Abstract
Melanoma skin cancer is the fastest growing cancer in the US [1]. A great need exists for improved formulations and mechanisms to prevent and protect human skin from cancers and other skin damage caused by sunlight exposure. Current efforts to prevent UV damage to human skin, which in many cases leads to melanoma and other skin cancers. The primordial melanocortin-1 receptor (MC1R) is involved in regulating skin pigmentation and hair color, which is a natural prevention from UV damage. The endogenous melanocortin agonists induce pigmentation and share a core pharmacophore sequence “His-Phe-Arg-Trp”, and it was found that substitution of the Phe by D-Phe results in increasing melanocortin receptor potency. To improve the melanocortin 1 receptor (MC1R) selectivity a series of tetra-peptides with the moiety of Ac-Xaa-Yaa-Nle-Trp-NH2, and structural modifications to reduce electrostatic ligand-receptor interactions have been designed and synthesized. It is discovered that the tetrapeptide Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2 resulted in a potent and selective hMC1R agonist at the hMC1R (EC50: 10 nM). Lizard anolis carolinensis pigmentation study shows very high potency in vivo. NMR studies revealed a reversed β turn structure which led to the potency and selectivity towards the hMC1R.
Keywords: α-MSH, Melanocortin peptide, Selectivity, hMC1R, Tetra-peptide, Skin pigmentation
1. Introduction
Melanoma is among the utmost prevalent cancer diseases [1]. It is estimated that over 80% of malignant melanomas express higher levels of melanocyte stimulating hormone (α-MSH) receptors, human melanocortin 1 receptor (hMC1R) [2], a member of the melanocortin receptor family, which belongs to 7-transmembrane, G protein-coupled receptors (GPCRs) that control various physiological functions that are critical for survival [3]. In particular, the hMC1R is associated with skin pigmentation. Upon activation, the hMC1R in melanocytes and keratinocytes will form the pigmentation to block the UV radiation to prevent skin damage [4–19]. The endogenous melanocortin peptides are all agonists to hMCRs and include α-melanocyte stimulate hormone, α-MSH; β-melanocyte stimulate hormone, β-MSH; and γ-melanocyte stimulate hormone, γ-MSH. They all have a core pharmacophore structure of tetra-peptide (His-Phe-Arg-Trp) sequence [20]. Our numerous previous studies have demonstrated that the tetra-peptide -His-Phe-Arg-Trp- is a minimum sequence which has the capability of activating all hMCRs [21–27]. Malignant melanoma is the most fatal form of skin cancer. The involvement of MC1 receptor during the proliferation of melanoma cells suggests that α-MSH and its analogues may be candidates for melanoma prevention [18,19,28–31]. The current marketed drug for skin pigmentation disorder is an α-MSH analogue NDP-α-MSH called melanotan I (MT-I). However, NDP-α-MSH is a 13 amino acid peptide with no selectivity towards all the other hMCRs subtypes. Therefore, development of novel analogues with higher MC1R selectivity, a shorter sequence and more druggable properties are needed. Our novel designed tetra peptides show highly selective hMC1R agonist activity and skin pigmentation capability, which can be used to protect against melanoma.
2. Design of novel tetrapeptides
It was previously discovered that the tetrapeptide Ac-His-D-Phe-Arg-Trp-NH2, which contains the tetrapeptide pharmacophore sequence of NDP-α-MSH, is the shortest melanotropin peptide required for binding and activation of melanocortin receptors [32]. However, it has poor potency and selectivity to all subtypes of hMCRs. Thus, modifying the tetrapeptide structure will be of critical importance to improve the potency and selectivity to hMC1R while keeping the short sequence. Modifications were mainly focused on three different sites of the Ac-His-D-Phe-Arg-Trp-NH2 template: 1. His was substituted with Pro in peptide 8–12. 2. We used either D-Phe with different halogenation groups (F, CF3, Cl, Br) at the para position or D-Nal(2′) to substitute the D-Phe position, which has been found to improve potency and modulate selectivity in the Ac-His-D-Phe-Arg-Trp-NH2 template [33,34]. It was noticed that substitutions such as halogenated D-Phe and D-Nal(2′) at D-Phe position were shown to reduce the tetrapeptide's ability to activate MC3R and MC4R, leading to partial agonism or even antagonism at MC3R and MC4R [33,34]. 3. We envisioned that enhanced selectivity towards the MC1R can be reached with reduced electrostatic interaction between the tetrapeptide and the respective aspartic acids on the MC3R and MC4R receptors. Previous receptor mutagenesis studies demonstrated that the electrostatic interaction between the Arg8 of the NDP-α-MSH and the Asp122, Asp126 of the hMC4R is of critical importance to achieve receptor activation, as evidenced by a more than 400-folds increase in the EC50 value for the Asp126Asn mutant [35]. Similarly, a key interaction between the Arg8 of the NDP-α-MSH and the Asp154, Asp158 of the MC3R is necessary, as Asp158Ala mutation on MC3R led to more than 350-fold increase for the EC50 value [36]. In contrast, Asp117 and Asp121 play much less role for interactions between hMC1R and NDP-α-SH, as evidenced by only around 10-fold increase on the IC50 and EC50 values for the Asp117Ala and Asp121Ala mutants [37]. Therefore, switching the arginine in the tetrapeptide to the neutrally charged amino acid norleucine, which has similar shape and size as arginine, should reduce binding towards the hMC3R and the hMC4R. Herein, a series of Nle8 tetra-peptides, Ac-Xaa-Yaa-Nle-Trp-NH2, were designed and synthesized. (Table 1).
Table 1.
Sequences and the physicochemical properties of peptides.
| No. Sequence | m/z calcd [M+H] | m/z obsd [M+H]/[M+Na] | HPLCa System 1 | HPLCa System 2 | TLCb System 1 4: 1 | TLCb System 2 4: 1: 0.5 | |
|---|---|---|---|---|---|---|---|
| 1 | Ac-His-DPhe-Arg-Trp-NH2 | 686.3 | 686.3 | 27.8 | 32.5 | 0.11 | 0.17 |
| 2 | Ac-His-DPhe-Nle-Trp-NH2 | 643.2 | 643.3 | 27.6 | 32.2 | 0.31 | 0.36 |
| 3 | Ac-His-DNal(2′)-Nle-Trp-NH2 | 693.2 | 693.3 | 25.1 | 31.6 | 0.16 | 0.41 |
| 4 | Ac-His-DPhe(4-F)-Nle-Trp-NH2 | 661.3 | 661.3 | 25.1 | 31.3 | 0.19 | 0.4 |
| 5 | Ac-His-DPhe(4-CF3)-Nle-Trp-NH2 | 711.2 | 711.3 | 27.5 | 32.8 | 0.18 | 0.35 |
| 6 | Ac-His-DPhe(4-Cl)-Nle-Trp-NH2 | 677.3 | 677.2 | 27.3 | 32.1 | 0.16 | 0.35 |
| 7 | Ac-His-DPhe(4-Br)–Nle-Trp-NH2 | 721.4 | 721.2 | 27.4 | 31.9 | 0.20 | 0.45 |
| 8 | Ac-Pro-DPhe-Nle-Trp-NH2 | 625.3 | 625.3 | 25.2 | 30.4 | 0.50 | 0.76 |
| 9 | Ac-Pro-DPhe(4-F)-Nle-Trp-NH2 | 643.3 | 643.3 | 25.4 | 31.8 | 0.47 | 0.73 |
| 10 | Ac-Pro-DPhe(4-CF3)-Nle-Trp-NH2 (Na+) | 671.3 | 693.3 | 27.7 | 32.2 | 0.47 | 0.76 |
| 11 | Ac-Pro-DPhe(4-Cl)-Nle-Trp-NH2 | 637.2 | 637.7 | 27.2 | 32.3 | 0.52 | 0.78 |
| 12 | Ac-Pro-DPhe(4-Br)-Nle-Trp-NH2 (Na+) | 681.2 | 703.2 | 27.2 | 32.2 | 0.52 | 0.77 |
HPLC column, YMC-Pack ODS-AM 150 _ 4.6 mm, S-3 ím, 120 A. HPLC system 1: solvent A, 0.1% TFA in water; solvent B, 0.08% TFA in acetonitrile; gradient, 2–80% B in A over 30 min, flow rate 0.8 mL/min. HPLC system 2: solvent A, 1% formic acid in water; solvent B, 1% formic acid in methanol; gradient, 2–80% B in A over 40 min, flow rate 0.8 mL/min.
TLC system 1, CHCl3/MeOH (4:1); TLC system 2, CHCl3/MeOH/AcOH (4:1:0.5).
3. Results and discussion
3.1. Binding and cAMP studies
The biological activities of the newly designed tetra-peptides were analyzed by binding and cAMP assays using stable HEK293 cell lines which express the hMC1R, hMC3R, hMC4R and hMC5R(Table 2). Our first step was replacing Arg8 with Nle8. As we expected, peptide 2, the Nle8 replaced tetra-peptide, lost 50% binding efficiency for all subtypes of hMCRs compared with the parent tetra-peptide, peptide 1. However, Peptide 2 kept 100% cAMP efficacy for the hMC1R, and the binding affinity for Peptide 2 towards hMC3R was greater than 1.0 μM. Thus, Peptide 2 tends to be more selective for the hMC1R. Introducing the bulky amino acid D-Nal (2′)7 into the Nle8 replaced tetra-peptide (peptide 3) abolished the binding affinity to all of the hMCRs. Nevertheless, it still retains 100% cAMP activity at the hMC1R. In order to improve the binding affinity for the hMC1R we introduced halogenated group (F, CF3, Cl, Br) into the D-Phe6 in the Nle8 replaced tetrapeptides (Peptides 4–7). Table 2 shows that peptides 5–7 have increased binding affinities towards the hMC1R along with binding efficiencies that are greater than 50%. In addition, peptides 4–7 retain 100% cAMP activity at the hMC1R. Among these four peptides, Peptide 4, Ac-His-D-Phe(4-F)-Nle-Trp-NH2, and Peptide 5, Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2, resulted in selective hMC1R agonists at the hMC1R with EC50: 25 nM and 10 nM respectively. Among these four peptides, peptide 5 (Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2) is a potent hMC1R agonist (EC50: 10 nM) with the strongest selectivity of at least 25-fold to other MCR subtypes.
Table 2.
Competitive Binding and cAMP Accumulation Results at the hMCRs.
| Peptides | MC1R
|
MC3R
|
MC4R
|
MC5R
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | % BE | EC50(nM) | Act % | IC50(nM) | % BE | EC50(nM) | Act % | IC50(nM) | % BE | EC50(nM) | Act % | IC50(nM) | % BE | EC50(nM) | Act % | |
| 1 | 105 ± 16 | 100 | 4.3 ± 0.2 | 100 | 63 ± 5 | 100 | 7.1 ± 0.1 | 100 | 359 ± 52 | 76 | 548 ± 58 | 100 | 109 ± 8 | 100 | 3.9 ± 0.6 | 63 |
| 2 | 50 ± 9 | 42 | 52 ± 4 | 100 | >1000 | 15 | 120 ± 12 | 100 | 65 ± 6 | 58 | 928 ± 23 | 52 | 46 ± 4 | 44 | 37 ± 4 | 48 |
| 3 | >1000 | 65 | 418 ± 69 | 100 | >1000 | 80 | 255 ± 26 | 17 | >1000 | 44 | 133 ± 3 | 46 | NB | 0 | NA | 0 |
| 4 | 243 ± 36 | 33 | 25 ± 3 | 100 | >1000 | 13 | 315 ± 38 | 100 | >1000 | 13 | 280 ± 34 | 58 | NB | 0 | NA | 0 |
| 5 | 339 ± 31 | 49 | 10 ± 0.7 | 100 | 90 ± 7 | 62 | 252 ± 26 | 85 | 835 ± 119 | 43 | 407 ± 28 | 79 | 37 ± 4 | 37 | >1000 | 44 |
| 6 | 122 ± 16 | 81 | 6.5 ± 0.7 | 100 | >1000 | 45 | 116 ± 13 | 87 | >1000 | 8 | 17 ± 2 | 55 | NB | 0 | 433 ± 60 | 52 |
| 7 | 154 ± 9 | 61 | 11 ± 1 | 100 | 224 ± 25 | 51 | 57 ± 12 | 93 | 53 ± 10 | 46 | 36 ± 4 | 79 | NB | 0 | >1000 | 33 |
| 8 | NB | 0 | NA | 0 | >1000 | 65 | NA | 0 | NB | 0 | NA | 0 | NB | 0 | NA | 0 |
| 9 | 0.74 ± 0.26 | 28 | >1000 | 46 | 46.8 ± 0.7 | 35 | >1000 | 56 | NB | 0 | NA | 0 | NB | 0 | NA | 0 |
| 10 | NB | 0 | NA | 0 | >1000 | 33 | NA | 0 | 2.1 ± 0.4 | 22 | NA | 0 | NB | 0 | NA | 0 |
| 11 | 1.9 ± 0.01 | 18 | 3 ± 0.3 | 63 | >1000 | 35 | >1000 | 23.1 | NB | 0 | NA | 0 | NB | 0 | NA | 0 |
| 12 | 80 ± 15 | 30 | >1000 | 39 | >1000 | 76 | >1000 | 23.1 | 10 ± 1 | 52 | NA | 0 | NB | 0 | NA | 0 |
| MT-II | 1 ± 0.1 | 100 | 1 ± 0.4 | 100 | 2.0 ± 0.1 | 100 | 5.1 ± 0.3 | 100 | 2.3 ± 0.85 | 100 | 2 ± 0.6 | 100 | 4 ± 1 | 100 | 6 ± 2 | 100 |
MT-II, Ac-Nle-c[Asp-His-D-Phe-Arg-Trp-Lys]-NH2, is used as a control for the entire assays. IC50 = concentration of peptide at 50% specific binding (N = 3). NB = 0% of [125I]-NDP-α-MSH displacement observed at 10 μM %BE (binding efficiency) = maximal % of [125I]-NDP-α-MSH displacement observed at 10 μM. EC50 = Effective concentration of peptide that was able to generate 50% maximal intracellular cAMP accumulation (N = 3). Act% = % of cAMP produced at 10 μM ligand concentration, in relation to MT-II. NA = 0% cAMP accumulation observed at 10 μM. The peptides were tested at a range of concentrations from 10−10 to10−5 M.
In order to study the effects of modifying the MCR pharmacophore His-Phe-Arg-Trp sequence with respect to the introduction of Nle8 to achieve MC1R selectivity, His6 was replaced with Pro6 in Peptides 8–12. This was done to determine the influence of a more sterically constrained residue and its ability to further improve molecular recognition towards hMC1R. As shown in Table 2, Pro6 substituted tetra-peptides lost most of the binding and functional activities for all subtypes of the hMCRs, except for Peptide 11. Interestingly, Peptide 11 displayed a cAMP activity level of 63%, but only had a binding efficiency of 18%. Despite the fact that the partial agonism of cAMP levels, the poor binding efficiency suggests that the proline substitution for histidine in these tetrapeptide is not ideal for hMC1R selectivity.
3.2. Pigmentation studies
Pigmentation studies were performed on lizard anolis carolinensis to analyze the in vivo pigmentation effect of Peptide 5. Lizards were given an intraperitoneal injection of the vehicle or peptide 5 at 3 μg/g. Peptide 5 induced pigmentation of the lizard within 1 h of injection (Fig. 1), while injection of the vehicle did not produce any pigmentation effect (data not shown). The natural green color was able to resume in less than 24 h.
Fig. 1.
Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2 induced pigmentation on the lizard. Left, before injection; Right, after injection. Peptide samples were dissolved in saline at the concentration of 1 mM. The total amount of peptide was through i.p. injection with 3 μg/g of each lizard. The strategy follows previous publications [21,38–41].
3.3. NMR analysis
Biological studies revealed that the Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2 (Peptide 5) is a selective hMC1R agonist. We performed a comprehensive NMR study of Peptide 5.
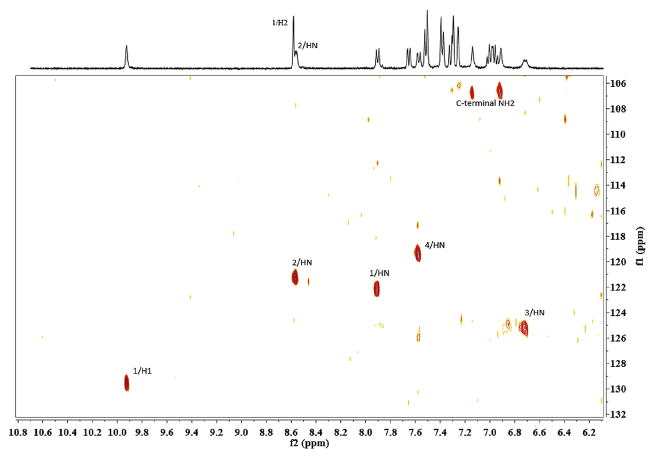
A complete assignment was achieved for all proton resonances based on the homonuclear 2D spectra protocol established by Wüthrich et al. (Table 3). The amide and aromatic proton resonances are well resolved in the 1D proton spectrum. The natural abundance 15N-HSQC facilitates differentiating the amide and aromatic proton resonances in the overlapping region (Fig. 2). Sequential NOE connectivities from N-terminal to C-terminal residues in the fingerprint region of 2D NOESY were assigned unambiguously. (Supporting Information) No chemical exchange or multiple spin systems due to minor conformations were observed in the spectra. The aromatic ring assignments were established by the intraresidual NOEs between beta protons to the spatially close protons in the aromatic rings. The 13C chemical shift of the Cα other than His1 Cα were also confirmed by the 13C-HSQC (His1 Hα overlapped with water peak).Chemical shift index method is routinely applied to identify the alpha-helices or beta-sheets using the 1Hα/13Cα chemical shifts variations relative to the values observed in the random coils. However, for the case of peptides with very short length, it is difficult to interpret the results of chemical shift index method. The lack of random coil reference values for non-natural amino acids is another hindrance. Nonetheless, the 1Hα/13Cα chemical shifts of His1 and Trp4 are very close to the values observed in random coils (His: Hα 4.73, random coil 4.63 ± 0.10; Trp Hα 4.58, Cα, 57.44 ppm, random coil 4.70 ± 0.10, 57.8 ± 0.7). The lack of significant up field shifts does exclude the possibility of the ring current effects due to the aromatic side chain stacking [42]. The inter-residual NOE connectivity's point to a β-turn structure. Other than the sequential dαN and dβN in the range of His1-Phe2- Leu3-Trp4, there exists the dαN (i, i+2) and dβN (i,i+2) between F2 and W4, and multiple dNN(i,i+2) and dNN (i,i+3) NOEs. The sequential HN-HN NOEs were observed along His1-Phe2-Leu3-Trp4. The direct HN-HN NOEs are also observed for His1-Trp4 and Phe2-Trp4. Another outward sign of the turn like structure is the long-range NOEs observed between N-terminal acetyl methyl group and Trp4 side chains, which results from the spatial closeness. The temperature coefficient for NH resonance of residue Trp4 is low (−Δδ/ΔT < 4.5 ppb/K), a hydrogen-bonding indicator [43]. The Nle3 NH is broad and weak, a sign that it is more exposed than other amide protons. Fig. 2.
Table 3.
Chemical shifts of Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2.
| NH | 3JHN-Hα | Cα | Hα | Hβ | Others | |
|---|---|---|---|---|---|---|
| His1 | 7.91 | 8.2 | – | 4.73 | 2.92, 3.05 | H2 8.58, H4 7.30; N-terminal CH3CO 1.92 |
| D-Phe(4-CF3)2 | 8.57 | 5.6 | 58.88 | 4.27 | 3.02 | H2 7.39, H3 7.52 |
| Nle3 | 6.72 | 8.2 | 56.70 | 3.86 | 0.72,1.06 | Hγ 0.34,0.24, Hδ 0.89, Hε 0.61 |
| Trp4 | 7.58 | 8.6 | 57.48 | 4.58 | 3.21, 3.35 | H1 9.93, H2 7.26, H4 7.66, H5 6.96, H6 7.00, H7 7.32; C-terminal NH2 6.92,7.14 |
pH = 5.5, 25°C, DSS as reference. His1 Cα was not assigned due to the Hα overlapped with water peak in the 13C-HSQC spectrum.
Fig. 2.
The natural abundance 15N-HSQC spectrum of peptide 5, sample concentration: 4.7 mM, number of scans: 128, F1 dimension: 128 data points, 40 ppm spectral width centered at 120 ppm, F2 dimension: 2048 data points, 14 ppm spectrum width centered at 120 ppm, relaxation delay:1.5s.
A consistent preference for the β-turn structure has emerged from the distance-restrained simulated annealing calculations of the peptide 5. The majority of the ensemble of the 300 structures generated by the distance-restrained molecular dynamics calculation shows that the His1 Cα-Trp 4 Cα distance is less than 7 Å, and the distance between the His1 CO and the amide hydrogen of Trp4 is less than 2.5 Å. An ensemble of 10 representative NMR structures were selected based on the criterion of low NOE derived distances violations and low potential energy Fig. 3. The summary of the RMSD of the structures and the distance violations are shown in Table 4. The side chains packing in the calculated structure ensembles is not well converged, showing that the peptide is flexible in solution without a single rigid conformation. It is in agreement with the observation that the 1Hα, 13Cα chemical shifts are close to the values in random coils. Meanwhile the peptide has the preference for the β-turn conformations in its free energy landscape, reflected in the consistent β-turn conformation in the ensemble of the NMR derived structures.
Fig. 3.
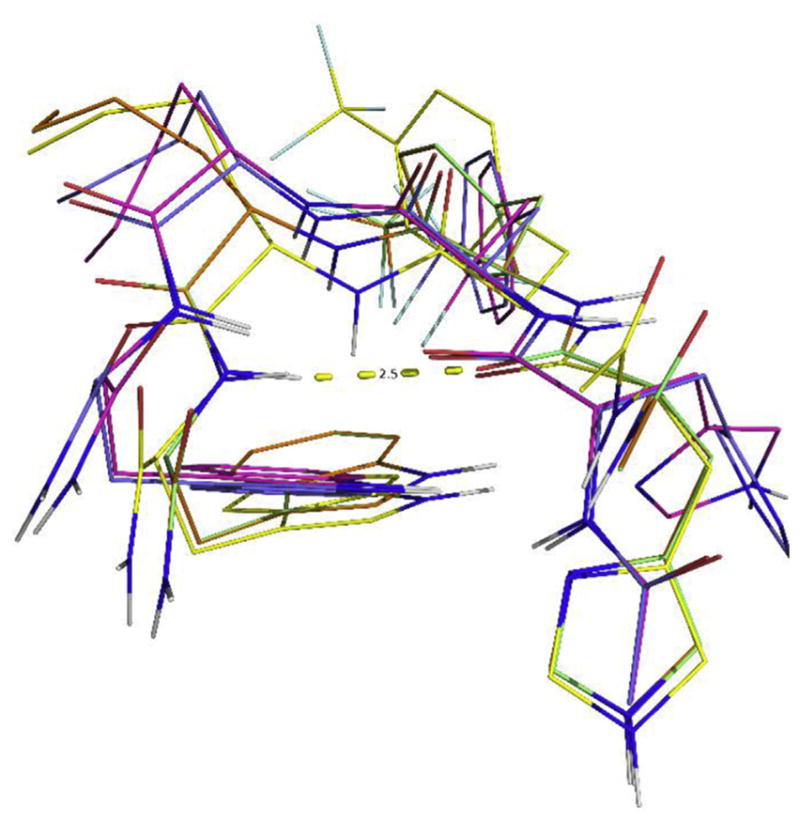
The ensemble of the 10 representative NMR derived structures of peptide 5, AcHis-D-Phe(4-CF3)-Nle-Trp-NH2. The hydrogen bond between the His1 CO and the Trp4NH is shown.
Table 4.
Summary of NMR structural statistics.
| Total distance restraints from NOEs | 70 |
|---|---|
| intraresidue | 19 |
| interresidue |i-j| = 1 | 43 |
| interresidue |i-j| 1 | 8 |
| NOE violations | |
| number of violations >0.2 Å | 1.2 ± 0.7 |
| RMSD of violations() | 0.176 ± 0.006 |
| Deviations from idealized geometry bond length (Å) | 0.008 ± 0.001 |
| bond angle (degree) | 0.70 ± 0.07 |
| Improper dihedral angle (degree) | 0.52 ± 0.16 |
| Averaged pairwise RMSD (Å) (heavy atoms) | 1.36 ± 1.54 |
3.4. Docking study of peptide 5 at the hMC1R
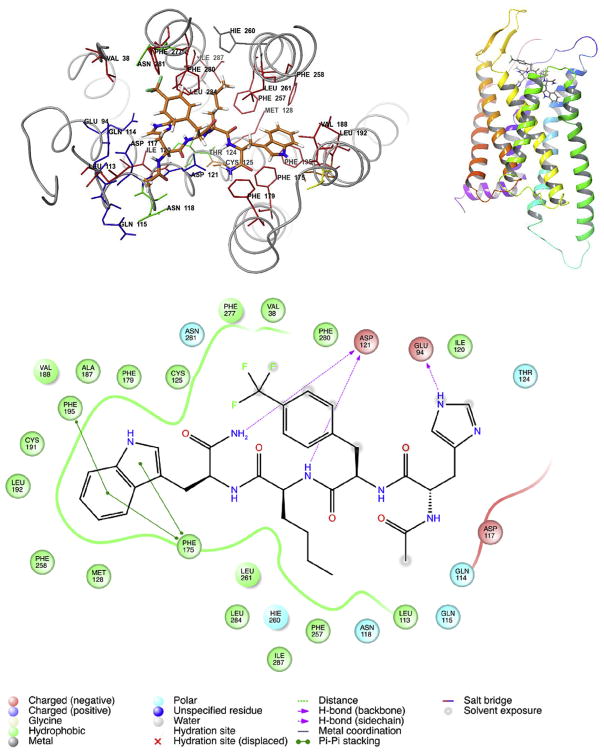
In addition, to investigate the peptide topology which might lead to new conformations of selective melanotropins for the hMC1R, a molecular docking study was performed for the peptide 5 with Glide (Schrodinger LLC, New York). The NMR structure of the Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2 was docked into the hMC1R structure which was generated from the Mosberg lab [44]. The MC1R-Peptide 5 interaction sites (3 Å cut-off) showed the binding pocket is hydrophobic comprising of a series of aromatic residues (Phe and Trp residues) spanning TM3 -7. These functionally attached receptor residues are involved in aromatic-aromatic interactions with residues D-Phe and Trp of the tetra-peptides. Fig. 4 shows that hydrophobic residues on the 7th transmembrane domain (TM) of the receptor contributed to the major force for binding; the D-Phe(4-CF3)6 forms a π-π stacking with the 7TM Phe280 and the 7TM Phe277 in the hMC1R. Also, the Trp9 of the tetra-peptide and the TM4 Phe175, Phe179 and TM5 Phe195, have π-π stacking interactions. Nle8 has frequent interactions with the 6TM Phe257 and Leu261. Multiple mutagenesis studies revealed that loss of a single aromatic receptor residue might be easily compensated by a network of aromatic-aromatic interactions and not induce any problematic effects on ligand binding or receptor activation. [35] [36], Our docking study directly observed the multiple effect of aromatic interactions supports the concept of a hydrophobic hMC1R binding pocket. The hMC1R can be potently activated by compound 5 in comparison with weak activation of MSHs tetra-peptides (His-Phe-Arg-Trp). This suggests that increasing hydrophobicity with the presence of D-Phe and Trp and lipophilic amino acid residue Nle, is very important for the potency and selectivity for the hMC1R. Introducing para-halogenated D-Phe residues in the tetrapeptides will increase the dipole moment of the ligand receptor interaction. As a result, this increases the binding potency towards the hMC1R.
Fig. 4.
Best docking pose for hMC1R selective ligand: Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2 into hMC1R. (Docking Score −11). The hMC1R-MCL interactions distance cut off 3 Å. Up Left: the selective molecule is highlight as tubing structure. The D-Phe(4-CF3) and Trp of the tetra peptide form π-π stacking interactions with the hMC1R transmembrane domains. Up Right: 3D view of selective molecule docking into hMC1R. Down: 2D structure of hMC1R-MCL interactions.
Finally, substitution of His with Pro was the initial goal in order to stabilize a β turn structure. However, the binding data show the loss of binding for all of these Peptides 8–12 to the hMC1R. Further investigation demonstrated that the position of Pro interferes with forming hydrogen bonding between the His1 CO and the amide hydrogen of Trp4. Analogues 5–6 indicate that the Nle position plays a critical role for the selectivity for the hMC1R, but does not enhance binding. To increase binding potency, halogenated groups were introduced on D-Phe6. It is known that in charge transfer compounds, halogens serve as acceptors and interact with a donor by transferring electronic charge. They can appear in biological systems amongst halogens. The π-electron clouds of benzene rings as well as with halogens and the delocalized pi;-electrons of peptide bonds of carboxy and amides stabilize π-π stacking. All of the halogen-containing compounds, including tetrapeptide 5, possess high lipophilicity, which improves in the following order: F < Cl < Br < CF3. The increased hydrophobicity of halogen-containing analogues with potent attractions between halogens and sulfur-containing receptor residues from transmembrane helices 3–6 may play an effective role in the stabilization of the firmly packed active receptor conformation. The hMC1R for halogen-containing peptides may be described with tighter packing of residues with the hMC1R binding pocket in comparison with other subtypes of human melanocortin receptors. Certainly, peptide 5 containing the bulky hydrophobic substituent p-CF3 increases stimulation activity only at the hMC1R, implying a more constrained geometry of its ligand binding pocket.
4. Conclusion
Structure-based drug design has become a useful approach to current drug discovery. In our long term peptide-based drug development, peptide truncation and amino acids scan have been used to discover the significant pharmacophore. Conformational constraints were applied to produce numerous stable and selective melanotropins, and the three-dimensional structure of ligands using NMR spectroscopy combined with computational based drug design have led to several selective compounds. In this research, we have combined previous knowledge on structure-activity relationships (SAR) of melanotropins as well as receptor mutagenesis studies to design tetrapeptide agonists selective to hMC1R. Our SAR results suggest that replacing the positively charged Arg residue with neutrally charged Nle is able to improve hMC1R selectivity with some sacrifice on potency in a tetrapeptide template, which is consistent with a recent discovery that replacing Arg residue with Leu in a γ-MSH template leads to a selective hMC1R agonist with only canonical amino acids [45]. Our further efforts to increase potency and binding affinity by adding parahalogenation groups to D-Phe led to the discovery of potent and hMC1R selective tetra-peptide peptide 5 (Ac-His-D-Phe(4-CF3)-Nle-Trp-NH2). Peptide 5 has an EC50 of 10 nM at hMC1R with at least 25-fold selectivity over other melanocortin receptors. An in vivo pigmentation study with lizards confirmed that peptide 5 can produce short-term skin pigmentation effect. Our NMR study revealed a β-turn conformation of peptide 5, which is stabilized by intramolecular hydrogen bond. Further docking studies identified extensive hydrophobic interactions between peptide 5 and hMC1R, which lead to hMC1R selectivity. Peptides are generally considered less toxic because they can be degraded into amino acids. As a result, we didn't measure or mention potential toxicity effects. We did use unnatural amino acids such as D-Phe with halogenations and Nle in this study, which could be potentially toxic to our body when metabolized. However, our peptide is potent in nanomolar range, which means that we can affect the physiological function in very low dose. Such dose is generally considered safe, even though it would require further experiments to confirm that. With its strong potency and selectivity to hMC1R as well as ease for synthesis, peptide 5 has great potential as a low side-effect product to induce skin pigmentation without sun for melanoma prevention.
5. Experimental section
5.1. Peptide synthesis
All peptides in this study were synthesized manually follow our previous published work [46–49]. See supporting Information for detail.
5.2. Bioassays
Binding and cAMP Assays followed our previously published work and the data Analysis [46–49]. IC50 and EC50 values represent the mean of two experiments performed in triplicate. IC50 and EC50 estimates and their associated standard errors were determined by fitting the data using a linear least-squares analysis, with the help of GraphPad Prism 5 (GraphPad Software, San Diego, CA).
5.3. Pigmentation study
Lizards were purchased from Carolina online. Peptide samples were dissolved in saline at the concentration of 1 mM. The total amount of peptide was given through i.p. injection with 3 μg/g for each lizard. The methods follow previous publications [21,38–41].
5.4. NMR spectroscopy
The micelle samples were prepared by dissolving the peptide and 50 equiv of perdeuterated SDS in 0.6 mL of acetate buffer (10 mM, pH5.5) containing 10% D2O. The pH of each sample was further measured and adjusted to 5.5 by using trace amount of DCl or NaOD as necessary. The peptide concentration used for the NMR experiments was 4.7 mM.
NMR spectra were recorded on a Varian INOVA 600 MHz spectrometer equipped with a z-gradient 5 mm HCN coldprobe. Homonuclear 1H 2D spectra were recorded at 25 °C and calibrated relative to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) internal reference. The water signal was suppressed by gradient echo or jump-return method. 2D DQF-COSY, TOCSY(70 ms mixing time), and NOESY spectra (100 ms mixing time) were recorded in the phase-sensitive mode with 4096 data points in t2 and 750 data points in t1. Shifted sine square window functions were applied in both dimensions. The 2D 15N HSQC and 13C HSQC at natural abundance were also recorded and referenced by the indirect method based on the gyromagnetic ratio. The 15N HSQC was recorded with 128 scans and 128 data points in F1 dimension, 40 ppm spectrum width centered around 120 ppm. The 13C HSQC was recorded with 128 scans and 300 data points in the F1 dimension, 160 ppm spectrum width centered around 80 ppm.
The assignment of the NMR spectra was done using the NMRFAM-Sparky software [50]. The NOE cross peak integrals were converted into upper distance bounds grouped as weak, medium and strong peaks. An ensemble of 300 structures was generated by the torsion-angle dynamics using the XPLOR-NIH simulated annealing protocol with the temperature range from 3500 K to 100 K [51], with NOE derived distance constraints (51 inter-residual and 19 intra-residual). A group of 10 structures were selected as the representative ensemble with the criterion of best fitted NOE derived distances (violations smaller than 0.20 Å) and lowest potential energy.
5.5. Docking studies
Molecular Docking Studies using the Glide programs (version 7.0, Schrodinger, LLC, New York, 2016). To analyze the docking results and execute the protocol, the Maestro user interface (version10.5, Schrodinger, LLC, New York, 2016) was employed. Docking was performed using the SP (Standard Precision Mode) protocol. This includes 1. Preparation of Protein. The protein was subjected to energy minimization using Schrodinger implementation of OPLS3 force field. 2. Preparation of Ligand. The ligand was prepared using the LigPrep 3.7 module of the Schrodinger suite using the standard protocol with OPLS3 force field. 3. Active Site Prediction. We employed Sitemap (version 3.8) to search for potential binding sites. Sitemap applies theoretical methods and predicts the most accurate binding site. Again, we used Sitemap after we had docked our ligand to evaluate the binding site. 4. Grid generation-docking calculation. Glide used a series of hierarchical filters to search for possible locations for the ligand in the active site region of the receptor. For the grid-based ligand docking, the receptor grid generation process was used. A grid box of 30 × 30 × 30 Å3 with a default inner box (10 × 10 × 10 Å3) was centered on the corresponding ligand. The receptor grid was defined as an enclosing box at the centroid of the ligand. Lastly, we performed a flexible docking calculation using the “Standard Precision” Glide algorithm and after the post-docking minimization we kept the pose with the best docking score.
Supplementary Material
Acknowledgments
Funding sources
These studies were supported in parts by grants from the U.S. Public Health Service, National Institutes of Health, DK017420, GM 108040 and DA06284.
Abbreviations
- SPPS
Solid Phase Peptide Synthesis
- Boc
Tert-butyloxycarbonyl
- Fmoc
Fluorenylmethoxycarbonyl
- Pbf
2,2,4,6,7-Pentamethyldihydrobenzofuran-5-sulfonyl
- Trt
Trityl
- Ac
Acetyl
- DNal(2′)
2′-Naphthylalanine
- D-Phe
D-Phenylalanine
- Nle
Norleucine
- Pro
Proline
- Tyr
Tyrosine
- CHCl3
Chloroform
- DCM
Dichloromethane
- DMF
N,N-Dimethylformamide
- MeOH
Methanol
- DIEA
Diisopropylethylamine
- HCTU
2-(6-Chlor-1H-benzotriazol-1-yl)-1,1,3,3-tetramethylammonium-hexafluorophosphate
- AcOH
Acetic Acid
- TFA
Trifluoroacetic Acid
- TIS
Triisopropylsilane
- DCl
Deuterium Chloride
- DMSO-d6
Dimethyl sulfoxide-d6
- D2O
Deuterium Oxide
- NaOD
Sodium Deuteroxide
- SDS-d25
Sodium Dodecyl-d25 Sulfate
- ACTH
Adrenocorticotropic Hormone
- AGRP
Agouti-Related Protein
- cAMP
cyclic Adenosine Monophosphate
- GPCRs
G Protein-Coupled Receptors
- hMCR
(hMC1R, hMC2R, hMC3R, hMC4R, and hMC5R) Human Melanocortin Receptor
- POMC
Pro-Opiomelanocortin
- MSH
Melanocyte-Stimulating Hormone
- TM
Transmembrane
- SAR
Structure–Activity Relationship
- m/z
mass-to-charge ratio
- NMR
Nuclear Magnetic Resonance
- RP-HPLC
Reversed-Phase High-Performance Liquid Chromatography
- TLC
Thin Layer Chromatography
- UV
Ultra-Violet
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ejmech.2018.04.021.
Footnotes
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Lowe GC, Saavedra A, Reed KB, Velazquez AI, Dronca RS, Markovic SN, Lohse CM, Brewer JD. Increasing incidence of melanoma among middle-aged adults: an epidemiologic study in Olmsted County. Minnesota Mayo Clin Proc. 2014;89:52–59. doi: 10.1016/j.mayocp.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engel J, Schlesinger-Raab A, Emeny R, Holzel D, Schubert-Fritschle G. Quality of life in women with localised breast cancer or malignant melanoma 2 years after initial treatment: a comparison. Int J Behav Med. 2014;21:478–486. doi: 10.1007/s12529-013-9334-x. [DOI] [PubMed] [Google Scholar]
- 3.Giuliani D, Minutoli L, Ottani A, Spaccapelo L, Bitto A, Galantucci M, Altavilla D, Squadrito F, Guarini S. Melanocortins as potential therapeutic agents in severe hypoxic conditions. Front Neuroendocrinol. 2012;33:179–193. doi: 10.1016/j.yfrne.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Zmijewski MA, Slominski AT. Is Mc1r an important regulator of non-pigmentary responses to UV radiation? Exp Dermatol. 2013;22:790–791. doi: 10.1111/exd.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolnicka-Glubisz A, De Fabo E, Noonan F. Functional melanocortin 1 receptor Mc1r is not necessary for an inflammatory response to UV radiation in adult mouse skin. Exp Dermatol. 2013;22:226–228. doi: 10.1111/exd.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Wan L, Hacker E, Dai X, Lenna S, Jimenez-Cervantes C, Wang Y, Leslie NR, Xu GX, Widlund HR, Ryu B, Alani RM, Dutton-Regester K, Goding CR, Hayward NK, Wei W, Cui R. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Mol Cell. 2013;51:409–422. doi: 10.1016/j.molcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadekaro AL, Chen J, Yang J, Chen S, Jameson J, Swope VB, Cheng T, Kadakia M, Abdel-Malek Z. Alpha-melanocyte-stimulating hormone suppresses oxidative stress through a p53-mediated signaling pathway in human melanocytes. Mol Canc Res MCR. 2012;10:778–786. doi: 10.1158/1541-7786.MCR-11-0436. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Malek Z, Suzuki I, Tada A, Im S, Akcali C. The melanocortin-1 receptor and human pigmentation. Ann N Y Acad Sci. 1999;885:117–133. doi: 10.1111/j.1749-6632.1999.tb08669.x. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki I, Im S, Tada A, Scott C, Akcali C, Davis MB, Barsh G, Hearing V, Abdel-Malek Z. Participation of the melanocortin-1 receptor in the UV control of pigmentation. J Invest Dermatol Symp Proc. 1999;4:29–34. doi: 10.1038/sj.jidsp.5640177. [DOI] [PubMed] [Google Scholar]
- 10.Scholzen TE, Brzoska T, Kalden DH, Hartmeyer M, Fastrich M, Luger TA, Armstrong CA, Ansel JC. Expression of functional melanocortin receptors and proopiomelanocortin peptides by human dermal microvascular endo-thelial cells. Ann N Y Acad Sci. 1999;885:239–253. doi: 10.1111/j.1749-6632.1999.tb08681.x. [DOI] [PubMed] [Google Scholar]
- 11.Pawelek JM. Approaches to increasing skin melanin with MSH analogs and synthetic melanins. Pigm Cell Res. 2001;14:155–160. doi: 10.1034/j.1600-0749.2001.140304.x. [DOI] [PubMed] [Google Scholar]
- 12.Scott MC, Wakamatsu K, Ito S, Kadekaro AL, Kobayashi N, Groden J, Kavanagh R, Takakuwa T, Virador V, Hearing VJ, Abdel-Malek ZA. Human melanocortin 1 receptor variants, receptor function and melanocyte response to UV radiation. J Cell Sci. 2002;115:2349–2355. doi: 10.1242/jcs.115.11.2349. [DOI] [PubMed] [Google Scholar]
- 13.Kadekaro AL, Kanto H, Kavanagh R, Abdel-Malek Z. Significance of the melanocortin 1 receptor in regulating human melanocyte pigmentation, proliferation, and survival. Ann N Y Acad Sci. 2003;994:359–365. doi: 10.1111/j.1749-6632.2003.tb03200.x. [DOI] [PubMed] [Google Scholar]
- 14.Kadekaro AL, Kavanagh R, Kanto H, Terzieva S, Hauser J, Kobayashi N, Schwemberger S, Cornelius J, Babcock G, Shertzer HG, Scott G, Abdel-Malek ZA. alpha-Melanocortin and endothelin-1 activate antiapoptotic pathways and reduce DNA damage in human melanocytes. Canc Res. 2005;65:4292–4299. doi: 10.1158/0008-5472.CAN-04-4535. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Malek ZA, Knittel J, Kadekaro AL, Swope VB, Starner R. The mela-nocortin 1 receptor and the UV response of human melanocytes–a shift in paradigm. Photochem Photobiol. 2008;84:501–508. doi: 10.1111/j.1751-1097.2008.00294.x. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Malek ZA, Ruwe A, Kavanagh-Starner R, Kadekaro AL, Swope V, Haskell-Luevano C, Koikov L, Knittel JJ. alpha-MSH tripeptide analogs activate the melanocortin 1 receptor and reduce UV-induced DNA damage in human melanocytes. Pigm Cell Melanoma Res. 2009;22:635–644. doi: 10.1111/j.1755-148X.2009.00598.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Borron JC, Abdel-Malek Z, Jimenez-Cervantes C. MC1R, the cAMP pathway, and the response to solar UV: extending the horizon beyond pigmentation. Pigm Cell Melanoma Res. 2014;27:699–720. doi: 10.1111/pcmr.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasti TH, Timares L. MC1R, eumelanin and pheomelanin: their role in determining the susceptibility to skin cancer. Photochem Photobiol. 2015;91:188–200. doi: 10.1111/php.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin K, Sturm RA, Smith AG. MC1R and NR4A receptors in cellular stress and DNA repair: implications for UVR protection. Exp Dermatol. 2014;23:449–452. doi: 10.1111/exd.12420. [DOI] [PubMed] [Google Scholar]
- 20.Cone RD. The Melanocortin System. New York Academy of Sciences; New York: 2003. [Google Scholar]
- 21.Sawyer TK, Hruby VJ, Darman PS, Hadley ME. [half-Cys4,half-Cys10]-alpha-Melanocyte-stimulating hormone: a cyclic alpha-melanotropin exhibiting superagonist biological activity. Proc Natl Acad Sci U S A. 1982;79:1751–1755. doi: 10.1073/pnas.79.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haskell-Luevano C, Miwa H, Dickinson C, Hadley ME, Hruby VJ, Yamada T, Gantz I. Characterizations of the unusual dissociation properties of melano-tropin peptides from the melanocortin receptor, hMC1R. J Med Chem. 1996;39:432–435. doi: 10.1021/jm950407s. [DOI] [PubMed] [Google Scholar]
- 23.Hadley ME, Marwan MM, al-Obeidi F, Hruby VJ, Castrucci AM. Linear and cyclic alpha-melanotropin [4–10]-fragment analogues that exhibit super-potency and residual activity. Pigm Cell Res. 1989;2:478–484. doi: 10.1111/j.1600-0749.1989.tb00242.x. Sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. [DOI] [PubMed] [Google Scholar]
- 24.Al-Obeidi F, Hruby VJ, Castrucci AM, Hadley ME. Design of potent linear alpha-melanotropin 4–10 analogues modified in positions 5 and 10. J Med Chem. 1989;32:174–179. doi: 10.1021/jm00121a032. [DOI] [PubMed] [Google Scholar]
- 25.Al-Obeidi F, Castrucci AM, Hadley ME, Hruby VJ. Potent and prolonged acting cyclic lactam analogues of alpha-melanotropin: design based on molecular dynamics. J Med Chem. 1989;32:2555–2561. doi: 10.1021/jm00132a010. [DOI] [PubMed] [Google Scholar]
- 26.Hruby VJ, Wilkes BC, Hadley ME, Al-Obeidi F, Sawyer TK, Staples DJ, de Vaux AE, Dym O, Castrucci AM, Hintz MF, et al. alpha-Melanotropin: the minimal active sequence in the frog skin bioassay. J Med Chem. 1987;30:2126–2130. doi: 10.1021/jm00394a033. [DOI] [PubMed] [Google Scholar]
- 27.Cody WL, Wilkes BC, Hruby VJ. Reversed-phase high-performance liquid chromatography studies of alpha-MSH fragments. J Chromatogr. 1984;314:313–321. doi: 10.1016/s0021-9673(01)97745-3. [DOI] [PubMed] [Google Scholar]
- 28.Jarrett SG, D'Orazio JA. Hormonal regulation of the repair of UV photoproducts in melanocytes by the melanocortin signaling axis. Photochem Photobiol. 2017;93:245–258. doi: 10.1111/php.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendt J, Rauscher S, Burgstaller-Muehlbacher S, Fae I, Fischer G, Pehamberger H, Okamoto I. Human determinants and the role of Melanocortin-1 receptor variants in melanoma risk independent of UV radiation exposure. JAMA Dermatol. 2016;152:776–782. doi: 10.1001/jamadermatol.2016.0050. [DOI] [PubMed] [Google Scholar]
- 30.Hacker E, Olsen CM, Kvaskoff M, Pandeya N, Yeo A, Green AC, Williamson RM, Triscott J, Wood D, Mortimore R, Hayward NK, Whiteman DC. Histologic and phenotypic factors and MC1R status associated with BRAF(V600E), BRAF(V600K), and NRAS mutations in a community-based sample of 414 cutaneous melanomas. J Invest Dermatol. 2016;136:829–837. doi: 10.1016/j.jid.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Swope V, Alexander C, Starner R, Schwemberger S, Babcock G, Abdel-Malek ZA. Significance of the melanocortin 1 receptor in the DNA damage response of human melanocytes to ultraviolet radiation. Pigm Cell Melanoma Res. 2014;27:601–610. doi: 10.1111/pcmr.12252. [DOI] [PubMed] [Google Scholar]
- 32.Haskell-Luevano C, Hendrata S, North C, Sawyer TK, Hadley ME, Hruby VJ, Dickinson C, Gantz I. Discovery of prototype peptidomimetic agonists at the human melanocortin receptors MC1R and MC4R. J Med Chem. 1997;40:2133–2139. doi: 10.1021/jm960840h. [DOI] [PubMed] [Google Scholar]
- 33.Proneth B, Pogozheva ID, Portillo FP, Mosberg HI, Haskell-Luevano C. Melanocortin tetrapeptide Ac-His-DPhe-Arg-Trp-NH2 modified at the para position of the benzyl side chain (DPhe): importance for mouse melanocortin-3 receptor agonist versus antagonist activity. J Med Chem. 2008;51:5585–5593. doi: 10.1021/jm800291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holder JR, Bauzo RM, Xiang Z, Haskell-Luevano C. Structure-activity relationships of the melanocortin tetrapeptide Ac-His-DPhe-Arg-Trp-NH(2) at the mouse melanocortin receptors: part 2 modifications at the Phe position. J Med Chem. 2002;45:3073–3081. doi: 10.1021/jm010524p. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, Cai M, Aprahamian CJ, Georgeson KE, Hruby V, Harmon CM, Yang Y. Contribution of the conserved amino acids of the melanocortin-4 receptor in [corrected] [Nle4,D-Phe7]-alpha-melanocyte-stimulating [corrected] hormone binding and signaling. J Biol Chem. 2007;282:21712–21719. doi: 10.1074/jbc.M702285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M, Aprahamian CJ, Celik A, Georgeson KE, Garvey WT, Harmon CM, Yang Y. Molecular characterization of human melanocortin-3 receptor ligand-receptor interaction. Biochemistry. 2006;45:1128–1137. doi: 10.1021/bi0521792. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Dickinson C, Haskell-Luevano C, Gantz I. Molecular basis for the interaction of [Nle4,D-Phe7]melanocyte stimulating hormone with the human melanocortin-1 receptor. J Biol Chem. 1997;272:23000–23010. doi: 10.1074/jbc.272.37.23000. [DOI] [PubMed] [Google Scholar]
- 38.Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB, Hadley ME. 4-Norleucine, 7-D-phenylalanine-alpha-melanocyte-stimulating hormone: a highly potent alpha-melanotropin with ultralong biological activity. Proc Natl Acad Sci U S A. 1980;77:5754–5758. doi: 10.1073/pnas.77.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castrucci AM, Hadley ME, Hruby VJ. Melanotropin bioassays: in vitro and in vivo comparisons. Gen Comp Endocrinol. 1984;55:104–111. doi: 10.1016/0016-6480(84)90134-5. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka S, Park MK, Takikawa H, Wakabayashi K. Comparative studies on the electric nature of amphibian gonadotropin. Gen Comp Endocrinol. 1985;59:110–119. doi: 10.1016/0016-6480(85)90425-3. [DOI] [PubMed] [Google Scholar]
- 41.Sugg EE, Castrucci AM, Hadley ME, van Binst G, Hruby VJ. Cyclic lactam analogues of Ac-[Nle4]alpha-MSH4-11-NH2. Biochemistry-US. 1988;27:8181–8188. doi: 10.1021/bi00421a029. [DOI] [PubMed] [Google Scholar]
- 42.Wishart DS, Sykes BD, Richards FM. The chemical-shift index - a fast and simple method for the assignment of protein secondary structure through Nmr-spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 43.Cierpicki T, Otlewski J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J Biomol NMR. 2001;21:249–261. doi: 10.1023/a:1012911329730. [DOI] [PubMed] [Google Scholar]
- 44.Chai BX, Pogozheva ID, Lai YM, Li JY, Neubig RR, Mosberg HI, Gantz I. Receptor-antagonist interactions in the complexes of agouti and agouti-related protein with human melanocortin 1 and 4 receptors. Biochemistry. 2005;44:3418–3431. doi: 10.1021/bi0478704. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, Mowlazadeh Haghighi S, Zoi I, Sawyer JR, Hruby VJ, Cai M. Design of MC1R selective γ-MSH analogues with canonical amino acids leads to potency and pigmentation. J Med Chem. 2017;60:9320–9329. doi: 10.1021/acs.jmedchem.7b01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai M, Cai C, Mayorov AV, Xiong C, Cabello CM, Soloshonok VA, Swift JR, Trivedi D, Hruby VJ. Biological and conformational study of beta-substituted prolines in MT-II template: steric effects leading to human MC5 receptor selectivity. J Pept Res Official J Am Peptide Soc. 2004;63:116–131. doi: 10.1111/j.1399-3011.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 47.Cai M, Stankova M, Pond SJ, Mayorov AV, Perry JW, Yamamura HI, Trivedi D, Hruby VJ. Real time differentiation of G-protein coupled receptor (GPCR) agonist and antagonist by two photon fluorescence laser microscopy. J Am Chem Soc. 2004;126:7160–7161. doi: 10.1021/ja049473m. [DOI] [PubMed] [Google Scholar]
- 48.Cai M, Mayorov AV, Cabello C, Stankova M, Trivedi D, Hruby VJ. Novel 3D pharmacophore of alpha-MSH/gamma-MSH hybrids leads to selective human MC1R and MC3R analogues. J Med Chem. 2005;48:1839–1848. doi: 10.1021/jm049579s. [DOI] [PubMed] [Google Scholar]
- 49.Cai M, Mayorov AV, Ying J, Stankova M, Trivedi D, Cabello C, Hruby VJ. Design of novel melanotropin agonists and antagonists with high potency and selectivity for human melanocortin receptors. Peptides. 2005;26:1481–1485. doi: 10.1016/j.peptides.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 50.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.