mechanisms will profoundly impact a variety of biological fields.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
Support is gratefully acknowledged from the NIH (R01GM084210 to JMG and R35GM119768 to WMH), American Diabetes Association (7‐13‐BS‐055 to JMG), American Heart Association (16GRNT27540010 to JMG), NSF (MCB1553334 to MLR), The Welch Foundation (I‐1936 to MLR and I‐1873 to WMH) and The Searle Foundation (SSP‐2016‐1482 to WMH).
The EMBO Journal (2018) 37: e98947
References
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol 192: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison AJ, Arsenault DJ, Ridgway ND (2015) Nuclear‐localized CTP: phosphocholine cytidylyltransferase alpha regulates phosphatidylcholine synthesis required for lipid droplet biogenesis. Mol Biol Cell 26: 2927–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, Cermelli S, Li Z, Kassan A, Bosch M, Sigua R, Huang L, Ouellette AJ, Pol A, Welte MA, Gross SP (2012) A novel role for lipid droplets in the organismal antibacterial response. eLife 1: e00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AP, Koster G, Guillermier C, Hirst EMA, MacRae JI, Lechene CP, Postle AD, Gould AP (2015) Antioxidant role for lipid droplets in a stem cell niche of Drosophila . Cell 163: 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su WM, Abreu S, Reggiori F, Carman GM, Siniossoglou S (2015) Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Biol Cell 26: 3641–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Siniossoglou S (2017) Function of lipid droplet‐organelle interactions in lipid homeostasis. Biochim Biophys Acta 1864: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li J‐L, Zhang Q, Wakelam MJO, Karpe F, Schulze A, Harris AL (2014) Fatty acid uptake and lipid storage induced by HIF‐1α contribute to cell growth and survival after hypoxia‐reoxygenation. Cell Rep 9: 349–365 [DOI] [PubMed] [Google Scholar]
- Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kuhnlein RP, Huang X (2012) Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci 125: 3568–3577 [DOI] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM (2006) An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster . Cell Metab 11: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette‐Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C (1995) Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res 36: 1211–1226 [PubMed] [Google Scholar]
- Bryant D, Liu Y, Datta S, Hariri H, Seda M, Anderson G, Peskett E, Demetriou C, Sousa S, Jenkins D, Clayton P, Bitner‐Glindzics M, Moore GE, Henne WM, Stanier P (2018) SNX mutations affect endoplasmic reticulum associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum Mol Genet 27: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Pearce EL (2015) T cell metabolism drives immunity. J Exp Med 212: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Honing S (2009) TIP47 functions in the biogenesis of lipid droplets. J Cell Biol 185: 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Zeimetz GM (1996) Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae . J Biol Chem 271: 13293–13296 [DOI] [PubMed] [Google Scholar]
- Cartwright BR, Goodman JM (2012) Seipin: from human disease to molecular mechanism. J Lipid Res 53: 1042–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM (2015) Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell 26: 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandak PG, Radovic B, Aflaki E, Kolb D, Buchebner M, Fröhlich E, Magnes C, Sinner F, Haemmerle G, Zechner R, Tabas I, Levak‐Frank S, Kratky D (2010) Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J Biol Chem 285: 20192–20201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Tsai TH, Li L, Saha P, Chan L, Chang BH (2017) PLIN2 is a key regulator of the unfolded protein response and endoplasmic reticulum stress resolution in pancreatic beta Cells. Sci Rep 7: 40855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U (2010) Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol 84: 6782–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A, Thiam AR (2018) An asymmetry in monolayer tension regulates lipid droplet budding direction. Biophys J 114: 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, Ojha N, Golden A, Prinz WA (2015) A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH (2008) Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci USA 105: 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Desselberger U (2016) Lipid droplets form complexes with viroplasms and are crucial for rotavirus replication. Curr Opin Virol 19: 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichlberger A, Schlager S, Maaninka K, Schneider WJ, Kovanen PT (2014) Adipose triglyceride lipase regulates eicosanoid production in activated human mast cells. J Lipid Res 55: 2471–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubots E, Cottier S, Peli‐Gulli MP, Jaquenoud M, Bontron S, Schneiter R, De Virgilio C (2014) TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS One 9: e104194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg‐Bord M, Mari M, Weill U, Rosenfeld‐Gur E, Moldavski O, Castro IG, Soni KG, Harpaz N, Levine TP, Futerman AH, Reggiori F, Bankaitis VA, Schuldiner M, Bohnert M (2018) Identification of seipin‐linked factors that act as determinants of a lipid droplet subpopulation. J Cell Biol 217: 269–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Zhong L, Ta MT, Shui G, Wenk MR, Yang H (2011) The size and phospholipid composition of lipid droplets can influence their proteome. Biochem Biophys Res Commun 415: 455–462 [DOI] [PubMed] [Google Scholar]
- Ferreira AV, Segatto M, Menezes Z, Macedo AM, Gelape C, de Oliveira Andrade L, Nagajyothi F, Scherer PE, Teixeira MM, Tanowitz HB (2011) Evidence for Trypanosoma cruzi in adipose tissue in human chronic Chagas disease. Microbes Infect 13: 1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Ohsaki Y (2006) Cytoplasmic lipid droplets: rediscovery of an old structure as a unique platform. Ann N Y Acad Sci 1086: 104–115 [DOI] [PubMed] [Google Scholar]
- Gao Q, Binns DD, Kinch LN, Grishin NV, Ortiz N, Chen X, Goodman JM (2017) Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J Cell Biol 216: 3199–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Chang YF, Jesch SA, Aregullin M, Henry SA (2017) Interaction between repressor Opi1p and ER membrane protein Scs2p facilitates transit of phosphatidic acid from the ER to mitochondria and is essential for INO1 gene expression in the presence of choline. J Biol Chem 292: 18713–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluchowski NL, Becuwe M, Walther TC, Farese RV Jr (2017) Lipid droplets and liver disease: from basic biology to clinical implications. Nat Rev Gastroenterol Hepatol 14: 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg IJ, Trent CM, Schulze PC (2012) Lipid metabolism and toxicity in the heart. Cell Metab 15: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire IP, Rabourdin‐Combe C, Faure M (2012) Autophagy and RNA virus interactomes reveal IRGM as a common target. Autophagy 8: 1136–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A, Buxo L, Mora G, Funaya C, Idrissi FZ, Mancuso F, Gomez R, Muntanya J, Sabido E, Carvalho P (2015) The seipin complex Fld1/Ldb16 stabilizes ER‐lipid droplet contact sites. J Cell Biol 211: 829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DA, Zhan C, Silver DL (2011) Direct binding of triglyceride to fat storage‐inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci USA 108: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubern A, Barcelo‐Torns M, Casas J, Barneda D, Masgrau R, Picatoste F, Balsinde J, Balboa MA, Claro E (2009) Lipid droplet biogenesis induced by stress involves triacylglycerol synthesis that depends on group VIA phospholipase A2. J Biol Chem 284: 5697–5708 [DOI] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J (2013) IRG and GBP host resistance factors target aberrant, “Non‐self” vacuoles characterized by the missing of “Self” IRGM proteins. PLoS Pathog 9: e1003414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T (2013) Autophagosomes form at ER‐mitochondria contact sites. Nature 495: 389–393 [DOI] [PubMed] [Google Scholar]
- Han GS, O'Hara L, Carman GM, Siniossoglou S (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem 283: 20433–20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Binns DD, Chang YF, Goodman JM (2015) Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(DeltaNterm) only in combination with Ldb16p. BMC Cell Biol 16: 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Rogers S, Ugrankar R, Liu YL, Feathers JR, Henne WM (2018) Lipid droplet biogenesis is spatially coordinated at ER‐vacuole contacts under nutritional stress. EMBO Rep 19: 57–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C, Herker E, Farese RV, Ott M (2011) Hepatitis C virus core protein decreases lipid droplet turnover: a mechanism for core‐induced steatosis. J Biol Chem 286: 42615–42625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M, Choudary V, Ojha N, Shin JJ, Han GS, Carman GM, Loewen CJ, Prinz WA, Levine T (2017) Fat storage‐inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes. Microb Cell 5: 88‐103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Zhu L, Balogi Z, Stefan C, Pleiss JA, Emr SD (2015) Mdm1/Snx13 is a novel ER‐endolysosomal interorganelle tethering protein. J Cell Biol 210: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Ott M (2012) Emerging role of lipid droplets in host/pathogen interactions. J Biol Chem 287: 2280–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A, Bosch M, Ariotti N, Reddy BJN, Fajardo A, Fernández‐Vidal A, Alvarez‐Guaita A, Fernández‐Rojo MA, Rentero C, Tebar F, Enrich C, Geli M‐I, Parton RG, Gross SP, Pol A (2013) Cell‐to‐cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr Biol 23: 1489–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C (2009) A class of dynamin‐like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Binns D, Reese ML (2017) The coccidian parasites Toxoplasma and Neospora dysregulate mammalian lipid droplet biogenesis. J Biol Chem 292: 11009–11020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunn JP, Feng CG, Sher A, Howard JC (2011) The immunity‐related GTPases in mammals: a fast‐evolving cell‐autonomous resistance system against intracellular pathogens. Mamm Genome 22: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R (2011) Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 124: 2424–2437 [DOI] [PubMed] [Google Scholar]
- Jiang S, Yan C, Fang QC, Shao ML, Zhang YL, Liu Y, Deng YP, Shan B, Liu JQ, Li HT, Yang L, Zhou J, Dai Z, Liu Y, Jia WP (2014) Fibroblast growth factor 21 is regulated by the IRE1alpha‐XBP1 branch of the unfolded protein response and counteracts endoplasmic reticulum stress‐induced hepatic steatosis. J Biol Chem 289: 29751–29765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan TX, Randall G (2017) Dengue virus activates the AMP kinase‐mTOR axis to stimulate a proviral lipophagy. J Virol 91: e02020‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan A, Herms A, Fernandez‐Vidal A, Bosch M, Schieber NL, Reddy BJ, Fajardo A, Gelabert‐Baldrich M, Tebar F, Enrich C, Gross SP, Parton RG, Pol A (2013) Acyl‐CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol 203: 985–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Kinameri A, Suzuki S, Senga S, Ke Y, Fujii H (2016) The cancer‐promoting gene fatty acid‐binding protein 5 (FABP5) is epigenetically regulated during human prostate carcinogenesis. Biochem J 473: 449–461 [DOI] [PubMed] [Google Scholar]
- Klemm RW, Norton JP, Cole RA, Li CS, Park SH, Crane MM, Li L, Jin D, Boye‐Doe A, Liu TY, Shibata Y, Lu H, Rapoport TA, Farese RV Jr, Blackstone C, Guo Y, Mak HY (2013) A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Rep 3: 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC (2008) Specialized perinodal fat fuels and fashions immunity. Immunity 28: 135–138 [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV Jr, Walther TC (2016) Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol 26: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt‐Supprian M, Vance DE, Mann M, Farese RV Jr, Walther TC (2011) Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP: phosphocholine cytidylyltransferase. Cell Metab 14: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto K, Okamura T, Yamaguchi T, Nakamura TY, Wakabayashi S, Morinaga H, Nomura M, Yanase T, Otsu K, Usuda N, Matsumura S, Inoue K, Fushiki T, Kojima Y, Hashimoto T, Sakai F, Hirose F, Osumi T (2012) Perilipin 5, a lipid droplet‐binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J Biol Chem 287: 23852–23863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto K, Sakai F, Yoshinori N, Nakamura TY, Wakabayashi S, Kojidani T, Haraguchi T, Hirose F, Osumi T (2014) Deficiency of a lipid droplet protein, perilipin 5, suppresses myocardial lipid accumulation, thereby preventing type 1 diabetes‐induced heart malfunction. Mol Cell Biol 34: 2721–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoeur H, Giraud E, Prevost MC, Milon G, Lang T (2013) Reprogramming neutral lipid metabolism in mouse dendritic leucocytes hosting live Leishmania amazonensis amastigotes. PLoS Negl Trop Dis 7: e2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi L, Lobo G, Doud MK, Lintig JV, Seachrist D, Tochtrop GP, Noy N (2013) Genetic ablation of the fatty acid–binding protein FABP5 suppresses HER2‐induced mammary tumorigenesis. Can Res 73: 4770–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thiel K, Thul PJ, Beller M, Kühnlein RP, Welte MA (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Current biology: CB 22: 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Luo X, Zhao S, Siu GK, Liang Y, Chan HC, Satoh A, Yu SS (2017) COPI‐TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J 36: 441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue J, Müller UB, Steinfeldt T, Howard JC (2013) Reciprocal virulence and resistance polymorphism in the relationship between Toxoplasma gondii and the house mouse. eLife 2: e01298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y‐C, Chang P‐F, Lin H‐F, Liu K, Chang M‐H, Ni Y‐H (2016) Variants in the autophagy‐related gene IRGM confer susceptibility to non‐alcoholic fatty liver disease by modulating lipophagy. J Hepatol 65: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr, Ory DS, Schaffer JE (2003) Triglyceride accumulation protects against fatty acid‐induced lipotoxicity. Proc Natl Acad Sci USA 100: 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, Bellen HJ (2015) Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Luo M, Fadeev EA, Groves JT (2005) Mycobactin‐mediated iron acquisition within macrophages. Nat Chem Biol 1: 149–153 [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Klemm RW, Junker M, Hannibal‐Bach HK, Ejsing CS, Rapoport TA (2014) An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep 6: 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A (2006) Transcriptional profiling of the human monocyte‐to‐macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177: 7303–7311 [DOI] [PubMed] [Google Scholar]
- Martinez‐Munoz GA, Kane P (2008) Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem 283: 20309–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManaman JL (2009) Formation of milk lipids: a molecular perspective. Clin Lipidol 4: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManaman JL (2014) Lipid transport in the lactating mammary gland. J Mammary Gland Biol Neoplasia 19: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC (2011) Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186: 3299–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Khaddaj R, Cottier S, Stradalova V, Jacob C, Schneiter R (2016) Mature lipid droplets are accessible to ER luminal proteins. J Cell Sci 129: 3803–3815 [DOI] [PubMed] [Google Scholar]
- Moldavski O, Amen T, Levin‐Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M (2015) Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev Cell 33: 603–610 [DOI] [PubMed] [Google Scholar]
- Nchoutmboube JA, Viktorova EG, Scott AJ, Ford LA, Pei Z, Watkins PA, Ernst RK, Belov GA (2013) Increased long chain acyl‐Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog 9: e1003401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, Olzmann JA (2017) DGAT1‐dependent lipid droplet biogenesis protects mitochondrial function during starvation‐induced autophagy. Dev Cell 42: 9–21.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan SJ, Romano JD, Coppens I (2017) Host lipid droplets: an important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog 13: e1006362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A (2011) Alternative macrophage activation and metabolism. Annu Rev Pathol 6: 275–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Fukasawa M, Satoh M, Hanada K, Saijo M, Uchiyama T, Ando S (2014) The intracellular pathogen Orientia tsutsugamushi responsible for scrub typhus induces lipid droplet formation in mouse fibroblasts. Microbes Infect 16: 962–966 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T (2006) Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell 17: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD (2009) Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem 284: 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, Burgess SC (2009) FGF21 induces PGC‐1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci USA 106: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CA, Nasser MW, Zhao H, Wochna JC, Zhang X, Shapiro C, Shilo K, Ganju RK (2015) Fatty acid binding protein 5 promotes metastatic potential of triple negative breast cancer cells through enhancing epidermal growth factor receptor stability. Oncotarget 6: 6373–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic M, Knittelfelder O, Cristobal‐Sarramian A, Kolb D, Wolinski H, Kohlwein SD (2013) The emergence of lipid droplets in yeast: current status and experimental approaches. Curr Genet 59: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott‐Schwartz J (2015) Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32: 678–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reali V, Mehdawy B, Nardacci R, Filomeni G, Risuglia A, Rossin F, Antonioli M, Marsella C, Fimia GM, Piacentini M, Di Sano F (2015) Reticulon protein‐1C is a key component of MAMs. Biochim Biophys Acta 1853: 733–745 [DOI] [PubMed] [Google Scholar]
- Renvoise B, Malone B, Falgairolle M, Munasinghe J, Stadler J, Sibilla C, Park SH, Blackstone C (2016) Reep1 null mice reveal a converging role for hereditary spastic paraplegia proteins in lipid droplet regulation. Hum Mol Genet 25: 5111–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Prados J‐C, Través PG, Cuenca J, Rico D, Aragonés J, Martín‐Sanz P, Cascante M, Boscá L (2010) Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185: 605–614 [DOI] [PubMed] [Google Scholar]
- Roingeard P, Melo RC (2017) Lipid droplet hijacking by intracellular pathogens. Cell Microbiol 19: e12688 [DOI] [PubMed] [Google Scholar]
- Romero‐Brey I, Merz A, Chiramel A, Lee J‐Y, Chlanda P, Haselman U, Santarella‐Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse‐Locker J, Bartenschlager R (2012) Three‐dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8: e1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magre J, Thiele C, Holtta‐Vuori M, Jokitalo E, Ikonen E (2016) Seipin regulates ER‐lipid droplet contacts and cargo delivery. EMBO J 35: 2699–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao‐Miranda I, Barbosa‐Lima G, Da Poian AT, Bozza PT, Gamarnik AV (2009) Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5: e1000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM (2008) Suppression of soybean oleosin produces micro‐oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, Lippincott‐Schwartz J (2017) AMPK and vacuole‐associated Atg14p orchestrate mu‐lipophagy for energy production and long‐term survival under glucose starvation. Elife 6: e21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010) Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker JP, Hoffman RV Jr, Huffman DG (1970) Trypanosoma cruzi: preference for brown adipose tissue in mice by the Tulahuen strain. Exp Parasitol 27: 403–407 [DOI] [PubMed] [Google Scholar]
- Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z (2015) Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J 34: 2117–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE (2009) Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem 284: 30941–30948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy DA, Giddings TH, Kirkegaard K (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy‐like origin for virus‐induced vesicles. J Virol 74: 8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd C, Brasaemle DL (2017) The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim Biophys Acta 1862: 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104: 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V, Johnsen L, Martinez‐Montanes F, Grippa A, Buxo L, Idrissi FZ, Ejsing CS, Carvalho P (2018) Regulation of lipid droplets by metabolically controlled Ldo isoforms. J Cell Biol 217: 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Farese RV Jr, Walther TC (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14: 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Foret L (2016) The physics of lipid droplet nucleation, growth and budding. Biochim Biophys Acta 1861: 715–722 [DOI] [PubMed] [Google Scholar]
- Tirinato L, Pagliari F, Limongi T, Marini M, Falqui A, Seco J, Candeloro P, Liberale C, Di Fabrizio E (2017) An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int 2017: 1656053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA (2013) Direct imaging reveals stable, micrometer‐scale lipid domains that segregate proteins in live cells. J Cell Biol 202: 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S, Rijo‐Ferreira F, Carvalho T, Pinto‐Neves D, Guegan F, Aresta‐Branco F, Bento F, Young SA, Pinto A, Van Den Abbeele J, Ribeiro RM, Dias S, Smith TK, Figueiredo LM (2016) Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host Microbe 19: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott‐Schwartz J (2017) Applying systems‐level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A (2006) Oxidative metabolism and PGC‐1beta attenuate macrophage‐mediated inflammation. Cell Metab 4: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez AP, Graef M (2016) Autophagy regulation depends on ER homeostasis controlled by lipid droplets. Autophagy 12: 1409–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M (2016) Lipid droplet‐mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol 212: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, Pon LA (2015) Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev Cell 35: 584–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T, von Landenberg P, Steinbuchel A (2005) Mechanism of lipid‐body formation in prokaryotes: how bacteria fatten up. Mol Microbiol 55: 750–763 [DOI] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV Jr (2017) Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33: 491–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong DW, Stanley WC, Sztalryd C (2011) Perilipin 5, a lipid droplet‐associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res 52: 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Miao YH, Chang YS (2014) Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci 127: 1214–1228 [DOI] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, Garg A, Olarte MJ, Lin Q, Frohlich F, Hannibal‐Bach HK, Upadhyayula S, Perrimon N, Kirchhausen T, Ejsing CS, Walther TC et al (2016) Seipin is required for converting nascent to mature lipid droplets. Elife 5: e16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA (2015) As the fat flies: the dynamic lipid droplets of Drosophila embryos. Biochim Biophys Acta 1851: 1156–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Gould AP (2017) Lipid droplet functions beyond energy storage. Biochim Biophys Acta 1862: 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr, Walther TC (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV Jr, Walther TC (2014) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3: e01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao‐Borengasser A, Rasouli N, Kern PA, Finck BN, Bickel PE (2006) OXPAT/PAT‐1 is a PPAR‐induced lipid droplet protein that promotes fatty acid utilization. Diabetes 55: 3418–3428 [DOI] [PubMed] [Google Scholar]
- Wolinski H, Hofbauer HF, Hellauer K, Cristobal‐Sarramian A, Kolb D, Radulovic M, Knittelfelder OL, Rechberger GN, Kohlwein SD (2015) Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim Biophys Acta 1851: 1450–1464 [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV Jr, Mak HY (2012) The FATP1‐DGAT2 complex facilitates lipid droplet expansion at the ER‐lipid droplet interface. J Cell Biol 198: 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li Y, Wu L, Li Y, Zhao D, Yu J, Huang T, Ferguson C, Parton RG, Yang H, Li P (2018) Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J Cell Biol 217: 975–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P (2012) The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 53: 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJ (2010) Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329: 1085–1088 [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD (2014) Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 25: 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]


