Abstract
Background
Wiskott-Aldrich syndrome (WAS), X-linked thrombocytopenia (XLT), and X-linked neutropenia (XLN), caused by WAS mutations affecting WASp expression or activity, manifest in immunodeficiency, autoimmunity, genomic-instability, and lymphoid-cancer. WASp supports filamentous-actin formation in the cytoplasm and gene-transcription in the nucleus. Although the genetic basis for XLT/WAS has been clarified, the relationships between mutant forms of WASp and the diverse features of these disorders remain ill-defined.
Objective
To define how dysfunctional gene transcription is causally linked to the degree of Thelper (Th) cell deficiency and genomic instability in XLT/WAS clinical spectrum.
Methods
In human Th1- or Th2-skewing cell culture systems, co-transcriptional R-loops (RNA:DNA duplex and displaced single-stranded DNA) and DNA double-strand breaks (DSBs) were monitored in multiple XLT and WAS patient samples, and in normal T cells depleted of WASp.
Results
WASp-deficiency provokes increased R-loops and R-loop-mediated DSBs in Th1-cells relative to Th2-cells. Mechanistically, chromatin-occupancy of Serine2-unphosphorylated-RNA Polymerase II is increased and that of topoisomerase-1, a R-loop preventing factor, is decreased at R-loop-enriched regions of IFNG and TBX21 (Th1 genes) in Th1-cells. These aberrations accompany increased unspliced (intron-retained) and decreased spliced mRNA of IFNG and TBX21 but not of IL13 (Th2-gene). Significantly, increased cellular load of R-loops and DSBs, which are normalized upon RNaseH1-mediated suppression of ectopic R-loops, inversely correlates with disease severity scores.
Conclusion
Transcriptional R-loop imbalance is a novel molecular defect etiologic in Th1-immunodeficiency and genomic-instability in WAS. The study proposes that cellular R-loop load could be used as a potential biomarker for monitoring symptom severity and prognostic outcome in the XLT-WAS clinical spectrum, and could be targeted therapeutically.
Keywords: Primary immunodeficiency, Wiskott-Aldrich syndrome, X-linked thrombocytopenia, Th1/Th2-differentiation, R-loops, DNA damage, Genomic instability
Graphical abstract
Introduction
WAS mutations result in X-linked thrombocytopenia (XLT), X-linked-neutropenia (XLN), or classic WAS.1-3 Although the genotype-phenotype association is imperfect, most manifest some combination of immunodeficiency, autoimmunity, genomic instability, and/or hematologic malignancy.4-8 WASp, encoded by WAS, is a multi-domain, adaptor protein that regulates actin-polymerization in the cytoplasm9 and RNA polymerase-II (RNAP2)-dependent transcription in the nucleus of cells of the hematolymphoid lineage.10-14 WASp-deficient CD4+Th-cells show decreased expression of Th1-genes in Th110 and Tfh (Th-follicular) cells.15 WASp is also required for the transcriptional-activation of the CD19 gene in human B-cells,16 and nuclear WASp partakes in murine T-cell development.17 WASp in human myeloid cells,18 and multiple other WASp-family proteins (Wave1, WASH) in different organisms,19-22 also support nuclear-functions or modulate transcription. Human WASp by modulating the activity of chromatin-remodeling complexes of the SWI/SNF-family influences promoter “readiness” and gene-activation.12 Murine WASH modulates chromatin activity of nucleosome-remodeling-factor (NURF) at the c-Myc promoter,20 and drosophila Wash influences global nuclear organization by influencing chromatin configuration.21 Yeast-Las17, a homolog of human WASp, is part of nucleolar rDNA-bound proteins involved in maintaining nucleolar integrity.23 Xenopus Wave1, like human WASp, facilitates RNAP2 transcription.19 Accordingly, multiple evidence links WASp-family proteins, well-known for their actin-polymerizing function, also to nuclear/transcriptional function(s) in both higher and lower organisms. As such, the transcriptional function of human WASp and xenopus Wave1, is uncoupled from its actin-polymerizing function,11,19 and the identification of the nuclear-localizing and nuclear-export sequences (NLS, NES) has allowed linking nuclear WASp, but not cytoplasmic WASp, with transcriptional activities.11 Together, these findings underscore the evolutionary pressure to preserve the nuclear/transcriptional role(s) of WASp-family proteins across different species, and for humans, propose WAS also as a disease of dysregulated transcription.
Regulated transcription is a multi-step process that includes safeguarding the newly synthesized, pre-mRNA (intron-containing) strand. Because DNA:RNA association is thermodynamically more stable compared to DNA:DNA association,24 such nascent RNA transiently hybridize to its template single-strand(ss) DNA, inducing the formation of R-loops, a 3-stranded nucleic-acid structure consisting of RNA:DNA duplex and a displaced non-template ssDNA.25 R-loops normally occur genome-wide, covering up to 5% of the human genome,26 where they function in gene transcription, immunoglobulin class-switching, DNA replication, and DNA repair.25-30 R-loops form both at 5′-TSS (promoter) and 3′-TTS (terminator) of a transcribing gene,31,32 where they are involved in remodeling the chromatin.33 These functions represent the physiologic roles of R-loops in transcription-initiation and -elongation. Conversely, unresolved or aberrant R-loops can retard elongation efficiency,34,35 and are detrimental to the genome by invoking activation of transcription-coupled nucleotide excision repair (TC-NER) factors that cleave R-loops to generate DNA single or double-strand breaks (SSBs or DSBs), which disrupts transcription.36-38 Accordingly, R-loops both activate or repress transcription, and hence can be beneficial or detrimental.39-43 To mitigate detrimental effects, cells employ multiple R-loop modulating factors to restrict aberrant R-loops. These are RNaseH1/H2 (degrade RNA moiety of RNA-DNA duplex),44,45 Aquarius and Senataxin (RNA/DNA-helicases),46,47 THO-complex and SRSF1 (mRNP-biogenesis factors),35,48-50 and topoisomerases (TOPs, resolve DNA-supercoiling).51,52 As such, TOP1 curbs R-loops both by relieving negative DNA-supercoils and facilitating mRNA-splicing.53,54 These factors are central to preventing R-loop-linked genome instability, since their loss/dysfunction results in human diseases including cancer.55-57
In this connection, we previously demonstrated that in WASp-deficient Th1-cells, decreased mRNA level of IFNG, a signature Th1-cytokine, accompanies reduction in the chromatin-enrichment of RNAP2-pCTD-Ser2 (S2P, a mark of transcription-elongation) but not of RNAP2-pCTD-Ser5 (S5P, a mark of transcription-initiation) in its gene-body.12 Because RNAP2-complexes that are Ser2-unphosphorylated (hereinafter S2unP) could still produce unspliced (intron-retaining, IR+) short-transcripts through non-productive elongation58 and because such IR+ transcripts could potentially reanneal to their template DNA-strand,59,60 we tested the possibility that WASp-deficiency might trigger the formation of aberrant R-loops at actively-transcribing genes in T cells. In this study, we demonstrate a link between WASp-deficiency, R-loop accumulation, and DNA-damage, defects that are more pronounced in Th1-cells relative to Th2-cells, which we propose contributes to the development of an aspect of WAS symptomatology. Furthermore, our data of R-loop imbalance rationalize the associated but less severe disorder, XLT, also caused by WAS mutations.
Methods
Cells, transfection, and pharmacological-modulation
CD4+ T helper (Th) cells were isolated from PBMCs by MACS-apparatus (Meltenyi) from 3 normal-donors: ND1 (hereinafter NIH001-WT), ND3, ND8, and 7 WAS-patients (using Transcript Variant NM_000377.2 based nomenclature): P1 (c.223G>A, p.V75M), P2 (c.116T>C, p.L39P), P3 (c.173C>G, p.P58R), P4 (c.570del, p.G191Vfs*70), P5 (c.450del, p.R151Gfs*110), P6 (c.1509A>C, p.*503ext*79 and P7 (c.23del, p.G8Efs*37), hereinafter NIH002-WAS).4-6,11,61 NIH001-WT and NIH002-WAS were immortalized by HTLV-1 (gift from Dr. Fabio Candotti).10 Patient samples were collected under the guidelines of the Institutional Review Board (IRB). NIH002-WAS was stably-transfected to express either Flag/Myc-dual tagged wild-type WASp (hereinafter wt-WASp) or different disease-causing WASp-mutants, as previously described.11-13 T cells were polarized into Th1 or Th2 cells using Human Th1- or Th2-Differentiation Kit from R&D Systems. Primary, non-immortalized, patient-derived Th cells (P1-P6) were propagated for multiple weeks in vitro in IL2-containing culture by adding irradiated, unrelated donor PBMCs (1×106/mL) and JY, an EBV-BLCL (0.1×106/mL), and stimulating with PHA (1μg/mL). For RNaseH1-expression, pEGFP-M27-RNaseH1 plasmid lacking mitochondrial-localizing sequence but containing a nuclear-localization sequence or its vector-only plasmid (gift from Dr. Robert Crouch) was transfected into Th1-cells by Amaxa®Nucleofector® (Lonza). RNaseH1-expressing Th1-cells were enriched by FACS-sorting the GFP+ subpopulation. For transcription inhibition assays, Th1-cells were treated with 5,6-Dichlorobenzimidazole-1-β-D-Ribofuranoside (DRB) at 25μM for 30min, 50μM for 1h or 2h, and 100μM for 2h, or with 5μg/ml α-amanitin for 16hr. For F-actin depolymerization, normal Th1-cells were treated with 2μM of Cytochalasin-D or Latrunculin-B for 1h or 2h.
CRISPR/Cas9-mediated knock-out and shRNA-mediated knock-down of WASp expression in human Th-cells
WASp-deficient Th-lines were generated from NIH001 and Jurkat (WT/WKO isogenic pairs) by CRISPR-mediated KO and from ND8 (WT/WKD isogenic pair) by shRNA-mediated KD (and scrambled-vector control KD). NIH001-WT and Jurkat-WT were transfected with 2ug of WASP-CRISPR/CAS9-GFP plasmid (sc-400712-KO-2) using Amaxa-nucleofaction. GFP+ cells were FACS-sorted, plated into 96-well plates by serial dilution, and screened by PCR to identify clones exhibiting biallelic (total) WAS-KO (NIH001-WKO and Jurkat-WKO). DNA-sequencing of gDNA, RT-PCR of mRNA and Western blot for WASp-expression were employed to verify successful WASp-depletion. For RNAi-mediated KD of WAS in primary Th-cells, lentiviral vectors encoding GFP and shRNA-sequences targeting WAS or a control (scrambled sequence) were used as described,62 and WASp expression assessed on sorted GFP+ Th-cells.
Coimmunoprecipitation and Mass-Spectrometry
Coimmunoprecipitatios/Western blots were performed as we described previously,10-13 using commercial reagents shown in Supplementary file-1. Multiple MS-assays on immunoprecipitated WASp-enriched protein-complexes were performed after pretreating normal donor, primary T cells samples with micrococcal nuclease (MNase),11 the latter known to mitigate false-positive protein-protein interaction by cleaving single- and double-stranded DNA and RNA.63
Confocal Microscopy and Flow cytometry
Immunofluorescence imaging was performed using Zeiss LSM710 integrated with Zen software, and z-stack images acquired at 0.3μM step-size at 63× magnification for analyses, as we described previously.10,64 FIJI-image analysis software (NIH) was used for all quantifications. Automated counting of S9.6+ foci/nucleus (nucleoplasm+nucleolus) was performed using the “particle analysis” function of Image-J software isolating the analyses to DAPI-stained regions. Signal saturation of γH2A.X in the nucleus was calculated using “3D-Object counter” and “Coloc-2” functions (Image-J software). Intracellular staining of proteins for flow was performed as we described previously.11-13
DNA-RNA immunoprecipitation (DRIP), ChIP-qPCR, and RT-PCR
Modified-DRIP was performed as described previously.65 Briefly, total nucleic-acid isolated from Th1-cells was sonicated to a size of 400-500 bp and pretreated with RNaseI to clear of contaminating single-stranded RNA. 3μg of eluted sample, after phenol/chloroform extraction, was incubated for 15hr with S9.6 antibody and immunoprecipitated RNA:DNA-hybrids were used in PCR-assays. All single-ChIP (1-round) and Re-ChIP (sequential 2-rounds of ChIP) assays were performed with MNase-digested chromatin as previously described.10-13 ChIP-grade and their isotype-Ig antibodies used to pull-down DNA:protein complexes are shown in Supplementary file-1. Non-specific signals obtained with control IgG-ChIP were subtracted from those obtained in the test samples. RT-PCR assays including qPCR analyses on RT2 Profiler™ PCR Array Human NF-κB-Signaling-Pathway-Plus (Qiagen) were performed as we described previously.11-13
Pre-mRNA estimation
Pre-mRNA was enriched using click-it nascent-RNA-capture-kit (Thermo-Fisher). Briefly, 10×106 cells were cultured with 5-ethynyluridine (EU) for 0.5h or 1h. 1ug/μl of RNA was biotinylated with 10mM biotin azide, 50μL of Dynabeads® MyOne™Streptavidin-T1 magnetic beads, and the 5-EU-labelled RNA used for qPCR assays (see Supplementary file-2 for primer/probe sequences).
Neutral comet assay
2.5 × 105 cells/ml mixed with 50μL of 1% low gelling agarose (Sigma) were pipetted onto CometSlide™ (Trevigen) and placed in the Sub-Cell® GT Agarose Gel Electrophoresis System (Bio-Rad) for 45 min at 30V in neutral electrophoresis buffer.66 Slides were stained with SYBR-Gold dye (Thermo-Fisher) and z-stack images acquired using a Zeiss-confocal microscope. Comet images were analyzed for tail-moment statistics using OpenComet software.
Results
WASp-deficiency triggers R-loop accumulation in human Th1-cells
Using S9.6 antibody (monitors RNA-DNA hybrids), we quantitated nuclear R-loops in NIH002-WAS Th-line (WAS patient-derived) and in 3 isogenic pairs of Th-lines (donor-derived), each either expressing or lacking WASp: 1) NIH001-WT and NIH001-WKO, 2) ND8-scrambled KD and ND8-shRNA WKD, and 3) Jurkat-WT and Jurkat-WKO (See Supplementary Figure 1 for phenotypic characteristics and Supplementary Figure 2 for WASp depletion). Th1-activation triggered high R-loops (S9.6+ foci) in NIH002-WAS and all 3 WASp-deficient Th-lines (NIH001-WKO, ND8-WKD, Jurkat-WKO) relative to their WT-control pairs, captured at a single-cell level by confocal microscopy (Figures 1A, B) and at population level by flow cytometry (Figure 1C). Overexpression of eGFP-tagged RNaseH1 (an endoribonuclease that degrades RNA in a RNA:DNA hybrid) into NIH001-WKO (Supplementary Figure 3), decreased the number of S9.6+foci/nucleus, which establishes S9.6+foci as authentic RNA:DNA hybrids. Moreover, treating NIH001-WKO with DRB or α-amanitin, both inhibitors of RNAP2-driven transcription, also decreased the number of S9.6+foci (Figures 1A, B). Disrupting F-actin assembly in normal (NIH001-WT) Th1-cells, by cytochalasin-D or latrunculin-B (Figure 1D), or stalling transcription-elongation by DRB (Supplementary Figure 4) did not provoke increased R-loop accumulation. The latter suggests that perturbing elongation alone is insufficient to trigger R-loops in human Th1-cells when WASp is present. Interestingly, Th2-skewing did not increase the number of S9.6+ foci in ND8-WKD relative to ND8-WT, but it did increase in NIH001-WKO relative to NIH001-WT (Supplementary Figure 5A). However, the number and size of S9.6+foci were significantly lower in NIH001-WKO Th2-cells compared to NIH001-WKO Th1-cells (Supplementary Figure 5B). Together, these findings demonstrate that R-loops accumulate to a greater degree in WASp-deficient Th1-cells as compared to WASp-deficient Th2-cells.
Figure 1. R-loops accumulate in WASp-deficient Th1-cells.
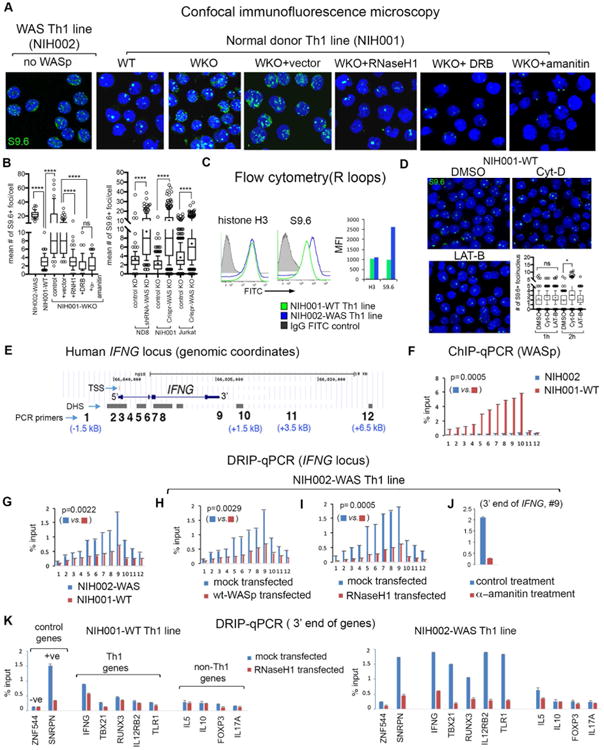
(A) confocal microscopy images of the indicated Th1-skewed cells under indicated experimental conditions dual-labelled with S9.6-antibody (R-loops, green) and DAPI (nucleus, blue). Displayed images are the collapsed composites of 20-30 z-stack images acquired at ×63 magnification. (B) Box-and-whisker plots (whiskers@10-90%, horizontal bar denotes median, “+” denotes mean) generated from 3D (z-stack) images of at least n=100 cells from n=3 independent experiments. (C) Flow cytometry. Histogram plots and bar graphs of their mean fluorescence intensity (MFI) for total histone-H3 (control) and S9.6 (R-loops) in the indicated Th cells. (D) confocal images of S9.6+ foci (R-loops) and their abundance (box-and-whisker plot) in normal donor Th1-skewed cells treated with Cytochalasin-D (Cyt-D), Latrunculin-B (LAT-B), or DMSO-control; n=100 cells from n=3 independent experiments. p-values: ****<0.0001; *=0.01; ns, not significant using Mann-Whitney 2-tailed nonparametric analyses. (E) Genomic locations of 12 PCR primer/probes across the human IFNG-locus (UCSC-Hg18) showing distances (kB) from the 5′TSS and 3′TTS. (F) ChIP-qPCR profile of WASp-enrichment across the IFNG-locus in indicated Th1-cells/conditions. (G-J) DRIP-qPCR profiles of R-loop frequency across the IFNG-locus in Th1-skewed, indicated Th-cells/conditions. (K) DRIP-qPCR: R-loop frequencies at 3′-ends of the indicated genes in the indicated Th1-cells, transfected with RNaseH1 or control vector. Displayed data is the mean of n=3 independent experiments, +SEM; shown are actual Mann-Whitney p-values that compare all data points for the entire IFNG-locus in one condition relative to other. Note: in this study, SNRPN is used as a positive-control for R-loops as previously described (primer coordinates, Chr15:25223714-2522381, in the SNRPN mRNA-sequence, region1-2).32 In another study, an upstream region of the SNRPN-SNURF locus (Chr 15: 25132924-25132943, not in the SNRPN mRNA-sequence) was used as a negative-control for R-loops. 87
To clarify the consequences of R-loops on Th1-differentiation, at the chromatin level, we next mapped R-loop frequency, in the presence or absence of WASp, on a subset of Th1-network genes (IFNG, TBX21, RUNX3, IL12RB2, TLR1). Using ChIP-qPCR, we first show that endogenous-WASp is recruited to multiple regions of the IFNG-locus (Figure 1E) in NIH001-WT, where its enrichment increases towards the 3′-region of the gene (Figure 1F), as shown previously using the ChIP-on-chip approach.10 This WASp-binding profile coincides with a similar 3′-biased accumulation of multiple other transcription factors.67,68 Using S9.6 antibody in DRIP-qPCR, we show in NIH001-WT Th1-cells that R-loops formed at a low level on the IFNG-locus, where the R-loop enrichment, like that of WASp, also increased towards the 3′-region of the gene (Figure 1G). This DRIP-profile is consistent with the physiologic role of R-loops in transcription elongation/termination of actively transcribing genes.27,31 By contrast, R-loops accumulated at a high level in NIH002-WAS Th1 cells, which were normalized upon transfecting either wild-type(wt)-WASp or RNaseH1 (Figures 1H, I), or treating with α-amanitin (Figure-1J), the latter established S9.6 DRIP-signals as representing co-transcriptional RNA:DNA hybrids. High R-loop accumulation was also captured at 3′-ends of multiple other Th1-genes in NIH002-WAS relative to NIH001-WT (Figure 1K). Notably, increased R-loops are not observed in Th1-cells at 3′-ends of IL5 (Th2-gene), IL10 (Treg-gene), FOXP3 (Treg-gene), IL17A (Th17-gene) or ZNF544 (R-loop-resistant locus)32 (Figure-1K).
WASp-deficiency provokes R-loop-linked DNA-damage in Th1-cells
Because unresolved R-loops cause DSBs,36,40 we investigated the effect of increased R-loops on genomic stability in WASp-deficient Th1- and Th2-cells. Using confocal microscopy and flow cytometry, we first quantified the signal-saturation of γH2A.X(pSer139)+ foci as a surrogate of DNA damage induction.69 This showed a significantly higher γH2A.X-signal saturations in both NIH002-WAS and NIH001-WKO Th1-cells relative to NIH001-WT Th1-cells by imaging (Figures 2A, B) and flow-cytometry (Figure 2C). Notably, overexpressing RNaseH1 into NIH001-WKO Th1-line or treating with DRB (or α-amanitin) both significantly decreased γH2A.X signal-saturation (Figures 2A, B), as did transfecting wild-type WASp into NIH002-WAS Th1-cells (Figure 2C). Furthermore, neutral comet assay, which reports on DSBs,66 showed that DSBs accumulated with higher frequency in all 3 WASp-deficient Th1-lines (NIH001-WKO, ND8-WKD, Jurkat-WKO) relative to their WT counterparts (Figure 2D), and the DSB-load is significantly reduced upon degrading R-loops (by RNaseH1) or inhibiting transcription (by DRB) (Figure 2E). Notably, like R-loops, the DSB load (tail-moment value) is also significantly higher in WASp-deficient Th1-cells compared to WASp-deficient Th2-cells (Supplementary Figure 5A).
Figure 2. Loss of WASp provokes R-loop-mediated DNA damage.
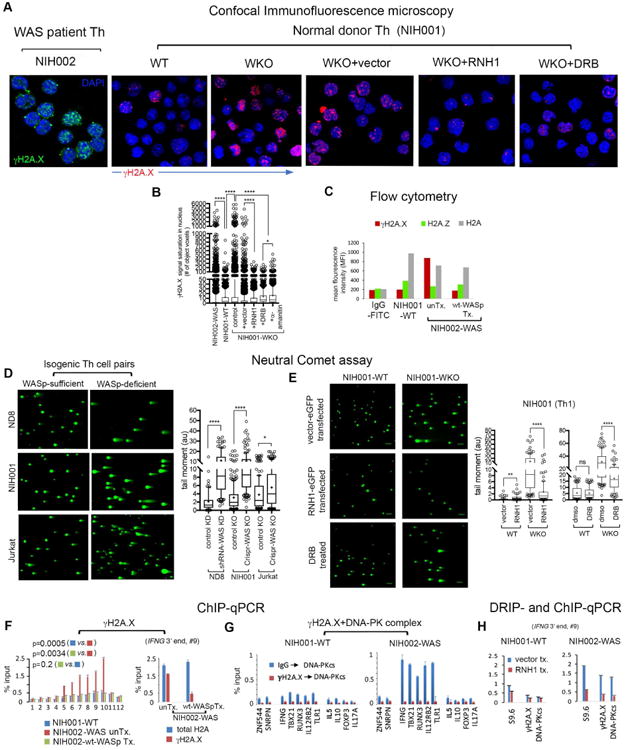
(A) Confocal images of indicated Th1-skewed cell lines under indicated conditions, dual-labelled with anti-γH2A.X antibody, an indicator of DNA damage and DNA replication stress (green for NIH002; red for NIH001) and DAPI (blue). (B) Box-and-whisker plots display γH2A.X signal saturation/nucleus in 3D images of the cell, n=100-120 cells, ****p<0.0001, *p=0.01 by Mann-Whitney. (C) Flow cytometry. Plot showing MFI values for γH2A.X, H2A.Z, and total-H2A (control) immunofluorescence in the indicated T cells/conditions; unTx, untransfected; Tx, transfected. (D and E) Neutral comet assay. Representative images of comet tails in indicated Th-line pairs/conditions. Tail moment statistics are displayed as box-and-whisker plots, n=100-150 cells from 3 independent experiments for each condition. ****p-value<0.0001, **p-value<0.001 *p-value=0.01 by Mann-Whitney. (F) ChIP-qPCR. Left, γH2A.X-enrichment at the IFNG-locus in indicated Th1-lines. Right, relative abundance of γH2A.X-modified histone-H2A at 3′-end of IFNG (point#9, Figure 1E). (G) Re-ChIP assay. In 2 sequential-rounds of ChIPs (1stChIP: γH2A.X or IgG, 2ndChIP: DNA-PKcs), co-enrichment profiles of γH2A.X and DNA-PKcs at the same 3′-ends of indicated genes in indicated Th1-lines. (H) ChIP- and DRIP-qPCR profiles of the indicated factors at 3′-end (point #9) of the IFNG-locus in indicated Th1-lines transfected with RNaseH1-eGFP (RNH1 tx.) or its empty-vector (vector tx.). The data in panels F-H are mean of n=3, +SEM; actual p-values by Mann-Whitney.
We next investigated whether DNA-damage occurs on Th1-genes at/or around the site(s) of R-loop formation. By ChIP-qPCR, we show that γH2A.X-enrichment at IFNG is significantly higher in NIH002-WAS relative to NIH001-WT Th1-cells (Figure 2F, left). At the 3′-region of IFNG, ∼80% of histone-H2A is γH2A.X-modified in NIH002-WAS compared to ∼20% in NIH001-WT Th1-cells (Figure-2F, right). Moreover, Re-ChIP assays (2-rounds of ChIPs: 1st:γH2A.X>2nd:DNA-PKcs) showed that γH2A.X and DNA-PKcs (targets DNA-breaks)70 co-enriched at a higher frequency at the same 3′-regions of multiple Th1-network genes in NIH002-WAS relative to NIH001-WT Th1-cells (Figure 2G), which is normalized by transfecting wt-WASp (Figure 2F) or RNaseH1 (Figure 2I). Interestingly, transfecting NIH002-WAS Th1-cells with ΔNES-WASp (nucleus-“only” WASp re-expression)11 reduced both R-loop- and γH2A.X-load, whereas, transfecting ΔNLS-WASp (cytosol-“only” WASp re-expression)11 did not (Supplementary Figure 6). We conclude that nuclear deficiency of WASp results in DNA-damage, in part through the accumulation of RNA:DNA-hybrids.
WASp-deficiency favors accumulation of intron-retaining, unspliced transcripts of IFNG and TBX21 but not IL13 in Th1-cells
In WASp-deficient Th1-cells, increased R-loops at IFNG and TBX21 loci occurs in the context of decreased mature mRNA-expression of the same genes (Figure-3A).10-13 Because R-loops are co-transcriptional products, these findings although counterintuitive, suggest that ongoing nascent transcription must be occurring at these Th1-genes to fuel R-loop formation. To test this possibility, we employed nonradioactive Click-iT® assay, in which Th1-cells were labelled with 5-ethynyluridine (EU) for 30min and 1h (Figure-3B). Using multiple qPCR primer-pairs covering all intron-exon boundaries of IFNG, TBX21 (Th1-genes), IL13 (Th2-gene) and TUBA1A (control-gene), we show a multifold increase in the expression of pre-mRNA-transcripts of IFNG and TBX21 in NIH001-WKO relative to NIH001-WT Th1-cells (Figure 3B). In contrast, there was a multifold decrease in the expression of mature, processed mRNA-transcripts (reported by primers covering coding-exons) of IFNG and TBX21, but not TUBA1B or IL13, in the same RNA-samples (Figure 3A).
Figure 3. Loss of WASp favors accumulation of intron-retained, nascent pre-mRNA transcripts of IFNG and TBX21 in Th1 cells.
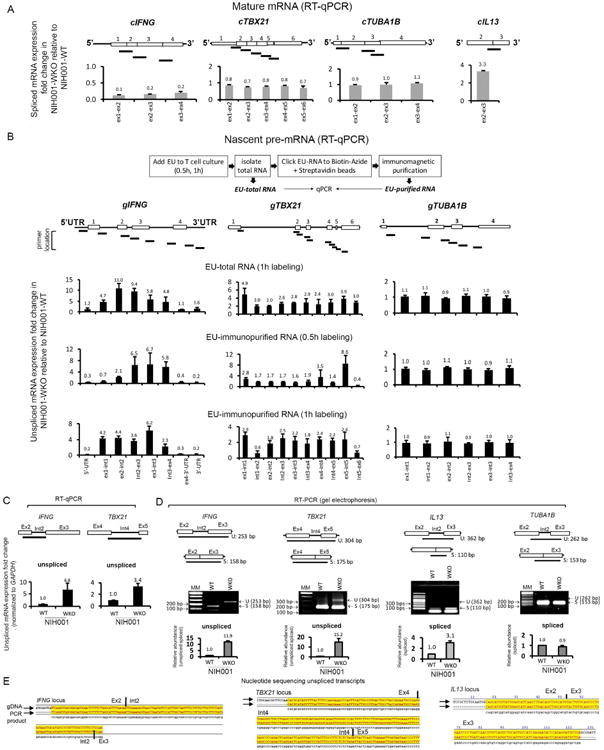
(A) Expression of processed (spliced, intron-removed) mRNA transcripts was quantified by generating multiple RT-qPCR primers that encompass indicated exon-exon junctions of all coding-exons of indicated genes, and is displayed as fold-change in WASp-deficient (WKO) relative to WASp-sufficient (WT) Th1-skewed lines (NIH001, isogenic pair). For IL13, only one exon-exon junction was tested. (B) Top, 5-ethynyluridine(EU)-labelling steps that yield EU-total RNA and EU-immunopurified RNA after 0.5h and 1h of EU-labelling. Bottom, expression of unprocessed (unspliced, intron-retained) pre-mRNA transcripts was quantified in both EU-total and EU-immunopurified RNA samples by generating multiple RT-qPCR primers that encompassed indicated intron-exon boundaries of indicated genes. Bar graphs depict mRNA fold-change in WKO Th1-cells relative to WT Th1-cells (NIH001, isogenic pair), after normalizing to their respective GAPDH mRNA levels. For fold-change calculations, mRNA-values obtained in WT were normalized as 1.0. Displayed values are mean+s.d., n=3 biological replicates, each with their separate, new EU-labeling. (C, D) Intron retention assayed by quantitative RT-qPCR (panel C) and semi-quantitative RT-PCR (panel D) for indicated introns displaying high splice-strengths (5′-acceptor and 3′-donor MaxEnt-scores) in WT and WKO Th1-line pairs. Data average of n=3, +s.d. In panel D, the relative abundance of unspliced (US) over spliced (S) transcripts was calculated using gel-densitometry, n=3 independent experiments. Note, the bar graphs display the abundance of “unspliced” mRNA for IFNG and TBX21, and “spliced” mRNA for IL13 and TUBA1B. (E) nucleotide sequences of the RT-PCR products, aligned to the indicated genomic-sequences (gDNA). Yellow shading denotes aligned sequences.
Because transcriptional defects that generate intron-retaining, pre-mRNAs also result in decreased level of mature mRNA, the latter by its degradation in the nonsense-mediated decay (NMD) pathway,71 we asked if WASp-deficiency triggers accumulation of pre-mRNA transcripts containing whole introns. Intron-retention was assayed by both quantitative RT-qPCR and semi-quantitative RT-PCR to monitor the relative abundance of unspliced versus spliced transcripts of IFNG, TBX21, IL13, and TUBA1B in 1h EU-labelled total-RNA from NIH001-WKO and NIH001-WT Th1-cells. MaxENT-scores (maximum-entropy-splicing-strength) were calculated for 5′-donor and 3′-acceptor splice-sites,72 and an intron associated with high splice-site strength, i.e., one that would be readily spliced-out co-transcriptionally, was selected. Accordingly, intron-2 of IFNG (MaxEnt 5′:9.85; 3′:8.68), intron-4 of TBX21 (5′:8.41; 3′:10.5), intron-2 of TUBA1B (5′:9.27; 3′:13.06), and intron-2 of IL13 (5′:9.33; 3′:9.88) were analyzed. We found a multi-fold increase in the frequency of intron-retained pre-mRNA transcripts (EU-labeled, 1h) with a high unspliced:spliced transcript ratio for IFNG and TBX21 in NIH001-WKO relative to NIH001-WT Th1-cells (Figure 3C-E). In contrast, TUBA1 and IL13 mRNA transcripts were fully-spliced, equally in NIH001-WKO and NIH001-WT Th1-cells, indicating that the splicing defect associated with WASp-deficiency is gene-specific. Together, the data demonstrates that transcription, albeit non-productive of mature mRNA, is still ongoing at these Th1-genes even when WASp is absent, and that WASp plays a yet to be defined role in pre-mRNA splicing.
WASp-deficiency favors accumulation of Ser2-unphosphorylated RNAP2-complexes at IFNG in Th1-cells
To further define the nature of transcriptional-dysfunction provoked by WASp-deficiency, we investigated the relationship between R-loops and RNAP2. Using ChIP-qPCR, we show decreased accumulation of Ser2-modified-RNAP2 (elongating-S2P) in the gene-body and at 3′-region of IFNG, this despite normal accumulation of Ser5-modified-RNAP2 (initiating-S5P) at its 5′-region, in NIH002-WAS Th1-cells compared to NIH001-WT Th1-cells (Figure-4A), which verifies our previous findings.12 Decreased S2P-occupancy was captured also at the 3′-regions of multiple other Th1-genes, but not at GAPDH, in NIH002-WAS Th1-line, in which re-expressing wt-WASp was sufficient to correct S2P-defect (Figures 4B, C).
Figure 4. Accumulation of Ser2-unphosphorylated (S2unP) RNAP2-complexes on the IFNG locus in WAS Th1-cells.
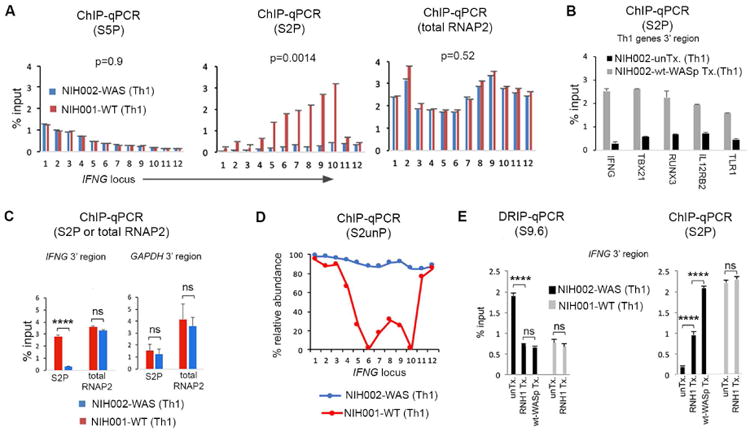
(A) ChIP-qPCR profiles of indicated RNAP2-modifications at the IFNG locus in indicated Th1-lines. S2P or S5P denotes ChIP performed with antibody specific for Ser2- or Ser5-phosphorylated RNAP2. (B) ChIP-qPCR profiles of S2P-enrichment at 3′-end of indicated genes in NIH002-WAS Th1-line (untransfected control, unTx.) and the same WAS Th1-line stably-transfected with wt-WASp. p-values are as described in the legend to Figure 1K. (C) ChIP-qPCR profiles of S2P- and total-RNAP2 (unmodified) at 3′-end of IFNG and GAPDH (control locus) in WAS and WT Th1-lines. (D) The ChIP-qPCR values for S2P and total-RNAP2 calculated from data in panel A show the relative abundance of S2unP-RNAP2 across the IFNG-locus, in WT and WAS Th1-lines. (E) Bar graphs show an inverse relationship between chromatin-enrichments of R-loops (S9.6 DRIP) and S2P (ChIP) at the 3′-end of IFNG, under indicated conditions (RNH1-transfected, wt-WASp-transfected, or untransfected control); data are an average of n=3 +SEM. ****p<0.0001; ns, nonsignificant, Wilcoxon nonparametric test.
Because R-loops could destabilize or remodel RNAP2-complexes,34,73-76 we investigated whether decreased S2P-occupancy is due to decreased occupancy of total-RNAP2 (i.e., destabilized) or from ectopic accumulation of Ser2-UNphosphorylated(S2unP)-RNAP2 (i.e., remodeled). ChIP-qPCR using pan-RNAP2-antibody, showed that total-RNAP2-occupancy was high (not low) at the IFNG-locus (Figure-4A). Accordingly, WASp-deficiency provokes increased chromatin-accumulation of S2unP-complexes (Figure-4D). Notably, RNaseH1-overexpression into NIH002-WAS Th1-cells resulted in ∼4.5-fold increase in S2P-occupancy over its untransfected control (Figure-4E). By comparison, re-expressing wt-WASp into NIH002-WAS Th1-cells increased S2P-occupancy by ∼10-fold. Because suppressing R-loops offered a partial S2P rescue, we postulate that loss of WASp impairs S2P-modification by altering multiple mechanisms, at least one of which is linked to R-loop accumulation.
WASp-deficiency disrupts TOP1 accumulation at Th1-genes
Because TOP1-recruitment to RNAP2 requires S2P-modification,74 it is possible that S2P-deficiency would disrupt TOP1-occupancy at Th1-genes, which could then provoke R-loop accumulation in WAS Th1-cells, as shown in other systems.51,52 To test this possibility, we queried our LC-MS/MS-dataset of WASp-interactors previously generated from human Th1-cells.11 These micrococcal nuclease (MNase)-treated, immunoprecipitation(IP)-based MS-assays63 captured polypeptides of DNA-topoisomerase-1, -2A, -2B (TOP1, TOP2A, TOP2B) from the nuclear-WASp-complexes (Figure 5A, Supplementary file 3), whereas control IgG-IP/MS did not recover TOP-peptides. The physiologic association between WASp and TOP1 was verified by co-immunoprecipitation/western-blot (Figure 5B) and MNase-treated Re-ChIP (1stChIP:TOP1→2ndChIP:WASp), which showed WASp:TOP1-complexes to co-enrich at 3′-regions of multiple Th1-genes in NIH001-WT Th1-cells (Figure 5C). In contrast, WASp-deficiency provoked a significant decrease in TOP1-occupancy, contemporaneously with increased R-loops, in the gene-body and at 3′-region of Th1-genes in NIH002-WAS Th1-line, both of which (TOP1, R-loops) were normalized by re-expressing wt-WASp (Figures 5D, E). We conclude that WASp plays an essential role, direct or indirect, in modulating TOP1-occupancy at certain Th1-genes, which we propose contributes to R-loop formation in WASp-deficient Th1 -cells.
Figure 5. Loss of WASp disrupts chromatin-occupancy of TOP1 at same cis-sites where R-loops accumulate in Th1 genes.
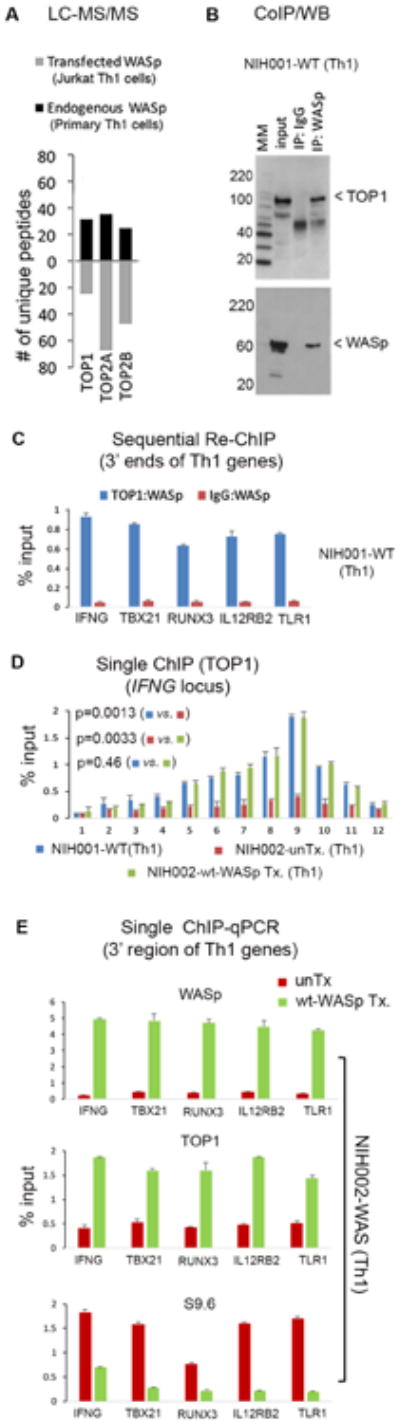
(A) LC-MS/MS. Number of unique polypeptides of indicated topoisomerases associated with WASp isolated from nuclear fraction of MNase-treated Th1-skewed, primary Th-cells (endogenous WASp-proteome) or Jurkat Th-cells (Flag/Myc-tagged, transfected wt-WASp-proteome) by immunoprecipitation (IP) with anti-WASp antibody (Ab) or anti-FLAG/Myc Abs (2-step immunopurification, 1stIP:Flag Ab, 2ndIP: Myc Ab). MultiConsensus reports of peptides/proteins were generated using the SEQUEST-platform from n=4-5 biological replicates after applying the filtering criteria previously described.11 MS data from IgG-IP conducted in parallel did not recover TOP peptides (not shown). (B) Coimmunoprecipitation (coIP)/Western blot. Protein-complexes isolated by coIP from nuclear extracts of WT Th1-cells resolved by sequentially immunoblotting the same gel with TOP1 and WASp. MM, Molecular marker (kDa). Input is 10%. (C). Re-ChIP assay. In 2 rounds of ChIP (1stChIP:TOP1/or IgG, 2ndChIP:WASp), co-enrichment profiles of TOP1 and WASp at the same 3′-ends of indicated genes in WT Th1-cells. (D) ChIP-qPCR. Enrichment profiles of TOP1 across the IFNG-locus (see Figure 1E) in indicated Th1-lines; unTx, untransfected; wt-WASp Tx, transfected with wt-WASp. (E) ChIP- and DRIP-qPCR. Enrichment profiles of indicted reagents at the 3′-ends of indicated genes in the same WAS Th1-line, unTx or wt-WASp Tx. For panels C, D, E, data is from n=3, mean +SEM; p-values in panel D by Wilcoxon nonparametric test.
Suppressing R-loops normalizes mRNA-levels of NF-κB-response genes in WASp-deficient Th1-cells
We previously showed,12,13 and verified here, that WASp-deficiency causes wide-spread decreases in steady-state mRNA-expression of multiple NF-κB-response genes in NIH002-WAS Th1-line (Figure 6A, Supplementary file 4). In contrast, expression of CSF2 and TNFAIP2, both pro-inflammatory cytokines, was elevated. (Figures 6A, B). To show a direct connection between R-loops and cell-function, we down-modulated R-loops in NIH002-WAS Th1-cells by RNaseH1, which resulted in normalization of mRNA-levels (e.g., ↑IFNG, STAT1, TBX21, IL12RB2; ↓CSF2, TNFAIP2) (Figures 6A-C). These results imply that the level of S2P-upregulation (∼4.5-fold) afforded by RNaseH1-mediated R-loop suppression (Figure 4E) is sufficient to restore Th1-cell functionality, at least of the immune-function genes examined. How RNaseH1-mediated R-loop suppression decreases mRNA-levels of CSF2 and TNFAIP2 in WASp-deficient Th1-cells is unclear, but we speculate that by decreasing R-loop-mediated DSBs, the expression of pro-inflammatory genes, known to be pathologically increased by chronic DDR-activation,77,78 will be also be suppressed.
Figure 6. R-loop load modulates the steady-state mRNA expression patterns in human Th1-cells.
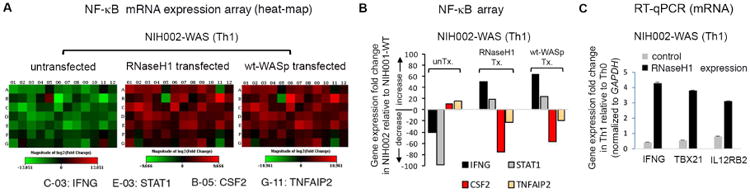
(A) Heat maps reporting on the RT-qPCR-based mRNA expression profiles of NF-κB response genes for WAS Th1-line, untransfected (control), or transfected with either RNaseH1 or wt-WASp using the 96-well plate format from Qiagen. The up/down gene modulation shown is in the context of transitioning from Th0-unstimulated to Th1-skewed state, in vitro. Red, gene up-regulation, Green, gene down-regulation, black, unchanged. Also see Supplementary file 4 for actual values. (B) Bar graphs display the actual values (fold-change) of the indicated 4 genes derived from the mRNA expression array (heat-maps) shown in panel B. IFNG is at C-03 and STAT1 at E-03 in the heat-map layout, and both become upregulated, whereas, CSF2 (GM-CSF) is at B05, TNFAIP2 is at G-11, both pro-inflammatory cytokine genes become downregulated in WAS Th1-cells upon expressing RNaseH1 or wt-WASp. (C) RT-qPCR. Fold change in the mRNA expression of 3 indicated Th1-genes in WAS Th-line (Th1-skewed) relative to the same WAS Th-line, untransfected (control) or transfected with RNaseH1. n=3, mean +SEM.
R-loop-mediated DNA-damage correlates with disease-severity in XLT/WAS clinical spectrum
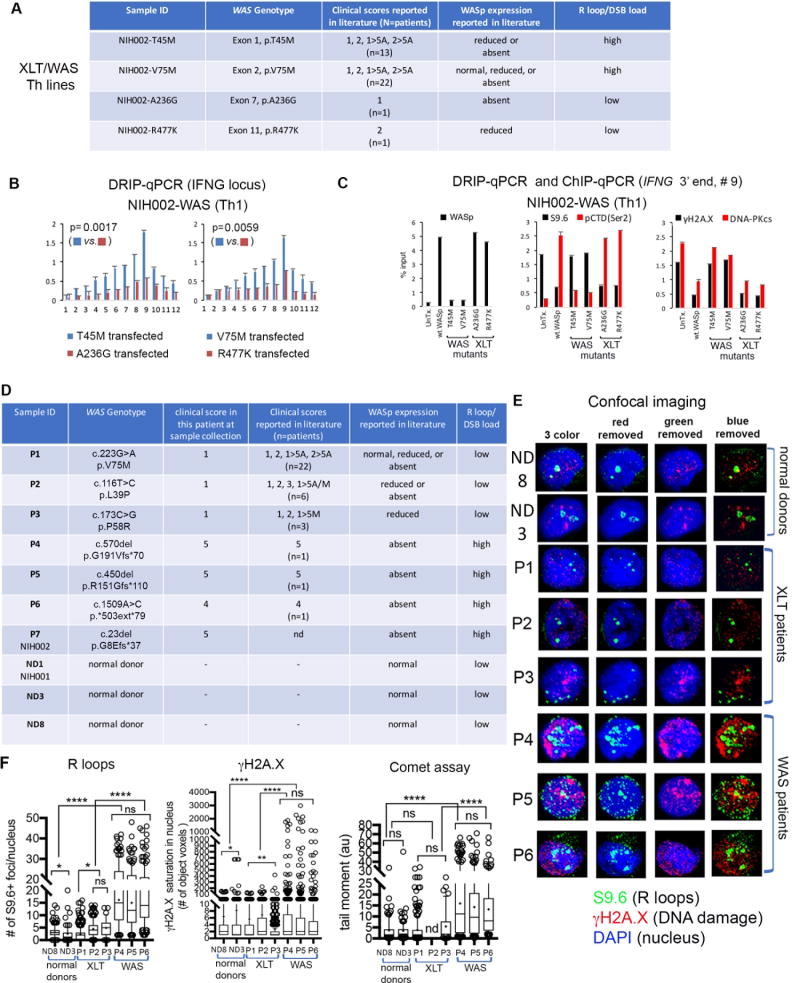
Finally, we asked if the R-loop defect in Th1-cells, uncovered here, correlates with the disease-severity scores of XLT/WAS.4-6,61 In a reconstitution study with engineered WASp-mutants, we show that mutations presenting in a subset of patients initially as XLT but may evolve subsequently into classic WAS with a score of 5 in the same patient (T45M, V75M)4-6 (Figure 7A) triggered R-loop accumulation and inscribed chromatin marks of DNA breaks at the IFNG locus, but not at non-Th1 gene loci in NIH002-WAS Th1-line stably transfected with these pathogenic mutants (Figures 7B, C; Supplementary Figure 7). In contrast, mutations that have been reported to always associate with XLT (A236G, R477K),4-6 did not manifest these aberrations at Th1 or non-Th1 gene loci. Significantly, in primary Th1-cells (not immortalized) from 6 patients (3 XLT [P1, P2, P3], 3 WAS [P4, P5, P6] and 2 normal-donors [ND8, ND3]), we show that the magnitude of R-loop and DSB-load in the patient Th1-cells with clinical-scores of 4-5 (i.e., WAS) is significantly higher than in the patient Th1-cells with clinical-scores of 0.5-1 (i.e., XLT) (Figure 7D-F). Together, the findings in primary Th cells (Figure 7D) and Th cell lines (Figures 1, 2, and 7A) support the conclusion that the degree of R-loop-linked DNA damage in Th1-cells correlates with the disease-severity scores of XLT-WAS for the pathogenic mutations tested.
Figure 7. R-loop and DNA-damage levels in Th1 cells correlate with disease-severity scores in WAS-XLT spectrum.
(A) The table shows 4 disease-causing WAS-mutations engineered in vitro for stable transfection into NIH002-WAS-null T-cell line to generate Th-lines representative of mutations that associate with mild or severe clinical phenotypes, as shown. Clinical-severity: score-1, 2: mild-XLT with eczema; score-5A, -5M, severe immunodeficiency with autoimmunity (A) or malignancy (M); 1, 2→5A/5M denotes disease progression to severe WAS after initially presenting as XLT in the same patient.4-6,86 (B) R-loop frequency across the IFNG-locus in NIH002-WAS Th1-line stably-expressing indicated WASp-mutants. p-values by Wilcoxon nonparametric test. (C) ChIP and DRIP (S9.6) enrichment profiles of indicated factors at 3′-end of IFNG (point #9, Figure 1E) in the indicated mutant. n=3, mean +SEM. (D) The table shows genotype-phenotype characteristics of pathogenic mutations in patients from whom primary Th cells (P1-P6) and a Th cell line (P7), as well as Th cells from normal donors (ND1, ND3, ND8) were generated.10,61(E) A representative confocal-image (a collapsed composite of multiple z-stack planes) for each patient and donor T-cell sample shows their respective R-loop load (S9.6+ foci) and DSB load (γH2A.X and neutral comet assay). (F) Box-and-whisker plots (whisker@ 10-90%), calculated on z-stacks (n=20-30 stacks/cell) from n=120-190 cells (exception: n=33 cells analyzed for patient-P2 due to small sample volume) for indicated read-outs. S9.6+ foci, γH2A.X signal-saturation in the nucleus, and comet tail moment were processed and quantified using ImageJ 2.0 and Prism7 programs. Entire multi-cell images were optimized equally by Adobe Photoshop to reduce any background fluorescence (“signal-noise”). ***p<0.0001, *p=0.01, ns= nonsignificant, p>0.1 by two-tailed non-parametric Mann-Whitney.
Discussion
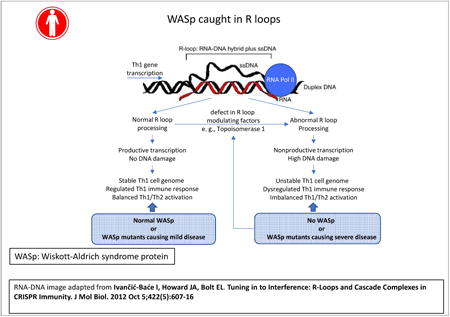
Our study uncovers R-loop-mediated genomic-instability as a new molecular defect in WAS Th1-cells, where the magnitude of defect correlates with clinical-severity scores in the XLT-WAS disease-spectrum. Accumulation of S2unP-RNAP2, decreased TOP1 chromatin-enrichment, and increased generation of unspliced pre-mRNA transcripts together manifest in increased R-loops, DNA-DSBs, and mature mRNA deficiency in a subset of Th1-genes. These aberrations manifest to a greater degree in WASp-deficient Th1-cells as compared to WASp-deficient Th2-cells, which may explain the observed Th1/Th2-imbalance10,79 contributing to eczema, hyper-IgE, and other atopic/clinical features of WAS.4-6 Based on our findings, we propose a model (Figure-8) in which the “platform” function of multi-domain WASp enables targeting of TOP1 and CDK9 to transcriptionally-active Th1-genes, and together with a similar “platform” function described for TDRD3 and BRCA1/BRAC2 in restricting R-loops,80-82 these observations support “adaptor-signaling” as a means to focally-concentrate anti-R-loop enzymology to transcribing- and replicating-chromatin. Accordingly, by providing new mechanistic clarification of how nuclear-WASp influences gene-transcription, this and previous studies together consolidate the idea that WAS is also a disease of dysregulated transcription.10-17,64,79
Figure 8. Model proposing WASp effect on proteins and pathways that modulate R loop balance.
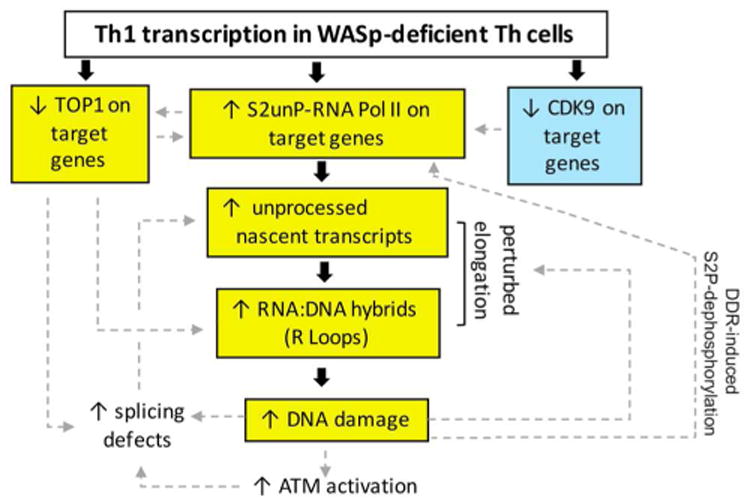
Event in blue-box denotes our previously published finding.12 Events in yellow-box denote our findings of this study. Unboxed events denote predicted consequences of our results. Bold black arrows connect proposed mechanistic pathway for the observed signaling outcomes based on our findings of this study. Hatched grey arrows indicate putative consequences of our findings on other related pathways, not tested in this study, but predicted from published results in other systems. Model: Loss of WASp disrupts chromatin-occupancy of CDK912 and TOP1. CDK9-deficiency contributes to impaired S2P-modification, the latter further compromising TOP1 recruitment to RNAP2.74 S2P-deficient RNAP2 can still generate short, unprocessed mRNA transcripts,58 which could fuel R-loop formation. TOP1 defect will also contribute to R-loop formation through its effects both on DNA-supercoiling and splicing factors.51-54 R-loops are known to incite DNA-damage, which via ATM-mediated spliceosome displacement,59 could further fuel R-loop formation. R-loop-mediated DDR-signaling activates S2P-phosphatases, e.g., protein phosphatase-1,88 which will dephosphorylate S2P contained within the elongation-complex in the gene-body. Together, a feed-forward causative loop of either R-loops⇒elongation-defect⇒R-loops (R-loop model) or elongation-defect⇒R-loops⇒elongation-defect (S2P model) will develop in WASp-deficient Th1-cells, resulting in R-loop-mediated DSBs. DSBs will elicit chronic-DDR, the latter known to perturb the global transcription programs of the affected cell, including provoking pro-inflammation,77,78 which we propose contributes to immunodeficiency and autoimmunity/autoinflammation in both cell-autonomous and non-cell autonomous fashion.
R-loops and S2P-deficiency: cause or consequence in WAS Th1-cell pathology?
R-loop-‘centric’ model (R-loop is a primary defect)
This model proposes that WASp-deficiency disrupts chromatin recruitment/retention of anti-R-loop factor(s) (e.g., TOP1), as a primary defect. Because TOP1 chromatin-recruitment is regulated by SWI/SNF-activity and H3K4me3,83 both of which we show are deficient in WAS Th1-cells,10,12 the model proposes that TOP1-defect could also arise from these chromatin/epigenetic aberrations. Because TOP1-defect impedes elongation and/or splicing,51-53,84 the latter causing accumulation of intron-retained (IR+) pre-mRNA, could further fuel R-loop formation (Figure-8), as shown recently in spinal muscular atrophy.60 Loss of yeast-TOP1 also results in accumulation of pre-rDNA transcripts, R-loops, and impeded RNAP1-transcription.51 Nonetheless, whether aberrant R-loops are a cause or consequence of splicing-defect in WAS Th-cells remains an open question.
S2P-‘centric’ model (S2unP is the primary defect)
Our findings of increased accumulation of S2unP-complexes present this alternative model, which offers an additional benefit of explaining a 3′-biased gradient for excessive R-loop formation (Figure-8). Our results in WASp-deficient Th1-cells demonstrating impaired chromatin-recruitment/retention of a S2P-kinase CDK912 and TOP1, the latter essential also for S2P-modification by BRD4,74 together support this model. Given NF-κB's role in S2P-elongation,58 a defect in NF-κB-activity seen in WASp-deficient lymphocytes,13,64 could also contribute towards S2P-deficiency.
Indeed, it is also possible that WASp exerts parallel, independent effects on both of these phenomena (R-loop and S2P models). Therefore, clarifying whether R-loops are a cause or consequence of defective S2P-elongation, which defect is a primary, disease-driving aberration in WAS, and how global are these aberrations in the WAS Th-cell genomes (Th1, Th2, etc.), will offer better definition of WASp-role in transcriptional R-loop metabolism. Similarly, since WASp-defect renders Th1 gene-promoters “inactive”,10,12-14 whether abnormal gene-body R-loops induce epigenetic-silencing of the 5′-promoter85 or an abnormal 5′-promoter chromatin-status induces abnormal gene-body R-loop formation33 in WAS is an open question, clarification of which would provide basis for modulating a specific chromatin-signaling pathway(s), potentially for therapy.
R-loops as a potential metric to monitor disease-severity in the XLT-WAS clinical spectrum
Of immediate clinical relevance are our findings that WAS-mutations that cause classic-WAS associate with high R-loops and DSBs, whereas, mutations that cause XLT do not, at least for mutations/patient-samples tested. Interestingly, for V75M, which has been reported to cause XLT or WAS in different patients, and in the same patient reported to evolve from XLT to WAS, R-loop profiles are instructive. Specifically, WASnull Th1-line (NIH002-WAS) stably-transfected with engineered V75M-WASp mutant shows high R-loop/DSB burden, whereas primary Th1-cells from a V75M-patient obtained at the time of mild-disease (score-1) show low R-loop/DSB burden. Such contrasting findings propose that for the same pathogenic-mutation, R-loop-mediated genomic-instability can be modulated by patient's genetic makeup and/or clinical context, two disease-modifiers that are challenging to test in any inbred, isogenic mouse-model of WAS, which underscores the necessity to conduct experimental studies in patient samples. Accordingly, serial monitoring of R-loop/DSB at different time-points in the same patient is likely to be more informative about disease-scores in XLT/WAS spectrum, especially because some XLT patients are known to subsequently evolve into serious WAS during their lifetime.4-6 Identification of R-loops as a potential biomarker of disease-severity is of clinical value because complete loss of WASp does not always correlate with severe WAS phenotypes.4-6,86 Nonetheless, reproducing the R-loop: disease-score correlation, identified here in 10 pathogenic-mutations, in a larger number of mutations, is required to bolster the diagnostic/prognostic value of monitoring R-loops in XLT/WAS spectrum. Certainly, we do not favor the idea that all aspects of WAS complications can be explained by the cell-autonomous consequences of R-loops in Th-cells alone. We anticipate the Th-cell-specific DNA-damage to have systemic consequences, including provoking autoimmunity/autoinflammation as a non-cell autonomous response to chronic DNA damage.77,78
Supplementary Material
Supplementary Figure 1. Phenotypic characterization of human CD4 Th cells. Flow cytometric analyses of the indicated primary Th cells (ND8) or Th lines (NIH001, Jurkat) stained with a panel of indicated antibodies or their isotype IgG control antibodies are shown as histogram plots. The data tabulated below indicates the Th subtype, i.e., effector, effector memory, naïve etc. based on the combination of cell receptor expression.
Supplementary Figure 2. Experimental depletion of WAS expression in normal (WT) Th cells. (A) For CRISPR/Cas9-mediated WAS KO: location of gRNA guides on the WAS gene showing primer pairs with the expected amplicon length. (B) representative gel images showing clones with PCR amplicon patterns that is consistent with no KO, partial KO, and complete KO of WAS. (C) Western blot sequentially reprobed with anti-WASp and α-tubulin loading control antibodies, to demonstrate complete WAS KO clones lacking WASp expression. (D) DNA sequencing of complete WAS KO clone show the expected gene region deletion. (E) RNA-interference-mediated depletion of WASp expression by shRNA vector. Shown is the western blot image of WASp expression in the indicated primary Th cells.
Supplementary Figure 3. RNaseH1 expression into WT and WKO Th1 cells. Flow cytometry (dot plot) of NIH001-WT and -WKO Th1 lines transfected with RNaseH1-eGFP construct or its empty vector control is shown as pre-sort (transfection efficiency) and post sort (purity) in 3 independent experiments. “X” denotes post-sort sample not tested for purity. The location of the gated population is shown, which was sorted for down-stream assays.
Supplementary Figure 4. R loop modulation in DRB-treated Th1 cells. Left, confocal images of WT Th1 cells dual-labelled with S9.6 antibody and DAPI. The image shown is representative of n=100 cells in DRB 100uM for 2 h treatment condition. Right, statistics of S9.6+ foci changes under the indicated DRB dose/duration conditions. The displayed data is from n=100-150 cells analyzed for each condition. p-value: ns= nonsignificant by Mann-Whitney nonparametric test.
Supplementary Figure 5. R-loop and DNA damage load in WASp-deficient Th2-cells. (A) Box-and-whisker plots comparing the magnitude of R-loop (S9.6+ foci) and DSB load (comet assay) in Th1 and Th2 cells, WASp-sufficient (WT) and WASp-deficient (WKO/WKD). The data is representative of analyzing 20-30 z-stack images of at least 100 cells each from 2-3 independent experiments. (B) shown are the indicated immunofluorescence images overlaid on the corresponding Nomarski (DIC) images for Th1 and Th2 cells. Right, plot showing the range of individual (single) S9.6+ foci size in Th1 and Th2 cells. ****p<0.0001, Mann-Whitney.
Supplementary Figure 6. Reconstituting nuclear but not cytosolic compartment of WASp-deficient Th1-cells is necessary to normalize R-loop frequency. Change in R-loop frequency in WAS Th1 line stably-transfected with NLS-deleted (ΔNLS) or NES-deleted (ΔNES) WASp mutants. Note, reconstituting only the nuclear compartment with WASp is sufficient to normalize the R-loop frequency (left panel), but reconstituting only the cytosolic compartment does not change the R-loop frequency (right panel). Also shown is the ChIP enrichment profile of γH2A.X. Note, the DNA damage as reported by γH2A.X enrichment is reduced by reconstituting only the nuclear compartment of WASp-deficient Th1 cells. Data is from n=3, mean+SEM.
Supplementary Figure 7. The magnitude of R-loop accumulation and DNA-damage at non-Th1 genes in WASp-deficient Th1 cells. (A) R loop frequency at the 3′-ends of the indicated Th1 and non-Th1 genes in WAS Th1 line (NIH002) untransfected (control) or transfected with wt-WASp or the indicated pathogenic mutant. (B) DNA damage frequency at the same cis-region in the same set of genes. Data is from n=3, mean+SEM.
Supplementary file 1. Antibodies and Reagents
Supplementary file 2. RT-qPCRPrimers and Probes for mature mRNA and nascent pre-mRNA estimation
Human Primary Th1 cells: nuclear fraction (IP with anti-WASp Ab)
Human Jurkat Th1 cells: nuclear fraction (2 step IP with anti-Flag and -Myc Abs)
Key Messages.
WASp deficiency triggers R-loop-mediated DNA damage in a subset of immune function genes.
R-loops accumulate to a greater degree in WASp-deficient Th1-cells as compared to WASp-deficient Th2-cells.
The magnitude of R-loop-linked DNA damage correlates with clinical severity scores in the XLT/WAS spectrum for a subset of mutations.
Acknowledgments
We thank R. Crouch (NIH/NICHD, Bethesda, USA) for providing RNaseH1-eGFP and its control vector, A. Galy (Généthon, Evry, France) for the ShRNA lentiviral vector, A. Selvakumar (Memorial Sloan-Kettering Cancer Center, New York, USA) for assistance with nucleotide blast searches, F. Candotti (University Hospital of Lausanne, Switzerland) and A. Aiuti (San Raffaele Telethon Institute for Gene Therapy, Milan, Italy) for patient samples, and the University of Iowa Dance Marathon (UIDM) for providing laboratory space. This work was supported by the NIH, National Institute of Allergy and Infectious Diseases (NIAID) grant R01AI084957 (to Y.M.V.), the Intramural Research Program of the NIH, National Institute on Aging (NIA) grant Z01AG000746-08 (to M.M.S.), and the DeJoria Wiskott-Aldrich Research Fund (to H.D.O).
Funding: This work was supported by the NIH grant R01AI084957 (to Y.M.V.), the Intramural Research Program of the NIH, National Institute on Aging grant Z01AG000746-08 (to M.M.S.), and the DeJoria Wiskott-Aldrich Research Fund (to H.D.O).
Abbreviations
- WAS
Wiskott-Aldrich syndrome
- XLT
X-linked thrombocytopenia
- XLN
X-linked neutropenia
- WASp
Wiskott-Aldrich syndrome protein
- RNAP2
RNA polymerase II
- pCTD
Phosphorylated C-terminal domain of RNAP2
- S2P or S5P
Phosphorylation of Serine 2 or Serine 5 residues of the CTD
- S2unP
Serine 2 unphosphorylated C-terminal domain of RNAP2
- Th1, Th2, Tfh cells
CD4+ T helper 1, T helper 2, T follicular helper lymphocytes
- RNaseH1, H2, I
Ribonuclease H1, H2, I
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- SRSF1
Serine/Arginine-Rich Protein-Specific Factor 1
- mRNP
Messenger ribonucleoprotein
- TOP
Topoisomerase
- TSS and TTS
transcription start site and transcription termination site
- PBMC
Peripheral blood mononuclear cells
- EBV-BLCL
Epstein-Barr virus immortalized B lymphoblastoid cell line
- PHA
Phytohemagglutinin
- EGFP
Enhanced green fluorescent protein
- HTLV-1
Human T-cell lymphotropic virus type 1
- LC-MS/MS
Laser capture tandem mass spectrometry
- TDRD3
Tudor Domain Containing 3
- BRCA1/BRAC2
Breast Cancer Susceptibility Protein Type 1/Type 2
- WKO/WKD
WAS knock out/knock-down
Footnotes
Conflict of interest: All authors declare no conflict of interest with the current study
Author contribution: K.S. performed ChIP, DRIP, coimmunoprecipitation/Western blot, WAS mutant and RNH1 transfections, flow cytometry, RT-qPCR assays, and assisted in comet assay. S.S.H. generated CRISPR/Cas9-mediated WAS KO cellular models, performed all nascent pre-mRNA assays, RNH1 transfection, and Western blot experiments. K.K.W. performed all confocal imaging, comet assays, assisted in RNH1 transfection, and performed statistical analyses for all images. H.D.O. and L.D., provided XLT/WAS patient samples and generated shRNA-mediated WAS KD cellular models. M.M.S. interpreted the data and wrote the paper; Y.M.V. conceived the study, designed the experiments, analyzed and interpreted the data, and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–44. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 2.Villa A, Notarangelo L, Macchi P, Mantuano E, Cavagni G, Brugnoni D, et al. X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet. 1995;9:414–7. doi: 10.1038/ng0495-414. [DOI] [PubMed] [Google Scholar]
- 3.Ancliff PJ, Blundell MP, Cory GO, Calle Y, Worth A, Kempski H, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–9. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- 4.Imai K, Morio T, Zhu Y, Jin Y, Itoh S, Kajiwara M, et al. Clinical course of patients with WASP gene mutations. Blood. 2004;103:456–64. doi: 10.1182/blood-2003-05-1480. [DOI] [PubMed] [Google Scholar]
- 5.Albert MH, Bittner TC, Nonoyama S, Notarangelo LD, Burns S, Imai K, et al. X-linked thrombocytopenia (XLT) due to WAS mutations: clinical characteristics, long-term outcome, and treatment options. Blood. 2010;115:3231–8. doi: 10.1182/blood-2009-09-239087. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, et al. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–9. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- 7.Westerberg LS, Meelu P, Baptista M, Eston MA, Adamovich DA, Cotta-de-Almeida V, et al. Activating WASP mutations associated with X-linked neutropenia result in enhanced actin polymerization, altered cytoskeletal responses, and genomic instability in lymphocytes. J Exp Med. 2010;207:1145–52. doi: 10.1084/jem.20091245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moulding DA, Blundell MP, Spiller DG, White MR, Cory GO, Calle Y, et al. Unregulated actin polymerization by WASp causes defects of mitosis and cytokinesis in X-linked neutropenia. J Exp Med. 2007;204:2213–24. doi: 10.1084/jem.20062324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–34. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 10.Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D'Silva S, et al. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci Transl Med. 2010;2:37ra44. doi: 10.1126/scitranslmed.3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J Immunol. 2014;193:150–60. doi: 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar K, Sadhukhan S, Han SS, Vyas YM. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood. 2014;124:3409–19. doi: 10.1182/blood-2014-07-587642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar K, Sadhukhan S, Han SS, Vyas YM. SUMOylation-disrupting WAS mutation converts WASp from a transcriptional activator to a repressor of NF -kappaB response genes in T cells. Blood. 2015;126:1670–82. doi: 10.1182/blood-2015-05-646182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teitell MA. Alternative control: what's WASp doing in the nucleus? Sci Transl Med. 2010;2:37ps31. doi: 10.1126/scitranslmed.3001336. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Dai R, Li W, Zhao H, Zhang Y, Zhou L, et al. Abnormalities of follicular helper T-cell number and function in Wiskott-Aldrich syndrome. Blood. 2016;127:3180–91. doi: 10.1182/blood-2015-06-652636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai X, Zhang Y, Huang L, Wang J, Li W, Niu L, et al. The early activation of memory B cells from Wiskott-Aldrich syndrome patients is suppressed by CD19 downregulation. Blood. 2016;128:1723–34. doi: 10.1182/blood-2016-03-703579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuznetsov NV, Almuzzaini B, Kritikou JS, Baptista MAP, Oliveira MMS, Keszei M, et al. Nuclear Wiskott-Aldrich syndrome protein co-regulates T cell factor 1-mediated transcription in T cells. Genome Med. 2017;9:91. doi: 10.1186/s13073-017-0481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Looi CY, Sasahara Y, Watanabe Y, Satoh M, Hakozaki I, Uchiyama M, et al. The open conformation of WASP regulates its nuclear localization and gene transcription in myeloid cells. Int Immunol. 2014;26:341–52. doi: 10.1093/intimm/dxt072. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science. 2013;341:1002–5. doi: 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, et al. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J Exp Med. 2014;211:2119–34. doi: 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verboon JM, Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM. Wash interacts with lamin and affects global nuclear organization. Curr Biol. 2015;25:804–10. doi: 10.1016/j.cub.2015.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM. Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn. 2012;241:608–26. doi: 10.1002/dvdy.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krol K, Jendrysek J, Debski J, Skoneczny M, Kurlandzka A, Kaminska J, et al. Ribosomal DNA status inferred from DNA cloud assays and mass spectrometry identification of agarose-squeezed proteins interacting with chromatin (ASPIC-MS) Oncotarget. 2017;8:24988–5004. doi: 10.18632/oncotarget.15332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–6. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 25.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 26.Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell. 2016;63:167–78. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–9. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–52. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 29.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 30.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–47. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–25. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chedin F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet. 2016;32:828–38. doi: 10.1016/j.tig.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–21. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–85. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, et al. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol Cell. 2015;60:351–61. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Rubio ML, Perez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–22. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costantino L, Koshland D. The Yin and Yang of R-loop biology. Curr Opin Cell Biol. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickramasinghe VO, Venkitaraman AR. RNA Processing and Genome Stability: Cause and Consequence. Mol Cell. 2016;61:496–505. doi: 10.1016/j.molcel.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–96. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–88. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim YW, Sanz LA, Xu X, Hartono SR, Chedin F. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife. 2015;4 doi: 10.7554/eLife.08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De I, Bessonov S, Hofele R, dos Santos K, Will CL, Urlaub H, et al. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat Struct Mol Biol. 2015;22:138–44. doi: 10.1038/nsmb.2951. [DOI] [PubMed] [Google Scholar]
- 47.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–78. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–75. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–30. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou JF, et al. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–2. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- 55.Richard P, Manley JL. R Loops and Links to Human Disease. J Mol Biol. 2017;429:3168–80. doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groh M, Gromak N. Out of balance: R-loops in human disease. PLoS Genet. 2014;10:e1004630. doi: 10.1371/journal.pgen.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chedin F, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife. 2016;5 doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–45. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–8. doi: 10.1038/nature14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jangi M, Fleet C, Cullen P, Gupta SV, Mekhoubad S, Chiao E, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci USA. 2017;114:E2347–E56. doi: 10.1073/pnas.1613181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Meester J, Calvez R, Valitutti S, Dupré L. The Wiskott-Aldrich syndrome protein regulates CTL cytotoxicity and is required for efficient killing of B cell lymphoma targets. J Leukoc Biol. 2010;88:1031–40. doi: 10.1189/jlb.0410197. [DOI] [PubMed] [Google Scholar]
- 62.Lafouresse F, Cotta-de-Almeida V, Malet-Engra G, Galy A, Valitutti S, Dupré L. Wiskott-Aldrich syndrome protein controls antigen-presenting cell-driven CD4+ T cell motility by regulating adhesion to intercellular adhesion molecule-1. Immunology. 2012;137:183–96. doi: 10.1111/j.1365-2567.2012.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen TN, Goodrich JA. Protein-protein interaction assays: eliminating false positive interactions. Nat Methods. 2006;3:135–9. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang W, Ochs HD, Dupont B, Vyas YM. The Wiskott-Aldrich syndrome protein regulates nuclear translocation of NFAT2 and NF-kappa B (RelA) independently of its role in filamentous actin polymerization and actin cytoskeletal rearrangement. J Immunol. 2005;174:2602–11. doi: 10.4049/jimmunol.174.5.2602. [DOI] [PubMed] [Google Scholar]
- 65.Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, P OB, Sato H, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–9. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 67.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–86. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 69.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–94. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–9. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–7. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 73.Belotserkovskii BP, Hanawalt PC. Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys J. 2011;100:675–84. doi: 10.1016/j.bpj.2010.12.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell. 2016;165:357–71. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Felipe-Abrio I, Lafuente-Barquero J, Garcia-Rubio ML, Aguilera A. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J. 2015;34:236–50. doi: 10.15252/embj.201488544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Broccoli S, Rallu F, Sanscartier P, Cerritelli SM, Crouch RJ, Drolet M. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol Microbiol. 2004;52:1769–79. doi: 10.1111/j.1365-2958.2004.04092.x. [DOI] [PubMed] [Google Scholar]
- 77.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–23. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawanishi S, Ohnishi S, Ma N, Hiraku Y, Murata M. Crosstalk between DNA Damage and Inflammation in the Multiple Steps of Carcinogenesis. Int J Mol Sci. 2017;18:1808. doi: 10.3390/ijms18081808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trifari S, Sitia G, Aiuti A, Scaramuzza S, Marangoni F, Guidotti LG, Martino S, Saracco P, Notarangelo LD, Roncarolo MG, Dupré L. Defective Th1 cytokine gene transcription in CD4+ and CD8+ T cells from Wiskott-Aldrich syndrome patients. J Immunol. 2006;177:7451–61. doi: 10.4049/jimmunol.177.10.7451. [DOI] [PubMed] [Google Scholar]
- 80.Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53:484–97. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. Livingston DM. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–47. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–65. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 83.Husain A, Begum NA, Taniguchi T, Taniguchi H, Kobayashi M, Honjo T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat Commun. 2016;7:10549. doi: 10.1038/ncomms10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, Pokharel S, Wang JT, Xu X, Liu Y. RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat Commun. 2015;6:6720. doi: 10.1038/ncomms7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ochs HD, Thrasher AJ. The Wiskott-Aldrich syndrome. J Allergy Clin Immunol. 2006 Apr;117(4):725–38. doi: 10.1016/j.jaci.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 87.Herrera-Moyano E, Mergui X, García-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014;28:735–48. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuntziger T, Landsverk HB, Collas P, Syljuasen RG. Protein phosphatase 1 regulators in DNA damage signaling. Cell Cycle. 2011;10(9):1356–62. doi: 10.4161/cc.10.9.15442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Phenotypic characterization of human CD4 Th cells. Flow cytometric analyses of the indicated primary Th cells (ND8) or Th lines (NIH001, Jurkat) stained with a panel of indicated antibodies or their isotype IgG control antibodies are shown as histogram plots. The data tabulated below indicates the Th subtype, i.e., effector, effector memory, naïve etc. based on the combination of cell receptor expression.
Supplementary Figure 2. Experimental depletion of WAS expression in normal (WT) Th cells. (A) For CRISPR/Cas9-mediated WAS KO: location of gRNA guides on the WAS gene showing primer pairs with the expected amplicon length. (B) representative gel images showing clones with PCR amplicon patterns that is consistent with no KO, partial KO, and complete KO of WAS. (C) Western blot sequentially reprobed with anti-WASp and α-tubulin loading control antibodies, to demonstrate complete WAS KO clones lacking WASp expression. (D) DNA sequencing of complete WAS KO clone show the expected gene region deletion. (E) RNA-interference-mediated depletion of WASp expression by shRNA vector. Shown is the western blot image of WASp expression in the indicated primary Th cells.
Supplementary Figure 3. RNaseH1 expression into WT and WKO Th1 cells. Flow cytometry (dot plot) of NIH001-WT and -WKO Th1 lines transfected with RNaseH1-eGFP construct or its empty vector control is shown as pre-sort (transfection efficiency) and post sort (purity) in 3 independent experiments. “X” denotes post-sort sample not tested for purity. The location of the gated population is shown, which was sorted for down-stream assays.
Supplementary Figure 4. R loop modulation in DRB-treated Th1 cells. Left, confocal images of WT Th1 cells dual-labelled with S9.6 antibody and DAPI. The image shown is representative of n=100 cells in DRB 100uM for 2 h treatment condition. Right, statistics of S9.6+ foci changes under the indicated DRB dose/duration conditions. The displayed data is from n=100-150 cells analyzed for each condition. p-value: ns= nonsignificant by Mann-Whitney nonparametric test.
Supplementary Figure 5. R-loop and DNA damage load in WASp-deficient Th2-cells. (A) Box-and-whisker plots comparing the magnitude of R-loop (S9.6+ foci) and DSB load (comet assay) in Th1 and Th2 cells, WASp-sufficient (WT) and WASp-deficient (WKO/WKD). The data is representative of analyzing 20-30 z-stack images of at least 100 cells each from 2-3 independent experiments. (B) shown are the indicated immunofluorescence images overlaid on the corresponding Nomarski (DIC) images for Th1 and Th2 cells. Right, plot showing the range of individual (single) S9.6+ foci size in Th1 and Th2 cells. ****p<0.0001, Mann-Whitney.
Supplementary Figure 6. Reconstituting nuclear but not cytosolic compartment of WASp-deficient Th1-cells is necessary to normalize R-loop frequency. Change in R-loop frequency in WAS Th1 line stably-transfected with NLS-deleted (ΔNLS) or NES-deleted (ΔNES) WASp mutants. Note, reconstituting only the nuclear compartment with WASp is sufficient to normalize the R-loop frequency (left panel), but reconstituting only the cytosolic compartment does not change the R-loop frequency (right panel). Also shown is the ChIP enrichment profile of γH2A.X. Note, the DNA damage as reported by γH2A.X enrichment is reduced by reconstituting only the nuclear compartment of WASp-deficient Th1 cells. Data is from n=3, mean+SEM.
Supplementary Figure 7. The magnitude of R-loop accumulation and DNA-damage at non-Th1 genes in WASp-deficient Th1 cells. (A) R loop frequency at the 3′-ends of the indicated Th1 and non-Th1 genes in WAS Th1 line (NIH002) untransfected (control) or transfected with wt-WASp or the indicated pathogenic mutant. (B) DNA damage frequency at the same cis-region in the same set of genes. Data is from n=3, mean+SEM.
Supplementary file 1. Antibodies and Reagents
Supplementary file 2. RT-qPCRPrimers and Probes for mature mRNA and nascent pre-mRNA estimation
Human Primary Th1 cells: nuclear fraction (IP with anti-WASp Ab)
Human Jurkat Th1 cells: nuclear fraction (2 step IP with anti-Flag and -Myc Abs)