Abstract
Research has largely reported that dog exposure is associated with reduced allergic disease risk. Responsible mechanism(s) are not understood. The goal was to investigate whether introducing a dog into the home changes the home’s dust microbiota. Families without dogs or cats planning to adopt a dog and those who were not were recruited. Dust samples were collected from the homes at recruitment and 12 months later. Microbiota composition and taxa (V4 region of the 16S rRNA gene) were compared between homes that did and did not adopt a dog. A total of 91 dust samples from 54 families (27 each, dog and no dog; 17 dog and 20 no dog homes with paired samples) were analyzed. A significant dog effect was seen across time in both unweighted UniFrac and Canberra metrics (both p=0.008), indicating dog introduction may result in rapid establishment of rarer and phylogenetically related taxa. A significant dog-time interaction was seen in both weighted UniFrac (p<0.001) and Bray-Curtis (p=0.002) metrics, suggesting that while there may not initially be large relative abundance shifts following dog introduction, differences can be seen within a year. Therefore, dog introduction into the home has both immediate effects and effects that emerge over time.
Keywords: microbiome, dog, allergy, asthma, microbiota hypothesis, hygiene hypothesis
BACKGROUND
Similar to how growing up on a livestock farm has been shown to be protective against the development of allergies and asthma in European children, living with a pet in early life has been investigated for its relationship with the subsequent development of allergies and asthma in numerous studies in the United States.1–9 Recent meta-analyses and additional studies have generally reported that pet exposure, particularly to dogs, decreases the risk of allergic diseases9–11, though some have reported an increased risk.12 With the reasonably consistent findings across many cohorts and populations, animal exposure—specifically to dogs—remains a promising avenue for identifying a prevention strategy for childhood allergic diseases.
The responsible mechanism(s), either wholly or in part, for the effects of dogs on allergic disease development, are not completely understood. Recent hypotheses posit that dogs change the environmental microbiome, which in turns alters the gut microbiome and subsequent immune system development of young children residing in homes with or in proximity to dogs. We and others have found that house dust microbiota communities vary with the presence of dogs and cats in the home (versus homes without dogs and cats).13–15 As this prior work was cross-sectional, there has been little evidence examining whether a dog changes the home’s dust microbiome. Additionally, dust sample collection in previous studies has typically not been targeted to coincide with or carefully measure timing of dog introduction. The goal of this work was to test the hypothesis that introducing a dog into the home has the capacity to change that home’s dust microbiota – both in terms of its overall composition and its relative abundance of specific taxa.
METHODS
Study Design
Participants were recruited from the metro-Detroit area. Eligible households had to have lived in their current home for at least one year, planned to live there for the next year and have not kept dogs or cats in their home for one week or more in the previous year. Keeping fish, birds, and other small caged/contained pets like snakes, turtles, hamsters, or insects was not an exclusion criterion. Other exclusions included smokers and household residents who work with, handle, or interact with pets or animals on a daily, weekly, or monthly basis. We recruited 2 groups of households: those that were adopting a new dog (“dog homes”), and those who were not planning to add a dog or other furred pet to their household over the subsequent year (“dog-free homes”). New dog owners were to have their newly added dog inside of their home for at least 12 hours per day with the dog permitted to roam freely about the home, and to not own another dog or cat for the duration of the study (12 months). Once a dog household was recruited, a dog-free household was recruited within 1 month. Participants were recruited from September 21, 2013 – July 09, 2014. This work was approved by the Henry Ford Health System IRB and written informed consent was provided by the study participants.
While participants were recruited through targeted radio broadcasts and Henry Ford Health System’s electronic communications and website, most “dog” households were recruited through the Michigan Humane Society’s local adoption center and their semi-annual adoption events at the local zoo. Trained interviewing staff identified participants that were eligible. Participants completed a baseline visit within several days of the dog’s introduction into their new home. Interviews about household characteristics, residents, and dog activities were conducted at recruitment, and six and twelve months later.
Dust Sample Collection
Dust samples were collected from participating dog homes by study staff within a few days of the dog’s introduction into their new home and again at 12 months after the introduction of the new dog. Dust samples were collected from dog-free homes at the time of recruitment and 12 months later. For families with a dog, the main floor area where the family and dog would spend time was vacuumed. For families without a pet, the main floor area where the family reported spending their time was vacuumed. Two areas of the same room were vacuumed. The first vacuuming was in a prefabricated rectangle measuring 1 meter by 2 meters for 2 minutes. Vacuum sample collection “socks” were then switched. The field staff then vacuumed all of the areas outside of that rectangle in the same room, including areas around baseboards, in corners and around and under furniture for 5 minutes. Participants were asked to maintain their typical cleaning practices prior to sampling.
Dust samples were stored at −80°C until initial processing. Dust socks were cut open in a sterile petri dish using sterile ethanol-flamed scissors and forceps. Dust was transferred from the sock into Lysing Matrix E tubes (MP Biomedicals, Santa Ana, CA) using a sterile single-use spatula. These tubes were again stored at −80°C until shipment on dry ice to our collaborating lab at the University of California – San Francisco (UCSF) for sequencing.
Microbiota Sequencing
The analyses of dust samples collected over 5 minutes are presented here. These sample volumes were generally larger than those collected from the premeasured rectangles and therefore provided a higher likelihood of having sufficient sample volume for microbial DNA sequencing. The methods for sequencing the microbial DNA are provided in the Supplemental Text.
Statistical Analysis
Differences in household characteristics between dog and dog-free homes were calculated using ANOVA for numerical covariates and the chi-square test for categorical covariates. Alpha diversity metrics of bacterial richness (number of unique OTUs present), Pielou’s evenness (relative distribution of OTUs in a community), Faith’s phylogenetic diversity, and Shannon’s (non-phylogenetic) diversity were estimated using QIIME16 and R vegan17. Differences in these metrics at each time point by group (dog versus dog-free homes) were calculated using ANOVA, while longitudinal effects were assessed using mixed effect models. Between-subject similarity (i.e., beta diversity) was defined using both weighted and unweighted UniFrac18, Canberra, and Bray-Curtis distance matrices. UniFrac matrices weight bacterial phylogenetic relationships in the calculation of between-sample distances, while Canberra and Bray-Curtis matrices are the non-phylogenetic correlates to the unweighted and weighted UniFrac matrices, respectively. Relationships between dust microbiota composition and variables measured cross-sectionally and for within-subject effects with repeated measures data (time effects, group*time interaction effects) were assessed using PERMANOVA19 as implemented in the R package vegan. Compositional tests for between-subject effects with repeated measures data (group effects) was performed using nested PERMANOVA in the BiodiversityR package20. Visual representations of compositional differences were plotted using non-metric multidimensional scaling (nMDS) in the R vegan package. Differences in OTU relative abundance (dog versus dog-free homes) were tested using a two-part zero-inflated beta regression model with the R package ZIBR, 21 which tests for differences in presence/absence, as well as relative abundance. To avoid testing overly sparse OTUs, only OTUs observed in 10% or more of homes were tested. OTUs were considered significant if the false discovery rate adjusted p-value for the joint test (i.e., test of both presence/absence and relative abundance) was less than 0.05.22
RESULTS
Sample Size and Description
At least one sample was included in the analysis from the homes of all 54 families (27 each, dog and no dog). These homes yielded 100 total 5-minute dust samples, with 99 successfully amplified for microbial sequencing. One of the 12-month samples was from a family that moved between baseline and 12-months—a protocol deviation—and was therefore excluded. Of the remaining 98 samples that met inclusion criteria, 7 samples were excluded due to unusually low sequencing depth (six with <11 sequence reads; one with <5,000 sequence reads). The OTU table was then rarefied to the minimum depth among the remaining 91 (minimum depth: 34,714 sequence reads).
Among dog homes, 17 (63%) had both baseline and 12-month dust samples, 9 (33%) only had a baseline dust sample included, and 1 (4%) family had only a 12-month dust sample included. Among dog-free homes, 20 (74%) homes had results from samples from both time points, 6 (22%) had dust sample results from baseline only, and 1 (4%) had only a 12-month sample included. Of the 26 dog home baseline dust samples included, 7 were collected on the same day or prior to the dog arriving (“early dog homes”, average number of days from collection of home dust until dog’s arrival into the home=2.1, median=0, range=0 to 9), 18 were collected a day or more after dog arrival (“late dog homes”, average number of days dog was in the home prior to dust collection=3.5, median=2.0, range 1 to 29), and one sample was missing information on timing of dog introduction.
Comparing Households
The dog and dog-free homes were very similar with respect to the household compositions and characteristics, including, but not limited to: number of residents in the home, the number of residents in the home under the age of 18 years, whether the home was a single-family dwelling, and the home’s square footage and room count (all p>0.05; Supplemental Table 1). Four homes without dogs did not have forced air heat while all other homes did (p=0.038; Supplemental Table 1). Homes without dogs also tended to have more children under the age of 5 years (p=0.002; Supplemental Table 1). Though there were differences in the month baseline dust was collected (p=0.003), this was strongly driven by May/June (large dog adoption event in May, followed by recruiting dog-free homes in June), which did not have an impact on seasonal differences between the two groups (p=1.00).
Alpha Diversity Measures
None of the alpha diversity metrics (richness, Pielou’s evenness, Faith’s phylogenetic diversity, and Shannon diversity) differed significantly between the dog homes and dog-free homes at baseline or at 12 months (Supplemental Figure 1). The results were unchanged after further separating the homes that adopted dogs by early and late dog introduction with respect to sample collection timing (Supplemental Table 2).
Dust microbiota compositional differences
Compositional differences between the homes with and without dogs were noted for the unweighted UniFrac and Canberra metrics at baseline and at 12-months, as well as only at 12-months for weighted UniFrac and Bray-Curtis (Table 1). At 12-months, having a dog in the home consistently explained a higher percentage of variation in bacterial dust composition (approximately 1.3-3.3 percent of variation explained), versus baseline. An nMDS plot of unweighted UniFrac distances visually confirmed the compositional distinction between dog and no dog homes (Figure 1).
Table 1.
Compositional differences between dog and dog-free homes, stratified by timing of sample collection.
| Metric | Baseline* | 12-Month† |
|---|---|---|
| p-value (R2) | p-value (R2) | |
| Unweighted UniFrac | 0.025 (0.028) | 0.021 (0.037) |
| Weighted UniFrac | 0.52 (0.017) | 0.049 (0.056) |
| Canberra | 0.043 (0.023) | 0.024 (0.031) |
| Bray-Curtis | 0.20 (0.023) | 0.015 (0.044) |
At baseline: N=26 dog-free samples; N=26 dog samples
At 12-months: N=21 dog-free samples; N=18 dog samples
Figure 1.
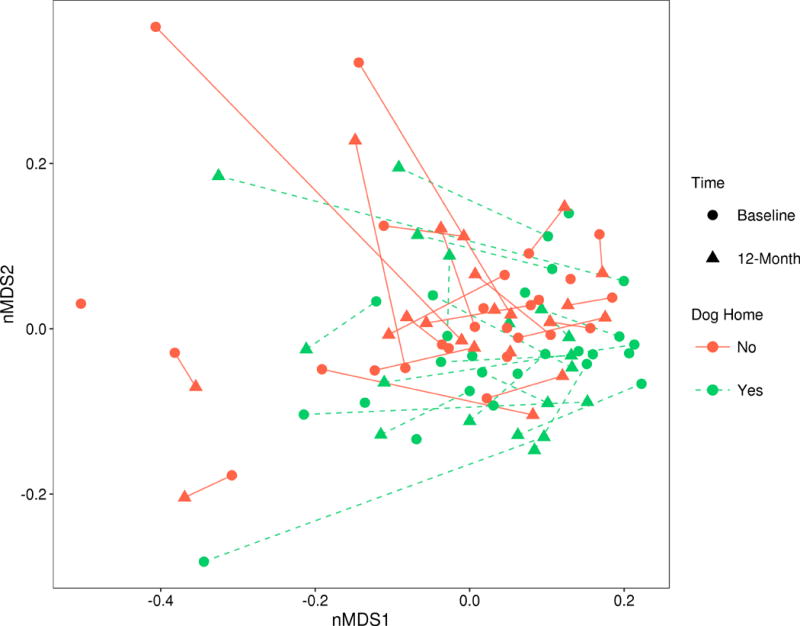
Non-metric multidimensional scaling (nMDS) plot of unweighted UniFrac distances, by dog introduction status and timing of sample collection. Samples from the same home are connected by a line (solid for dog-free homes and dashed for dog homes).
Whether these results were affected by the timing of the dust sample collection compared to the introduction of the dog into the home (early vs. late dog homes) was investigated using pairwise comparisons between the 3 groups (Table 2). When dog-free homes were compared to late dog homes at baseline, a significant difference was found using both unweighted metrics (unweighted UniFrac p=0.046, R2=0.031; Canberra p=0.056, R2=0.026). These effects persisted at 12-months (unweighted UniFrac p=0.025, R2=0.025; Canberra p=0.025, R2=0.033). Further, there were significant differences at 12-months in the weighted metrics for early dog vs. dog-free homes (weighted UniFrac p=0.004, R2=0.143; Bray-Curtis p=0.015, R2=0.072).
Table 2.
Compositional differences by dog introduction status and timing of sample collection.
| Metric | Overall* | Dog-free vs. Early Dog Home | Dog-free vs. Late Dog Home | Early Dog Home vs. Late Dog Home |
|---|---|---|---|---|
| p-value (R2) | p-value (R2) | p-value (R2) | p-value (R2) | |
| Baseline† | ||||
| Unweighted UniFrac | 0.12 (0.05) | 0.25 (0.03) | 0.05 (0.03) | 0.92 (0.04) |
| Weighted UniFrac | 0.69 (0.03) | 0.27 (0.04) | 0.79 (0.01) | 0.78 (0.02) |
| Canberra | 0.20 (0.04) | 0.39 (0.03) | 0.06 (0.03) | 0.92 (0.04) |
| Bray-Curtis | 0.30 (0.04) | 0.15 (0.04) | 0.33 (0.03) | 0.62 (0.04) |
| 12-Months‡ | ||||
| Unweighted UniFrac | 0.04 (0.06) | 0.14 (0.05) | 0.03 (0.04) | 0.30 (0.06) |
| Weighted UniFrac | 0.02 (0.11) | 0.004 (0.14) | 0.17 (0.04) | 0.14 (0.10) |
| Canberra | 0.02 (0.06) | 0.07 (0.05) | 0.03 (0.03) | 0.11 (0.06) |
| Bray-Curtis | 0.02 (0.08) | 0.02 (0.07) | 0.05 (0.04) | 0.17 (0.07) |
These p-values correspond to the overall test of the three group comparison (dog-free vs. early dog vs. late dog).
At baseline: N=26 dog-free samples; N=7 early dog samples; N=18 late dog samples (one baseline dog sample with unknown timing of dog introduction, excluded)
At 12-months: N=21 dog-free samples; N=3 early dog samples; N=15 late dog samples
These cross-sectional results were consistent with repeated measures analyses performed to test if the effect of dog varied over time (Table 3). For each metric, the dog-by-time multiplicative interaction term was not statistically significant. However, the main effect of dog was significant in 3 of the 4 metrics: unweighted UniFrac (p=0.008), Canberra (p=0.008), and Bray-Curtis (p=0.045), meaning dog and dog-free homes were compositionally distinct across the study period. From this, we hypothesized that the lack of a dog-by-time interaction may be due to the fact that many baseline dust samples from dog homes were collected shortly after the dog was introduced (late dog homes), which would result in baseline and 12-month dog dust samples being more similar to one another. Therefore, we subsequently recomputed this model after removing all baseline samples from late dog homes (Table 3). In this case, the interaction term was statistically significant for the weighted UniFrac (p<0.001) and Bray-Curtis metrics (p=0.002), while the main effect of dog remained statistically significant for unweighted UniFrac (p=0.047) and approached, but did not reach, statistical significance for Canberra dissimilarity (p=0.081).
Table 3.
Compositional differences by dog introduction status, across time using repeated measures analysis.*
| Metric | p-value for Dog Effect | p-value for Time Effect | p-value for Interaction Term between Dog and Time |
|---|---|---|---|
| Overall (N=91) | |||
| Unweighted UniFrac | 0.01 | 0.23 | 0.27 |
| Weighted UniFrac | 0.11 | 0.07 | 0.55 |
| Canberra | 0.01 | 0.15 | 0.32 |
| Bray-Curtis | 0.05 | 0.13 | 0.56 |
| After removing baseline dust samples collected in late dog homes from the analyses (N=73) | |||
| Unweighted UniFrac | 0.05 | 0.21 | 0.34 |
| Weighted UniFrac | 0.16 | 0.03 | <0.001 |
| Canberra | 0.08 | 0.24 | 0.39 |
| Bray-Curtis | 0.25 | 0.31 | 0.002 |
Nested PERMANOVA models were used for the repeated measures analyses.
Taxonomic Differences
Dust OTU differences between dog and dog-free homes at the 12-month time point were then tested. Figure 2 shows a total of 109 OTUs at 12-months that were associated with differences between dog and dog-free homes (joint FDR-adjusted p-value<0.05; N=48 where presence/absence differed significantly, N=51 where relative abundance differed significantly, and N=10 where both significantly differed; Supplemental Table 3). The majority of differential OTUs were more likely to be detectable in dog homes or have greater relative abundance in dog homes, compared to dog-free homes. Specifically, of the 58 OTUs where presence/absence significantly differed, 51 (88%) were more likely to be present in dog homes compared to dog-free homes. Similarly, of the 61 OTUs with significant differential relative abundance, 46 (75%) had higher relative abundance in dog homes compared to dog-free homes. OTUs that were more likely to be detected in dog homes at 12-months included specific members of the genera Moraxella, Porphyromonas, Capnocytophaga, Fusobacterium, Streptococcus, and Treponema. OTUs that had higher relative abundance in dog homes at 12-months included specific members of the genera Anaerococcus and Prevotella.
Figure 2.
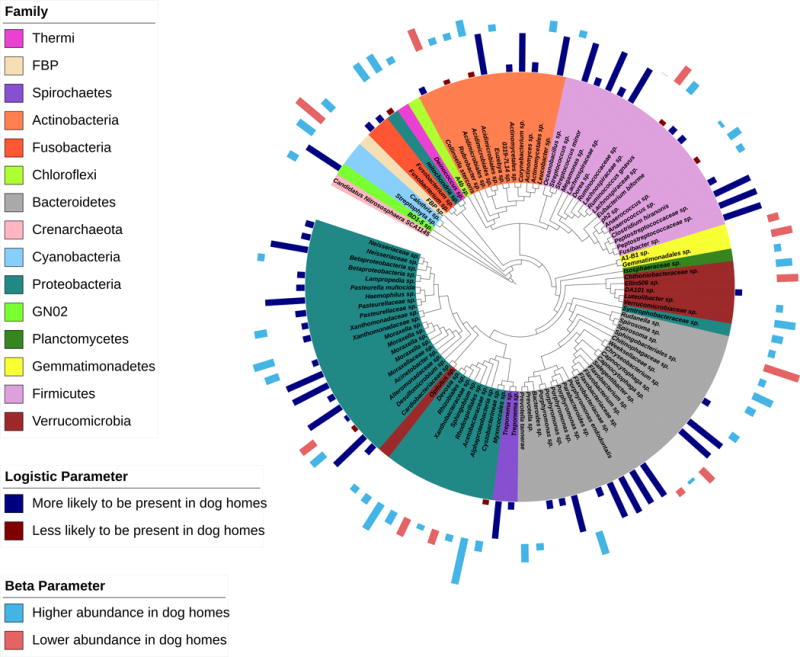
12-month OTUs associated with differences between dog and no dog homes, (joint FDR-adjusted p-value<0.05), identified by a two-part zero-inflated beta regression model.
DISCUSSION
These data suggest that introducing a dog into the home has both immediate effects on dust bacterial composition as well as effects that emerge over time. The effects seen in both unweighted UniFrac and Canberra—which do not take into account relative abundance, and therefore have increased power to detect shifts in rare lineages23—indicate that the introduction of a dog in the home may result in the rapid establishment of rarer and phylogenetically related taxa into the dust microbiota. The significant dog-by-time interaction seen in both the weighted UniFrac and Bray-Curtis metrics—which take relative abundance into account, and therefore have increased power to detect shifts in more common lineages—further suggests that while there may not initially be large relative abundance shifts following dog introduction, these differences may be seen with time. Given that this dog-by-time interaction is only significant in the early but not the late dog homes, this suggests that the time course for this relative abundance shift may begin within the first week following dog introduction, dampening the ability to detect any effect in the late dog homes.
The immediate observed changes in dust microbiota after dog introduction may have been expected given other studies showing rapid shifts in the built environment. For example, Lax et al. recently examined changes in microbial colonization patterns in a newly opened hospital, and found that several hospital surfaces began to resemble human skin microbiota immediately after opening.24 In a study of house dust embedded in carpet coupons, bacteria growth appeared after a week (with 100% equilibrium relative humidity levels).25 Similar quick shifts have also been observed in gut microbial communities in response to new dietary exposures 26–27, and in vaginal microbial communities in response to bacterial vaginosis antibiotic treatment.28
Our results appear to be consistent with what we and others have previously shown regarding the effect of dog ownership on house dust microbiota. A previous study from our group examined a small set of house dust samples drawn from a birth cohort and revealed that dust from dog-keeping homes had higher relative abundances of specific Treponema, Capnocytophagta, and Moraxella taxa compared to dust from no-pet homes13. Additionally, Barberán et al. recently demonstrated in a sample of approximately 1,200 homes across the US that house dust in homes with dogs had higher relative abundances of Porphyromonas and Moraxella compared to house dust in homes without dogs.15
While the current study indicates that dogs alter dust microbiota composition, understanding the specific mechanisms of this effect require further investigation. A limitation of this study is that we did not measure the microbiota of dogs themselves (e.g., oral, gut, skin) nor of their outdoor environments (e.g., soil microbiota). Therefore, it is unclear if the bacteria introduced into dog homes was sourced from the dogs directly or, alternatively, if they were outdoor environment-associated “bacterial hitchhikers” brought into the homes by dogs. However, many of the taxa enriched in dog homes have previously been identified as common members of the canine oral microbiota (Porphyromonas, Fusobacterium, Capnocytophaga, Moraxella) 29–30, as well as the canine gastrointestinal tract microbiota (Fusobacterium, Prevotella, Streptococcus)31–32. The microbiota of the skin of the dog has also been investigated in cross-sectional studies.33–35 While between dog variability of microbiota is high, with sampling site, sex of the dog, and skin health status contributing to the variability, all skin samples from a series of pure-breed dogs included the following families: Corynebateriaceae (Actinobacteria), Streptococcaceae and Lachnospiraceae (Firmicutes); Fusobacteriaceae (Fusobacteria); and Comamonadaceae, Oxalobacteraceae and Neisseriaceae (Proteobacteria).35 Another cross-sectional study of dogs of varying breeds reported Oxalobacteraceae and Proteobacteria were the most abundant phylum and family identified, respectively.33
Many of the key taxa found to be significantly different by dog (Moraxella, Porphyromonas, Capnocytophaga, Fusobacterium, Streptococcus, and Treponema) 36–39 have also emerged in recent studies of the microbiota and its associations with allergies and asthma.36, 38, 40–42 However, the evidence from these reports is conflicting when comparing results across studies – possibly because the data were derived from various types of samples (e.g., stool, sputum, airway, etc.). It should be noted that though these specific taxa were found to differentiate by dog ownership in house dust, they may not necessarily also differentiate by dog ownership in the gut microbiota of at-risk children. Alternatively, it is possible that they alter the gut environment in such a way that other microbes may thrive or decline, and these other microbes could directly affect the risk of allergies and asthma. Additional studies are needed to understand how dog-exposed house dust alters gut microbiota in early life.
These results should be viewed as further evidence in support of the evolving “Microbiota Hypothesis” related to allergy and asthma risk. 43 The hypothesis is that the environmental microbial diversity influences the assemblage process of an infant’s gut microbiota ecosystem which subsequently, in partnership with exposure to allergens and microbes, influences the child’s immune development and risk for allergies and asthma.43–45 Evidence supporting this hypothesis continues to increase. Children with dogs in homes in early life have been shown to have less allergy and asthma.6, 9–11, 46–47
Other work has shown that children with dogs in the home in the first year of life have greater microbial diversity in their stool, 48 while other studies have shown that children with greater microbial diversity in their stool in early life have less allergy and asthma. 43, 49–53 The present work demonstrates that dogs change the home dust microbiota by increasing the types and relative abundances of specific genera. Home dust bacterial composition and allergen levels have been found to be associated with subsequent allergic sensitization and wheezing54. However, the precise mechanisms by which a child’s gut microbiota can be influenced by their home dust microbiota are not specifically known. Additionally, it is not known if a specific species or a network of species are necessary to impact the immune system’s development. It is possible that many different combinations of bacteria in early life could yield better health in the child, but perhaps the optimal combinations depend on to what the child has already been exposed. It is also not known if gut microbial composition and immune function changes can be induced after the immune system has been educated in early life.
A limitation of the study is that we did not conduct repeated sampling over shorter intervals that would have allowed us to more precisely identify when significant changes in taxa presence and relative abundance occurred. Further, due to the difficulty in identifying and recruiting dog families prior to receiving their new dog, many of the baseline dog home samples were collected after the dog was introduced, adding variation to dog home samples at baseline. However, given that we observed differences by dog at baseline among those who had only been resident for a short time—which were not merely explained by dog-associated demographic or household characteristics—dogs may affect bacterial house dust composition more rapidly than anticipated.
Further investigation is required to determine if these particular taxonomic changes are generalizable or are specific to metropolitan Detroit. Additionally, specific dog characteristics such as size, fur type or indoor/outdoor activity were not examined due to the small sample size. However, by including in this project homes that had not been keeping furred animals for a period of approximately one year or more, as well as a comparison population similar in terms of household characteristics and demographics, we think the strengths of this work reinforce its importance.
In summary, this work demonstrates that dogs can rapidly alter the home dust microbiota, primarily by increasing the presence and relative abundance of specific bacteria and that this alteration is more easily seen after one year. This evidence, partnered with epidemiological studies relating early dog exposure to the diminished occurrence of allergic disorders, supports the Microbiota Hypothesis for impacting asthma and allergies. Future studies must identify which taxon or combinations of taxa will be most beneficial for the developing gut microbiota for the long-term health of children.
Supplementary Material
Supplemental Table 1. Dog vs. dog-free household comparisons
Supplemental Table 2. Household dust alpha diversity metrics for bacterial microbes, by dog (grouped by dust collection time) vs. dog-free households – baseline only
Supplemental Table 3. OTUs with significant differences in dog vs. dog-free homes at 12 months (N=109, joint FDR-adjusted p-value<0.05)
Supplemental Figure 1. Household dust alpha diversity metrics for bacterial microbes, by dog vs. dog-free households
Practical Implications.
In many studies, living with dogs has been shown to decrease the subsequent risk of allergies and asthma; however, the mechanisms are not well understood. Recent and current investigations are examining the role of the gut microbiome in allergic disease development. As part of this hypothesis, dogs could alter the environmental microbiome which could, in turn, modify the gut microbiome. This study was conducted to investigate how the introduction of a dog into a home changes the home’s environmental microbiota. Dog introduction into the home appears to immediately introduce low abundant bacterial taxa, and by 12-months post-introduction, results in large relative abundance shifts. Operation taxonomic units (OTUs) that were more likely to be detected in dog homes at 12-months included specific members of the genera Moraxella, Porphyromonas, Capnocytophaga, Fusobacterium, Streptococcus, and Treponema. OTUs that had higher relative abundance in dog homes at 12-months included specific members of the genera Anaerococcus and Prevotella.
Acknowledgments
This work was completed with the efforts of Mary Ann Aubuchon, Andrew Bossick, Mark Kolar and Kole Lynch.
This work was funded by NIAID.
Footnotes
DR. GANESA WEGIENKA (Orcid ID : 0000-0001-7281-1193)
References
- 1.Von Ehrenstein OS, Von Mutius E, Illi S, Baumann L, Böhm O, von Kries R. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy. 2000;30(2):187–93. doi: 10.1046/j.1365-2222.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 2.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 2002;288(8):963–72. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs O, Genuneit J, Latzin P, Buchele G, Horak E, Loss G, et al. Farming environments and childhood atopy, wheeze, lung function, and exhaled nitric oxide. J Allergy Clin Immunol. 2012;130(2):382–8 e6. doi: 10.1016/j.jaci.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 4.Genuneit J. Exposure to farming environments in childhood and asthma and wheeze in rural populations: a systematic review with meta-analysis. Pediatr Allergy Immunol. 2012;23(6):509–18. doi: 10.1111/j.1399-3038.2012.01312.x. [DOI] [PubMed] [Google Scholar]
- 5.Campbell BE, Lodge CJ, Lowe AJ, Burgess JA, Matheson MC, Dharmage SC. Exposure to ‘farming’ and objective markers of atopy: a systematic review and meta-analysis. Clin Exp Allergy. 2015;45(4):744–57. doi: 10.1111/cea.12429. [DOI] [PubMed] [Google Scholar]
- 6.Bufford JD, Reardon CL, Li Z, Roberg KA, DaSilva D, Eggleston PA, et al. Effects of dog ownership in early childhood on immune development and atopic diseases. Clin Exp Allergy. 2008;38(10):1635–43. doi: 10.1111/j.1365-2222.2008.03018.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen CM, Tischer C, Schnappinger M, Heinrich J. The role of cats and dogs in asthma and allergy–a systematic review. Int J Hyg Environ Health. 2010;213(1):1–31. doi: 10.1016/j.ijheh.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Gern JE, Reardon CL, Hoffjan S, Nicolae D, Li Z, Roberg KA, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol. 2004;113(2):307–14. doi: 10.1016/j.jaci.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Pelucchi C, Galeone C, Bach JF, La Vecchia C, Chatenoud L. Pet exposure and risk of atopic dermatitis at the pediatric age: a meta-analysis of birth cohort studies. J Allergy Clin Immunol. 2013;132(3):616–22 e7. doi: 10.1016/j.jaci.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Lodrup Carlsen KC, Roll S, Carlsen KH, Mowinckel P, Wijga AH, Brunekreef B, et al. Does pet ownership in infancy lead to asthma or allergy at school age? Pooled analysis of individual participant data from 11 European birth cohorts. PLoS One. 2012;7(8):e43214. doi: 10.1371/journal.pone.0043214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters RL, Allen KJ, Dharmage SC, Lodge CJ, Koplin JJ, Ponsonby AL, et al. Differential factors associated with challenge-proven food allergy phenotypes in a population cohort of infants: a latent class analysis. Clin Exp Allergy. 2015;45(5):953–63. doi: 10.1111/cea.12478. [DOI] [PubMed] [Google Scholar]
- 12.Pyrhönen K, Näyhä S, Läärä E. Dog and cat exposure and respective pet allergy in early childhood. Pediatr Allergy Immunol. 2015;26(3):247–55. doi: 10.1111/pai.12369. [DOI] [PubMed] [Google Scholar]
- 13.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–2. 2 e1–3. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2016;26(2):179–92. doi: 10.1111/ina.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barberán A, Dunn RR, Reich BJ, Pacifici K, Laber EB, Menninger HL, et al. The ecology of microscopic life in household dust. Proc Biol Sci. 1814;2015:282. doi: 10.1098/rspb.2015.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin P, O’Hara RB, et al. vegan: Community ecology package. R package version 2.0-9. 2013 [Google Scholar]
- 18.Lozupone CA, Knight R. UnifFrac: A new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 20.Kindt R, Coe R. Tree diversity analysis: A manual and software for common statistical methods for ecological and biodversity studies. World Agroforestry Centre (ICRAF); Nairobi: 2005. [Google Scholar]
- 21.Chen EZ, Li H. A two-part mixed-effects model for analyzing longitudinal microbiome compositional data. Bioinformatics. 2016;32(17):2611–7. doi: 10.1093/bioinformatics/btw308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini YHY. A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57(1):289–300. [Google Scholar]
- 23.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–13. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lax S, Sangwan N, Smith D, Larsen P, Handley KM, et al. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med. 2017;9:eeah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dannemiller KC, Wechsler CJ, Peccia J. Fungal and bacterial growth in floor dust at elevated relative humidity levels. Indoor Air. 2017;27:354–363. doi: 10.1111/ina.12313. [DOI] [PubMed] [Google Scholar]
- 26.Gaulke CA, Barton CL, Proffitt S, Tanguay RL, Sharpton TJ. Triclosan exposure is associated with rapid restructuring of the microbiome in adult zebrafish. PLoS ONE. 11(5):e0154632. doi: 10.1371/journal.pone.0154632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer BT, Srinivasan S, Fielder TL, Marrazzo JM, Fredricks DN, Schiffer JT. Rapid and profound shifts in the vaginal microbiota following antibiotic treatment for bacterial vaginosis. J Infect Dis. 2015;212:793–802. doi: 10.1093/infdis/jiv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturgeon A, Stull JW, Costa MC, Weese JS. Metagenomic analysis of the canine oral cavity as revealed by high-throughput pyrosequencing of the 16S rRNA gene. Vet Microbiol. 2013;162(2–4):891–8. doi: 10.1016/j.vetmic.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Oh C, Lee K, Cheong Y, Lee SW, Park SY, Song CS, et al. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PLoS One. 2015;10(7):e0131468. doi: 10.1371/journal.pone.0131468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hand D, Wallis C, Colyer A, Penn CW. Pyrosequencing the canine faecal microbiota: breadth and depth of biodiversity. PLoS One. 2013;8(1):e53115. doi: 10.1371/journal.pone.0053115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middelbos IS, Vester Boler BM, Qu A, White BA, Swanson KS, Fahey GC., Jr Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One. 2010;5(3):e9768. doi: 10.1371/journal.pone.0009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman AR, Patterson AP, Diesel A, Lawhon SD, Ly SJ, et al. The skin microbiome in health and allergic dogs. PLOS One. 2014;9:e83197. doi: 10.1371/journal.pone.0083197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuscó A, Bellanger JM, Gershony L, Islas-Trejo A, Levy K, et al. Individual signatures and environmental factors shape skin microbiota in healthy dogs. Microbiome. 2017;5:139. doi: 10.1186/s40168-017-0355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuscó A, Sanchez A, Altet L, Ferrer L, Francino O. Individual signatures define canine skin microbiota composition and variability. Frontiers Vet Sci. 2017;4:1–12. doi: 10.3389/fvets.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–95. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 37.Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol. 2017;139(3):826–834. doi: 10.1016/j.jaci.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 38.Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140(1):63–75. doi: 10.1016/j.jaci.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbes SJ, Jr, Matsui EC. Can oral pathogens influence allergic disease? J Allergy Clin Immunol. 2011;127(5):1119–27. doi: 10.1016/j.jaci.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131(2):346–52. e1–3. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2015 doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 42.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129(2):434–40. 40 e1–2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 43.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62(11):1223–36. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 44.Johnson CC, Ownby DR. Allergies and Asthma: Do Atopic Disorders Result from Inadequate Immune Homeostasis arising from Infant Gut Dysbiosis? Expert Rev Clin Immunol. 2016;12(4):379–88. doi: 10.1586/1744666X.2016.1139452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson CC, Ownby DR. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res. 2017;179:60–70. doi: 10.1016/j.trsl.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegienka G, Johnson CC, Havstad S, Ownby DR, Nicholas C, Zoratti EM. Lifetime dog and cat exposure and dog- and cat-specific sensitization at age 18 years. Clin Exp Allergy. 2011;41(7):979–86. doi: 10.1111/j.1365-2222.2011.03747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wegienka G, Johnson CC, Havstad S, Ownby DR, Zoratti EM. Indoor pet exposure and the outcomes of total IgE and sensitization at age 18 years. J Allergy Clin Immunol. 2010;126(2):274–9. 9 e1–5. doi: 10.1016/j.jaci.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. 2016;6:31775. doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. J Allergy Clin Immunol. 2013 doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 50.Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39(4):518–26. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 51.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187–91. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132(3):601–7 e8. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 53.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134(3):593–601 e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3(2):168–78. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 56.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31(21):3476–82. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 61.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 64.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Dog vs. dog-free household comparisons
Supplemental Table 2. Household dust alpha diversity metrics for bacterial microbes, by dog (grouped by dust collection time) vs. dog-free households – baseline only
Supplemental Table 3. OTUs with significant differences in dog vs. dog-free homes at 12 months (N=109, joint FDR-adjusted p-value<0.05)
Supplemental Figure 1. Household dust alpha diversity metrics for bacterial microbes, by dog vs. dog-free households


