Abstract
Purpose of review
To synthesize, integrate, and comment on recent research developments to our understanding of the molecular basis of ependymoma, and to place this in context with current treatment and research efforts.
Recent findings
Our recent understanding of the histologically defined molecular entity ependymoma has rapidly advanced through genomic, transcriptomic, and epigenomic profiling studies.
Summary
These advancements lay the groundwork for development of future ependymoma biomarkers, models, and therapeutics. Our review discusses these discoveries and their impact on our clinical understanding of this disease. Lastly, we offer insight into clinical and research areas requiring further validation, and open questions remaining in the field.
Keywords: brain tumor, ependymoma, genetics, epigenetics, tumour heterogeneity
INTRODUCTION
Ependymoma is a histologically defined tumor of the central nervous system (CNS) that occurs in children and adults. Ependymomas of childhood arise most commonly within the supratentorial (ST) brain (i.e. cerebral hemispheres) or posterior fossa (PF, i.e. cerebellum and brainstem). Spinal ependymomas occur most often in adulthood. Treatment for ependymoma to this date remains maximal-safe surgical resection followed by conformal radiation[1]. The effectiveness of chemotherapy for treatment of ependymoma is still highly debated, and continues to be evaluated in numerous ongoing clinical trials (United States - ACNS0831, Europe - SIOP Ependymoma II). There are no targeted therapies in clinical use or being evaluated in multi-center clinical trials for ependymoma. As a result, survival rates have seen limited improvement (~80%) in the last decade, with survivors suffering from the debilitating side effects of treatment-related surgery and radiation.
Historically, histopathologic features have been used to diagnose and risk-stratify ependymoma. However, there has been a failure to develop consistent and reliable criteria to predict ependymoma patient survival, outside of World Health Organization (WHO) Grade I tumors (i.e. subependymoma myxopapillary ependymoma)[2]. Frustration with a lack of consistency with respect to ependymoma histopathologic grading, have motivated clinicians and researchers to leverage unbiased, sensitive, and molecular approaches to both develop reliable prognostic markers of ependymoma, and to decipher the molecular biology of this disease to identify the first set of targeted therapies[3–9].
This review provides a synthesis of recent molecular research developments in the field of ependymoma. We describe the integration and impact of molecular tools and knowledge on our understanding of the biology of ependymoma. We comment upon how these findings may be interpreted with respect to clinical treatment and variables, and propose research areas that require potential future investment.
Ependymoma is comprised of at least nine different subgroups
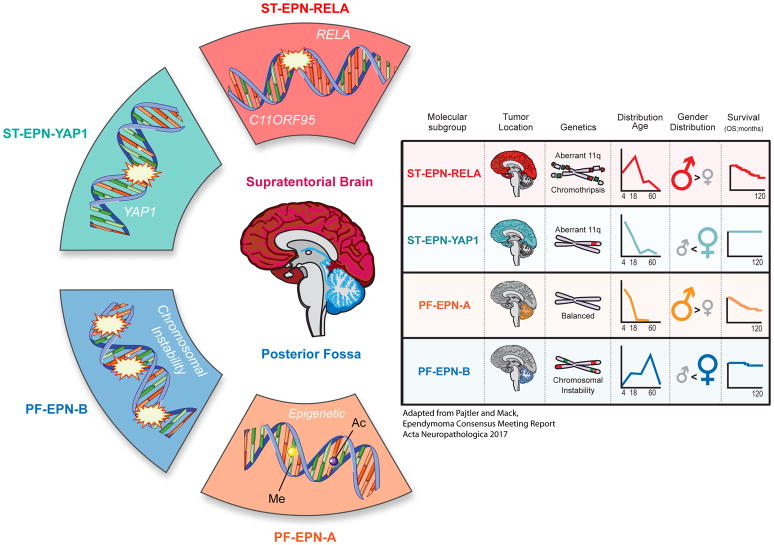
Through independent analysis of multiple ependymoma cohorts, using transcriptomics, genomics, and epigenomics (termed ‘-omics’) technologies, we now recognize that ependymomas are divided into multiple clinically and biologically distinct subgroups. The most recent large-scale analysis reveals that ependymomas are segregated into at least nine molecular subgroups[9]. ST ependymomas are divided into tumors defined by predominant gene fusions between C11ORF95-RELA (72% of ST ependymomas), and less frequently fusions of the YAP1 oncogene with other gene partners, denoted ST-EPN-RELA and ST-EPN-YAP1 subgroup, respectively [4,9] (Figure 1). PF ependymomas are divided into frequent PF-EPN-A tumors (74% of PF ependymomas) with a poor outcome, and relatively balanced genome, versus PF-EPN-B tumors with increased genomic instability and a favorable outcome[7,8,11] (Figure 1). Within both ST and PF compartments exist WHO Grade I classified subependymomas[2]. Spinal ependymomas frequently harbor loss-of-function NF2 mutations or deletions, traditionally classified as Grade I or II, and further composed of subependymoma and myxopapillary ependymoma variants[12,13]. These ‘-omics’ profiling studies of ependymoma reveal, in the largest series of cohorts analyzed to date, that ependymomas are comprised of at least 9 molecularly, demographically, and clinically distinct disease entities[9]. They reinforce that, outside of WHO Grade I ependymomas (i.e. subependymomas and myxopapillary ependymoma), histopathologic grading has no prognostic utility. Therefore we recommend that outside of current clinical trials, histopathologic grading of Grade II or III ependymoma should not be used to risk stratify future patients[10] (Figure 1).
Figure 1.
Summary of intracranial ependymoma subgroups and recommendations for future ependymoma clinical trials. Adapted from the Ependymoma Consensus Meeting manuscript[10].
Clinical integration of ependymoma subgroups
Molecular profiling of multiple large ependymoma cohorts (namely through Illumina DNA methylation analysis) has allowed for analyses to be made on the impact of molecular subgroup and clinical variables[5,9,11]. A study of four distinct PF ependymoma cohorts revealed that while use radiotherapy is effective in the case of PF-EPN-A tumors that were gross-totally resected, it had limited impact in patients harboring PF-EPN-A ependymoma with a sub-total resection[5]. In the same vein, current clinical trial cohorts that are addressing important questions related to other modalities such as the role of chemotherapy (United States - ACNS0831) need to be analyzed in the context of molecular subgroup. In the case of PF-EPN-B, there potentially exists a population of patients that may be cured with surgery alone, and in the case of recurrence, may be salvaged with radiotherapy[5]. This raises a possibility of future clinical efforts to study therapy de-escalation for PF-EPN-B patients[10]. Similar molecular and multi-cohort analysis of ST ependymoma is needed to validate the incidence of C11ORF95-RELA and YAP1 fused tumors in other ependymoma cohorts, and their correlation with survival and other clinical parameters[10]. In contrast to ST-EPN-RELA tumors, which have been shown to have a poor outcome, ST-EPN-YAP1 tumors perform favorably, thus suggesting potentially distinct treatment modalities for this distinct patient population in the future. We therefore recommend that future ependymoma clinical trials should stratify patients by their molecular features, or at very minimum collect frozen tissue for future molecular diagnostics[10].
Frequent fusion oncoproteins define supratentorial ependymoma
72% of ST ependymomas harbor a gene fusion between C11ORF95 and RELA, now recognized by WHO as a distinct clinical entity[2,4]. This event leads to sequestration of the C11ORF95-RELA fusion protein within the nucleus causing aberrant NF-κB activity[4]. Viral over-expression of the C11ORF95-RELA fusion protein in murine embryonic day 14.5 radial glial cells (candidate cell of origin of ependymoma) is capable of cellular transformation and tumor initiation, when transduced cells are implanted in immunodeficient mice[4]. Although C11ORF95-RELA is the most commonly observed fusion, other fusion events have been observed[4]. Recurrent fusion of YAP1 with other fusion partners is thought to induce a distinct subgroup of ST ependymoma, labeled ST-EPN-YAP1 [9]. Unlike ST-EPN-RELA tumors, which have been reported to have a poor survival, ST-EPN-YAP1 tumors carry a favorable prognosis and respond well to current treatments[9]. There remain several important questions to be validated, that similar to PF ependymoma, would benefit from an expanded molecular based analysis of multiple ST ependymoma cohorts. For example, the incidence of these fusion events in ST ependymoma needs to be independently validated. Are ST-EPN-YAP1 fused tumors substantially over-represented from other non-C11ORF95-RELA fusion events? In other cohorts, do YAP1 fused ependymomas carry a favorable prognosis? More broadly, do all ST ependymomas carry a gene fusion? It will be important that current and future ST ependymoma clinical trial studies examine the incidence, and clinical associations with clinical parameters of onco-fusion protein events[10]. While Illumina DNA methylation provides an indirect (yet highly reliable) measure of molecular subgroup associated with a particular fusion, break apart fluorescence in situ hybridization can be used to identify and verify samples harboring the C11ORF95-RELA fusion. RNA-seq also provides an approach to identify C11ORF95-RELA fusions and provides a discovery method to identify novel or less obvious fusion events. Therapeutically targeting these fusion oncogenic events is currently a challenge as this class of transcriptional machinery proteins (i.e. Transcription factors and regulators) has been challenging in the past to target with small molecules. There may be potential in evaluation of NF-κB or YAP1 inhibitors in pre-clinical models where available[14,15]. An important direction moving forward will be to determine if these fusion proteins reveal potential druggable binding partners and synthetic lethal cancer cell dependencies[16,17].
Global epigenomic rewiring defines posterior fossa ependymoma
PF ependymoma are divided into two principle molecular subgroups, PF-EPN-A and PF-EPN-B. PF-EPN-B tumors arise in older children through to adulthood, carry a favorable prognosis, and also demonstrate widespread chromosomal aneuploidy[8]. This is in direct contrast to the most frequent PF-EPN-A subgroup (74% of PF cases), which despite in-depth genomic characterization, has yet to reveal a highly recurrent focal driver alteration[10,18]. Despite lacking a clear genetic driver mutation, PF-EPN-A tumors demonstrate widespread epigenomic alterations in the form of DNA CpG island hypermethylation and global DNA hypomethylation, a phenotype well described in many solid cancers[10,18–20]. Furthermore, PF-EPN-A tumors demonstrate global H3K27me3 loss, a histone modification important for regulating bivalent gene expression and maintaining cell identity[21,22]. Gene targets of DNA and H3K27 hypermethylation overlap significantly with patterns of DNA and histone H3K27 methylation seen in embryonic stem cells. Perhaps a necessary component of neoplastic transformation of ependymoma cells involves acquisition of features found in the embryonic stem cell state. Interestingly, diffuse intrinsic pontine gliomas (DIPGs) that carry recurrent K27M mutations exhibit similar patterns of global H3K27me3 and DNA methylation loss, suggesting potentially convergent mechanisms of neoplastic transformation and/or similar cell types or states that give rise to these distinct brain tumor entities[23,24]. Although very rare, a small subset of PF-EPN-A ependymomas harbor K27M mutations, raising the question as to what is the cause of global DNA and H3K27me3 loss in non-histone mutated ependymoma[25]. This also raises the opportunity for potential shared therapeutic paradigms to exploit dysregulated epigenomes. As seen in ependymoma and DIPG, the residual H3K27me3 in tumors is important for tumor cell maintenance, because inhibition with EZH2 inhibitors is sufficient to block tumor cell growth[11,18,19,26,27]. Research efforts in the future may benefit from exploring the potential for exploiting tumors that demonstrate global H3K27me3 loss for potential synthetic lethal pathways[16,17].
Models desperately needed
Understanding the genetic basis of ependymoma has led to the development of ex vivo models through over-expression of the C11ORF95-RELA fusion protein, and implantation of transduced radial glia into immunodeficient mice[3,6,28]. This has allowed for the mechanistic dissection of the biology of C11ORF95-RELA fusion protein in initiating and maintaining ependymoma tumorigenesis in mouse models. These mouse isogeneic models offer opportunities to identify ST-EPN-RELA cell dependencies through mechanistic study, and genetic and chemical screens. They also provide a foundation for development of transgenic ependymoma models, which would be useful for mapping the cellular origins of the disease. Still widely lacking is the availability of human patient derived models. In the case of C11ORF95-RELA tumors, the model EP1-NS harbors the RELA fusion and shares similar transcriptional and DNA methylation signatures with human primary ST-EPN-RELA tumors (data unpublished)[29]. Given the number of ependymoma subgroups, additional models are desperately needed for pre-clinical therapeutic development. This is especially the case for PF-EPN-A ependymoma where primary cultures can be maintained for a few passages before they senescence or begin to exhibit patterns of genetic and epigenetic drift. Given the rarity of these tumors, collaborative efforts are essential to establishing, sharing, and characterizing new ependymoma models in order for biological advancements to proceed[10].
Conclusion: The future of ‘-omics’ to study ependymoma
‘-omics’ based discovery approaches identified the main driver of ST ependymoma. Can additional applications such as epigenomics, proteomics, single cell analysis, and metabolomics reveal other oncogenic drivers, and offer further insight into the mechanisms of ependymoma-genesis? Such analysis may proof useful in the case of PF-EPN-A and PF-EPN-B ependymomas where frequent drivers are unknown. For example, the repressive landscape has been defined through histone methylation mapping, but what about other epigenetic modifications in particular active epigenomic marks, such as H3K27 acetylation? Could these actively transcribed domains reveal recurrent regulatory programs that lead to oncogenic activation as reported in other brain tumors?[30] Will metabolic approaches performed in primary tumors help us determine the optimal conditions for primary cell culture, such that more models are established, and platforms can be developed for therapeutic screening? Have we missed cryptic lesions or mutations that might be revealed with advanced genomic sequencing? Have we underestimated the full degree of heterogeneity within subgroups of ependymoma, and that these tumors - like medulloblastoma - are composed of many more subtypes?[31,32] These questions may be answered readily with continued partnerships between researchers and clinicians, working towards international collaboration and shared access to samples, models, and data.
KEYPOINTS.
Outside of clinical trials histopathologic grading II/III of ependymoma is not useful
Ependymoma is composed of at least nine different diseases
Frequent gene fusions define supratentorial ependymoma, namely C11ORF95-RELA
Global epigenomic rewiring defines posterior fossa ependymoma
‘-omics’ based tumor characterization will continue to unravel the molecular basis of ependymoma and its subgroups
Acknowledgments
FINANCIAL
This work was supported by an Alex’s Lemonade Stand Young Investigator Award (SCM) and The CIHR Banting Fellowship (SCM). MDT is supported by The Garron Family Chair in Childhood Cancer Research, and grants from the Pediatric Brain Tumour Foundation, Grand Challenge Award from CureSearch for Children’s Cancer, the National Institutes of Health (R01CA148699 R01CA159859), The Terry Fox Research Institute, and Brainchild. MDT is also supported by a Stand Up To Cancer − St. Baldrick’s Pediatric Dream Team Translational Research Grant (SU2C-AACR-DT1113). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. We thank Susan Archer for technical writing expertise.
Footnotes
ACKNOWLEDGEMENTS
None
CONFLICTS OF INTEREST
None
REFERENCE LIST
- 1.Merchant TE, Li C, Xiong X, Kun LE, Boop FA, Sanford RA. Conformal radiotherapy after surgery for paediatric ependymoma: A prospective study. The lancet oncology. 2009;10(3):258–266. doi: 10.1016/S1470-2045(08)70342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 who classification of tumours of the central nervous system. Acta neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C, Hogg T, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, White E, et al. Nature. 2014. C11orf95-rela fusions drive oncogenic nf-kappab signalling in ependymoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW, Jones DT, Luu B, Cavalli FM, Aldape K, Remke M, et al. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: A retrospective multicohort analysis. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016 doi: 10.1200/JCO.2015.65.7825. An analysis of four independent posterior fossa ependymoma cohorts correlating ependymoma molecular subgroup with clinical outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J, Macdonald T, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer cell. 2005;8(4):323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Wani K, Armstrong TS, Vera-Bolanos E, Raghunathan A, Ellison D, Gilbertson R, Vaillant B, Goldman S, Packer RJ, Fouladi M, Pollack I, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta neuropathologica. 2012;123(5):727–738. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, Jones DT, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, et al. Molecular classification of ependymal tumors across all cns compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. doi: 10.1016/j.ccell.2015.04.002. An unsupervised analysis demonstrating that ependymomas consist of at least 9 molecular subgroups using both epigenomic and transcriptomic data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A, Hansford JR, von Hoff K, Wright KD, Hwang E, Frappaz D, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta neuropathologica. 2017;133(1):5–12. doi: 10.1007/s00401-016-1643-0. The current molecular consensus of ependymoma molecular subgroups, impact on clinical parameters, and consensus recommendations for future clinical trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, Zhang X, et al. Epigenomic alterations define lethal cimp-positive ependymomas of infancy. Nature. 2014 doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Mack SC, Agnihotri S, Bertrand KC, Wang X, Shih DJ, Witt H, Hill N, Zayne K, Barszczyk M, Ramaswamy V, Remke M, et al. Spinal myxopapillary ependymomas demonstrate a warburg phenotype. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-14-2650. An analysis of the genomic landscape of spinal ependymomas highlighting the importance of metabolic alterations in distinct subsets of spinal myxopapillary tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert C, von Haken M, Meyer-Puttlitz B, Wiestler OD, Reifenberger G, Pietsch T, von Deimling A. Molecular genetic analysis of ependymal tumors. Nf2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am J Pathol. 1999;155(2):627–632. doi: 10.1016/S0002-9440(10)65158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiDonato JA, Mercurio F, Karin M. Nf-kappab and the link between inflammation and cancer. Immunological reviews. 2012;246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R, Halder G. The two faces of hippo: Targeting the hippo pathway for regenerative medicine and cancer treatment. Nature reviews Drug discovery. 2014;13(1):63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson JM, Shelat AA, Carcaboso AM, Kranenburg TA, Arnold LA, Boulos N, Wright K, Johnson RA, Poppleton H, Mohankumar KM, Feau C, et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell. 2011;20(3):384–399. doi: 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Mohankumar KM, Currle DS, White E, Boulos N, Dapper J, Eden C, Nimmervoll B, Thiruvenkatam R, Connelly M, Kranenburg TA. An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. 2015;47(8):878–887. doi: 10.1038/ng.3323. An in vivo gain and loss of function screen of ependymoma oncogenes and tumor suppressor genes identified by recurrent copy number alterations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mack SC, Witt H, Wang X, Milde T, Yao Y, Bertrand KC, Korshunov A, Pfister SM, Taylor MD. Emerging insights into the ependymoma epigenome. Brain pathology (Zurich, Switzerland) 2013;23(2):206–209. doi: 10.1111/bpa.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mack SC, Taylor MD. The genetic and epigenetic basis of ependymoma. Child’s nervous system: ChNS: official journal of the International Society for Pediatric Neurosurgery. 2009;25(10):1195–1201. doi: 10.1007/s00381-009-0928-1. [DOI] [PubMed] [Google Scholar]
- 20.Mack SC, Hubert CG, Miller TE, Taylor MD, Rich JN. An epigenetic gateway to brain tumor cell identity. 2016;19(1):10–19. doi: 10.1038/nn.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C, Yip S, Hukin J, Sun Y, Schipper MJ, Chavez L, et al. Immunohistochemical analysis of h3k27me3 demonstrates global reduction in group-a childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017 doi: 10.1007/s00401-017-1752-4. Correlation of depleted H327me3 levels with PF-EPN-A status of ependymomas as a potential diagnostic and prognostic markers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, Pekmezci M, et al. Lowered h3k27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Science translational medicine. 2016;8(366):366ra161. doi: 10.1126/scitranslmed.aah6904. Identification of a global loss of H3K27me3 levels in PF-EPN-A ependymomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, et al. Driver mutations in histone h3. 3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 24.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, Easton J, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nature genetics. 2014;46(5):444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryall S, Guzman M, Elbabaa SK, Luu B, Mack SC, Zapotocky M, Taylor MD, Hawkins C, Ramaswamy V. H3 k27m mutations are extremely rare in posterior fossa group a ependymoma. 2017;33(7):1047–1051. doi: 10.1007/s00381-017-3481-3. [DOI] [PubMed] [Google Scholar]
- 26.Mack SC, Hubert CG, Miller TE. An epigenetic gateway to brain tumor cell identity. 2016;19(1):10–19. doi: 10.1038/nn.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, Tvardovskiy A, et al. Ezh2 is a potential therapeutic target for h3k27m-mutant pediatric gliomas. 2017;23(4):483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 28.Parker IM, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, White E, et al. C11orf95-rela fusions drive oncogenic nf-kappab signalling in ependymoma. Nature. 2014;506(7489):451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milde T, Kleber S, Korshunov A, Witt H, Hielscher T, Koch P, Kopp HG, Jugold M, Deubzer HE, Oehme I, Lodrini M, et al. A novel human high-risk ependymoma stem cell model reveals the differentiation-inducing potential of the histone deacetylase inhibitor vorinostat. Acta neuropathologica. 2011;122(5):637–650. doi: 10.1007/s00401-011-0866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, Jones DT, et al. Enhancer hijacking activates gfi1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, Agnihotri S, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer cell. 2017;31(6):737–754. e736. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, et al. Medulloblastoma comprises four distinct molecular variants. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]