Summary
Meiotic double-strand breaks (DSBs) are generated and repaired in a highly regulated manner to ensure formation of crossovers (COs) while also enabling efficient non-CO repair to restore genome integrity. We use Structured-Illumination Microscopy to investigate the dynamic architecture of DSB repair complexes at meiotic recombination sites in relationship to the synaptonemal complex (SC). DSBs resected at both ends are converted into inter-homolog repair intermediates harboring two populations of BLM helicase and RPA, flanking a single population of MutSγ. These intermediates accumulate until late pachytene, when repair proteins disappear from non-CO sites and CO-designated sites become enveloped by SC-central region proteins, acquire a second MutSγ population, and lose RPA. These and other data suggest that the SC may protect CO intermediates from being dismantled inappropriately and promote CO maturation by generating a transient CO-specific repair compartment, thereby enabling differential timing and outcome of repair at CO and non-CO sites.
Keywords: Meiosis, DNA repair, synaptonemal complex, super-resolution, recombination
ETOC
Structured-illumination microscopy of meiotic recombination reveals how compartmentalization and differential timing impact the outcome of repair at crossover and non-crossover sites.
Introduction
During meiosis in most organisms, each pair of homologous chromosomes must receive at least one crossover (CO) in order to undergo reliable segregation during the first meiotic division. COs are generated by a specialized form of recombinational repair of double-strand DNA breaks (DSBs), introduced by the meiosis–specific SPO-11 protein. In order to ensure that every homolog pair receives at least one CO, a surplus of DSBs is introduced during meiotic prophase. Only a subset of these DSBs mature into COs, while the rest are repaired by alternative homologous recombination-based mechanisms that do not yield COs. Despite an excess of DSBs, COs exhibit wide spacing along chromosomes, and many plant and animal models typically undergo only a single CO per chromosome arm. Thus, CO formation is both ensured and limited (for reviews: (Mercier et al., 2015; Zickler and Kleckner, 2016)).
Several conserved meiosis-specific proteins, such as the MutSγ (MSH4-MSH5) complex or the cyclin-related protein COSA-1/CNTD1, were identified as being essential for controlled CO formation. These pro-CO factors localize to sites of ongoing meiotic recombination and modulate the outcome of DNA repair to yield COs as final DSB repair products at “CO-designated” sites (for review: (Hillers et al., 2017; Zickler and Kleckner, 2016). Nevertheless, how these proteins promote the CO repair outcome, and how different modes of DSB repair can operate at other sites on the same chromosomes in the same cells to yield non-CO products, are far from being understood.
Controlled CO formation further depends on components of the synaptonemal complex central region (SC-CR). The SC-CR is composed of predicted coiled-coil proteins and assembles between aligned homologous chromosomes, maintaining them at a distance of roughly 100–150 nm along their lengths throughout the pachytene stage of meiotic prophase. The SC-CR is dispensable for initial homolog pairing and DSB formation, but when absent, meiotic COs are reduced or eliminated, indicating that SC-CR components play a role in promoting CO formation (for review: (Hillers et al., 2017; Page and Hawley, 2004)). The SC is a long known cytological hallmark of meiosis (Moses, 1956) that has been implicated previously in multiple aspects of CO regulation, and recent studies have revealed dynamic properties of the SC and provided insight regarding how changes in SC dynamics might contribute both to nucleus-wide and chromosome-autonomous signaling systems that limit DSBs and inhibit excess COs (Machovina et al., 2016; Nadarajan et al., 2017; Pattabiraman et al., 2017; Rog et al., 2017; Subramanian et al., 2016). However, how the SC-CR functions in promoting CO formation remains poorly understood.
In the nematode C. elegans, in contrast to many other model systems, assembly of the SC-CR is not dependent on recombination (for review: Keeney et al., 2014). This makes it possible to differentiate experimentally between the contributions of DNA repair factors and those of SC components to the process of meiotic recombination. In the current work, we exploit this and other features of the C. elegans system to investigate how meiotic DNA repair proteins and meiotic chromosome structure collaborate to bring about a robust outcome of meiosis, i.e., formation of a single CO per each pair of homologous chromosomes (bivalent) and restoration of chromosome integrity prior to the meiotic cell divisions.
In particular, we employed a cytological preparation that affords greatly improved detection of recombination sites to establish a comprehensive time course of the dynamic localization of meiosis-specific recombination factors and DSB repair proteins during wild-type and mutant meiosis. Further, we used 3D-Structured Illumination microscopy (SIM) to reveal spatial organization of DSB repair complexes at recombination sites and changes in the architecture of these sites during the course of meiotic prophase, allowing us to make inferences regarding the timing of recombination progression and the nature of the underlying DNA intermediates. Moreover, we discovered a unique spatial arrangement of SC-CR components surrounding CO-designated recombination sites that provides a new framework for thinking about how distinct outcomes of DSB repair can be accomplished at different sites within a single cell.
Results
Progressive differentiation of meiotic recombination sites during C. elegans meiosis
We re-examined the dynamic localization of multiple DSB repair proteins at the sites of ongoing recombination during the course of meiotic prophase, using a nuclear spreading protocol that enables improved visualization of these events (Loidl et al., 1991; Pattabiraman et al., 2017). Chromosome axis components and chromosome-bound repair proteins are retained in these preparations, but nucleoplasmic protein pools that obscure recombination foci are washed out, resulting in superior detection relative to previously-used in situ methods (Fig S1A). At the same time, the relative spatial organization of nuclei within a gonad is maintained largely intact, preserving the spatial/temporal gradient of nuclei progressing through meiotic prophase.
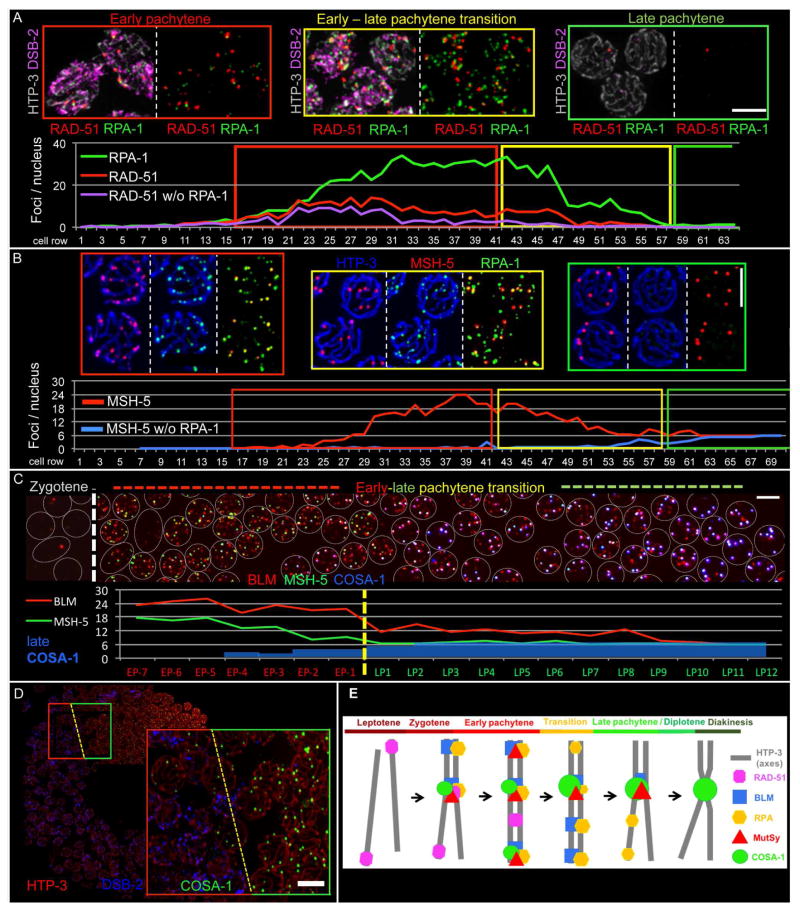
Fig 1A and Fig S1B show simultaneous visualization of RAD-51, the sole C. elegans recombinase, and RPA-1, a subunit of the eukaryotic ssDNA binding protein RPA. Similar to previous reports (Alpi et al., 2003; Colaiacovo et al., 2003), RAD-51 foci are detected beginning in zygotene, rise in abundance and reach a plateau during early pachytene before declining and then disappearing during late pachytene (identified based on loss of the early prophase marker DSB-2 (Fig S1B; (Rosu et al., 2013)). RPA foci initially rise in abundance in parallel with RAD-51, but RPA foci continue to increase in abundance after RAD-51 foci have peaked and begun to decline, and they accumulate to much higher levels, plateauing at 30 ± 4 foci per nucleus (Fig 1B and S2C). RPA foci decline markedly at the early-to-late pachytene transition, but RPA remains detectable at a substantial number of sites before eventually being lost from all sites in late pachytene.
Fig. 1. Localization of RAD-51, RPA, BLM, MSH-5 and COSA-1 during meiotic prophase (see also Figs S1–S2).
(A) Top: Representative images of meiotic stages in a spread WT gonad (also see Fig S1B). DSB-2 indicates nuclei in early prophase, and HTP-3 marks chromosome axes. (B) Top: Representative nuclei from a spread gonad. (A and B) Bottom: Quantitative displays of appearance, disappearance and co-localization of indicated foci in a single gonad (distinct from the examples depicted). (C) Top: Image of the early-to-late pachytene transition region of a spread gonad, with individual nuclei represented by white circles. Bottom: Quantitative display of BLM, MSH-5 and late COSA-1 foci in the portion of the gonad shown above. (D) Left: WT gonad expressing GFP::COSA-1. Right: enlargement of the early (red outline) to-late (green) pachytene transition, coinciding with the appearance of six late COSA-1 foci. Scale bars (in A–D): 5 μm. (E) Schematic of appearance and disappearance of recombination factors at recombination sites during meiotic prophase.
MSH-5 and MSH-4 make up the heterodimeric MutSγ complex (for review: Manhart and Alani, 2016) that concentrates at CO-designated sites in late pachytene and is essential for CO formation in C. elegans (Kelly et al., 2000; Yokoo et al., 2012; Zalevsky et al., 1999). MSH-5 foci first appear in zygotene and peak near the end of early pachytene at an average of 22 ± 4 MSH-5 foci per nucleus (Fig 1B and S2C). At this stage, MSH-5 foci nearly always colocalize with RPA, with fewer than one RPA-negative MSH-5 focus per nucleus. MSH-5 foci represent interhomolog (IH) recombination intermediates, as they are usually not detected on chromosome segments that are unable to engage a (non-sister) homologous chromosome segment as a template for HR (Fig S1C–D). At the early-to-late pachytene transition, MSH-5 is lost from the majority of repair sites while being retained at a single CO-designated repair site on each bivalent. These late MSH-5 foci are brighter than the earlier foci, suggesting an increased concentration of MutSγ at CO-designated sites. RPA foci also decrease in number following the transition to late pachytene, but in contrast to MSH-5, RPA resides longer at (MSH-5-negative) NCO sites and vanishes from (MSH-5-positive) future CO sites, suggesting a transition at CO sites to a recombination intermediate that lacks ssDNA.
Throughout prophase, MSH-5-marked sites also harbor BLM helicase (Fig 1C and S2). BLM (HIM-6 in worms) is required both for efficient CO maturation and for dismantling NCO repair intermediates (for review: (Hatkevich and Sekelsky, 2017). Essentially all BLM foci in early prophase localize with RPA; in contrast, a few RPA foci lacking BLM are detected, predominantly at the early-to-late pachytene transition. Similar to RPA, BLM temporarily remains at NCO repair sites after removal of MSH-5. However, BLM is retained at CO sites throughout late pachytene (like MSH-5), while RPA is lost (e.g.: Fig S1B and S2A).
Previous work had identified COSA-1 as a marker COs (Yokoo et al., 2012). Our spread preparations reveal that in addition to becoming highly concentrated together with MSH-5 at CO sites in late pachytene, GFP::COSA-1 also decorates IH recombination sites in early prophase as very faint foci, co-localizing with MSH-5, BLM and RPA (Fig 1C and S2). These early foci are more numerous and fainter than the six brighter COSA-1 foci detected at CO sites in late pachytene (Fig S2A). Loss of COSA-1 and MSH-5 from NCO sites and their accumulation on CO sites in late pachytene occurs concurrently (Fig 1C). Further, loss of early prophase nuclear marker DSB-2 strongly coincides with the presence of 6 late COSA-1 foci, and conversely, if nuclei retain DSB-2, they invariably have fewer than 6 (if any) late COSA-1 foci (Fig 1D). This indicates an abrupt change in the state of recombination sites as germ cells transition to the late pachytene stage.
Based on these and previous observations, the following picture emerges (Fig 1E): DSBs are introduced, resected and load RAD-51 throughout early prophase. RAD-51-decorated DNA ends engage in homology search and strand exchange and turn over into RAD-51-free intermediates that contain RPA and BLM. A subset of IH repair sites additionally recruits MutSγ (and COSA-1) to create intermediates with the potential to become COs. Multiple such sites accumulate until each bivalent has received at least one such IH intermediate before nuclei transition to late pachytene. (Rosu et al., 2011; Woglar et al., 2013). A single DSB repair site on each bivalent is designated to become the future CO, either before or during a rapid transition to late pachytene. Upon transition to late pachytene, recombination sites designated to mature as COs or NCOs become differentiated from each other, with COSA-1 and MSH-5 simultaneously becoming enriched at future CO sites and lost from NCO sites. BLM and RPA remain associated with NCO sites briefly after the transition to late pachytene but disappear soon thereafter, presumably reflecting earlier completion of NCO repair at these sites. Concurrently, CO-designated sites lose RPA, indicating completion of repair DNA synthesis at those sites; these differentiated CO sites persist throughout the late pachytene stage and vanish by diakinesis, when the presence of a chiasma connecting each homolog pair reflects the presence of a completed CO event.
The architecture of recombination sites during early prophase
Having established this comprehensive time course of dynamic localization of meiotic DNA repair proteins, we combined nuclear spreading with 3D Structured Illumination Microscopy (SIM) to better understand how recombination sites are organized spatially and how this organization changes during meiotic progression. As these spread preparations both greatly improve signal-to-noise ratio and decrease sample thickness, they optimize the specimens for SIM. This enabled us to visualize the relative spatial organization of different recombination proteins present at CO and NCO sites and their relationships to chromosome axes and the SC-CR.
RAD-51
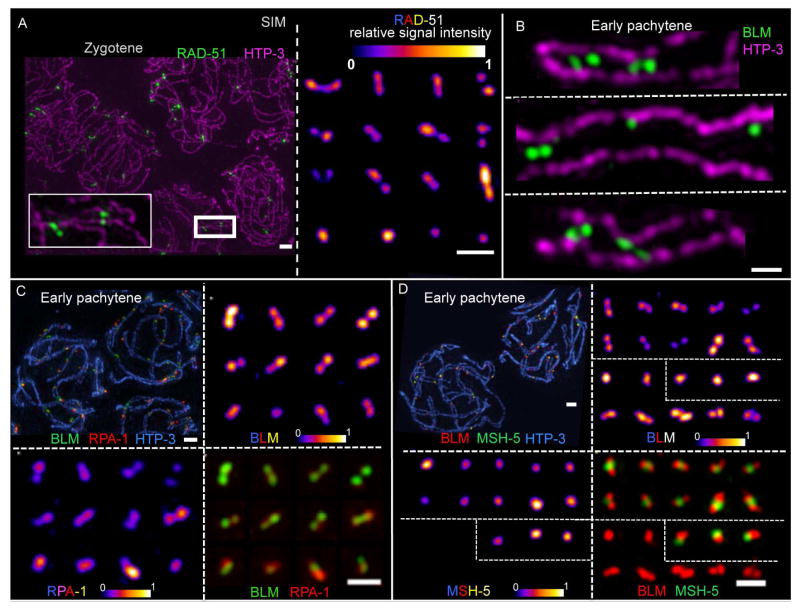
RAD-51 signals are readily resolved as doublets (57%) or elongated structures (14%) in the majority of cases (Fig 2A); in the remaining cases, RAD-51 is detected as a single point focus. RAD-51 doublets are readily detected regardless of the number of DSB sites present in a nucleus and also when chromosomes are strongly spread, and we never detect more than two foci at a single site. Thus, we infer that RAD-51 doublets represent single DSB events (rather than two adjacent DSBs) and that both sides of a DSB are resected and load RAD-51. This suggests that both ends are potentially capable of promoting homology search and strand invasion during C. elegans meiosis, as was recently shown for budding yeast (Brown et al., 2015).
Fig. 2. Structural features of DNA repair complexes at recombination sites in early prophase.
Images of nuclei, chromosome segments, and individual recombination sites generated using 3D-SIM and Z-stack projection. (A) Left: HTP-3 and RAD-51 in zygotene nuclei. Scale bar: 1 μm. Right: Individually cropped RAD-51 signals from the same field. To illustrate the structural organization of foci, relative signal intensities are displayed using an 8-bit color look-up table (“LUT Fire” in ImageJ). Scale bar: 500 nm. (B) Individually-cropped early pachytene SC segments in frontal view, illustrating the roughly parallel orientation of BLM doublet foci between the aligned axes. Scale bar: 500 nm. (C+D) Individually-cropped early pachytene recombination foci (from the same field as the representative nuclei at top left) illustrating both structural organization for individual components and relative spatial relationships of distinct components. Scale bars: 1 μm (top left) and 500 nm (bottom right).
BLM
BLM was frequently detected as dual foci aligned longitudinally with the chromosome axis in pachytene nuclei analyzed in situ in whole-mount preparations of C. elegans gonads (Jagut et al., 2016). We confirm and extend these findings for BLM using SIM on nuclear spreads (Fig 2B). In well-spread early prophase nuclei, we can resolve BLM signals as two populations (doublets; 46.8%) or elongated structures (9.5%) at the majority of repair sites; these BLM doublets or elongated structures are oriented longitudinally along the chromosomes, and when the axes can be resolved as two parallel tracks, the BLM signals are located between the two axial tracks. Further, multiple BLM doublets and single foci can be observed between the axes along the length of a single bivalent during early pachytene.
RPA
Like BLM, RPA is also frequently detected as doublets or elongated structures oriented longitudinally between the chromosome axes during zygotene and early pachytene (Fig 2C) (34.0% doublets, 17.5% elongated structures). However, BLM and RPA SIM signals don’t strictly coincide, as RPA sometimes localizes as a single focus in the middle of a BLM doublet. Further, when both RPA and BLM are detected as doublets, their relative orientation is similar, yet slightly oblique. This suggests that ssDNA (marked by RPA) and BLM helicase complexes occupy nearby but distinct positions within recombination intermediates.
MSH-5
At IH recombination sites marked by MSH-5 during zygotene and early pachytene, MSH-5 is consistently detected as a single focus, located either in the middle of a BLM/RPA doublet or side-by-side with a BLM/RPA singlet (Fig 2D). BLM is detected as doublets or elongated foci much more frequently at MSH-5-positive recombination sites (61%, n=389) than at MSH-5-negative sites, (33%, n=117) (p< 0.0001, Fisher exact test), suggesting that MSH-5-positive and negative sites represent structurally distinct classes of recombination intermediates.
The architecture of CO sites during late prophase
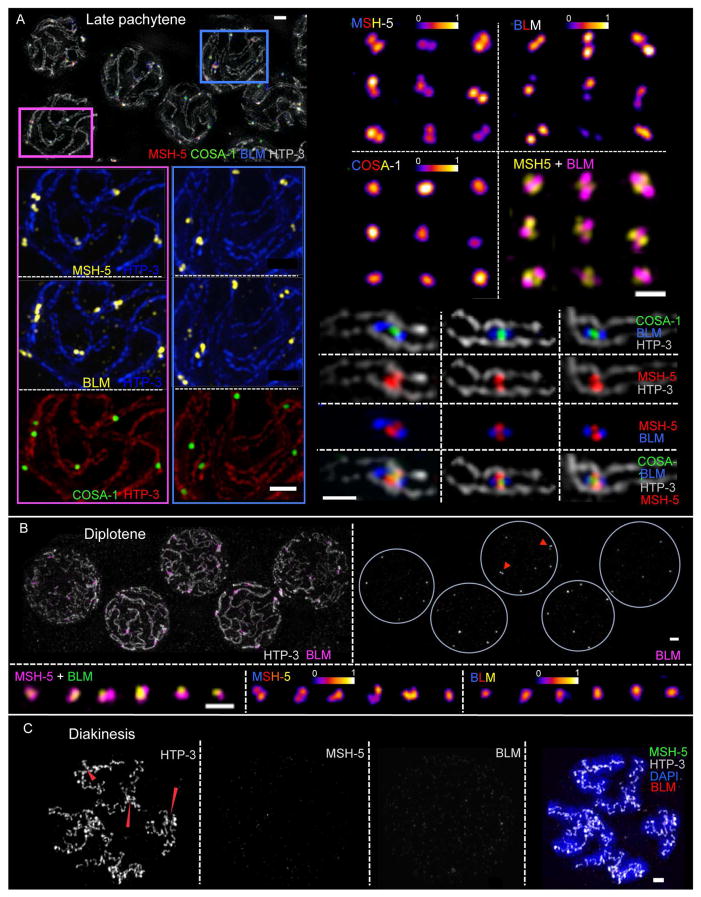
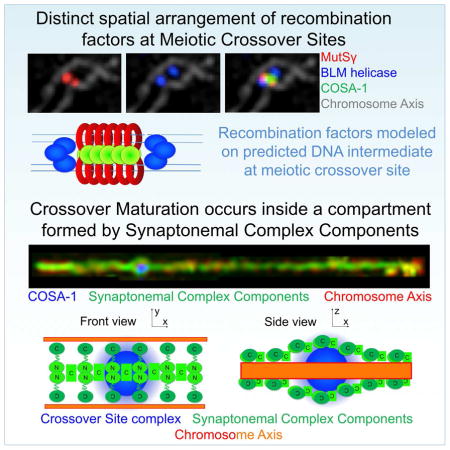
CO-designated recombination sites acquire a striking new architecture following transition to late pachytene. CO sites not only lose RPA (presumably reflecting completion of repair synthesis), they also change the spatial organization of proteins that remain and/or become concentrated at these sites (Fig 3A and S3B–D). BLM is still present at each CO site and is detected as doublet foci aligned roughly in parallel with and between the homolog axes at nearly all CO sites in well-spread samples. In contrast to the MSH-5 singlets present in early pachytene, most MSH-5 signals at CO sites are now resolved as doublets (70%) or elongated foci (15%) that are oriented orthogonally to the BLM doublets and span the distance between the aligned homolog axes. COSA-1 is detected at the center of the cruciform structure defined by the orthogonally-oriented MSH-5 and BLM doublets. We infer that this organization reflects the presence of a late CO-specific recombination intermediate at these sites. Based on measurements of the mean distances between the centroids of paired BLM signals (240 ± 34 nm) and paired MSH-5 signals (168 ± 24 nm), we derive a lower-limit estimate of 290–300 nm, or about 800–900 bp, for the average length of these underlying CO-specific DNA intermediates (see Methods and Discussion).
Fig. 3. SIM images of late prophase CO-designated recombination sites.
(A) Left: Representative late pachytene nuclei (top) and enlargements of portions of this field (below). Right: Individually-cropped recombination sites. Examples displayed show CO-designated sites at chromosome segments imaged in frontal aspect, illustrating two populations of BLM (oriented in parallel with the chromosome axes), two populations of MSH-5 (oriented perpendicular to the axes) and a single population of COSA-1 at the center. Scale bars: 1 μm (left) and 500 nm (right). (B) Top: diplotene nuclei; most BLM signals are no longer reliably resolved as doublets; arrowheads highlight rare examples of BLM doublets. Scale bar: 1 μm. Bottom: Individually-cropped diplotene recombination sites. Scale bar: 500 nm. (C) Diakinesis nucleus. Red arrowheads indicate chiasmata, temporary connections between desynapsed homologs that reflect the formation of COs; BLM and MSH-5 are no longer detected. Scale bar: 1 μm.
The architecture of CO sites changes once more upon transition to diplotene, when homologs start to desynapse The fraction of BLM signals (12%) and MSH-5 signals (29%) resolvable as doublets diminishes substantially, and the cohorts of proteins no longer exhibit any consistent arrangement relative to each other or to the chromosome axes (Fig 3B and S3E). Finally, by diakinesis, when the bivalents are completely de-synapsed, BLM and MSH-5 are no longer detected at the CO sites, which are now visible as chiasmata (Fig 3C).
CO-designated recombination sites are encased by SC-CR proteins
Since meiotic recombination events progress and mature in the context of the SC, we investigated the spatial relationships between the SC-CR and the cohorts of DNA repair proteins present at recombination sites (Fig 4 and S4). When spreading is conducted using physiological or hypertonic conditions (“Hanks”), SC destabilization is minimized compared to hypotonic spreads (“H20”) and a distance more similar to in situ spacing between the axes is maintained, reflecting better preservation of the SC-CR (Fig S3B). In early pachytene nuclei analyzed under such conditions, most signals for DNA repair proteins are found adjacent to, but not directly co-localizing with SC-CR proteins; i.e., repair foci appear laterally associated with the SC-CR and only rarely appear embedded in it (Fig 4A and S4A).
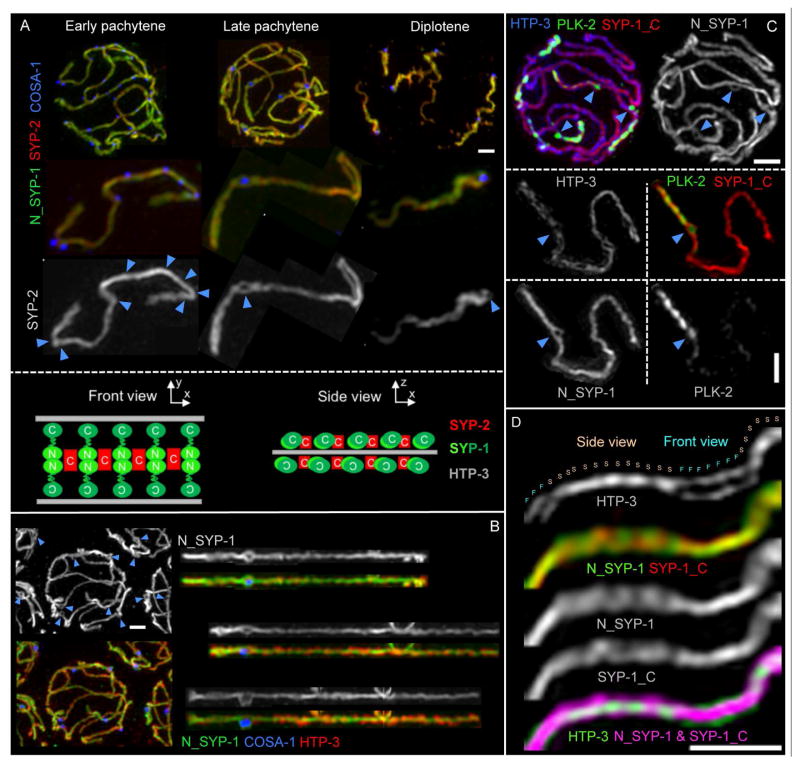
Fig. 4. The SC-CR engulfs CO-designated sites as a bubble-like structure.
(A) Top: Representative images of nuclei from the indicated stages. Middle: one bivalent, individually cropped from each nucleus above. Blue arrowheads indicate the positions of multiple COSA-1-marked recombination sites in early pachytene and the single CO-designated site at late prophase. Bottom: schematics depicting frontal and lateral views of SC organization. (B) Representative late pachytene nucleus (left) and individually straightened bivalents (right), illustrating that SC-CR bubbles are detected specifically at CO-designated sites marked by GFP::COSA-1 (blue arrowheads). (C) Representative late pachytene nucleus and one individually-cropped SC, illustrating that the SC-CR bubble is detected by antibodies against both the N- and C-termini of SYP-1. (D) Segment of a late pachytene SC, illustrating both the frontal (F) and lateral (side; S) aspects of the SC. Scale bars: 1 μm.
In contrast, late pachytene CO-designated sites are frequently associated with bubble-like structures in which the SC-CR proteins fully or partially surround the cohort of DNA repair proteins concentrated at the CO site (Fig 4A–C). These SC-CR bubbles appear following the transition to late pachytene and can be detected using antibodies against both the SYP-1 N-terminus, which localizes at the center of the C. elegans SC (Schild-Prufert et al., 2011), and the SYP-1 C-terminus, which localizes more laterally within the SC, closer to the chromosome axes, and with antibodies against the small SYP-2 protein, which localizes at the center of the SC (Fig 4 and S4). SC-CR bubbles are present throughout late pachytene and are strongly correlated with CO sites: 89% of the bubble-like structures identified when SIM images were screened using only the SC-CR channel were found to correspond to CO sites, despite such sites comprising less than 5% of the total SC length. Conversely, SC-CR bubbles could be detected at nearly half of all late pachytene CO sites (47%); since the ability to detect SC-CR bubbles is dependent on the orientation of the SC relative to the plane of imaging, it is likely that SC-CR bubbles form at most, if not all, late pachytene CO sites. Correspondence of SC-CR bubbles and CO sites is further reinforced by SIM analysis of chromosome spreads from worms homozygous for the mnT12(IV;X) fusion chromosome, as we could detect fusion bivalents harboring two CO sites that were both encased by SC-CR bubbles (Fig S4).
Previous work showed that polo-like-kinase PLK-2 is recruited to the SCs and becomes concentrated on the SC segment extending from the CO-designated site to the nearest chromosome end (referred to as the “short arm”) at the onset of late pachytene (Harper et al., 2011; Labella et al., 2011; Pattabiraman et al., 2017). However, we find that PLK-2 is not part of the structure that encases the future CO site upon transition to late pachytene. Rather, PLK-2 localizes within the SC-CR bubble together with the recombination proteins (in addition to its localization on the SC short arm; Fig 4C and S4B). Further, while the CO-site SC-CR bubbles persist throughout late pachytene, they disappear concomitantly with disassembly of the SC. By mid-diplotene, remnants of the SC-CR bubble can be detected only on the side of the CO site adjacent to the short arm of the bivalent, where the SC-CR proteins remain specifically enriched at this stage (Nabeshima et al., 2005); Fig 4A).
Our SIM imaging of SC-CR proteins together with CO-site markers and chromosome axis marker HTP-3 (Goodyer et al., 2008; MacQueen et al., 2005) provides additional information regarding the structural organization of the C. elegans SC. As homolog pairs are twisted around each other during pachytene, SC stretches can be visualized either in a frontal view, where two parallel HTP-3 tracks are visible, or in a lateral view, in which HTP-3 appears as a single line (Fig 4D). When the SC is visualized in frontal views, SYP-2 and both the N- and C-termini of SYP-1 all localize between the aligned chromosome axes, as expected. When the SC is visualized in lateral aspect, the SYP-1 N- and C-termini and SYP-2 are all detected on both sides of the single HTP-3 track, indicating the presence of SC-CR proteins both above and below the plane defined by the chromosome axes (Fig 4D).
Together, our data suggest that the C. elegans SC-CR is composed of (at least) two layers, similar to the 3D structures recently reported for the Drosophila and mouse SCs (Cahoon et al., 2017; Schucker et al., 2015), and the CO intermediates become encased between these layers in a bubble-like structure, flanked on either side by the chromosome axis.
SC-CR proteins play a role in promoting CO formation that is distinct from the roles of other pro-CO factors
SC-CR proteins have been implicated in promoting CO formation in multiple organisms, and all meiotic COs in C. elegans depend on the SC-CR proteins (MacQueen et al., 2002). However, the nature of the CO-promoting activity(ies) of the SC-CR is not well understood, and thus it has been unclear whether the SC-CR makes distinct contributions from other conserved pro-CO factors such as orthologs of MutSγ, ZHP-3 and COSA-1. Therefore, we analyzed and compared the dynamic localization of multiple DNA repair factors in two classes of mutants that fail to form COs: 1) null mutant that lack SC-CR proteins and thus fail to form SCs (syp-1 and syp-3) (MacQueen et al., 2002; Smolikov et al., 2007); and 2) mutants that are proficient for SC assembly, DSB formation and DSB repair, but lack conserved pro-CO factors (msh-4, cosa-1 and zhp-3, termed “recombination mutants” from here on; (Jantsch et al., 2004; Yokoo et al., 2012; Zalevsky et al., 1999).
In both SC-CR and recombination mutants, DSB repair sites marked by RAD-51 and/or RPA appear throughout early prophase and accumulate, as in WT. As early prophase is prolonged in these mutants (Woglar et al., 2013) the numbers of detectable DSB repair sites are higher and peak at a more proximal region of the gonad (Figure 5A). Similar to WT, BLM localizes to a subset of RPA sites during early prophase (Fig S5A). In both classes of mutants, MSH-5 foci can also be detected beginning in zygotene and accumulate until the transition to late pachytene (Figs 5B–C and S5B), albeit they appear fainter than in WT. Further, as in WT, these early MSH-5 foci appear to be largely dependent on the ability of chromosomes to engage in interactions with their homologs (Fig S5C).
Fig. 5. Dynamic localization of recombination factors during mutant meiosis.
(A) Top left: Schematic depicting the positions of nuclei just prior to the early-to-late prophase transition (*), which is delayed in mutants deficient in crossing over. Top right: Quantification of RPA-1 and RAD-51 foci in nuclei from the positions indicated in the schematic; foci levels for all mutants are significantly higher than WT (p < 0.0001). Bottom: Representative examples of such nuclei. (B) Early-to-late prophase transition region of WT (top) and cosa-1 mutant (bottom) gonad, stained for MSH-5, HTP-3 and early prophase marker DSB-1. MSH-5 foci are not retained after the transition to late prophase in the cosa-1 mutant (see also Fig S4B). (C) Early prophase (red outline), early-to-late prophase transition (yellow) and late prophase (green) nuclei from a syp-1 mutant gonad. Throughout the prolonged early prophase of syp-1 mutants, MSH-5 colocalizes with BLM foci (left). At the transition to late prophase, MSH-5 foci reduce in number, and BLM is still present (middle). Upon transition to late prophase, BLM is lost from MSH-5-marked sites (right). Scale bar: 5 μm. (D) Left: late prophase nuclei from WT (gfp::cosa-1) and the syp-3 mutant (syp-3; gfp::cosa-1). Right: Quantification of late prophase MSH-5/COSA-1 foci. (E) Representative SIM images of early prophase (red outline), early-to-late-prophase transition (yellow) and late prophase (green) nuclei from syp-1; blm::HA worms (scale bar: 1 μm; with enlarged individually cropped DNA repair sites from the same fields shown below (scale bar: 500 nm). In late prophase nuclei of the syp-1 mutant, MSH-5 doublets are association with and oriented along single unpaired axes without BLM being present, as indicated in the schematic.
However, in late prophase these two classes of mutants differ markedly from each other with respect to localization of pro-CO factors. In cosa-1 and zhp-3 mutants, MSH-5 foci are not stabilized, and they disappear concurrently with other DNA repair markers following transition to late pachytene (Fig 5B and Fig S5B). In contrast, in SC-CR mutants, MSH-5 foci can be detected following transition to a late pachytene-like state (Fig 5C and 5D). COSA-1 and ZHP-3 are also recruited to these late MSH-5-positive sites, and as in WT, RPA is lost from these late COSA-1/MSH-5/ZHP-3-marked sites (Fig 5D and Fig S5B). Further, as in WT, late prophase nuclei in SC-CR mutants consistently display no more than six foci (average of 4.2 foci per nucleus in the syp-3 mutant), and these foci are non-randomly distributed, such that two foci rarely occur on the same chromosome axis (p<0.0001). However, in contrast to WT, where BLM is maintained at CO-designated sites during late pachytene, BLM becomes undetectable at late COSA-1/MSH-5 marked sites soon after RPA is lost (Fig 5C).
SIM imaging revealed that as in WT, MSH-5 signals in SC-CR mutants are detected as singlets in early prophase, and then can be resolved as doublets from the early-to-late prophase transition and throughout the remainder of the late pachytene like-stage. In contrast to WT, BLM is rarely detected as doublets in early prophase. At the early-to-late prophase transition, BLM is transiently detected as doublets associated with MSH-5 doublets; however, these structures do not consistently adopt the specific cruciform arrangement observed at CO-designated sites during WT meiosis, and they are not detected between parallel-aligned chromosome axes (Fig 5E). In late prophase when BLM can no longer be detected at MSH-5 marked sites, the doublet MSH-5 foci in SC-CR mutants are both tightly associated with a single unpaired chromosome axis (Fig 5E). These structures may potentially reflect non-productive remnants of earlier unstable IH repair intermediates, and/or they may represent the formation of CO-like intermediates between sister chromatids rather than between homologs.
Thus, mutants lacking the SC-CR are able to concentrate pro-CO factors at late prophase repair sites and generate intermediates that share some features in common with CO-specific intermediates present in WT meiosis. However, these intermediates exhibit abnormal architecture and protein composition and never yield COs. Together the data show that SC-CR proteins play a role in promoting normal formation, architecture and stability of CO recombination complexes that is distinct from the roles of conserved CO-promoting factors such as COSA-1 or MutSγ that actually become concentrated at the CO sites.
Discussion
Simultaneous visualization of multiple recombination markers in germline spreads illuminates the progression of meiotic recombination
Our analysis of localization of multiple recombination proteins together, in a context with greatly improved signal-to-noise ratio, has been informative in several ways:
First, we detect abundant RPA foci arising and accumulating during meiotic progression through the end of early pachytene. Based on timing, numbers, distinct localization relative to RAD-51 and colocalization with MSH-5 and BLM, we can infer that the majority of these RPA foci represent post-strand-exchange recombination intermediates. Second, based on analysis of both wild-type meiosis and nuclei where specific chromosome segments cannot engage in homologous synapsis, we infer that MSH-5/RPA/BLM co-foci mark sites of IH recombination, and conversely, that chromosome segments that cannot engage the homolog as a repair template accumulate earlier RAD-51-marked intermediates. These observations and inferences strongly parallel findings from cytological analyses examining pairwise combinations of recombination proteins in mouse meiosis (e.g. (Moens et al., 2002; Walpita et al., 1999), indicating broad conservation of multiple early steps in the meiotic recombination. Thus, new insights gained from our simultaneous analyses of multiple recombination factors in C. elegans are relevant for extending our understanding of mammalian reproduction.
Third, simultaneous immunolocalization of specific combinations of three recombination proteins (i.e. RAD-51 and RPA together with either BLM, MSH-5 or COSA-1) enables improved accounting of the total number of meiotic recombination events. By the end of early pachytene, after DSBs have been converted into repair intermediates over many hours, the total number of active repair sites detected peaks at 25–40 per nucleus before declining upon transition to late pachytene. Accordingly, we infer that each of the six bivalents receives an average of at least 4–7 breaks over the course of meiotic prophase. This estimate of DSB number is compatible with the average number of IR-induced DSBs (3.9 per bivalent) required to reliably restore COs in the absence of endogenous DSBs.
We propose that the aforementioned three-marker combinations enable visualization of virtually all ongoing meiotic recombination events in a given nucleus at a given time. Accordingly, we hypothesize that DSB-dependent intermediates generated during early prophase accumulate until the transition to late pachytene, but repair is not completed until after this transition. Further, as recombination markers disappear from NCO sites after this transition while being retained at CO sites, we infer that completion of NCO repair likely occurs earlier than completion of CO repair. Differential timing for completion of NCO and CO repair is reminiscent of the differential timing of appearance of NCO and CO products in budding yeast meiosis (Allers and Lichten, 2001). However, in contrast to budding yeast, where NCOs (but not COs) can be generated prior to a programmed transition to late pachytene, in C. elegans, completion of both NCO and CO events appears to be dependent on an analogous developmental transition.
Inferring the architecture of underlying recombination intermediates
SIM images of recombination proteins at DSB repair sites enable us to make inferences and hypotheses regarding the types of recombination intermediates that may be present, their relative timing of appearance and key transitions in the process.
First, our finding that RAD-51 foci occur as doublets indicates that following DSB formation, both DNA ends undergo resection and loading of strand-exchange proteins, paralleling the finding that strand-exchange proteins occupy both ends of a DSB during S. cerevisiae meiosis (Brown et al., 2015). Thus, we infer that dual end resection is a conserved feature of the meiotic program and that both DSB ends may have the potential to participate in homology search.
Second, we infer that resected DSBs rapidly give way to post-strand-exchange intermediates. Such intermediates accumulate throughout early prophase and are typically decorated by two populations each of BLM and RPA, frequently flanking a single population of MSH-5; however, RAD-51 is notably absent at most of these sites. Given the initial presence of RAD-51 on both DSB ends, its absence from most BLM/RPA-marked intermediates implies that the behavior of the two DSB ends must be coupled. This may involve temporally coupled invasion of the same DNA duplex by both ends and/or rapid second-end capture following initial invasion by one end. Alternatively, the two ends might be coordinated in a regulatory manner, e.g. strand invasion by the first end could be coupled to RAD-51 filament removal and replacement by RPA on the ssDNA tail of the second end, as has been proposed to occur during synthesis-dependent strand annealing (SDSA) mediated DSB repair in S. cerevisiae (Liu et al., 2017; Mitchel et al., 2013). Further, the fact that BLM doublets are more readily detected during early pachytene at sites also harboring MSH-5 supports a role for MutSγ in stabilizing post-strand-exchange intermediates.
Changes in organization of recombination intermediates following transition to late pachytene presumably reflect changes in the underlying structure of recombination intermediates. One intermediate per bivalent converts into a CO-specific intermediate, now marked by a MSH-5 doublet and lacking appreciable ssDNA, reflecting completion of repair DNA synthesis at those sites. The estimated lengths of the underlying DNA intermediates (800–900 bp) is compatible with the average lengths of CO-associated gene conversion tracts in budding yeast (1.6 – 2 kb) and mammals (500–600 bp) (Cole et al., 2014; Mancera et al., 2008; Martini et al., 2011). Moreover, the organization of the recombination factors present at these sites is consistent with the interpretation that these likely represent double Holliday junction (dHJ) recombination intermediates, which have been demonstrated to be CO-specific intermediates during yeast meiosis (Schwacha and Kleckner, 1995). Accordingly, we envision that the two population of MSH-5 represent two cohorts of MutSγ sliding clamp complexes (Snowden et al., 2004) that embrace and accumulate on the DNA duplexes that span the distance between the two junctions, and that the two populations of BLM localize near or at the junctions themselves (Jagut et al., 2016). Thus, the MutSγ complexes could serve as a roadblock inhibiting branch migration, thereby preventing conversion of such intermediates into NCO products via dHJ dissolution.
Structure of the SC-CR and roles in promoting the formation of meiotic COs
Our imaging of the SC-CR in relation to recombination sites, in combination with our analysis of recombination sites in syp mutants, provides new insight into the potential functions of the SC-CR in promoting the formation of meiotic COs.
First, as late prophase MSH-5/COSA-1 marked sites appear non-randomly distributed on now unpaired chromosome axes in syp mutants, we suggest that C. elegans chromosomes retain a capacity to communicate along and/or between each other to limit the number of “CO-like” sites even when the SC-CR is absent, as reported for CO site markers during yeast meiosis (Fung et al., 2004). However, syp mutants display significantly fewer “CO-like” sites than the six CO sites consistently observed in WT meiosis. Thus, we envision that the previously-described CO-limiting function of the SYP proteins (Hayashi et al., 2010; Libuda et al., 2013) is needed to counteract a strong impetus promoting CO designation and maturation that operates when the SYP proteins are present.
Second, our data suggest a model in which the SC-CR helps to create spatially-segregated compartments that could enable distinct biochemistry to yield different outcomes of repair at CO and NCO sites within the same nucleus. Upon transition to late pachytene, CO designated sites become encased by SC “bubbles” in which CO intermediates appear sandwiched between two layers of SC-CR proteins and embraced laterally by the chromosome axes. This organization effectively creates two distinct compartments, i.e., inside and outside the bubble. We propose that by segregating CO and NCO intermediates into spatially distinct compartments, the SC-CR can protect the CO-designated sites from NCO-promoting activities exerted at the same time at all other repair sites within the same cell (such as SDSA, dHJ dissolution and/or single HJ cleavage followed by branch migration). Conversely, the “inside” environment could favor local recruitment and retention of pro-CO factors that promote maturation of CO intermediates.
This idea that the SC-CR may help to prevent CO-designated intermediates from becoming dismantled prematurely is reinforced by our analysis of recombination site structures in mutants lacking SC-CR components. Some structures that bear resemblance to CO-designated sites in the WT do form in these mutants, but these sites lose BLM prematurely, concurrently with the disappearance of DNA repair markers from all other repair sites.
In order to form a CO from a dHJ precursor (see above) the two junctions must be consistently resolved in “opposite sense” (Szostak et al., 1983), but the basis of such biased resolution is unknown. One possibility is that pre-existing asymmetry in the DNA of the intermediate (e.g. nicks) could be used to direct asymmetric cutting by resolvases. Alternatively, architecture of the CO intermediate in the context of chromosome structure might provide cues needed for biased resolution. Our images revealing changes in CO-site architecture at diplotene, the (likely) time of CO resolution, may provide insight into this unresolved issue. Remnants of the SC-CR bubble are specifically retained only at one side of the CO site. CO sites concurrently undergo a change in organization, as BLM foci are no longer detected as doublets. One possible explanation for this change is that dHJ resolution may occur in two temporally distinct steps, with CO-biased resolution resulting from transient presence of SC-CR proteins in close proximity to only one of the two junctions. Another possible scenario is that large-scale reorganization of chromosomes at the diplotene stage enables a 3D reconfiguring of dHJ intermediates that results in the two HJs being brought into close spatial proximity, thereby enabling coupled resolution.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Anne M. Villeneuve (annev@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans culture conditions
For most figures, worms were grown on E. coli (OP50) seeded NG agar plates at 20°C according to the standard method (www.wormbook.org). For experiments involving strains carrying balancer chromosomes, homozygous mutant worms (lacking markers associated with the balancer chromosome) were selected as L4 larvae. For homozygous strains, worms were synchronized at the L1 larval stage by bleaching (www.wormbook.org). Worms were processed for cytological analysis (below) at 24–36 hours post L4 stage. For Figures S5A and S4D: AV962 (him-14(it44); him-6(jf93[him-6::HA] IV) or AV695 (gfp::cosa-1 II; mnT12 (X; IV), worms were grown at 25°C and analyzed 16 hours post L4. For experiments using the mIn1 inversion on chromosome II (Fig S1C), mnIn1/+ worms were generated by crossing VC1474 (top-2(ok1930) / mIn1[mls14 dpy-10(e128) II]) hermaphrodites with N2 males and were processed at 24 hours post L4.
C. elegans strains used in this study
N2 (wild type)
AV776: spo-11(me44) IV / nT1[qIs51 let-?] (IV;V)
AV596: cosa-1(tm3298) / qC1[qIs26] III
AV115: msh-5(me23) IV / nT1[unc-?(n754) let-?] (IV;V)
AV818: meIs8[unc-119(+); Ppie-1::gfp::cosa-1] II; cosa-1(tm3298) III
AV831: meIs8[unc-119(+); Ppie-1::gfp::cosa-1] II; cosa-1(tm3298) III; spo-11(me44) / nT1[qIs51 let-?] (IV;V)
CA258: zim-2(tm574) IV
AV307: syp-1(me17) V / nT1[unc-?(n754) let-?] (IV;V)
AV842: meIs8[unc-119(+); Ppie-1::gfp::cosa-1] II; cosa-1(tm3298) III; him-6(jf93[him-6::HA] IV
WS458: unc-119(ed3) III; opls 263[rpa-1p::rpa-1::YFP + unc-119]
AV687: syp-3(ok758) I / hT2[bli-4(e937) let-?(q782 qls48] (I;III); meIs8[unc-119(+); Ppie-1::gfp::cosa-1] II
CV2: syp-3(ok758) I / hT2[bli-4(e937) let-?(q782 qls48] (I;III)
VC1474: top-2(ok1930) / mIn1[mls14 dpy-10(e128)] II (for experiments described VC1476 was mated with N2 males and F1 worms heterozygous for mIn1 were analyzed)
UV116: him-6(jf93[him-6::HA] IV
AV959: him-6(jf93[him-6::HA] IV; syp-1(me17) V / nT1[unc-?(n754) let-?] (IV;V)
AV960: cosa-1(tm3298) / qC1[qIs26] III; him-6(jf93[him-6::HA] IV
AV961: zhp-3(me95) I / hT2[bli-4(e937) let-?(q782 qls48] (I;III); him-6(jf93[him-6::HA] IV
AV962: him-14(it44); him-6(jf93[him-6::HA] IV
AV963: him-8(me4) IV; syp-1(me17) V / nT1[unc-?(n754) let-?]
AV695: meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)] II; mnT12 (X; IV)
METHOD DETAILS
Cytological preparations
Spreading of C. elegans gonads was performed as in (Pattabiraman et al., 2017). The gonads of 20–100 adult worms were dissected in 5μl dissection solution (see below) on an ethanol-washed 22×40mm coverslip. 50μl of spreading solution (see below) was added and gonads were immediately distributed over the whole coverslip using a pipette tip. Coverslips were left to dry at room temperature (approximately 1 hour) and post-dried for two more hours at 37°C, washed for 2 0 minutes in methanol at −20°C and rehydrated by washing 3 times for 5 minutes in PBS-T. A 20-minute blocking in 1% w/v BSA in PBS-T at room temperature was followed by overnight incubation with primary antibodies at 4°C (antibodies diluted in: 1% w/v BS A in PBS-T supplied with 0.05% w/v NaN3). Coverslips were washed 3 times for 5 minutes in PBS-T before secondary antibody incubation for 2 hours at room temperature. After PBS-T washes, the nuclei were immersed in Vectashield (Vector) and the coverslip was mounted on a slide and sealed with nail polish. Dissection solution: For most experiments, gonads were dissected in 0.1% v/v Tween-20 in H20. For nearly all experiments in which the SC-CR was visualized, gonads were dissected in 85% v/v Hank’s Balanced Salt Solution (HBSS, Life Technology, 24020-117) with 0.1% v/v Tween-20; the exceptions were for images shown in Figures S1C and S3D, where gonads were dissected in H20 as above. Results similar to dissection in HBSS were obtained by dissecting gonads in increasing concentrations of PBS, Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma-Aldrich, D6546), or similar. Spreading solution: (for one coverslip, 50μl): 32μl of Fixative (4% w/v Paraformaldehyde and 3.2–3.6% w/v Sucrose in water), 16μl of Lipsol solution (1% v/v/Lipsol in water), 2μl of Sarcosyl solution (1% w/v of Sarcosyl in water).
Additional remarks: Conditions used for dissection should be chosen based on the goals of the experiment, considering the following issues: For visualization of recombination foci (or meiotic axes marked by HTP-3, HTP1-2 or HIM-3), spreading in low or no salt conditions works ideally in our hands. However, the lower the salt concentration, the fewer gonads stick to the slide, and while many individual well-spread nuclei are obtained under low salt conditions, it is more difficult to obtain whole gonads or large gonad fragments. To increase the fraction of gonads retained, we separate the gonads (behind the spermatheca) from the worm carcasses to release free-floating gonads, which adhere better to the cover slip upon spreading. We also find that the lower the salt concentration, the more the chromosomes are spread apart and the more the SC-CR is washed out. Furthermore, early stages spread more efficiently than late stages in the gonad under all conditions, with mitotic nuclei being the easiest to spread and the oocytes being the most recalcitrant (little or no spreading).
Antibodies used in this study
The following primary antibodies were used at the indicated dilutions: Chicken anti-HTP-3 (1:500) (MacQueen et al., 2005), guinea pig anti-HTP-3 (1:500) (MacQueen et al., 2005), rabbit anti-MSH-5 (1:10000) (SDI/Novus), chicken anti-GFP (1:2000) (Abcam), mouse anti-GFP (1:500) (Roche), rabbit anti-GFP (1:500) (Yokoo et al., 2012), mouse anti-HA (1:1000) (Covance), guinea pig anti-ZHP-3 (1:500) (Bhalla et al., 2008), rabbit anti-PLK-2 (Nishi et al., 2008), guinea pig anti-SYP-1 (1:200) (MacQueen et al., 2002), rat anti-RAD-51 (1:200) (Rosu et al., 2013), rabbit anti-RPA-1 (1:500) (Lee et al., 2010), mouse anti-H3K4me2 (1:2000) (Merck), rabbit anti-SYP-1 (1:2000) (MacQueen et al., 2002) and rabbit anti-SYP-2 (1:200) (Colaiacovo et al., 2003).
Secondary antibodies conjugated to Alexa dyes 405, 488, 555 or 647, obtained from Molecular Probes, were used at 1:500 dilution. In cases where antibodies raised in mouse and guinea pig were used on the same sample, we used highly cross-absorbed goat anti-mouse secondary antibodies, obtained from Biotum (conjugated to CF488, or CF555 respectively) for secondary detection of the mouse primary antibody in order to avoid cross-reaction against antibodies raised in guinea pig.
Imaging
Imaging, Deconvolution and 3D-SIM reconstruction were performed as in (Pattabiraman et al., 2017). Spreading results in squashing of C. elegans germline nuclei from 5 to 1–2 μm in thickness. Wide field (WF) images were obtained as 200 nm spaced Z-stacks, using a 100× NA 1.40 objective on a DeltaVison OMX Blaze microscopy system, deconvolved and corrected for registration using SoftWoRx. Subsequently, gonads were assembled using the “Grid/Collection” plugin (Preibisch et al., 2009) in ImageJ. For display, assembled gonads were projected using maximum intensity projection in ImageJ. 3D-Structured Illumination microscopy images were obtained as 125 nm spaced Z-stacks, using a 100× NA 1.40 objective on a DeltaVison OMX Blaze microscopy system, 3D-reconstructed and corrected for registration using SoftWoRx. For display, images were projected using maximum intensity projection in ImageJ or SoftWoRx. For display in figures, contrast and brightness were adjusted in individual color channels using ImageJ. For Figure 4B and Figure S3C, SCs were computationally straightened using ImageJ 2D-straightening tool on well-spread chromosomes (250–1250 nm maximum Z-stack thickness).
QUANTIFICATION AND STATISTICAL ANALYSIS
Definition and quantification of recombination foci in wide-field images (related to Figure 1A–C, S2A, S2C, 5A and 5D)
Recombination foci were counted manually on deconvolved, stitched and projected 32-bit images. For these quantifications, we considered a signal to be a recombination focus when it co-localized with HTP-3 signals. Furthermore, to be counted as recombination focus, a signal must be at least 5 times brighter than background for the imaged nucleus (offside HTP-3 axes). To estimate “brightness” of recombination foci, we measured signal intensities for a 9-by-9 pixel (80.1 × 80.1 nm/pixel in wide-field raw data) area (which equals the maximum expansion in X and Y of a recombination focus signal)) encompassing each of 20 recombination sites per meiotic stage (ep, ep-to-transition, lp and diplo) per gonad and compared it to background measurements (on and off HTP-3 signals). Both, the maximum and mean brightness values were considered and both met the above-mentioned criterion.
To quantitatively display the appearance, disappearance and co-localization of multiple types of recombination foci in a single representative spread gonad (Figure 1A–C), all nuclei within the assayed portion of the gonad that did not overlap with other nuclei were considered (usually 6–12 nuclei per cell row); the total number of nuclei scored for Figure 1A was 448. For Figure 1B, an additional 6 rows of late pachytene nuclei were included (containing 4–8 nuclei each; in total 478 nuclei were scored). For the early-to late pachytene transition region depicted in Figure 1C (same gonad as in Figure S2A) all nuclei were considered (n = 91). Meiotic entry was defined by the appearance of HTP-3 as continuous tracks along chromosomes in the majority of nuclei in the cell row. Transition of stages was defined by the appearance of the defining event in the majority of nuclei in the cell row.
Reproducibility of maximum numbers of recombination foci / nucleus (related to Figure 1 and Figure S2C)
To assess reproducibility in quantitation of MSH-5, BLM and RPA foci, we quantified maximum numbers of MSH-5 and BLM foci in three gonads (Gonads 1–3 in Figure S2C) and maximum numbers of MSH-5 and RPA-1 foci in another three gonads (Gonads 4–6 Figure S2C). For each gonad, foci were counted in 20 non-overlapping nuclei in the 2–3 rows prior to the first row in which a late pachytene nucleus was observed. The error bars in Figure 2C represent standard deviation; one-way ANOVA did not detect significant differences among the means for MSH-5 foci (6 gonads), for BLM foci (3 gonads), or RPA foci (3 gonads). However, numbers of both BLM and RPA foci exceeded numbers of MSH-5 foci (Figure S2C).
Definition of early and late COSA-1 foci (related to Figure 1C–E, S2A and S2B)
Categorization of GFP::COSA-1 foci (detected by immunofluorescence) as early vs. late was validated by measuring peak fluorescence intensities for 20 9-by-9 pixel areas, each encompassing a single COSA-1 focus scored as “early” or “late”, for each of the stages indicated in Figure S2C and for 20 control background areas, using 32-bit projected images of deconvolved and stitched gonads. “Early” GFP::COSA-1 foci scored in early pachytene nuclei just prior to the early-to-late pachytene transition region (average brightness value 60,431; SD=17,096) were 16 times brighter on average than background areas (3,653; SD=1,623). Further, “late” GFP::COSA-1 foci in late pachytene nuclei located in the early-to-late pachytene transition region (196,683; SD=33,552) were 3 times brighter than early foci, and in the late pachytene region, the COSA-1::GFP signal intensity further increased (285,446; SD=19,910) and plateaued until diplotene (274,678; SD=16,085). Moreover, the ranges of intensities for early and late foci did not overlap (Figure S2), indicating that early and late COSA-1 foci can be distinguished as separate classes.
Measurement/estimation of distances between meiotic chromosome axes and paired recombination foci (related to figure S3A and S3B)
The centroids of recombination foci, or axial signals, respectively were determined as the brightest pixel(s) within the signal (in SIM data, one pixel in XY represents 40 × 40 nm). As chromosome pairs twist around their longitudinal axes, the fullest extent of separation between individual HTP-3 signals (in the XY plane) is observed when the SC is visualized in a fully frontal aspect. Therefore, to determine the separation between HTP-3-marked chromosome axes, we measured the distances at 7–9 different sites along a pair of aligned homologous chromosomes in segments that displayed separation of HTP-3 axis signals, with the three highest values being recorded. The distances between the centroids of paired recombination foci (MSH-5-MSH-5 or BLM-BLM) were measured (in nuclei spread using hypotonic conditions) at sites where parallel axes were visualized in single XY plane in a fully frontal view, as indicated by at least 190 nm of axial separation.
Measurement of the angle of BLM doublets with respect to the chromosome axes (related to Figure S3A and S3D)
18 CO-designated repair sites were selected based on the following criteria: 1) the SC was visualized in frontal aspect at the CO site, and 2) all signal maxima analyzed were contained within a depth of ≤ 2 Z planes (total: 250 nm). The centroids of each of the two BLM foci at each selected CO site were connected by a straight line, and the angle of this line relative to each of the flanking chromosome axes at the CO site was assessed as depicted in Figure S3D. No measurement exceeded an angle of 23°, and for 80% of measurements, the angle did not exceed 12°. For Figure S3D, the two measurements taken at each site were averaged; the values for all investigated sites are all depicted in the Figure.
For estimation of DNA length underlying late recombination structures
We used the Pythagorean theorem to estimate the length of the DNA underlying late pachytene cohorts of recombination factors based on the mean measured MSH-5-MSH-5 (m = 168) and BLM-BLM (b =240) distances, using the formula: d = 2[ (m/2)2 + (b/2)2 ]1/2, yielding a value of 293 nm. As a dHJ or similar recombination intermediate of this length is expected to be composed primarily of B-form DNA, and as 1 kb of B-form DNA has a length of 340 nm, we estimate that the average length of DNA underlying late pachytene CO-site structures is on the order of 800–900 bp.
Quantification of foci classes for SIM images of recombination sites (related to Figures 2, 3 and S3)
For each recombination protein analyzed, the occurrence of doublets, elongated foci and/or singlets was quantified in 20 nuclei for early pachytene,25 nuclei for late pachytene, and 20 nuclei for diplotene. Foci were scored in 3D image stacks of spread nuclei from the centers of two SIM fields. Doublets were identified based on close proximity of signals with two resolvable maxima in signal intensity (no further apart than 500 nm). Total numbers of foci analyzed: BLM (early pachytene): 506; RPA (early pachytene): 509; RAD-51 (early pachytene): 112; MSH-5 (late pachytene): 150; BLM (late pachytene): 150; COSA-1 (late pachytene): 150; BLM (diplotene): 113 and MSH-5 (diplotene): 120.
Quantification of SC-CR bubbles (related to Figure 4, S4)
SC-CR bubbles were analyzed in nuclei prepared by spreading in 85% Hanks + 0.1%Tween, using SYP-1 as a marker for the SC-CR. All nuclei in one late pachytene SIM field (n = 35 nuclei, with 210 total COSA-1/MSH-5 marked CO sites) and 20 early pachytene nuclei from 2 SIM fields (with a total 454 MSH-5 marked recombination sites) were analyzed.. SIM images were first screened in 3D-SIM reconstructed image stacks using only the SYP-1 cannel to identify bubble-like structures in an unbiased manner. 111 bubble-like structures were observed in 35 late pachytene nuclei containin (average of 3.2 per nucleus), and 89% of these corresponded to CO sites. In contrast, only 5 possible bubble-like structures were detected in 20 early pachytene nuclei, and none of these corresponded to sites of recombination foci (despite MSH-5 foci in these early pachytene nuclei being 3.8-fold more numerous than CO sites in late pachytene nuclei).
Quantification of maximum numbers of recombination foci / nucleus in recombination and SC-CR defective mutants (related to Figure 5A)
Numbers of nuclei scored were: WT (n=34), syp-1 (n=23), cosa-1 (n=20) and zhp-3 (n=20). For all three mutants, total numbers of recombination intermediates detected (RPA-1 and RAD-51 foci combined) were significantly higher in the mutants than in WT (p < 0.0001; two-tailed student’s t-tests).
GFP::COSA-1/MSH-5 at late recombination sites in SC-CR mutants (related to Figure 5D)
Quantification of MSH-5/COSA-1 foci in syp-3; gfp::cosa (syp-3) and gfp::cosa-1(WT). For WT, average = 6 (n=35); for syp-3, average = 4.2 (n=45,). All data points are represented in the stacked bar graph. A two-tailed Mann-Whitney test indicated a highly significant difference (p < 0.0001) between WT and syp-3. We further showed that these foci were non-randomly distributed among the 12 chromosome axes in the syp-3 mutant by using a Chi-square test to compare the observed distribution of axes with 0, 1 or ≥2 foci to that expected based on the Poisson distribution (p <0.0001).
Statistical analyses
Statistical analyses including one-way ANOVA, t-tests, Mann-Whitney tests, Chi-square and Fisher-exact tests (as indicated above and in results and figure legends) were conducted using GraphPad Prism software.
Supplementary Material
Recombination proteins adopt distinct spatial architecture at meiotic crossover sites
Recombination site architecture undergoes dynamic changes during meiotic progression
The Synaptonemal complex envelops crossover-designated recombination intermediates
Crossover and non-crossover repair are completed in spatially distinct compartments
Acknowledgments
We thank the Caenorhabditis Genetics Center, M. Colaiacovo, A. Dernburg, A. Gartner, Y. Kim, D. Libuda, and R. Lin for strains and antibodies. We are grateful to V. Jantsch for allowing usage of the him-6::HA transgene prior to publication, to F. Klein and J. Loidl for sharing reagents and experience in cytological preparations, to J. Mulholland for technical assistance and to A. MacQueen for inspiration. We thank C. Akerib, C. Girard, V. Jantsch, and B. Roelens for comments on the manuscript. This work was supported by the FWF Erwin Schrödinger Fellowship (J-3676), NIH grants R01GM53804, R01GM67268, and U54 HD068158, American Cancer Society Research Professor Award RP-15-209-01-DDC and grant 1S10OD01227601 from the NCRR.
Footnotes
Author contributions:
A.W. designed and conducted the experiments, A.W. and A.M.V. analyzed the data and wrote the manuscript.
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS genetics. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Grubb J, Zhang A, Rust MJ, Bishop DK. Small Rad51 and Dmc1 Complexes Often Co-occupy Both Ends of a Meiotic DNA Double Strand Break. PLoS genetics. 2015;11:e1005653. doi: 10.1371/journal.pgen.1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon CK, Yu Z, Wang Y, Guo F, Unruh JR, Slaughter BD, Hawley RS. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E6857–E6866. doi: 10.1073/pnas.1705623114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Cole F, Baudat F, Grey C, Keeney S, de Massy B, Jasin M. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nature genetics. 2014;46:1072–1080. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Developmental cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Developmental cell. 2011;21:934–947. doi: 10.1016/j.devcel.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T, Sekelsky J. Bloom syndrome helicase in meiosis: Pro-crossover functions of an anti-crossover protein. BioEssays: news and reviews in molecular, cellular and developmental biology. 2017;39 doi: 10.1002/bies.201700073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Mlynarczyk-Evans S, Villeneuve AM. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics. 2010;186:45–58. doi: 10.1534/genetics.110.115501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ, Jantsch V, Martinez-Perez E, Yanowitz JL. Meiosis. WormBook. 2017:1–43. doi: 10.1895/wormbook.1.178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagut M, Hamminger P, Woglar A, Millonigg S, Paulin L, Mikl M, Dello Stritto MR, Tang L, Habacher C, Tam A, et al. Separable Roles for a Caenorhabditis elegans RMI1 Homolog in Promoting and Antagonizing Meiotic Crossovers Ensure Faithful Chromosome Inheritance. PLoS biology. 2016;14:e1002412. doi: 10.1371/journal.pbio.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Molecular and cellular biology. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annual Review of Genetics. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Wojcik M, Xu K, Dernburg AF. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4734–E4743. doi: 10.1073/pnas.1702312114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Developmental cell. 2011;21:948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Gartner A, Hyun M, Ahn B, Koo HS. The Caenorhabditis elegans Werner syndrome protein functions upstream of ATR and ATM in response to DNA replication inhibition and double-strand DNA breaks. PLoS genetics. 2010;6:e1000801. doi: 10.1371/journal.pgen.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 2013;502:703–706. doi: 10.1038/nature12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ede C, Wright WD, Gore SK, Jenkins SS, Freudenthal BD, Todd Washington M, Veaute X, Heyer WD. Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase delta-extending D-loops. eLife. 2017;6 doi: 10.7554/eLife.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- Machovina TS, Mainpal R, Daryabeigi A, McGovern O, Paouneskou D, Labella S, Zetka M, Jantsch V, Yanowitz JL. A Surveillance System Ensures Crossover Formation in C. elegans. Current biology: CB. 2016;26:2873–2884. doi: 10.1016/j.cub.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & development. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst) 2016 doi: 10.1016/j.dnarep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS genetics. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Mezard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annual review of plant biology. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS genetics. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. Journal of cell science. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- Moses MJ. Chromosomal structures in crayfish spermatocytes. J Biophys Biochem Cytol. 1956;2:215–218. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Villeneuve AM, Colaiacovo MP. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. The Journal of cell biology. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Lambert TJ, Altendorfer E, Gao J, Blower MD, Waters JC, Colaiacovo MP. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife. 2017;6 doi: 10.7554/eLife.23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y, Rogers E, Robertson SM, Lin R. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development. 2008;135:687–697. doi: 10.1242/dev.013425. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pattabiraman D, Roelens B, Woglar A, Villeneuve AM. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS genetics. 2017;13:e1006670. doi: 10.1371/journal.pgen.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Dernburg AF. A Family of Zinc-Finger Proteins Is Required for Chromosome-Specific Pairing and Synapsis during Meiosis in C. elegans. Developmental cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelens B, Schvarzstein M, Villeneuve AM. Manipulation of Karyotype in Caenorhabditis elegans Reveals Multiple Inputs Driving Pairwise Chromosome Synapsis During Meiosis. Genetics. 2015;189:1363–1379. doi: 10.1534/genetics.115.182279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Kohler S, Dernburg AF. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. eLife. 2017;6 doi: 10.7554/eLife.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–1289. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS genetics. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prufert K, Saito TT, Smolikov S, Gu Y, Hincapie M, Hill DE, Vidal M, McDonald K, Colaiacovo MP. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics. 2011;189:411–421. doi: 10.1534/genetics.111.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schucker K, Holm T, Franke C, Sauer M, Benavente R. Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2029–2033. doi: 10.1073/pnas.1414814112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Smolikov S, Eizinger A, Schild-Prufert K, Hurlburt A, McDonald K, Engebrecht J, Villeneuve AM, Colaiacovo MP. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Molecular cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Stergiou L, Eberhard R, Doukoumetzidis K, Hengartner MO. NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell death and differentiation. 2011;18:897–906. doi: 10.1038/cdd.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook; 2006. www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, MacQueen AJ, Vader G, Shinohara M, Sanchez A, Borde V, Shinohara A, Hochwagen A. Chromosome Synapsis Alleviates Mek1-Dependent Suppression of Meiotic DNA Repair. PLoS biology. 2016;14:e1002369. doi: 10.1371/journal.pbio.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Walpita D, Plug AW, Neff NF, German J, Ashley T. Bloom’s syndrome protein, BLM, colocalizes with replication protein A in meiotic prophase nuclei of mammalian spermatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5622–5627. doi: 10.1073/pnas.96.10.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS genetics. 2013;9:e1003335. doi: 10.1371/journal.pgen.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Seminars in cell & developmental biology. 2016;54:135–148. doi: 10.1016/j.semcdb.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.