Abstract
Viruses from the Coronaviridae, Togaviridae, and Hepeviridae families all contain genes that encode a conserved protein domain, called a macrodomain; however, the role of this domain during infection has remained enigmatic. The recent discovery that mammalian macrodomain proteins enzymatically remove ADP-ribose, a common post-translation modification, from proteins has led to an outburst of studies describing both the enzymatic activity and function of viral macrodomains. These new studies have defined these domains as de-ADP-ribosylating enzymes, which indicates that these viruses have evolved to counteract antiviral ADP-ribosylation, likely mediated by poly-ADP-ribose polymerases (PARPs). Here, we comprehensively review this rapidly expanding field, describing the structures and enzymatic activities of viral macrodomains, and discussing their roles in viral replication and pathogenesis.
Keywords: macrodomain, ADP-ribosylation, poly-ADP-ribose polymerase (PARP), interferon (IFN), replication, pathogenesis, Coronaviridae, Togaviridae, Hepeviridae
Highlights
Macrodomains were discovered in Togaviruses, Coronaviruses, and the hepatitis E virus over 25 years ago. They were called the ‘X’ domain because they had no known function.
About 10 years later, several macrodomain structures were determined. They consist of central β-sheets surrounded by α-helices and bind to ADP-ribose and its derivatives. They were named after the structurally homologous domain of the MacroH2A histone.
Originally described as ADP-ribose-1″-phosphatases, both cellular and viral macrodomains enzymatically remove mono- and poly-ADP-ribose from proteins, supporting the notion that protein ADP-ribosylation is a component of the antiviral response.
Chikungunya virus and hepatitis E virus macrodomains are critical for replication, while the Coronavirus macrodomain both blocks the innate immune response and separately promotes in vivo replication.
ADP-Ribosylation
ADP-ribosylation is a post-translational modification (PTM) that was identified serendipitously over 50 years ago. While studying RNA synthesis, Chambon and colleagues discovered a large increase in 32P incorporation when adding nicotinamide mononucleotide (NMN), a precursor of NAD+, into liver nuclear extracts in the absence of other nucleotides [1]. The product was determined to be polymers of ADP-ribose, opening up a whole new field of investigation.
ADP-ribosylation is a ubiquitous modification present across all domains of life [2]. ADP-ribose is added to proteins by ADP-ribosyl transferases (ARTs) – a large family of homologous proteins with similarity to diphtheria and cholera toxins [3]. Based on these similarities, ARTs can be classified as ARTDs (diphtheria toxin-like) and ARTCs (cholera toxin-like), respectively [4]. These enzymes use nicotinamide adenine dinucleotide (NAD+) as a substrate to transfer ADP-ribose onto proteins. The best understood group of ARTs are poly-ADP-ribose polymerases (PARPs), which, in humans, consists of 17 members. Modification by PARPs occurs largely on acidic residues (glutamate and aspartate), but serines, lysines, arginines, and cysteines can also act as acceptors [2]. PARPs are the only ART group that can synthesize poly-ADP-ribose (PAR) on proteins (chains of repeating ADP-ribose units), but only some PARPs have PARylating activity (PARPs 1/2/5a/5b). Thus, most PARPs are only known to transfer a single ADP-ribose to their target protein [5].
ADP-ribosylation has a large array of effects on proteins, including modification of enzymatic activity, promoting the ubiquitination and degradation of proteins, and acting as a platform for protein interactions. Several protein domains are known to bind to ADP-ribose, including the WWE, PAR-binding motif (PBM), PAR-binding zinc finger (PBZ), and macrodomains [6]. ADP-ribosylation is well-known for its role in the DNA damage response, mediated by PARP1-dependent ADP-ribosylation of histones and other proteins which recruits repair proteins to sites of DNA damage [7]. In addition, the Tankyrases (PARP5a/5b) control Wnt signaling [8]. Recently, more defined roles for the MARylating PARPs have been described, including a prominent role in the cellular stress response. PARP16 regulates the ER-stress response [9], and several PARPs are present in cytoplasmic stress granules (PARPs 12,13,14,15, and 5a) [10]. ADP-ribosylation of Argonaute proteins in stress granules by an unknown PARP inhibits RNA interference 10, 11. Finally, PARPs have both antiviral and proviral activities (Box 1 ).
Box 1. The Interplay between PARPs and Virus Infections.
Several studies have found PARPs to have both proviral and antiviral activities. Using PARP inhibitors, several DNA viruses, such as herpesviruses, poxviruses, and adenoviruses, have been found to require ADP-ribosylation for optimal replication 56, 57, 58. This is not surprising given the importance of PARPs in cellular DNA replication and repair. The HSV-1 ICP0 protein enhances PARylation by degrading PAR-glycohydrolase, which removes PAR from proteins [56]. Furthermore, TiPARP was shown to ADP-ribosylate TBK-1 and decrease IFN-β production, leading to enhanced replication of several RNA and DNA viruses [59]. The best known antiviral PARP, PARP13, also known as ZAP (zinc-antiviral protein) restricts the replication of several RNA viruses by targeting viral or host RNA for degradation [60]. PARP13 contains mutations in its PARP domain, making it enzymatically inactive, indicating that its antiviral activity does not require ADP-ribosylation [61]. However, PARP13 is known to be important for stress granule ADP-ribosylation, and it also contributes to the ADP-ribosylation of the overexpressed PA and PB2 proteins of Influenza A virus which leads to their ubiquitination and proteasomal degradation 10, 11, 62. Thus, while it does not directly ADP-ribosylate proteins, it appears that PARP13 participates in this process, possibly by binding to active PARPs and recruiting them to target proteins. Similarly, PARPs 7, 10, and 12 directly inhibit alphavirus replication, but in a PARP-independent manner, as catalytically inactive mutants still blocked replication [63]. Overexpression of PARP12 has also been shown to inhibit the replication of several negative-strand RNA viruses, including vesicular stomatitis virus (VSV) [64]. PARP1 represses the transcription of integrated retroviruses in a chicken lymphoblastoid cell line, again in a PARP-independent manner [65]. In addition, multiple PARPs are interferon-stimulated genes (ISGs), and in silico analyses indicate that PARPs 4, 9, 14, and 15 are rapidly evolving, a hallmark of antiviral defense proteins [66]. However, despite these data, antiviral ADP-ribosylation of cellular or viral targets have yet to be identified.
Alt-text: Box 1
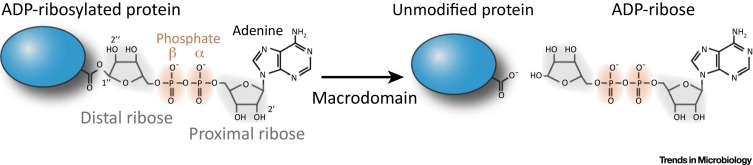
As with any PTM, ADP-ribose must be removed in a timely manner to prevent an untoward response and to recycle ADP-ribose components (Figure 1 ). The cellular enzymes that remove PAR from proteins are PAR glycohydrolase (PARG), and to a lesser extent ADP-ribosyl hydrolase 12, 13. The importance of removing PAR is demonstrated by the embryonic lethality of mice with genetic deletion of PARG [14]. However, PARG only cleaves between the unique glycosidic bonds of PAR [15], and thus it was unclear how the terminal ADP-ribose moiety was removed from the modified protein. This conundrum was resolved by the discovery that cellular macrodomain-containing proteins, including terminal ADP-ribose glycohydrolase 1 (TARG1), MacroD1, and MacroD2 removed this terminal ADP-ribose 16, 17, 18. The human genome contains at least 11 macrodomain-containing genes, but the substrate specificity and physiological roles of these proteins are largely unknown.
Figure 1.
Removal of ADP-Ribose from Protein by Macrodomains. A schematic representation of de-ADP-ribosylation reaction of proteins modified on acidic residues. Proximal and distal ribose are shaded in gray, α and β phosphate groups are shaded in orange.
Almost three decades ago, bioinformatics approaches identified a unique conserved gene domain within three viral families, the Togaviridae, Coronaviridae, and Hepeviridae 19, 20, 21. All of these families are positive-sense RNA viruses and cause disease in humans and animals (Box 2 ). The domain was named the ‘X’ domain as its structure and function was unknown. These domains were defined as ‘macrodomains’ after crystal structures of both cellular and viral macrodomains revealed a core fold homologous to the nonhistone part of the macroH2A protein 22, 23, 24. These macrodomains were shown to have phosphatase activity and were named ADP-ribose-1″-phosphatases (ADRPs) 23, 25, 26, 27, 28.
Box 2. Pathogenesis of Macrodomain-Containing Viruses.
Hepatitis E virus (HEV) is an emerging pathogen and is probably the most common cause of acute viral hepatitis in the world, with as many as 3 million infections per year. Mortality rates for acute infections are ∼3% for normal adults but as high as 30% for pregnant women in their third trimester, also often resulting in stillbirths of the fetus. HEV, like HCV, can also lead to chronic hepatitis and cirrhosis [67]. In developing countries, HEV is spread by the fecal–oral route, while in developed countries HEV is generally acquired through animal reservoirs, most likely pigs. Treatment for HEV infection includes either monotherapy with ribavirin or combination therapy of pegylated IFN and ribavirin. Additionally, two vaccines have been developed for HEV that have shown to be effective [68].
The Togaviridae are divided into two genera, the Alphaviruses and the Rubiviruses. Rubella virus, a Rubivirus, causes a congenital syndrome characterized by multiorgan birth defects. However, this disease has largely disappeared in the USA and other developed countries since an effective vaccine was developed. Alphaviruses, which include Chikungunya virus and Ross River virus, are transmitted by mosquitoes and can infect both vertebrate and invertebrate species. In humans, these infections can lead to a number of different pathologies, the most common being severe arthritis or rash 69, 70. There are no alphavirus vaccines currently available.
Coronaviruses (CoVs) cause a wide variety of diseases in both humans and mammals. CoVs such as porcine epidemic diarrhea virus (PEDV), bovine CoV, and infectious bronchitis virus (IBV) cause severe disease in veterinary animals. While originally only thought to be a cause of the common cold in humans, recent epidemics have demonstrated that CoVs can also cause severe human disease. Severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) have emerged in the last 15 years, causing serious respiratory disease in humans with high mortality rates (10–40%). Both viruses probably originated in bats and were transmitted to humans through animal reservoirs. MERS-CoV is endemic in camels in the Middle East and will likely continue to cause lethal disease for many years [71]. There are no approved vaccines or antiviral agents for human CoVs; however, several vaccines have been licensed for CoV-mediated veterinary diseases [72].
Alt-text: Box 2
Despite the demonstrated ADRP activity, it was unclear why viruses would devote a gene product to converting ADP-ribose-1″-phosphate to ADP-ribose. The discovery that macrodomains de-ADP-ribosylate proteins strongly suggested that their major role was in reversing antiviral ADP-ribosylation. However, this hypothesis has yet to be proven. As all three viral families that contain macrodomains include established human pathogens, understanding how macrodomains contribute to replication and pathogenesis is important. Here we comprehensively review the structures and enzymatic activities of these viral macrodomains, describe their potential functions, and discuss future challenges in understanding how macrodomains impact virus biology.
Viral Macrodomain Structure and Enzymatic Activity
Macrodomains are conserved, ∼150 amino acid protein domains present in all kingdoms of life. Based on phylogeny, macrodomains are divided into different classes. Most RNA virus macrodomains fall into the MacroD-like family, which includes human macrodomains MacroD1 and MacroD2 [29]. The exceptions are the SARS Coronavirus (SARS-CoV) unique macrodomains (Mac2/Mac3 – formely known as SUD domains), which we will not further discuss as they are distinct from the conserved macrodomain (Mac1).
Viral Macrodomain Structure
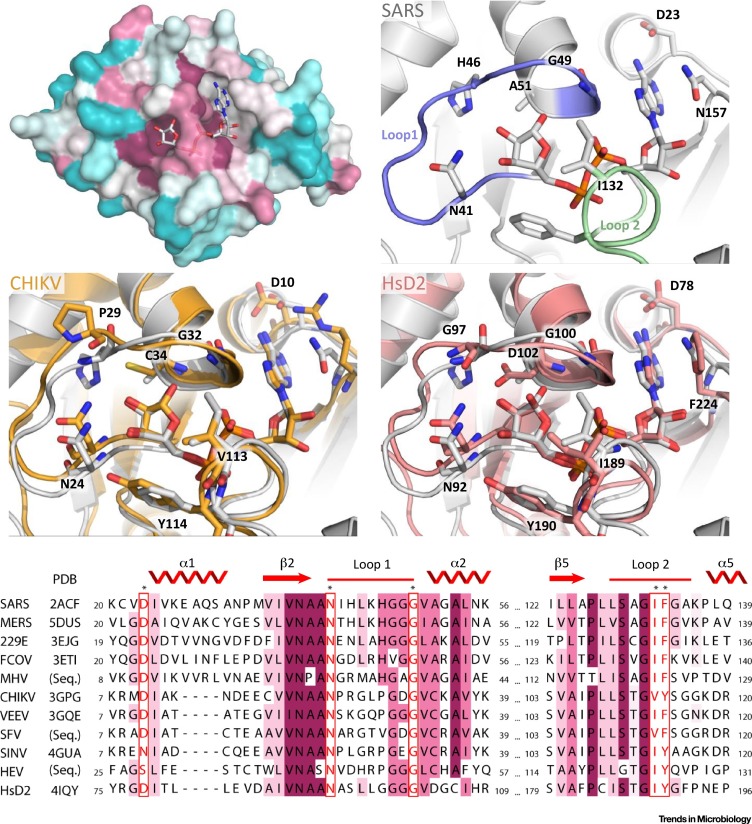
Macrodomains adopt a globular, mixed α/β/α sandwich fold, first shown when the structure of the archaeal macrodomain AF1521 [22] was determined. Many viral macrodomain structures, including those of Alphaviruses and Coronaviruses (CoVs), but not from Hepatitis E virus, have been determined, with 23 PDB entries available (www.rcsb.org). The globular macrodomain contains a conserved cleft that has been shown to bind ADP-ribose [30], a feature that has been confirmed for viral macrodomains by numerous biochemical studies 27, 31, 32, 33. Some of the most conserved residues of macrodomains are located at the surface and near the ADP-ribose binding cleft (Figure 2 ).
Figure 2.
Structures and Multiple Sequence Alignment of Viral Macrodomains. (Top left) A conserved surface representation of SARS macrodomain (PDB:2FAV) based on ConSurf analysis. The most conserved residues are coloured in magenta, while the least conserved residues are coloured in cyan. ADP-ribose is shown in stick representation. (Top right) ADP-ribose binding cleft of SARS macrodomain alone (gray) (PDB:2FAV), aligned with CHIKV macrodomain (Middle left, orange) (PDB:3GPO), or human MacroD2 macrodomain (Middle right, salmon) (PDB:4IQY). The proteins are shown as cartoon, with key residues shown as sticks. Loop 1 and loop 2 are coloured in slate and pale-green, respectively (SARS-CoV macrodomain only). (Bottom) Sequence alignment of selected viral and human macrodomains. The name of the virus is indicated on the left. Structure availability is indicated by a Protein Data Bank code; for viral macrodomains where no structural information is available, UniProt retrieved sequences of the macrodomains were used and are indicated as (Seq.). Secondary structure elements are schematically depicted above the alignment, and the numbering is for a generic macrodomain (i.e., not including additional helices or sheets present in some but not all macrodomains). Asterisks and red boxes indicate highly conserved, mostly substituted or key catalytic determinant residues of viral macrodomains. Magenta-shaded boxes depict the degree of conservation.
Based on ADP-ribose-macrodomain complex structures, these conserved residues provide high specificity and affinity for ADP-ribose. The distal ribose moiety is especially tightly coordinated by key residues found in loop 1 (between β-sheet 2 and α-helix 2) and loop 2 (between β-sheet 5 and α-helix 5) (Figure 2). Loop 1 contributes backbone contacts to the ADP-ribose α-phosphate and 1″ and 2″ OH groups of the distal ribose. In addition, the conserved asparagine residue (N41 in SARS-CoV) makes a hydrogen bond to the 2″ OH group. In CoV macrodomains, the conserved histidine residue (H46) contributes to ADP-ribose binding through a coordinated water. Alphavirus macrodomains have a conserved cysteine (C34) that could fulfil a similar role, whereas human MacroD2 contains an aspartic acid (D102) that makes a hydrogen bond with the 2″ OH group. Loop 2 contributes backbone contacts primarily to the β-phosphate of ADP-ribose. In addition, isoleucine (I132) and phenylalanine (F133) residues provide van der Waals contacts and are thought to direct orientation of the distal ribose 16, 24, 27, 34.
In some macrodomains, tyrosine replaces phenylalanine and forms a hydrogen bond to the 3″ OH of ADP-ribose. Macrodomains with a threonine before the G131 residue contribute an additional hydrogen bond to the 3ʹ OH of the proximal ribose (e.g., T111 in Chikungunya virus (CHIKV)). Besides the two loops, other notable ADP-ribose binding residues are D23 (N10 in Sindbis virus (SINV) and S28 in hepatitis E virus (HEV)), which contacts the amino group of adenine, and N157 (R144 in CHIKV and F224 in MacroD2), which interacts with the adenine ring via π-charge or π-π interactions. Depending on the ADP-ribose contacting residues, the affinities for ADP-ribose by different macrodomains vary, greatly with dissociation constants ranging from 0.1 to above 20 μM 16, 27, 31, 32.
Viral Macrodomain Enzymatic Activity
Binding of ADP-ribose is not the sole property of macrodomains. The first enzymatic activity ascribed to macro-D macrodomains was the dephosphorylation of ADP-ribose-1″-phosphate (a by-product of tRNA splicing reactions) identified through a biochemical-genomic screen in yeast [25]. This phosphatase activity was also detected for viral macrodomains 24, 27, 28, 31, 32. However, the enzymatic activity of macrodomains is low, the activities vary across different virus species, and no easy explanation exists as to why ADP-ribose-1″-phosphate hydrolysis would benefit these viruses. Therefore, other functions of viral macrodomains were considered. They include but are not limited to: binding ADP-ribosylated proteins, poly-ADP-ribose, RNA or other nucleic acids [31].
The discovery that macroD-like macrodomains are de-ADP-ribosylating enzymes 16, 17 suggested that viral macrodomains could have the same activity. Indeed, Li et al. were the first to show both de-MAR-ylating and de-PAR-ylating activities for macrodomains from members of all three macrodomain-containing virus families [35]. Fehr et al. confirmed and additionally tested the effects of several mutations on the SARS-CoV macrodomain de-ADP-ribosylating activity [36]. Eckei et al. demonstrated de-ADP-ribosylating activity of macrodomains from different Alphaviruses, addressing both de-MAR-ylating and de-PAR-ylating activities [37]; however, it is still unknown which features of the macrodomain limit its activity to de-MAR-ylation [16] and which are required for de-PAR-ylation 35, 37. Finally, McPherson et al. showed de-MAR-ylating activity for the Chikungunya virus macrodomain and determined that this macrodomain removes ADP-ribose from acidic residues but not from lysines [38]. Together, these four studies strongly suggest that de-ADP-ribosylation is likely the biochemical function of viral macrodomains. ADP-ribosylated proteins are linked to ADP-ribose through the 1″ position, reminiscent of ADP-ribose-1″-phosphate, explaining how macrodomains could have both activities. These latest studies have shifted the focus to de-ADP-ribosylation, but other potential macrodomain functions should not be neglected, as the catalytic mechanism of de-ADP-ribosylating macrodomains is not fully understood and the residues impacting de-ADP-ribosylation often affect ADP-ribose binding and other catalytic activities (e.g., ADP-ribose-1″-phosphate hydrolysis).
Macrodomain mutations experimentally shown to disrupt catalytic function are summarized in Table 1 . The corresponding residues in different viral macrodomains are identified by sequence alignment and their relation to the ADP-ribose substrate are visualized in Figure 2. The mutational approaches interfere with macrodomain function by (i) reducing ADP-ribose binding affinity (e.g., D23A (SARS-CoV), T111A (CHIKV), Y114F (CHIKV)), (ii) introducing steric hindrance (e.g., G131V (SARS-CoV), G123A (HEV), G112E (CHIKV)), or (iii) displacing potential catalytic water and reducing conformational strain on the distal ribose (e.g., G32E+V113R+Y114N (CHIKV)).
Table 1.
Representative Mutations That Disrupt Viral Macrodomain Function and Their Effects In Vivo
| Family | Virus | Mutations | ARH act.a, c | ADRP act.b, c | In vivo effect | Refs |
|---|---|---|---|---|---|---|
| Coronaviridae | SARS-CoV | Wild type (wt) | ++ | ++ | 27, 35, 36 | |
| D23A | + | ↓ Virulence | [36] | |||
| N38A | +/− | [27] | ||||
| N41A | − | − | ↓ Virulence | 27, 36 | ||
| H46A | +/− | +/− | ↓ Virulence | 27, 36 | ||
| G131V | − | ↓ Virulence | [36] | |||
| G47A+G48A | +/− | 27, 36 | ||||
| F133A | − | [27] | ||||
| HCoV229Ed | wt | ++ | ++ | 28, 35 | ||
| N28A | ++ | [28] | ||||
| N37A+N40A | − | [28] | ||||
| N37A | − | [28] | ||||
| N40A | − | [28] | ||||
| H45L | − | [28] | ||||
| G47A | V | +/− | − | [28] | ||||
| G48A | V | +/− | − | [28] | ||||
| FIPVe | wt | ++ | [37] | |||
| MHV | N30A | ↓ Virulence & replication | 49, 53 | |||
| Hepeviridae | HEV | wt | ++ | ++ | 27, 35, 37 | |
| N38A | ↓ Replication | [42] | ||||
| N42A | +/− | Not viable|↓ Replication | 35, 42 | |||
| H45A | Not viable | [42] | ||||
| G48A | V | ↓ Replication|no effect | [42] | ||||
| G49A | V | Not viable | [42] | ||||
| G50A | V | +/− | ↓ Replication|not viable | 35, 42 | |||
| G48S+G49S | − | ↓ Replication | [35] | |||
| G48S+G49S+G50A | − | ↓ Replication | [35] | |||
| G123A | +/− | ↓ Replication | [35] | |||
| I124A | + | [35] | ||||
| Y125F | + | [35] | ||||
| Y125A | − (misfold) | [35] | ||||
| Togaviridae | CHIKV | wt | ++ | ++ | 31, 37, 38 | |
| D10A | +/− | + | Reverted to WT | 31, 38 | ||
| N24A | +/− | +/− | 31, 37 | |||
| N24R | Y | − | − | [37] | ||||
| G32E | D | Q | − | Reverted to WT (G32E) | [38] | |||
| G32A | S | + | +/− | ↓ Virulence & replication | [38] | |||
| V33A | F | +/− | [37] | ||||
| V33E | − | [37] | ||||
| T111A | + | ↓ Virulence & replication | [38] | |||
| G112E | +/− | Reverted to WT | [38] | |||
| Y114A | +/− | − | ↓ Virulence & replication | 31, 37, 38 | ||
| Y114V | W | +/− | [37] | ||||
| R144A | ++ | Reverted to WT | [38] | |||
| G32E+V113R+ Y114N | − | Reverted to G32A+V113R+Y114N or V113R+V114N | [38] | |||
| G32A+V113R+Y114N | +/− | [38] | ||||
| V113R+Y114N | ++ | [38] | ||||
| SFV | wt | + | [32] | |||
| D10A | − | [32] | ||||
| N21A+N24A | − | [32] | ||||
| D31G | +/− | [32] | ||||
| G32Y | − | [32] | ||||
| G112Y | − | [32] | ||||
| SINV | wt | ++ | [37] | |||
| N10A | ↓ Virulence & replication | [44] | ||||
| N24A | ↓ Virulence | [44] | ||||
| N10A+N24A | ↓ Virulence & replication | [44] | ||||
| VEEVf | wt | ++ | 35, 37 | |||
| ONNVg | wt | ++ | [37] |
ADP-ribosyl hydrolase activity.
ADP-ribose-1″-phosphatase activity.
− no activity; +/− minimal activity; + modest activity; ++ robust activity.
HCoV-229E − human coronavirus 229E.
FIPV − feline infectious peritonitis virus.
VEEV − Venezuelan equine encephalomyelitis virus.
ONNV − O’nyong’nyong virus.
The most common studied mutation is substitution of the highly-conserved asparagine to alanine (N41A in SARS-CoV). This asparagine coordinates the 2″ OH of distal ribose and was proposed to be essential for catalysis. While the N41A mutation abolishes the catalytic activity of SARS-CoV macrodomain 27, 28, 36, it reduces but does not abolish the catalytic activity of other cellular and viral macrodomains 16, 37, 38. Such apparent discrepancy could be explained by reduced ADP-ribose binding by the asparagine-to-alanine mutant since viral macrodomains, especially the CoVs, have a lower affinity for ADP-ribose (20 μM in SARS-CoV compared to 0.15 μM in human MacroD2). The loss of a single hydrogen bond between the CoV macrodomain and ADP-ribose could reduce its affinity to a point where the macrodomain enzymatic activity is nondetectable. An alternative explanation is that different catalytic mechanisms exist among macro-D type macrodomains. Despite these uncertainties, the strict conservation of this asparagine and the effects of its mutation on in vitro activity and virus fitness make it a valuable tool for studying viral macrodomains.
The fact that viral macrodomains are generally required for virulence (see section below) suggests that small-molecule inhibition of macrodomains could be a novel therapeutic approach to preventing severe Alphavirus, hepatitis E virus, or Coronavirus-induced disease. One concern in considering the development of such molecules is the potential difficulty in specifically targeting the viral macrodomains but not the cellular MacroD1 or MacroD2 proteins. While such concern is warranted, there is potential for exploiting differences between human and viral macrodomains. Residues contributing to distal ribose 2″ OH coordination differ between human and viral macrodomains. In addition, the space around the proximal ribose and the surface residues contacting the adenine-ring might be additional areas of interest for specific small-molecule design. The potential for virus escape by mutating at these differing residues could hamper the feasibility of inhibitor design and usability, but these concerns could be addressed experimentally. The observation that even a 50% reduction in viral macrodomain activity greatly reduced virus fitness suggests that viruses would struggle to escape potential inhibitors [38]. If the risk of off-target effects in humans is too high, such inhibitors could be used in veterinary medicine.
Role of Viral Macrodomains During Infection
In recent years, significant progress has been made in deciphering the role of viral macrodomains, largely due to the advent of reverse genetic methods for these viruses. In this section, we review the known and potential roles for viral macrodomains.
Hepatitis E Virus (HEV) Macrodomain
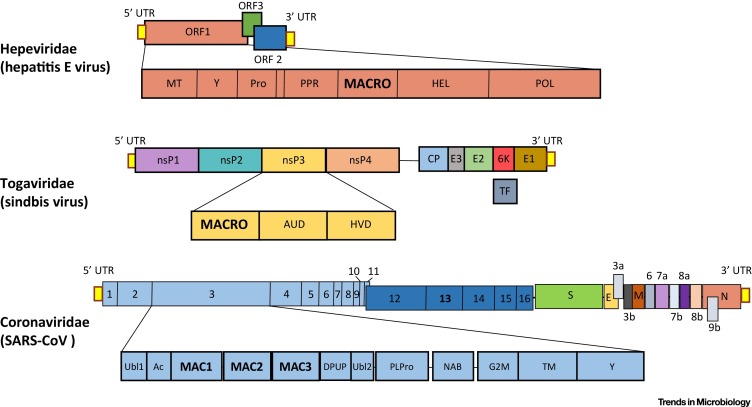
HEV is a nonenveloped positive-sense RNA virus with a genome of 7.2 kb that contains three open reading frames: ORF1, ORF2, and ORF3. The HEV macrodomain is a part of the multidomain ORF1 gene product of HEV (Figure 3 ) [39]. In a study on the ORF1 gene products of HEV, Nan et al. found that ORF1 inhibited poly I:C-dependent induction of interferon-β (IFN-β) [39]. Using overexpressed proteins, only the papain-like cysteine protease and the macrodomain could block IFN-β expression. Importantly, the HEV macrodomain inhibited both IKKε overexpression-induced IFN-β expression and interferon regulatory factor-3 (IRF-3) phosphorylation. Since the protein kinase IKKε phosphorylates IRF-3, these data suggest that the macrodomain targets IRF-3. However, macrodomain inhibition of IKKε-induced IFN-β expression was significantly less than its inhibition of poly I:C-induced IFN-β expression, suggesting that other mechanisms are involved. In a study by Ojha and Lole, the HEV macrodomain was found to interact with the light-chain subunit of human ferritin (FTL), which led to a reduction of secreted ferritin [40]. Ferritin is an iron-binding protein that helps the cell store free iron. Ferritin complexed with iron can activate NF-κB-dependent IFN production, suggesting that this interaction could be a mechanism of innate immune suppression [41]. Neither of these studies addressed whether the ability of the HEV macrodomain to bind or hydrolyze ADP-ribose was important for these observations.
Figure 3.
Genomic Location of Viral Macrodomains. Diagram depicting the genomic locations of macrodomains in the Hepeviridae, Togaviridae, and Coronaviridae. All viral macrodomains are a distinct domain of a larger nonstructural protein, ORF1 in Hepeviridae and nsP3 in both the Togaviridae and Coronaviridae. For Coronaviridae there are multiple macrodomains, listed Mac1/2/3. Mac1 is analogous to macrodomains found in other viruses.
Utilizing full-length replicon-based reverse genetic systems with either a GFP or luciferase reporter, Parvez et al. and Li et al. tested the role of the macrodomain in HEV replication 35, 42. In these studies, several amino acids in the active site were mutated (N806, N809, H812, G815, G816, G817) and tested for their ability to replicate. In the Parvez study, all mutations produced wild-type levels of viral RNA, but only N806 and G815 mutants were viable as measured by GFP expression. Li et al. found that replication strongly correlated with the enzymatic activity of each mutation they created. These results demonstrate that the HEV macrodomain is important for replication and acts after RNA replication, likely prior to translation of viral proteins. Another study found that the HEV macrodomain interacted with the viral methyltransferase and ORF3 using yeast-2-hybrid and coimmunoprecipitation methods [43]. It will be of interest to determine if these proteins are ADP-ribosylated and whether these interactions are important for replication.
Togavirus Macrodomain
Togaviruses are positive-sense enveloped viruses with 10–12 kb RNA genomes. The first two-thirds of the genome encodes four nonstructural proteins while the final one-third of the genome encodes structural genes. The macrodomain is located at the N-terminus of nsP3 (Figure 3) [44]. In 2009, Park and Griffin published a study focusing on two asparagine mutants (N10A and N24A) in the ADP-ribose binding pocket of the SINV macrodomain [44]. These mutant viruses replicated normally in BHK-21 cells but were modestly attenuated in neuronal cells and significantly attenuated in 2-week-old mice. Upon replication in neurons, the N10A mutation changed to threonine or aspartic acid, and a second site mutation of a glutamic acid at position 31 to a glycine appeared. Most macrodomains have a glycine at this position, suggesting that this mutation may enhance macrodomain activity. It was unclear how these mutations affected this protein, as neither mutant affected PAR binding and it was unknown whether alphaviruses’ macrodomains had de-ADP-ribosylating activity. However, recently it was demonstrated that CHIKV contains these activities 37, 38, making it likely that these mutations within the ADP-ribose-binding pocket of the SINV macrodomain attenuated its enzymatic activity.
As opposed to SINV, mutations in the active site of the CHIKV macrodomain had severe effects on virus replication [38]. Three recombinant viruses with mutations that severely affected both ADP-ribose binding and hydrolase activity were unrecoverable as they quickly reverted to wild type (D10A, G32E, G112E). The G32E mutation even reverted when transfected into an Aedes albopictus cell line, suggesting that antiviral ADP-ribosylation also occurred in mosquito cells. Other mutations that partially ablated hydrolase activity and ADP-ribose binding (T111A, G32S, G32A) replicated at 1 to 2 logs lower efficiency than wild-type virus in a neuronal cell line (NSC-34). Finally, one mutation, Y114A, was identified that had enhanced ADP-ribose binding but decreased hydrolase activity. This mutation was also significantly attenuated in NSC-34 cells, demonstrating that ADP-ribosyl hydrolase activity alone is important for replication. In this study, de-ADP-ribosylating activity, replication, and virulence did not perfectly correlate, suggesting a role for ADP-ribose binding in virulence. Of note, an additional mutant, R144A, which had normal levels of ADP-ribose binding and hydrolase activity in vitro, quickly reverted in tissue culture cells. This mutation could affect alternative functions of the macrodomain. For example, studies have found that mutations in the ADP-ribose binding domain of the VEEV macrodomain could compensate for mutations in other regions of the genome; however, it is unclear whether these activities were related to de-ADP-ribosylating activity or other functions 45, 46, 47. Additionally, Lulla et al. found that C-terminal amino acids of the Semliki Forest Virus (SFV) macrodomain are required in the processing of the nsP2/3 cleavage site, which could explain the defect of the R144A mutation [48].
Coronavirus Macrodomain
CoVs are large, positive-sense enveloped RNA viruses with ∼30 kb genomes. The N-terminal two-thirds of the CoV genome contains 16 nonstructural proteins (NSPs) while the C-terminal one-third of the genome contains the structural and accessory genes. The conserved macrodomain is located at the N-terminus of the multidomain nsp3 protein (Figure 3) [49]. In SARS-CoV and possibly other CoVs, two additional macrodomains (Mac2/Mac3) immediately follow the conserved macrodomain (Mac1) [50]. Mac2 and Mac3 do not bind or hydrolyze ADP-ribose; in contrast, they bind G quadruplexes [51]. Using a SARS-CoV replicon system, Kusov et al. found that Mac3 was essential for replication [52].
Most studies of the conserved CoV macrodomain (Mac1) function have used recombinant viruses with the highly-conserved asparagine mutated to an alanine. This mutation has small to no effects on CoV replication in cell culture 28, 36, 49, 53, 54. Also, deletion of the macrodomain had no effect on replication of a SARS-CoV replicon [52].
In marked contrast, several studies have found that the CoV macrodomain is essential for viral pathogenesis. Eriksson et al. first showed that the macrodomain was required for murine hepatitis virus (MHV) strain A59-induced hepatitis [53], while Fehr et al. demonstrated that mutation of the macrodomain of an encephalitic strain of MHV and of mouse-adapted (MA) SARS-CoV resulted in attenuation in vivo 36, 49. In all cases, the mutation was associated with reductions in viral load. It was also shown by both Eriksson et al. and Kuri et al. that the macrodomain could repress IFN production; however, it was unclear if this occurred in vivo and whether it was important for pathogenesis 53, 54. Fehr et al. found that the SARS-CoV macrodomain repressed IFN and cytokine production both in mice and in a bronchial epithelial cell line [36]. A coinfection with wild type and the macrodomain mutant SARS-CoV demonstrated that the enhanced interferon response was partially protective in mice. Also, the reduced viral load observed in SARS-CoV-infected mice was independent of the IFN response, as viral loads were not increased in IFNAR−/− mice infected with macrodomain mutant virus. In combination, these studies led to the conclusion that the SARS-CoV macrodomain promotes replication in vivo and independently suppresses the IFN response, with both functions playing a role in pathogenesis.
Concluding Remarks
Despite the discovery that viral ‘X’ domains closely resemble cellular macrodomains, the function of these domains remained an enigma until recent discoveries showed that viral macrodomains remove ADP-ribose from proteins. While this activity has not been demonstrated during infection, the publications discussed in this review strongly suggest that this is the case. As such, the primary focus of macrodomain research will likely shift from basic functional and biochemical studies to identifying antiviral PARPs and the protein targets of viral macrodomains (see Outstanding Questions). While identifying ADP-ribosylated proteins has proven difficult in the past, new methods for detecting ADP-ribosylation should lead to important discoveries in this field [55]. These studies will significantly enhance our understanding of these proteins, which could lead to new approaches for treating and preventing disease caused by macrodomain-expressing pathogens.
Outstanding Questions.
Do viral macrodomains counteract antiviral ADP-ribosylation? Which PARP(s) is responsible for antiviral ADP-ribosylation? Do invertebrates have antiviral PARP activity that is counteracted by macrodomains?
What are the ADP-ribosylated protein targets of viral macrodomains? Are targets mono- or poly-ADP-ribosylated? How does ADP-ribosylation impact these targets? What are the determinants of mono- versus poly-ADP-ribose binding/removal? Do all viruses require macrodomain enzymatic activity or is MAR/PAR binding sufficient to promote virus fitness/virulence? What effects do mutations in viral macrodomains have on de-ADP-ribosylation kinetics?
How do viral macrodomains acquire specificity? Do regions outside of the ADP-ribose binding pocket contribute to interactions with targets? Can a macrodomain from one virus family substitute for a different one?
Why are macrodomains encoded by some but not all virus families? Does antiviral ADP-ribosylation occur in other virus infections? Do other viruses interfere with PARP activity, and by what mechanisms?
Is the development of small-molecule inhibitors specific for viral macrodomains to repress viral pathogenesis feasible? Could viruses escape these inhibitors? Would these inhibitors also affect mammalian enzymes, leading to significant side effects?
Acknowledgments
This research is supported by a Wellcome Trust grant (101794), and a Cancer Research UK grant (C35050/A22284) to Ivan Ahel, an NRSA award to Anthony R. Fehr (F32-113973), and National Institute of Health grants to Stanley Perlman (R01 AI129269; RO1 NS36592; P01 AI060699).
References
- 1.Chambon P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 2.Palazzo L. ADP-ribosylation: new facets of an ancient modification. FEBS J. 2017;284:2932–2946. doi: 10.1111/febs.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holbourn K.P. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 4.Hottiger M.O. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Vyas S. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014;5 doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teloni F., Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44:993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messner S., Hottiger M.O. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011;21:534–542. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Yang E. Wnt pathway activation by ADP-ribosylation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jwa M., Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. Nat. Cell Biol. 2012;14:1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung A.K. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo G.J. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashimo M., Moss J. Functional role of ADP-ribosyl-acceptor hydrolase 3 in poly(ADP-ribose) polymerase-1 response to oxidative stress. Curr. Protein Pept. Sci. 2016;17:633–640. doi: 10.2174/1389203717666160419144603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana P. Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife. 2017;6 doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh D.W. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slade D. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jankevicius G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenthal F. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 18.Sharifi R. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez G. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya A.E. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E.V. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen M.D. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 23.Kumaran D. Structure and mechanism of ADP-ribose-1ʹʹ-monophosphatase (Appr-1ʹʹ-pase), a ubiquitous cellular processing enzyme. Protein Sci. 2005;14:719–726. doi: 10.1110/ps.041132005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saikatendu K.S. Structural basis of severe acute respiratory syndrome Coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martzen M.R. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 26.Shull N.P. A highly specific phosphatase that acts on ADP-ribose 1ʹʹ-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:650–660. doi: 10.1093/nar/gki211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egloff M.-P. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putics A. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rack J.G. Macrodomains: structure, function, evolution, and catalytic activities. Annu. Rev. Biochem. 2016;85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- 30.Karras G.I. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malet H. The crystal structures of Chikungunya and Venezuelan Equine Encephalitis Virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y. Crystal structures of two Coronavirus ADP-ribose-1″-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 2009;83:1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barkauskaite E. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Li C. Viral macro domains reverse protein ADP-ribosylation. J. Virol. 2016;90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehr A.R. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome Coronavirus infection. mBio. 2016;7:e01721–16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckei L. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017;7:41746. doi: 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPherson R.L. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nan Y. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J. Virol. 2014;88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojha N.K., Lole K.S. Hepatitis E virus ORF1 encoded macro domain protein interacts with light chain subunit of human ferritin and inhibits its secretion. Mol. Cell. Biochem. 2016;417:75–85. doi: 10.1007/s11010-016-2715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoll J. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-kappa B. J. Virol. 2002;76:9664–9672. doi: 10.1128/JVI.76.19.9664-9672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parvez M.K. The hepatitis E virus ORF1 ‘X-domain’ residues form a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene. 2015;566:47–53. doi: 10.1016/j.gene.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anang S. Identification of critical residues in hepatitis E virus macro domain involved in its interaction with viral methyltransferase and ORF3 proteins. Sci. Rep. 2016;6:25133. doi: 10.1038/srep25133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E., Griffin D.E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology. 2009;388:305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foy N.J. Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J. Virol. 2013;87:7569–7584. doi: 10.1128/JVI.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michel G. Adaptation of Venezuelan equine encephalitis virus lacking 51-nt conserved sequence element to replication in mammalian and mosquito cells. Virology. 2007;362:475–487. doi: 10.1016/j.virol.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De I. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 2003;77:13106–13116. doi: 10.1128/JVI.77.24.13106-13116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lulla A. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J. Virol. 2012;86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fehr A.R. The nsp3 macrodomain promotes virulence in mice with Coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan J. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kusov Y. A G-quadruplex-binding macrodomain within the ‘SARS-unique domain’ is essential for the activity of the SARS-coronavirus replication–transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eriksson K.K. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1ʹʹ-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuri T. The ADP-ribose-1ʹʹ-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 55.Daniels C.M. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J. Proteome Res. 2014;13:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grady S.L. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J. Virol. 2012;86:8259–8268. doi: 10.1128/JVI.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Child S.J. Inhibition of vaccinia virus replication by nicotinamide: evidence for ADP-ribosylation of viral proteins. Virus Res. 1988;9:119–132. doi: 10.1016/0168-1702(88)90027-5. [DOI] [PubMed] [Google Scholar]
- 58.Tempera I. Regulation of Epstein–Barr virus OriP replication by poly(ADP-ribose) polymerase 1. J. Virol. 2010;84:4988–4997. doi: 10.1128/JVI.02333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada T. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 2016;17:687–694. doi: 10.1038/ni.3422. [DOI] [PubMed] [Google Scholar]
- 60.Guo X. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuny C.V., Sullivan C.S. Virus–host interactions and the ARTD/PARP family of enzymes. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C.H. Battle between influenza A virus and a newly identified antiviral activity of the PARP-containing ZAPL protein. Proc. Natl. Acad. Sci. U. S. A. 2015;112:14048–14053. doi: 10.1073/pnas.1509745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atasheva S. Interferon-stimulated poly(ADP-ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 2014;88:2116–2130. doi: 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atasheva S. New PARP gene with an anti-alphavirus function. J. Virol. 2012;86:8147–8160. doi: 10.1128/JVI.00733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bueno M.T. Poly(ADP-ribose) polymerase 1 promotes transcriptional repression of integrated retroviruses. J. Virol. 2013;87:2496–2507. doi: 10.1128/JVI.01668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daugherty M.D. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host–virus conflicts. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nan Y., Zhang Y.-J. Molecular biology and infection of hepatitis E virus. Front. Microbiol. 2016;7:1419. doi: 10.3389/fmicb.2016.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamar N. Hepatitis E virus infection. Clin. Microbiol. Rev. 2014;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parkman P.D. Togaviruses: Rubella virus. In: Baron S., editor. Medical Microbiology. 4th edn. University of Texas Medical Branch at Galveston; 1996. Chapter 55. [PubMed] [Google Scholar]
- 70.Schmaljohn A.L., McClain D. Alphaviruses (Togaviridae) and flaviviruses (Flaviviridae) In: Baron S., editor. Medical Microbiology. 4th edn. University of Texas Medical Branch at Galveston; 1996. Chapter 54. [PubMed] [Google Scholar]
- 71.Vijay R., Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr. Opin. Virol. 2016;16:70–76. doi: 10.1016/j.coviro.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song D. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]