Abstract
The rice blast fungus, Magnaporthe oryzae, causes one of the most destructive diseases of cultivated rice in the world. Infections caused by this recalcitrant pathogen leads to the annual destruction of approximately 10–30% of the rice harvested globally. The fungus undergoes extensive developmental changes to be able to break into plant cells, build elaborate infection structures, and proliferate inside host cells without causing visible disease symptoms. From a molecular standpoint, we are still in the infancy of understanding how M. oryzae manipulates the host during this complex multifaceted infection. Here, we describe recent advances in our understanding of the cell biology of M. oryzae biotrophic interaction and key molecular factors required for the disease establishment in rice cells.
Keywords: Rice blast, appressorium, cytoplasmic effectors, apoplastic effectors
The impact of rice blast disease on rice production
Rice (Oryza sativa) is by far the most important staple food for more than half of the human population, providing approximately 19% of the daily calories consumed worldwide [1]. By 2050, the human world population is predicted to increase from nearly 7.6 to 9.8 billioni. Thus, to keep up with future rice demands, rice producers must increase global rice yields by 25% before 2030ii. To support this increase, rice crops will face several future challenges that will seriously jeopardize its annual production. Among these challenges are fungal diseases threatening rice production that cause decreasing annual yields and increasing cultivation costs [2]. Rice blast disease represents a significant threat to rice production worldwide. The causal agent of this disease is the hemibiotroph (see Glossary) filamentous fungus Magnaporthe oryzae (anamorph Pyricularia oryzae), which annually contributes to the loss of enough rice to feed 60 million people [3]. Unfortunately, neither traditional breeding nor chemical approaches have been able to contain this disease, due to the fact that the fungus can rapidly adapt and mutate to evolve resistance to multiple rice varieties [3]. Despite outstanding efforts to understand M. oryzae biology and its interactions with the host, this pathogen continues to be a major threat to global food security [3]. Additionally, rice is not the only plant susceptible to this pathogen. Several M. oryzae pathotypes are also able to infect a wide variety of annual and perennial grass species including economically important cereal crops, such as wheat (Box 1), barley, and millet [2]. In this review, we focus on recent advances in our understanding of how M. oryzae undergoes different morphological changes in order to penetrate and invade rice cells. In particular, to gain a better understanding of how the fungus colonizes rice tissues, we highlight several aspects of the Magnaporthe life cycle and major key factors required for the establishment of rice blast disease. We do not discuss the biology of avirulent effector proteins in detail, because these have been reviewed recently [4].
Box 1: Wheat Blast - potential threat to global wheat production.
The Magnaporthe oryzae Triticum pathotype (MoT) is the causal agent of wheat blast or ‘brusome’ disease. This disease was first identified infecting wheat plants in the Paraná State of Brazil in 1985 [63]. Soon after, the pathogen spread to other wheat-producing regions of Brazil, and neighboring countries such as Bolivia, Paraguay and Argentina (see review [64]). The first wheat blast outbreak outside of South America was detected in Bangladesh in February 2016 [65]. The disease originates from a M. oryzae subpopulation that infects Triticum species (wheat) but not rice plants. This fungus can infect all above-ground tissues of wheat plants. However, the most obvious and damaging symptom of wheat blast infections is the bleaching of the spike. It occurs when the fungus attacks rachis at the base of the wheat heads, thereby blocking the transport of nutrients to the upper head parts resulting in the bleached appearance and hindrance of seed development [66]. Moreover, MoT is recognized as a seedborne pathogen, meaning that it can spread through infected seeds [64]. Therefore, wheat blast disease is a major threat to wheat production because the pathogen can severely infect wheat heads and result in serious yield losses-up to 100% in susceptible varieties [67]. The continued spreading of MoT to new wheat-growing regions is particularly worrisome because this disease is a potential threat to global wheat production and there is lack of effective disease control. Therefore, the need for a better understanding of the wheat blast biology and epidemiology is a top priority for the blast research community. Recently, researchers sought to understand the origin of this aggressive pathogen in 11 districts in Bangladesh, confirming that the outbreak resembles that of the South American wheat blast isolates [68, 69]. Moreover, a recent study revealed the key reasons why the blast fungus jumped hosts, from oat or perennial ryegrass to wheat, in Brazil. Genetic studies identified two Avr effectors, PWT3 and PWT4, from ryegrass and oat fungal isolates, which elicit a rapid host response in wheat plants containing resistance genes rwt3 and rwt4 [70]. In the early 1980s, a new high-yield wheat cultivar lacking the cognate resistance gene to Avr effector PWT3 was introduced into Brazil, allowing certain fungal isolates to colonize wheat plants. Wheat cultivars still carrying the cognate resistance pwt3 gene in nearby areas imposed selective pressure that shaped the current genetics of M. oryzae PWT3-containing isolate, resulting in a new nonfunctional PWT3 strain [70]. These events prompted the emergence of a wheat blast outbreak a few years thereafter in Brazil. Thus, the use of wheat cultivars possessing both the Rwt3/Rwt4 is essential for reoccurrence of host jumps or for wheat blast disease prevention.
Infection Stage, Part I: Host surface recognition and appressorium formation
Host cell contact
To be a successful pathogen, M. oryzae faces extensive developmental changes to colonize the plant tissues and complete its life cycle. Unlike other plant pathogens, M. oryzae has a hemibiotrophic life style, in which the fungus undergoes an initial biotrophic stage during which the plant immune system is suppressed, and then switches to a necrotrophic stage that promotes plant cell death. Once the three-celled asexual spores (known as conidia) settle on the leaf cuticle, the conidium releases an adhesive substance called spore tip mucilage from the conidial apex and attaches itself tightly to the hydrophobic plant surface (Figure 1) [5]. Furthermore, upon attachment, the conidium germinates and forms a polarized germ tube at one of the apical cells, and grows across the leaf surface (Figure 1). When the germ tube perceives physical cues, such as surface hardness and hydrophobic surface, the tip of the germ tube differentiates into a dome-shaped infection structure called the appressorium (Figure 1) [6–8]. As the appressorium matures, a thick layer of melanin deposition forms on the inner side of the appressorium cell wall providing an impermeable barrier to prevent the efflux of comparable solutes. At the same time, the conidium contents are recycled into the appressorium contributing to the next stages of appressorium development. Subsequently, a substantial internal turgor pressure of up to 8.0 MPa is generated in the appressorium, which is translated into a physical force acting at the base of the appressorium forming a penetration peg to rupture the rice cell cuticle (Figure 1) [9, 10]. During appressorial morphogenesis, M. oryzae accumulates high levels of endogenous reactive oxygen species (ROS) hence strengthening the appressorium cell wall [11]. This ROS accumulation is regulated by two M. oryzae NADPH oxidases, Nox1 and Nox2. Mutation on any of these Nox genes result defects in appressorium-mediated cuticle penetration. Once the appressorium matures, it remodels its actin cytoskeleton to form a toroidal F-actin network at the base of the cell, scaffolded by septins GTPases to promote the penetration peg emergence to invade plant tissue [12]. Remodeling of the F-actin cytoskeleton also requires the action of Nox2 NADPH oxidase complex [13]. Following penetration of the leaf cuticle, the fungus rapidly colonizes and destroys the plant cells, generating necrotic lesions filled with spores to spread the diseases. To achieve this, M. oryzae utilizes different mechanisms to rapidly response to nutrient fluctuations and redox conditions inside the plant cell during infection (Box 2).
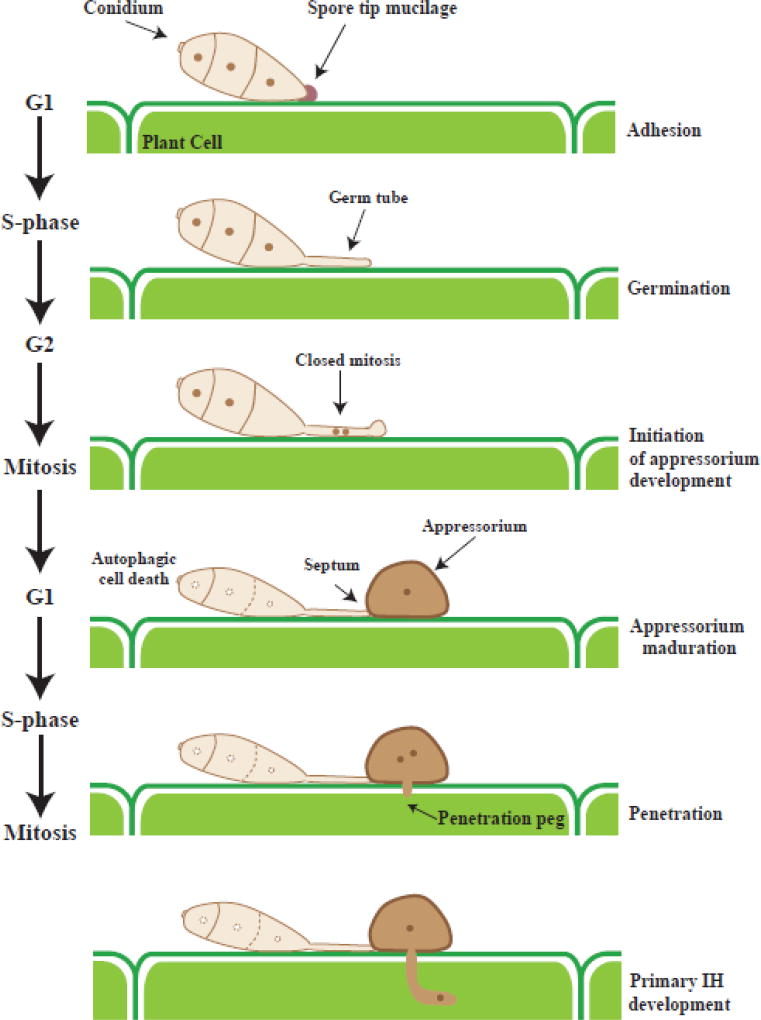
Figure 1.
Schematic representation of cell cycle progression during appressorium development in the rice blast fungus M. oryzae. The fungus three-celled conidium adheres to the plant hydrophobic cuticle and germinates. Each cell contains one nucleus. One of the nuclei migrates to the germ tube and goes through a single mitotic event, in which two daughter nuclei are generated in a closed mitosis. On daughter nucleus migrates into the immature appressorium and a septum is formed, while the other nucleus migrates back to the conidium, which then goes through an autophagic cell death process. Appressorium remains mitotically active after maturation. Brown circles: nucleus, dashlines: conidium degradation, HI: invasive hyphae.
Box 2. Metabolic strategies employed by M. oryzae in planta.
Once M. oryzae spores germinates on the leaf cuticle, the germ tube constantly monitors its environment for physical cues to generate infection structures to colonize plant tissues. At this point of infection, the fungus actively moves from the nutrient-poor leaf surface conditions to the nutrient-rich conditions inside the plant cell. How does M. oryzae cope with nutrient fluctuations during plant infection? Sugar sensing may enable the fungus to respond rapidly to nutrient availability in the environment. Indeed, trehalose-6-phosphate synthase (TPS1) plays a pivotal role in glucose-6-phosphate (G6P) sensing. Tps1 is a biosynthetic enzymes required for the production of the non-reducing disaccharide trehalose from G6P and uridine disphosphate (UDP)-glucose [71–73]. Additionally, Tps1 regulates gene expression in response to G6P availability during M. oryzae infection [72, 74]. Previous studies demonstrated that Tps1 is required for M. oryzae pathogenesis due to G6P binding independent of its trehalose production [71, 75]. In response to G6P, Tps1 activates glucose-6-phosphate dehydrogenase (G6PDH) to generate high levels of NADPH in the oxidative stage of pentose phosphate pathway (PPP). This in turn leads to the activation of different NADPH requiring genes, some of which are involve in nitrogen metabolism, pathogenicity via NADPH metabolism, and repression of genes required for alternative carbon sources [72]. Furthermore, NADPH levels lead to the activation of several GATA factors including Nut1, a nitrogen regulator in M. oryzae, and to G6P displacement from Tps1 active side [72]. Notably, Tps1 acts as a central regulator of G6P sensing to manipulate NADPH levels, leading to rapid genetic responses that will enable M. oryzae to adapt and grow in such a harsh environment like the host cell. A recent study provided evidence that G6P/Tps1 sensing and NADPH production are essential for both glutathione and thioredoxin antioxidant systems during M. oryzae biotrophic growth [76]. Another study demonstrated how glucose metabolism through the transketolase enzyme, encoded by TKL1, is essential for M. oryzae invasive growth and cell-to-cell movement during infection [77]. Loss of TKL1 in M. oryzae has severe effects on ATP levels, IH growth and cell cycle progression in planta [77]. Addition of exogenous ATP restores IH growth and reverses mitotic delay in Δtkl1 mutant strains during infection. This supports the notion that glucose metabolism through the activation of the non-oxidative PPP to generate ATP and NADPH is an important process for M. oryzae biotrophic growth and infection. Interestingly, connections have been made between primary metabolism and suppression of host plant defenses during rice blast infections [78]. The sugar sensor Tsp1 regulates the expression of nitronate monooxygenases 2, encoded by the NMO2 gene, which catalyzes the oxidative denitrification of nitroalkanes. Loss of NMO2 resulted in strains severely attenuated in IH growth, unable to grow under nitrate and nitrite containing media, and highly susceptible to nitooxidative stress conditions and plant immune responses [78]. In general, M. oryzae uses NMO2 to protect itself against nitrooxidative stress conditions, and to maintain redox balance by suppressing the first line of plant defenses to avoid plant recognition during rice blast infection.
Translating the environmental cues via cyclic AMP and Pmk1 MAP kinase signaling pathways
The formation of a functional appressorium is an essential step in the infection cycle of M. oryzae. The infection begins when the conidium germinates on the leaf surface and the germ tube recognizes physical cues leading the fungus to develop an appressorium. The recognition of the physical cues is primarily regulated by highly conserved G-protein/cAMP-dependent signaling and the Pmk1 MAP kinase pathways (Figure 2) [8, 14, 15]. Upon conidia germination, Pth11 a bona fide G-protein-coupled receptor senses a hydrophobic surface and interacts with the G-proteins complex to mediate the activation of the cAMP signaling pathway (Figure 2) [20]. The Pth11 protein contains seven transmembrane regions and a CFEM (Common in several Fungal Extracellular Membrane protein) domain [21, 22]. Pth11 functions upstream of the cAMP signaling pathway in M. oryzae [20]. Interestingly, the Pth11 CFEM domain is required for the formation of appressorium and pathogenicity, implying that CFEM is the putative surface sensing function of Pth11 [23]. Moreover, ROS accumulation during appressorium formation is impaired in the pth11 mutant strain. However, exposure to antioxidants induces appressorium formation in the pth11 mutant strain on hydrophobic and in planta surface. M. oryzae contains three distinct G-α subunits: MagA, MagB and MagC as well as an adenylate cyclase, Mac1 [14, 16, 17]. Genetically, Rgs1 (Regulator of G-protein Signaling) modulates the G-α subunit MagA to enable M. oryzae to perceive and response to physical cues during appressorium formation [18]. PKA activity also is essential for vegetative growth and appressorium formation in M. oryzae. Two forms of PKA, cPKA and cPK2, have overlapping functions in cAMP cascade [14]. The generation of cpkA cpk2 double mutants showed severe defects in growth and sporulation and failed to develop infection-related structures in M. oryzae [19]. Moreover, spontaneous suppressor of cpkA cpk2 restores hyphal growth and appressorium formation on hydrophobic surface but not pathogenesis in M. oryzae. Interestingly, loss of function mutations in the transcription factor, MoSfl1, can bypass PKA activity to restores growth and appressorium formation in the cpkA cpk2 mutant [19]. MoSfl1 interacts with the conserved Cyc8-Tup1 transcriptional co-repressor to block the transcription of target genes required for growth [19]. In general, phosphorylation of MoSfl1 by PKA disrupts MoSfl1-Cyc8-Tup1 interaction to regulate the expression of growth and infection-related genes [19].
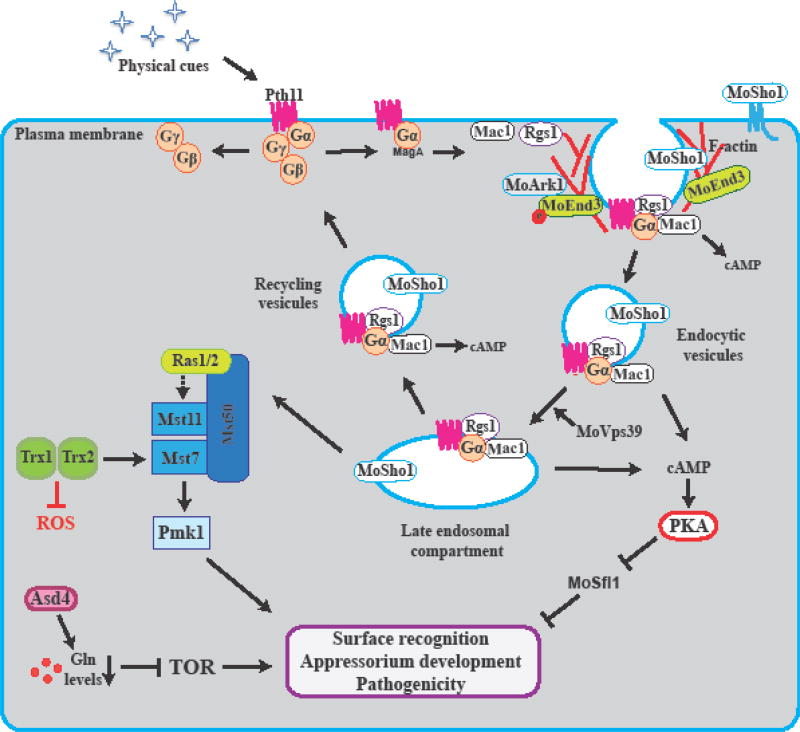
Figure 2.
Schematic representation of the spatio-temporal cell dynamics of the G-protein/Pth11 and cAMP-signaling pathway during surface sensing and appressorium morphogenesis. Upon conidia germination, the key activators and regulators of G protein/cAMP (Pth11, membrane sensor MoSho1, G-proteins, Rgs1, and Mac1) are internalized to endosomal compartments leading to the accumulation of cAMP and subsequently PKA activity. MoEnd3 mediates the receptors Pth11 and MoSho1 endocytosis. MoArk1 negatively regulates MoEnd3 through protein phosphorylation. Mst50 interacts with Mst11/Mst7 and Ras1/2 proteins to activate the Pmk1 MAPK cascade and regulate appressorium development, penetration and pathogenicity. Asd4 acts downstream to cPKA, maintains intracellular glutamine levels in the cell, and promote appressorium formation. TOR signaling pathway inhibits the appressorium formation under conditions of high glutamine levels or in the presence of glucose. Diagram modified from [24
A recent study identify endosomal compartments to function as signaling scaffolds that anchor the components of the G-protein/cAMP signaling in M. oryzae [22]. Upon surface sensing, the key activators and regulators, including Pth11, MagA, Mac1 and Rgs1, localized in the dynamic tubulo-vesicular endosomal compartments (Figure 2) [22]. Vps39 is a key member of the homotypic fusion and vacuole protein-sorting complex, essential for the conversion of early endosome into late endosomal compartments. Loss on MoVPS39 function disrupts the endosomal localization of the Pth11, MagA, Mac1, and Rgs1, cAMP signaling and inhibit appressoriun development (Figure 2) [22]. Exogenous cAMP treatments restore appressorium formation on vps39 mutant strain suggesting that functional Vps39 is essential for proper cAMP signaling and pathogenesis. Late endosomal scaffolding and trafficking of G-protein signaling components are vital for robust cAMP signaling leading to appressorium development in M. oryzae (Figure 2).
Mass spectrometry analysis identified a M. oryzae Ark1 (actin regulating kinase) interacting protein, MoEnd3, that is required for endocytosis transport and F-actin assembly (Figure 2). MoEnd3 function is linked to the internalization of Pth11 and the membrane sensor MoSho1 to the endosomal compartments [24]. Deletion of MoEnd3 gene resulted in delays in endocytic transport and appressorium development [24]. Moreover, the authors demonstrate how MoEnd3 is involved in multiple cellular processes including autophagy, MAP kinase pathway and might has additional role in effector secretion during plant infection [24].
The Pmk1 MAP kinase pathway regulates late stages of appressorium formation, penetration and invasive growth (Figure 2) [14, 25]. Pmk1 is orthologous to the terminal kinase in the MAP kinase cascade, Fus3/Kss1 MAP kinases (MAPKs), found in the budding yeast Saccharomyces cereviciae [26]. Loss in Pmk1 function results in an appressorium defective mutant that is non-pathogenic. Upstream to Pmk1, MAPK kinase (MEK) Mst7, MEK kinase (MEKK) Mst11 and an adaptor protein Mst50 have been identified [14, 27, 28]. Mst7 and Mst11 interact with Mst50, to activate the Pmk1 MAPK cascade. As expected, deletion of any of these genes upstream of Pmk1 result in impairments of appressorium formation and plant infection [27–29].
New evidence has revealed two cell-surface signaling mucins, Msb2 and Cbp1, promote the activation of Pmk1 MAPK pathway through the GTP-binding proteins Ras2 interaction [30]. The extracellular mucin domain region of Msb2 is required for appressorial formation whereas the cytoplasmic domain is essential for M. oryzae penetration and invasive growth [30]. Previously that it was demonstrated two Ras GTP-binding proteins, Ras1 and Ras2, play an essential role in Pmk1 activation by interacting with Mst11 and Mst50 (Figure 2) [28]. Contrary to Ras1 that displays no obvious phenotype, Ras2 has been proven essential for M. oryzae appressorium morphogenesis [29]. Interestingly, the generation of dominant active RAS2 protein in M. oryzae resulted in improper activation of both Pmk1 MAPK and cAMP signaling pathways, and the formation of appressoria on non-inductive surfaces [29]. Therefore, constitutive expression of Ras2 enables the fungus to bypass the physical cues and attachment required for initial appressoria development. Moreover, the interaction of MEKK Mst11 with activated Ras2 and two phosphorylation events releases Mst11 from its self-inhibitory binding and activates the Pmk1 pathway [31].
Additionally, two other M. oryzae thioredoxin genes, TRX1 and TRX2, have been found to play an important role in pathogenesis [32]. TRX2 interacts with the Mst7 kinase and regulates the activation of the Pmk1 MAPK pathway for appressorium formation (Figure 2) [32]. Interestingly, both TRX1 and TRX2 are implicated in another signaling pathway that involves intra-cellular reactive oxygen species (ROS) signaling [32].
Both cAMP PKA and Pmk1 MAPK signaling pathways positively regulate appressorial morphogenesis and maturation. However, recent evidence has revealed a negative regulatory pathway of cAMP PKA and Pmk1 MAPK signaling that prevents appressorium formation in M. oryzae [33]. Marroquin and Wilson have demonstrated that the activation of Target of Rapamycin (TOR) signaling pathway in response to the accumulation of intracellular glutamine trigger the inhibition of appressorium development (Figure 2) [33]. The GATA transcription factor, Asd4, regulates intracellular glutamine levels and promotes appressorium formation. Δasd4 mutant strains are unable to develop an appressorium on inductive surface, and contain high levels of glutamine [33]. Interestingly, the inhibition of TOR pathway by rapamycin restores appressorium formation in both Δasd4, ΔcPKA mutants but not in the MAP Kinase ΔPmk1 mutant [33]. Therefore, TOR inactivation requires a functional Asd4 to regulate intracellular glutamine levels and develop an appressorium in M. oryzae.
Cell cycle progression during appressorium formation
In addition to physical cue responses by cAMP and MAPK signaling pathways, M. oryzae utilizes essential cellular processes to ensure functional appressorial formation and maturation. M. oryzae conidia undergo a sequential cell cycle progression and autophagic cell death for appressoria development [34, 35]. The conidium has three nuclei, one of which migrates into the developing appressorium where it undergoes a single mitotic division (Figure 1). This process generates two daughter nuclei, of which one nucleus migrates into the developing appressorium and the other one migrates back to the conidium. Once the daughter nucleus translocates to the appressorium, a septum is formed between the germ tube and the appressorium structure [36]. Therefore, this daughter nucleus is the source of all genetic material during in planta infection. The initial appressorium formation is controlled by a cell cycle progression at the germ tube, in which DNA replication in the S-phase is required to initiate swelling at the germ tube tip [35]. Inhibition of DNA replication by either hydroxyurea (HU) or generation of a temperature sensitive nim1 mutant prevent the germ tube tip from differentiating into appressorium [35]. It is now known that M. oryzae requires two independent S-phase checkpoints that ensure proper timing of initial appressorium morphogenesis and appresorium repolarization [37]. The first Sphase checkpoint is regulated by the protein kinase Cds1 involving the DNA damage response (DDR) pathway. HU treatments on cds1 mutant result in defects on conidium cell death, and the formation of unmelanized appressorium. The second S-phase checkpoint take place at the appressorium, where it is vital for penetration peg formation and plant infection. This novel S-phase checkpoint is independent of DDR pathway and is regulated by turgor sensing and linked to melanin biosynthesis. Following S-phase, G2/M cell cycle checkpoints are required for maturation of the appressorium and subsequent plant cell infection [35]. Indeed, an inactivated TOR pathway is required to arrest the cell cycle at G2 and initiate both autophagy and appressorial development. Nutrients released from autophagy re-activate TOR signaling to reinitiate cell cycle progression through mitosis. Then the appressorial nuclei arrest at G1 because TOR signaling is inactivated again [38]. After appressorium formation, the three remaining nuclei in the conidium degrade, and the conidium dies via an autophagy process [34]. During this nuclear degradation, M. oryzae employs a nonselective macroautophagy mechanism to successfully infect the plant cell [39]. Subsequently, the conidium contents are recycled into the appressorium contributing to the next stages of appressorium development.
In a recent report, a transcription factor for polarity control, TPC1, was identified and shown to be a key regulator of vegetative growth and appressorium-mediated plant infection processes in M. oryzae [40]. Tpc1 function is associated with the activation of the Pmk1 MAP kinase and Atg1 kinase signaling pathways. Interestingly, Tpc1 activation leads to the transcription of target genes required for autophagy, glycogen/lipid degradation and septin-mediated asymmetric reorganization of F-actin cytoskeleton to facilitate plant cell invasion [40].
Infection stage, part II: Under the radar and inside the leaf
Biology of the invasive growth
Once the melanized appressorium matures, the fungus breaches the host surface and develops a narrow penetration peg to enter into the first host cell (Figure 3, Key Figure). After penetration, the peg differentiates into thin filamentous primary invasive hyphae (IH) and grows inside the live rice cell. At this stage of infection, M. oryzae and the rice cell establish an intimate biotrophic association, in which the fungus switches from filamentous primary IH to bulbous intracellular IH that are surrounded by a plant-derived extrainvasive hyphae-membrane (EIHM) (Figure 3) [41]. This biotrophic association creates an enclosed apoplastic compartment between the intracellular IH and the rice cell membrane [41]. The bulbous IH grows inside the first cell and switches back to primary IH to move into uninfected adjacent cells, potentially through the host’s plasmodesmata [41].
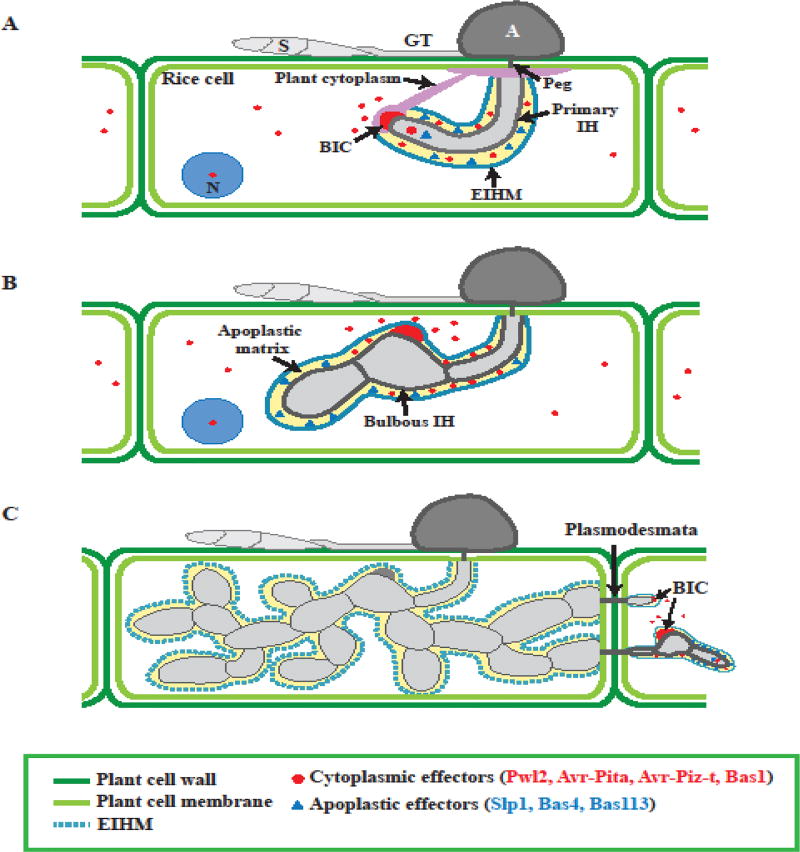
Figure 3.
BIC development and effector secretion during biotrophic growth. These illustrations represent the growth of the invasive hyphae (IH) from 22–40 hour post inoculation inside rice cells. (A) BIC develops at the tip of the primary filamentous invasive hyphae (IH). (B) BIC localizes at a subapical position in the first bulbous IH, and remains behind as the fungus grow in the rice cell. (C) New BICs are formed in the IH tip in the adjacent cells. Cytoplasmic effectors (red circles), including Pwl2, Avr-Pita, Avr-Piz-t and Bas1, accumulate at the BIC and are secreted into the host cytoplasm. In contract, apoplastic effectors (blue triangles), including Slp1, Bas4 and Bas113 are secreted into the apoplastic matrix between EIHM and fungal cell wall. Abbreviations: S, Spores; GT, Germ tube; A, Appressorium; N, Nucleus; BIC, Biotropic interface complex; IH, Invasive hyphae; EIHM, Extrainvasive invasive hyphae membrane
In planta live-cell imaging with fluorescently labeled proteins have become a valuable tool to track and visualize M. oryzae biotrophic invasion and the host cellular events occurring during pathogen colonization. For instance, high spatio-temporal resolution imaging studies demonstrate how M. oryzae appressorium remains mitotically active post-penetration and during IH proliferation [42]. Once the appressorial mitosis occur, a one new nucleus migrates into the swollen apex of the first bulbous IH, while the other nucleus remains in the appressorium (Figure 1) [43]. This cellular event leads to a septum formation in the filamentous region of the first IH [43]. The mitotic nucleus displays a remarkable constriction and elongation during migration through penetration peg [42]. Furthermore, a second round of appressorial mitosis was observed during second IH formation implying that the appressorium viability contributes the genetic material for IH proliferation. However, it remains to be elucidated whether the appressorial mitotic activity persists throughout the IH proliferation in the first infected cell and the adjacent cells. Another study developed a live cell imaging method using two florescent vital dyes, fluorescein diacetate (FDA, indicating green viable cells) and propidium iodide (PI, indicating dead red cells), in rice cells to gain new insights into the dynamics of host cell death during rice blast infection [44]. The authors demonstrated how the first infected rice cell loses viability once M. oryzae advances into adjacent cells [44]. Additionally, during biotrophic growth M. oryzae IH invaginates the host vacuole membrane causing vacuolar shrinkage [45]. However, early collapse of the host vacuole during early infection with M. oryzae has an effect on hyphal growth. This study elegantly demonstrates that the host vacuolar integrity in the first infected rice cell is important for M. oryzae early stages of infection.
As the primary IH develops, a plant membrane-rich structure known as biotrophic interfacial complex (BIC) appears at the tip of the hyphae, where BIC is positioned at the differentiated bulbous IH (Figure 3A,B). As the fungus proliferates inside the first rice cell, BIC remains behind the bulbous IH and then appears again at the tip of the filamentous IH that will move into adjacent cells (Figure 3B, C) [46, 47]. At this biotrophic stage of infection, M. oryzae secretes a repertoire of effector proteins into rice cells to suppress plant immunity and manipulate host cell physiology for the next stages of infection (Figure 3) [48]. To date, many M. oryzae effectors have been identified, although the precise function of most of these effectors has not yet been determined (Table 1). Most fungal effectors are small-secreted proteins without known conserved domains or motifs and presumably have functional redundancy due to the lack of visible virulence phenotypes in single deletion strains [49]. Therefore, it remains as a challenge to predict their function inside the host cell. Despite the lack of sequence commonalities among fungal effectors, recent efforts to understand fungal effector function has revealed that 50% of the M. oryzae avirulence effectors and other fungal effectors belong to a new family of structurally conserved MAX effectors (Magnaporthe Avrs and ToxB like) [50]. Interestingly, the vast majority of the M. oryzae MAX effectors are expressed during the biotrophic stage of infection.
Table 1.
M. oryzae effector proteins identified to date.
| Effector name | ID number | Size a | Function/Expression/Localizationb | Rc | References |
|---|---|---|---|---|---|
| Avirulent effectors | |||||
| AVR-Pita | AF207841 | 224 | Zinc metalloprotease/ In planta*/ BIC; Cytoplasm | Pita | [47,79–81] |
| AVR-Piz-t | HE578813 | 108 | Target rice U3 ubiquitin ligase; reduces Flg-22 and chitin induced ROS production/In planta/Cytoplasm | Piz-t | [53] |
| AVR-CO39 | AF463528 | 89 | Interact with RGA4/RGA5/In planta/Cytoplasm | CO39 | [82,83] |
| AVR-Pia | AB498873 | 85 | -/In planta/Cytoplasm | Pia | [84] |
| AVR-Pii | AB498874 | 70 | C2H2 Zinc finger motif/in planta/Cytoplasm | Pii | [84] |
| AVR-Pik/km/kp | AB498875-AB498879 | 113 | -/In planta/Cytoplasm | Pik | [84] |
| AVR-Pib | KM887844 | 74 | -/-/Cytoplasm predicted | Pib | [85] |
| AvrPi9 | MGG_12655 | 91 | -/In planta/BIC; Cytoplasm | Pi9 | [86] |
| PWL1 | AB480169 | 147 | -/In planta/BIC; Cytoplasm | - | [87] |
| PWL2 | MGG_04301 | 145 | -/In planta/BIC; Cytoplasm | - | [52,87–89] |
| ACE1 | AJ704622 | 4035 | Polyketide synthase-peptide synthetase/Appressorium penetration/Appressorium; Cytoplasm | Pi33 | [90] |
| AVRPi54 | MGG_01947 | 153 | -/In planta/Appressorium | Pi54 | [91] |
| Secreted proteins | |||||
| SLP1 | MGG_10097 | 162 | LysM domains, binds to chitin oligosaccharides; suppresses chitin-induced immunity in rice/In planta/Apoplast | - | [55] |
| MC69 | MGG_02848 | 54 | -/in planta/Apoplast | - | [54] |
| MSP1 | MGG_05344 | 137 | Cerato-platanin family; triggers autophagic cell death and elicits host defense responses/In plant/Apoplast | - | [92] |
| Biotrophy-associated secreted proteins | |||||
| BAS1 | MGG_04795 | 115 | -/In planta/BIC; Cytoplasm | - | [46,52] |
| BAS2 | MGG_09693 | 102 | -/In planta/ BIC; Cell wall crossing points | - | [46,52] |
| BAS3 | MGG_11610 | 113 | -/In planta /BIC; Cell wall crossing points | - | [46,52] |
| BAS4 | MGG_10914 | 102 | -/In planta/EIHM | - | [46,52] |
| BAS 107 | MGG_10020 | 132 | -/In planta /BIC; Cytoplasm | - | [89] |
| Group of predicted effector proteins | |||||
| IUGs | Isolate unique genes; Suppression of SA and ET signaling/In planta/BIC; Cytoplasm | [93] | |||
| MoHEGs | M. oryzae hypothetical effector genes; suppress plant defenses; virulence function in Barley plants/In planta/Apressorium | [94] | |||
| MoCDIPs | Induced plant cell death in rice protoplasts/In planta/Apoplast | [57] | |||
| SDPs | Suppression of plant host defenses/In planta/Apoplast and cytoplasm | [58] | |||
Indicates the number of amino acids residues of the predicted protein.
Inside the host plant cell upon rice blast infection.
Cognate resistance protein in rice plants.
In planta: expression during biotrophic growth; - unknown function or host interactor.
All localization matter: cytoplasmic versus apoplastic effectors
Live-cell imaging of fluorescently labeled effector proteins has proven to be an outstanding tool for monitoring secreted proteins in living rice cells during early M. oryzae infections with minimal perturbation of their normal function [47, 51]. Indeed, fluorescently labeled blast effectors reveal two groups of effectors based on their localization in planta. Some effectors such as the known avirulent (Avr) effectors Avr-Pita, Avr-Pizt, Pwl1, Pwl2 as well as the secreted protein MC69 and the biotrophic-associated secreted (BAS) protein Bas1, are first accumulated in the BIC structure at the primary IH before their translocation into the rice cytoplasm (Figure 3) [52–54]. The native promoter region of these cytoplasmic effectors is responsible for BIC localization, but this activity is not linked to any obvious sequence motifs. Interestingly, once cytoplasmic effectors are translocated into the rice cells, some effectors accumulate around the host cell wall crossing points or they move into uninfected neighbor cells, presumably preparing the adjacent uninvaded host cell for further infection (Figure 3A, B). Fluorescence recovery after photobleaching has revealed that fluorescent cytoplasmic effectors such as Pwl2 and Bas1 are continuously secreted into the BIC while the bulbous IH grows elsewhere in the rice cell [52]. Intriguingly, BIC structures appear in every newly infected cell and accumulate cytoplasmic effectors. How this accumulation at the BIC mediates the delivery of M. oryzae effectors into the rice cytoplasm remains to be elucidated.
In contrast to cytoplasmic effectors, apoplastic effectors are not associated with the BIC structure. The apoplastic effectors such as the secreted LysM protein 1 (Slp1), Bas4 and Bas113 accumulate at the enclosed apoplastic matrix between the fungal cell wall and EIHM outlining the entire IH (Figure 3) [47, 51, 55]. For instance, Slp1 directly binds to chitin oligosaccharides and competes with the plant receptor CEBiP to evade host recognition and subsequent chitin-triggered immune responses [55]. To achieve this, an α-1,3-mannosyltransferase, Alg3 mediates the N-glycosylation of the Slp1 effector, and this posttranslational modification is required to prevent host innate immunity [56]. In addition, the apoplastic effector Bas4 was observed to uniformly outline the bulbous IH, and also N-glycosylated by Alg3 [52, 56].
Using a transient expression assay, five apoplastic effectors in M. oryzae, named as MoCDIP1–5, were shown to induce plant cell death in rice protoplasts [57]. Applying similar transient expression approaches in Nicotiana benthamiana leaves, a recent study identified 11 suppressors of plant cell death (SPD) effectors from M. oryzae that were able to suppress plant cell death induced by necrosis-inducing protein 1 (Nep1) or the apoptosis regulator BAX protein [58]. The biological function of a M. oryzae specific gene, Required-for-Focal-BIC-Formation 1 (RBF1) was identified as a virulence gene essential for focal BIC formation during biotrophic growth [59]. Similar to cytoplasmic effectors, RBF1 accumulates in the BIC structure and translocates into rice cytoplasm. However, loss of RBF1 function resulted in dispersed BIC formation and low effector translocation efficiency [59]. In addition, the lack of RBF1 caused not only a pathogenicity defect but also the inability to suppress host immune responses [59]. Notably, the focal BIC structure plays a critical role in the establishment of biotrophic growth to evade or suppress host defenses responses by secreting effectors into plant cell. However, understanding the molecular mechanism by which M. oryzae Rbf1 mediates the focal BIC formation will bring new clues on the mode of action of BIC and host immune suppression.
Secretion mechanisms during biotrophic stage
To understand effector secretion, a recent study demonstrated that M. oryzae has two distinct secretory pathways for effector proteins. In the first, cytoplasmic effectors preferentially accumulate at the BIC and are subsequently delivered into the plant cytoplasm (Figure 3) [51]. Targeted deletion of two genes in the exocyst complex, SEC5 and EXO70, in M. oryzae revealed significant loss of pathogenicity and inefficient secretion of these cytoplasmic effectors. In addition, a t-SNARE called Sso1 in M. oryzae is required for BIC development and pathogenesis [51]. This study provides evidence that cytoplasmic effectors require an unconventional pathway in the BIC for secretion that involve the exocyst complex and t-SNARE components. Moreover, live-cell imaging of cytoplasmic effectors fused to fluorescent proteins demonstrated that some effectors are not only translocated into the infected cell but also into neighboring uninfected cells [52].
Perturbations in the aforementioned pathway had no effect on the secretion of apoplastic effectors, indicating that these effectors use a second secretion pathway for delivery into the apoplastic matrix during biotrophic invasion. Indeed, treatments with brefeldin A, an inhibitor of conventional endoplasmic reticulum (ER)-Golgi secretion pathway, blocked apoplastic effector secretion into the apoplastic space without having an impact on BIC-mediated accumulation of cytoplasmic effectors [51]. These findings provide evidence that the fungus uses distinct pathways to secrete effector proteins during early stages of rice cell colonization.
Hormonal mimicking to modulate plant metabolism
Plant pathogens employ sophisticated strategies to disrupt hormonal homeostasis in order to create a suitable niche and facilitate nutrient uptake and colonization in plant tissues [60]. To achieve this many pathogens either target components of the phytohormone metabolism or synthesize compounds that can act as phytohormone mimics during infection to modulate plant physiology. For instance, M. oryzae secretes a monooxygenase, Abm, which hydroxylases endogenous free jasmonic acid (JA) into 12OH-JA to disables JA-based host innate immunity to facilitate pathogenesis [61, 62]. Moreover, the fungus also secretes the metabolite 12OH-JA during host penetration to subvert the plant immune responses [62]. In fact, it has been shown that loss of ABM function in the fungus induces host immune responses and inhibits plant colonization [62]. Fungi also produce hormonal compounds similar to plant hormones in order to hijack plant development and nutrient allocation process to promote sustained colonization and dissemination [60]. For example, a secreted cytokinin synthase 1, Cks1, has been shown to be essential for cytokinin (CK) biosynthesis and full virulence in M. oryzae [61]. Fungus-derived CKs likely contribute to nutrient fluxes and rice defense inhibition. The cks1 mutants induce an early and strong transcriptional response of rice defense-related genes during infection [61]. Moreover, M. oryzae Cks1 was clearly implicated with nutrient fluxes due to the inability of the cks1 mutants to maintain key nutrient levels at the infection site [61]. Remarkably, in the presence of cks1 mutant, the relative expression of rice CK-responsive genes were altered in planta, suggesting that M. oryzae CKs are likely perceived by the plant system during infection [61].
Concluding Remarks and Future Perspectives
Undoubtedly, rice blast disease remains a threat to global rice production and food security. With its remarkable biology, M. oryzae actively secretes a large repertoire of small-secreted proteins to suppress or evade host pathogen surveillance. Upon spore-leaf attachment, the fungus undergoes distinct morphological changes to develop an appressorium, generate IH and subsequently move the IH to adjacent cells. These changes are intrinsic to rice blast disease progression, but how the fungus maintains a biotrophic association and switches to a necrotrophic behavior remains unclear. Many questions remain open regarding M. oryzae biotrophic biology (see Outstanding Questions). Significant advances have been made in recent years to understand effector secretion during biotrophic interaction, revealing how M. oryzae deploys repertoires of effectors to suppress plant immunity and allow the fungus to propagate inside the host cell. Despite the outstanding discoveries on M. oryzae effector biology, our understanding on how these effectors are translocated into host cell is unknown. Elucidating the precise composition of apoplastic matrix between the IH and the plant membrane will improve our knowledge of the effector translocation mechanisms. Furthermore, the mechanism underlying effector-host defense suppression is not yet understood, and with this, the question of how M. oryzae biotrophic effectors suppress plant immune defenses to promote intracellular colonization is of particular interest. The use of biochemical approaches to detect protein-protein associations and protein complexes in living cells may facilitate the identification of host targets. Answers to these outstanding questions will provide us with clues to the general understanding of how pathogenic fungi successfully colonize plant tissues and subvert plant immunity.
Outstanding questions.
What other processes are involved in conidia cell death and degradation during appressorial development?
Once inside the plant cell, how does M. oryzae maintain host cell membrane integrity during early biotrophic infection stage?
How do M. oryzae effectors translocate from the IH into the host cytoplasm?
Does M. oryzae deliver the effector proteins by extracellular vesicles such as exosomes into apoplastic interface? How is this accomplished? How is effector secretion regulated?
How does the fungus manipulate the plant plasmodesmata channels to move from cell-to-cell?
How are cytoplasmic effectors delivered into the BIC structures? Is the translocation mechanism BIC-effector accumulation dependent?
How do M. oryzae effectors suppress basal plant defenses during biotrophic growth?
What are the main signaling pathways that effectors proteins target, and how does this contribute to the infection process?
What are the intracellular cues that promote the fungus to switch from biotrophic growth to necrotrophic behavior
Highlights.
The internalization and transport of the G-protein signaling components to late endosomal compartments plays a critical role during appressoria development in rice blast disease.
TOR pathway activation by intracellular glutamine levels negatively regulate appressorium development.
Rice blast fungus has evolved two secretion mechanisms to deliver effector proteins into the host cell during biotrophic growth.
M. oryzae disrupts hormonal homeostasis to evade or suppress host defenses during plant cell invasion.
Acknowledgments
This work was supported by the Welch Foundation grant I-1561 and Once Upon a Time…Foundation. JF was funded by the Microbiology Training (grant T32 AI007520-17) and the Once Upon a Time… Foundation. K.O. is a Burroughs Welcome Investigator in Pathogenesis of Infectious Disease, a Beckman Young Investigator, and a W. W. Caruth, Jr., Biomedical Scholar and has an Earl A. Forsythe Chair in Biomedical Science. We thank the members of the Orth lab for thoughtful discussion and revision of this manuscript. We are very thankful to Dor Salomon and Richard A. Wilson for their critical review of our manuscript.
Glossary
- Apoplastic (also known as apoplast)
Extracellular space outside of the plant cell membrane. It facilitates transport of water and solutes across the cell wall to different plant tissues.
- Appressorium
A specialized infection structure formed at the tip of the germ tube that many plant pathogenic fungi use to penetrate the plant cuticle and colonize the host cell.
- Avirulence
Effector proteins that are recognized by host cognate resistance (R) proteins
- Biotrophic Interface Complex (BIC)
A membrane rich complex in which the fungus accumulates a specific group of secreted proteins.
- Biotrophic
Refer to a fungal pathogen that has a close relationship with the host and requires living plant tissue to survive. Biotrophs suppress host immune systems and derive nutrients from living host cells.
- Extrainvasive hyphal membrane (EIHM)
A plant membrane that surrounds the fungal cell wall.
- Hemibiotroph
A pathogenic organism that initially establishes a close biotrophic association with the host without causing any symptoms that later turns into necrotrophic association, resulting in death of the plant cells.
- Invasive hyphae (IH)
Fungal cells that develops during host infection, and grows and proliferates inside the plant cell.
- Necrotrophic
Refer to a plant pathogen that colonizes host tissues by secreting toxins and enzymes that kill the living plant cells, followed by acquisition of nutrients from the dead and dying host tissues.
- Pathotypes
A fungal pathogen that is distinguished from others of the same species by its specificity for a specific host.
- Penetration Peg
A specialized fungal hyphae that the fungus uses to pierce the host plant surface. It is formed at the base of the mature appressorium.
- Plasmodesmata
Small channels that connect the plant cytoplasm of neighboring cells. These channels facilitate the movement of molecules and communication between the cells
- Target of Rapamycin (TOR)
Signaling pathway responsible for cell metabolism, growth, proliferation and survival under favorable conditions.
- Turgor pressure
A hydrostatic force generated by the accumulation of solutes in a cell.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elert E. Rice by the numbers: A good grain. Nature. 2014;514:S50–S51. doi: 10.1038/514s50a. [DOI] [PubMed] [Google Scholar]
- 2.Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends in Biotechnology. 2009;27:141–150. doi: 10.1016/j.tibtech.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi E. Armed and dangerous. Science. 2010;327:804–5. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 4.Yan X, Talbot NJ. Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae. Current Opinion in Microbiology. 2016;34:147–153. doi: 10.1016/j.mib.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Hamer JE, et al. A Mechanism for surface attachment in spores of a plant pathogenic fungus. Science. 1988;239:288. doi: 10.1126/science.239.4837.288. [DOI] [PubMed] [Google Scholar]
- 6.Jelitto TC, et al. Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus, Magnaporthe grisea. Planta. 1994;194:471–477. [Google Scholar]
- 7.Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Micro. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 8.Ryder LS, Talbot NJ. Regulation of appressorium development in pathogenic fungi. Current Opinion in Plant Biology. 2015;26:8–13. doi: 10.1016/j.pbi.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumley FG, Valent B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Molecular Plant Microbe Interactions. 1990;3(3):135–143. [Google Scholar]
- 10.de Jong JC, et al. Glycerol generates turgor in rice blast. Nature. 1997;389:244–244. [Google Scholar]
- 11.Egan MJ, et al. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proceedings of the National Academy of Sciences. 2007;104:11772–11777. doi: 10.1073/pnas.0700574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagdas YF, et al. Septin-Mediated Plant Cell Invasion by the Rice Blast Fungus, Magnaporthe oryzae. Science. 2012;336:1590–1595. doi: 10.1126/science.1222934. [DOI] [PubMed] [Google Scholar]
- 13.Ryder LS, et al. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3179–3184. doi: 10.1073/pnas.1217470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, et al. Genetic control of infection-related development in Magnaporthe oryzae. Current Opinion in Microbiology. 2012;15:678–684. doi: 10.1016/j.mib.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes & Development. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 16.Choi W, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. The Plant Cell. 1997;9:1973. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi K, Hamer JE. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. The Plant Cell. 1998;10:1361–1374. doi: 10.1105/tpc.10.8.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, et al. Rgs1 regulates multiple Gα subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. The EMBO Journal. 2007;26:690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. PKA activity is essential for relieving the suppression of hyphal growth and appressorium formation by MoSfl1 in Magnaporthe oryzae. PLOS Genetics. 2017;13:e1006954. doi: 10.1371/journal.pgen.1006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeZwaan TM, et al. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. The Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni RD, et al. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biology. 2005;6:R24–R24. doi: 10.1186/gb-2005-6-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanujam R, et al. The late endosomal HOPS complex anchors active G-protein signaling essential for pathogenesis in Magnaporthe oryzae. PLOS Pathogens. 2013;9:e1003527. doi: 10.1371/journal.ppat.1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kou Y, et al. Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytologist. 2017;214:330–342. doi: 10.1111/nph.14347. [DOI] [PubMed] [Google Scholar]
- 24.Li X, et al. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLOS Pathogens. 2017;13:e1006449. doi: 10.1371/journal.ppat.1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J-R, et al. The CPKA Gene of Magnaporthe grisea is essential for appressorial penetration. Molecular Plant-Microbe Interactions. 1997;10:187–194. [Google Scholar]
- 26.Chen RE, Thorner J. Function and regulation in mapk signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochimica et biophysica acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, et al. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. The Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park G, et al. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. The Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, et al. Bypassing both surface attachment and surface recognition requirements for appressorium formation by overactive ras signaling in Magnaporthe oryzae. Molecular Plant-Microbe Interactions. 2014;27:996–1004. doi: 10.1094/MPMI-02-14-0052-R. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, et al. Activation of the signalling mucin MoMsb2 and its functional relationship with Cbp1 in Magnaporthe oryzae. Environmental Microbiology. 2015;17:2969–2981. doi: 10.1111/1462-2920.12847. [DOI] [PubMed] [Google Scholar]
- 31.Gong X, et al. pFPL vectors for high-throughput protein localization in fungi: Detecting cytoplasmic accumulation of putative effector proteins. Mol Plant Microbe Interact. 2015;28:107–121. doi: 10.1094/MPMI-05-14-0144-TA. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, et al. Thioredoxins are involved in the activation of the PMK1 MAP kinase pathway during appressorium penetration and invasive growth in Magnaporthe oryzae. Environmental Microbiology. 2016;18:3768–3784. doi: 10.1111/1462-2920.13315. [DOI] [PubMed] [Google Scholar]
- 33.Marroquin-Guzman M, Wilson RA. GATA-dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR Inhibition of cAMP/PKA signaling. PLoS Pathog. 2015;11:e1004851. doi: 10.1371/journal.ppat.1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veneault-Fourrey C, et al. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 35.Saunders DGO, et al. Cell cycle–mediated regulation of plant infection by the rice blast fungus. The Plant Cell. 2010a;22:497. doi: 10.1105/tpc.109.072447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders DGO, et al. Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. The Plant Cell. 2010b;22:2417. doi: 10.1105/tpc.110.074492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osés-Ruiz M, et al. Two independent S-phase checkpoints regulate appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Proceedings of the National Academy of Sciences. 2017;114:E237–E244. doi: 10.1073/pnas.1611307114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marroquin-Guzman M, et al. Glucose-ABL1-TOR signaling modulates cell cycle tuning to control terminal appressorial cell differentiation. PLoS Genet. 2017;13:e1006557. doi: 10.1371/journal.pgen.1006557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He M, et al. Infection-associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non-selective macro-autophagy. PLOS ONE. 2012;7:e33270. doi: 10.1371/journal.pone.0033270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galhano R, et al. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLOS Pathogens. 2017;13:e1006516. doi: 10.1371/journal.ppat.1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kankanala P, et al. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. The Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkinson CB, et al. The appressorium of the rice blast fungus Magnaporthe oryzae remains mitotically active during post-penetration hyphal growth. Fungal Genet Biol. 2017;98:35–38. doi: 10.1016/j.fgb.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Shipman EN, et al. Nuclear and structural dynamics during the establishment of a specialized effector-secreting cell by Magnaporthe oryzae in living rice cells. BMC Cell Biol. 2017;18:11. doi: 10.1186/s12860-017-0126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones K, et al. Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biology. 2016;16:69. doi: 10.1186/s12870-016-0756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochizuki S, et al. Live-cell imaging of rice cytological changes reveals the importance of host vacuole maintenance for biotrophic invasion by blast fungus, Magnaporthe oryzae. MicrobiologyOpen. 2015;4:952–966. doi: 10.1002/mbo3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosquera G, et al. Interaction transcriptome analysis identifies Magnaporthe oryzae Bas1-4 as biotrophy-associated secreted proteins in rice blast disease. The Plant Cell. 2009;21:1273. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khang CH, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22:1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi M, Valent B. Communication between filamentous pathogens and plants at the biotrophic interface. Annual Review of Phytopathology. 2013;51:587–611. doi: 10.1146/annurev-phyto-081211-172916. [DOI] [PubMed] [Google Scholar]
- 49.Rafiqi M, et al. Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Current Opinion in Plant Biology. 2012;15:477–482. doi: 10.1016/j.pbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 50.de Guillen K, et al. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLOS Pathogens. 2015;11:e1005228. doi: 10.1371/journal.ppat.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giraldo MC, et al. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun. 2013;4:1996. doi: 10.1038/ncomms2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khang CH, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. The Plant Cell. 2010;22:1388. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park C-H, et al. The Magnaporthe oryzae effector AvrPiz-t Targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. The Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saitoh H, et al. Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLOS Pathogens. 2012;8:e1002711. doi: 10.1371/journal.ppat.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mentlak TA, et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. The Plant Cell. 2012;24:322–335. doi: 10.1105/tpc.111.092957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X-L, et al. N-Glycosylation of effector proteins by an α-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. The Plant Cell. 2014;26:1360–1376. doi: 10.1105/tpc.114.123588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, et al. Identification and characterization of in planta–expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Molecular Plant-Microbe Interactions. 2012;26:191–202. doi: 10.1094/MPMI-05-12-0117-R. [DOI] [PubMed] [Google Scholar]
- 58.Sharpee W, et al. Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Molecular Plant Pathology. 2017;18:850–863. doi: 10.1111/mpp.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimura T, et al. Magnaporthe oryzae Glycine-Rich Secretion Protein, Rbf1 Critically Participates in Pathogenicity through the Focal Formation of the Biotrophic Interfacial Complex. PLOS Pathogens. 2016;12:e1005921. doi: 10.1371/journal.ppat.1005921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma K-W, Ma W. Phytohormone pathways as targets of pathogens to facilitate infection. Plant Molecular Biology. 2016;91:713–725. doi: 10.1007/s11103-016-0452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chanclud E, et al. Cytokinin Production by the Rice Blast Fungus Is a Pivotal Requirement for Full Virulence. PLOS Pathogens. 2016;12:e1005457. doi: 10.1371/journal.ppat.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patkar RN, et al. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nature Chemical Biology. 2015;11:733. doi: 10.1038/nchembio.1885. [DOI] [PubMed] [Google Scholar]
- 63.Igarashi S, et al. Pyriculariaemtrigo. 1. Ocorrência de Pyricularia sp. no estado do Paraná. Fitopatol Bras. 1986;11 [Google Scholar]
- 64.Cruz CD, Valent B. Wheat blast disease: danger on the move. Tropical Plant Pathology. 2017;42:210–222. [Google Scholar]
- 65.Callaway E. Devastating wheat fungus appears in Asia for first time. Nature. 2016;532:421–422. doi: 10.1038/532421a. [DOI] [PubMed] [Google Scholar]
- 66.Kohli MM, et al. Pyricularia blast — a threat to wheat cultivation. Czech J Genet Plant Breed. 2011;47 [Google Scholar]
- 67.Goulart ACP, et al. Damages in wheat caused by infection of Pyricularia grisea. Summa Phytopathol. 2007;33 [Google Scholar]
- 68.Islam MT, et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biology. 2016;14:84. doi: 10.1186/s12915-016-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malaker PK, et al. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Disease. 2016;100:2330–2330. [Google Scholar]
- 70.Inoue Y, et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science. 2017;357:80. doi: 10.1126/science.aam9654. [DOI] [PubMed] [Google Scholar]
- 71.Wilson RA, et al. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. The EMBO Journal. 2007;26:3673. doi: 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson RA, et al. An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proceedings of the National Academy of Sciences. 2010;107:21902–21907. doi: 10.1073/pnas.1006839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandez J, et al. Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1-3, and a MATE–family pump regulate glucose metabolism during infection. PLOS Genetics. 2012;8:e1002673. doi: 10.1371/journal.pgen.1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernandez J, Wilson RA. The sugar sensor, trehalose-6-phosphate synthase (Tps1), regulates primary and secondary metabolism during infection by the rice blast fungus: Will Magnaporthe oryzae's “sweet tooth” become its “Achilles’ heel”? Mycology. 2011;2:46–53. [Google Scholar]
- 75.Foster AJ, et al. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. The EMBO Journal. 2003;22:225. doi: 10.1093/emboj/cdg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fernandez J, Wilson RA. Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLOS ONE. 2014;9:e87300. doi: 10.1371/journal.pone.0087300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandez J, et al. Evidence for a transketolase-mediated metabolic checkpoint governing biotrophic growth in rice cells by the blast fungus Magnaporthe oryzae. PLOS Pathogens. 2014;10:e1004354. doi: 10.1371/journal.ppat.1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marroquin-Guzman M, et al. The Magnaporthe oryzae nitrooxidative stress response suppresses rice innate immunity during blast disease. Nat. Microbiology. 2017;2:17054. doi: 10.1038/nmicrobiol.2017.54. [DOI] [PubMed] [Google Scholar]
- 79.Orbach MJ, et al. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. The Plant Cell. 2000;12:2019. doi: 10.1105/tpc.12.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chuma I, et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLOS Pathogens. 2011;7:e1002147. doi: 10.1371/journal.ppat.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jia Y, et al. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. The EMBO Journal. 2000;19:4004. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ribot C, et al. The Magnaporthe oryzae effector AVR1–CO39 is translocated into rice cells independently of a fungal-derived machinery. The Plant Journal. 2013;74:1–12. doi: 10.1111/tpj.12099. [DOI] [PubMed] [Google Scholar]
- 83.Cesari S, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. The Plant Cell. 2013;25:1463. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshida K, et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. The Plant Cell. 2009;21:1573. doi: 10.1105/tpc.109.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang S, et al. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Scientific Reports. 2015;5:11642. doi: 10.1038/srep11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu J, et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytologist. 2015;206:1463–1475. doi: 10.1111/nph.13310. [DOI] [PubMed] [Google Scholar]
- 87.Kang S, et al. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 1995;8:939–948. doi: 10.1094/mpmi-8-0939. [DOI] [PubMed] [Google Scholar]
- 88.Sweigard JA, et al. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 1995;7:1221–1233. doi: 10.1105/tpc.7.8.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giraldo MC, Valent B. Filamentous plant pathogen effectors in action. Nat Rev Micro. 2013;11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 90.Böhnert HU, et al. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. The Plant Cell. 2004;16:2499. doi: 10.1105/tpc.104.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ray S, et al. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Frontiers in Plant Science. 2016;7:1140. doi: 10.3389/fpls.2016.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, et al. Magnaporthe oryzae-secreted protein MSP1 induces cell death and elicits defense responses in rice. Molecular Plant-Microbe Interactions. 2016;29:299–312. doi: 10.1094/MPMI-12-15-0266-R. [DOI] [PubMed] [Google Scholar]
- 93.Dong Y, et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLOS Pathogens. 2015;11:e1004801. doi: 10.1371/journal.ppat.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mogga V, et al. Magnaporthe oryzae effectors MoHEG13 and MoHEG16 interfere with host infection and MoHEG13 counteracts cell death caused by Magnaporthe-NLPs in tobacco. Plant Cell Reports. 2016;35:1169–1185. doi: 10.1007/s00299-016-1943-9. [DOI] [PubMed] [Google Scholar]