Abstract
At the nexus of specialized cellular responses are localized enrichments of protein activity. The localization of messenger RNA (mRNA) coupled with translational control often plays a crucial role in the generation of protein concentrations at defined subcellular domains. Although mRNA localization is classically associated with large specialized cells, such as neurons and embryos, RNA localization is a highly conserved paradigm of post-transcriptional regulation observed in diverse cellular contexts. Functions of localized mRNAs extend far beyond the well-studied examples of neuronal polarization and developmental patterning. Since the initial discovery of the intracellular localization of cytoskeletal mRNAs within migrating cells, hundreds of mRNAs are now known to be enriched at specific organelles where they contribute to cell function. In this short review, we discuss basic principles regulating RNA localization and consider the contribution of localized mRNA to several essential cellular behaviors. We consider RNA localization as a mechanism with widespread implications for cellular function.
Keywords: RNA localization, translational control, focal adhesions, mitochondria, endoplasmic reticulum, centrosomes, RNA-binding proteins
Introduction
The spatial distribution of messenger RNAs (mRNAs) contributes to the compartmentalized organization and function of intracellular processes. While numerous proteins are targeted to distinct subcellular domains, the localization of some proteins requires the prior localization of their cognate mRNAs. Since RNA localization is often coupled to translational control, the localization of a given mRNA often corresponds to the site of enrichment for the corresponding protein. Understanding which RNAs localize within cells, the mechanisms used for localization, the translational status of such RNAs, and the functional significance for each targeted transcript remain active areas of investigation. Indeed, RNA localization is a conserved paradigm observed in bacteria, fungi, plants, and animal cells.
Why do some RNAs localize, while others do not? In general, RNA localization functions to generate enrichments of specific proteins at subcellular domains. In cases where particular proteins are required at defined domains (or in cases where such proteins would be deleterious elsewhere), RNA localization coupled to translational control efficiently enriches protein only where it is needed1. Instead of localizing individual protein molecules, each mRNA may serve as a template for repeated rounds of translation when and where the protein functions. Thus, RNA localization effectively generates cellular domains of gene activity.
Generally speaking, RNAs can become enriched at subcellular compartments by a variety of mechanisms: active transport, diffusion and anchoring, and local protection from degradation (reviewed by St. Johnston1). Studies from several model transcripts have uncovered general principles that dictate how mRNAs become localized within cells. Conserved cis-acting sequences, usually within the mRNA 3′-untranslated region (3′UTR), are recognized by trans-acting RNA-binding proteins (RBPs) to regulate mRNA transport, translation, stability, and degradation2. Each RBP can interact with few or many target mRNAs on the basis of primary sequence and/or secondary structure. Most localized mRNAs interact with multiple RBPs throughout their lifetime, and the localization of multiple mRNAs to the same subcellular domain is often regulated by the same RBPs2.
One key function of RBPs is to coordinate the active transport of mRNAs to subcellular targets via molecular motors that move along cytoskeletal networks (reviewed by Gagnon and Mowry3 and Bullock4). All three families of molecular motors – myosins, kinesins, and dynein – are known to transport mRNAs3. Individual motors are responsible for RNA localization to defined domains, such as the role for kinesins in regulating transport of mRNAs to axonal compartments in neurons5. Localization specificity is typically conferred by molecular motors in association with RBPs and adaptor proteins. For example, the localization of several mRNAs with divergent localization motifs (e.g., gurken and hairy mRNAs) to the minus-ends of microtubules requires the association of the dynein-Bicaudal-D (BicD) complex to the RBP Egalitarian (Egl)6. To maintain a steady state of concentrated mRNA at a subcellular domain, localized RNAs must be either anchored in place or undergo continuous active transport. Adding further complexity, patterns of RNA localization are often dynamic and subject to reorganization in response to cellular signals or developmental cues.
Of the various models used to investigate RNA localization, the best understood are large, highly polarized cells, such as oocytes, embryos, and neurons, where subcellular enrichments of RNAs are readily observed1. Subcellular RNA localization is well known to play an important role within the extended projections of neurons (Fig. 1A) and in embryonic patterning. Numerous excellent reviews detail the extensive work characterizing the mechanism and function of RNA localization within these systems.
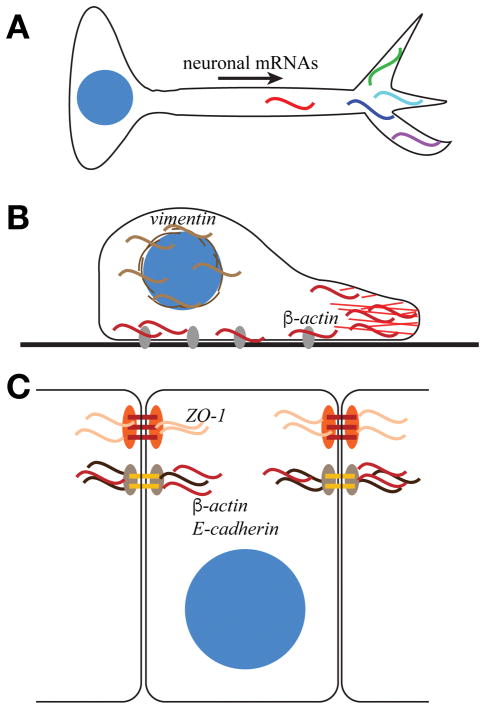
Figure 1. RNA localization within polarized cells.
(A) Multiple mRNAs (colored sinusoidal lines) localize to neuronal processes and contribute to neuronal polarization and function. (B) In migrating fibroblasts, β-actin mRNA (red) localizes to the leading edge of cells and to FA plaques (grey ovals), while vimentin mRNA (brown) is enriched at the perinuclear domain. β-actin localization and translation at focal adhesions contributes to the regulation of adhesion and migration. (C) In epithelial cells, β-actin and E-cadherin mRNAs localize to adherens junctions, whereas zonula occludens-1 mRNA (ZO-1) localizes to tight junctions to regulate cell adhesion.
Recent technological advances leveraging high-throughput imaging7, next-generation sequencing modalities, such as RNA sequencing8, as well as advances in RNA detection and microscopy (e.g., single molecule fluorescence in situ hybridization (smFISH)9) continue to uncover intriguing subcellular patterns of RNA localization in diverse cell types. For example, a high-throughput screen in Drosophila early embryos revealed that more than 70% of transcripts were localized in discrete subcellular patterns, such as the cell division machinery, cell junctions, nuclear or perinuclear domains, and apical or basal membranes7. This work by Lécuyer and colleagues demonstrated the prevalence of localized RNA as a regulatory mechanism and spurred investigation of these mechanisms in other cell types.
In this short review, we will highlight examples of localized RNA participating in diverse pathways with acute cellular responses: cell migration and adhesion, intracellular membrane trafficking, and cell division. Our goal is to revise the conception of RNA localization from a process unique to specialized or polarized cell types to a widespread mechanism with far-reaching consequences. For readers well versed in the RNA field, the recurring importance of RNA localization in basic cellular function may not come as a surprise. For the more general audience, however, the diversity and specificity of RNA localization patterns and functions are noteworthy and ever expanding, encompassing a multitude of cellular processes.
The Contribution of RNA Localization to Acute Cellular Responses
RNA localization regulates cellular adhesion
The first example of localized RNA was the identification that actin mRNA partitions to specific cytoplasmic domains within ascidian embryos and concentrates within mesodermal cell lineages10. This pioneering work in an embryonic system paved the way for the discovery that cytoskeletal mRNAs are localized in migratory fibroblasts where, in contrast to bulk poly(A) mRNA, β-actin is notably enriched at the leading edge and vimentin is found at a perinuclear domain (Fig. 1B)11. The recognition that mRNAs are localized to subcellular domains was a major breakthrough in cell biology. Specific RNAs showed unique patterns of localization, the prediction being that such RNA enrichments may contribute to protein enrichments, thereby influencing cellular morphology, polarity, and behavior.
In addition to its enrichment in cytoplasmic domains of ascidian embryos and at the leading edge of migrating fibroblasts, β-actin mRNA is also enriched at focal adhesions (FAs) (Fig. 1B)12. Work by Singer and colleagues confirmed that localized β-actin regulates the adhesive properties of migrating fibroblasts and epithelial cells (Fig. 1B and 1C). Moreover, β-actin displays dynamic patterns of processivity that contribute to the adhesive behaviors of the cell. Recent work using advanced single molecule tracking shows particles of β-actin transiently move more slowly and even become stationary within the vicinity of FAs13. Pharmacological studies and live imaging of ribosomes indicate that the increased residence time and reduced motility of β-actin at FAs is modulated by its association with the translation machinery12,13. These data are consistent with the ideas that mRNAs associated with ribosomes exhibit reduced motility and β-actin is locally translated at FAs. The RBP Zipcode-binding protein 1 (ZBP1) is required for β-actin localization to FAs12. This localization contributes to cell motility and adhesion, as ZBP1-knockout cells have reduced FA stability12. In addition, artificially tethering β-actin to FAs via Vinculin produces larger FA plaques, longer adhesion lifetimes, more extensive actin stress fibers, and a corresponding reduction in cell migration velocity12. These studies indicate that the regulation of cellular adhesion and motility is modulated, in part, through the regulation of RNA localization and translational control at FAs.
In epithelial cells, which must form dynamic adhesive contacts, localized β-actin is required for assembly of adherens junctions (Fig. 1C). Recent work shows the disruption of β-actin localization in confluent MDCK cells prevents the assembly of adherens junctions but does not affect established adhesive contacts14. These data suggest the localization of β-actin to nascent junctions represents an early step in adherens junction assembly. It is likely that local synthesis of β-actin monomers contributes to the polymerization or remodeling of the actin cytoskeleton.
These examples in fibroblasts and epithelial cells support a role for regulation of cell adhesion by localized β-actin mRNA. Increasing evidence demonstrates β-actin is not the only localized mRNA that regulates cell adhesion. Knockdown of ZBP1, for example, results in the diminished localization of E-cadherin mRNA and protein at adherens junctions, effectively increasing the invasive properties of MDA231 breast cancer cells15. A recent unbiased screen identified more than 500 mRNAs that associate with an adherens junction protein, PLEKHA7, including components of signaling pathways known to locally regulate adherens junctions16. These findings suggest that localization of mRNAs to adherens junctions contributes to the dynamic regulation of adhesion and proliferation.
Finally, localized mRNAs contribute to epithelial cell adhesion beyond adherens junctions. For example, localization of zonula occludens (ZO-1) mRNA to the apical surface of epithelial cells is required for tight junction assembly and epithelial cell polarity (Fig. 1C)17. These studies are consistent with a role for RNA localization and local protein synthesis in regulating diverse aspects of cellular adhesion and motility with broad implications for developmental and disease processes.
RNA localization to intracellular membranes contributes to organelle specialization and the host-pathogen response
Extensive analyses of mRNA localization patterns in yeast reveal a propensity for many mRNAs to associate with membrane-bound organelles. For example, the mitochondrion is a depot for numerous mRNAs. The majority of mitochondrial proteins are encoded by nuclear genes, which must be transported to mitochondria for proper function. The canonical pathway for mitochondrial import is cytosolic translation of nuclear-encoded mitochondrial proteins followed by post-translational translocation through the outer or inner membranes (reviewed by Backes and Herrmann18). An alternative pathway relies upon the localization of mitochondrial mRNAs to the mitochondrial membrane or vicinity prior to translation and import. For example, the ATP2 mRNA, which encodes the β subunit of F1-ATPase, localizes to mitochondria in yeast and rat hepatocytes, where it is required for proper respiratory function19–21. A 50 nucleotide consensus motif in the 3′UTR of ATP2 is necessary and sufficient for this sorting process22. Mitochondrial localization of mRNA is not unique to ATP2. An estimated 50% of nuclear-encoded mitochondrial mRNAs associate with cytoplasmic ribosomes that biochemically co-purify with mitochondria23, suggesting that mRNA localization to mitochondria contributes to the localization of a large proportion of mitochondrial proteins. The localization of mitochondrial RNAs to the mitochondrial outer membrane raises the question of co-translational import. Indeed, cytoplasmic ribosomes have long been detected in biochemical preparations of mitochondria, but were recently visualized directly at the mitochondrial surface using cryo-EM coupled to sensitive direct electron detectors24. These data demonstrate RNA localization precedes mitochondrial import for a significant subset of mitochondrial proteins. The conserved localization of mRNAs to mitochondria suggests this is an ancient paradigm that contributes to mitochondrial function25.
Another membrane-bound organelle enriched in localized mRNAs is the endoplasmic reticulum (ER). The ER is the major site of biosynthesis of integral membrane and secreted proteins. Translation of mRNAs encoding these proteins is initiated in the cytosol and paused after translation of a signal sequence (SS) or transmembrane domain (TMD). The signal recognition peptide binds the SS or TMD and mediates transport of the ribosome-mRNA-nascent polypeptide complex to the ER where the nascent polypeptide is then translocated through ER membrane and translation continues26. Recent data supports a second targeting pathway to the ER that enriches mRNAs for cytosolic proteins at this organelle (for a review, see Cui and Palazzo26). Ribosome profiling from biochemical fractionations of the ER reveal that many cytosolic mRNAs are bound by ER-associated ribosomes and that ribosome density is higher in ER fractionations than the cytosol, suggesting increased translational efficiency at the ER27. In mammalian cells, the ER membrane protein p180 contains a lysine-rich cytoplasmic domain that binds multiple mRNAs and is required for their localization to the ER membrane28. While p180 is only present in metazoans, overexpression of p180 in yeast is sufficient to increase the ER membrane association and stability of mRNAs, resulting in ER proliferation, suggesting the mechanism by which p180 binds and stabilizes mRNAs at the ER is functionally conserved29. Since p180 binds multiple mRNAs non-specifically, other ER-localized RBPs likely participate in this pathway to confer specificity to the process of targeting mRNAs to defined ER subdomains.
Multiple, non-exclusive functions have been described for these ER-localized mRNAs. In budding yeast, the coordinated movement of mRNA encoding the cytoplasmic cell-fate determinant ASH1 with ER membranes is required for proper segregation of ASH1 to the yeast bud tip, although a biochemical association of ASH1 with ER membranes has not been demonstrated30. This coordinated movement of ASH1 mRNA and ER membranes requires the type V myosin motor Myo4P, although this motor is not required for the association of ASH1 with ER membranes30. During the stress response, many cytoplasmic transcripts are sequestered into stress granules, which are translationally silent31. ER-associated mRNAs encoding cytosolic proteins often avoid sequestration into stress granules and thereby escape translational inhibition32. These examples demonstrate that regulated RNA localization to the ER is conserved in eukaryotes, while the functions of these localized RNAs vary. Some RNAs partition with ER as cargo during cell division; in other cases, RNA localization to the ER influences translation status.
The demonstration of RNA localization to mitochondria and the ER raises the question if mRNAs are a component of other membrane-bound organelles. The ability of RNA viruses to co-opt intracellular membranes for replication demonstrates that RNA can localize to and even remodel diverse membranes within mammalian cells (for a recent review, see Altan-Bonnet33). For example, HIV-1 has two viral replication pathways: a direct pathway via the plasma membrane and an indirect pathway via endosomes34. Inhibition of endosomal trafficking disrupts vesicular budding and increases levels of HIV-1 RNA at endosomes, demonstrating that this virus co-opts components of the host membrane trafficking machinery35. Additionally, negative-sense RNA viruses in the Paramyxoviridae and Orthomyxoviridae familes, which include the parainfluenza, measles, mumps, and influenza A viruses, synthesize viral ribonucleotide protein (vRNP) complexes that are transported via Rab11 endosomes36. In the case of influenza A virus, the viral polymerase subunit PB2 is required for attachment to Rab11-positive endosomes, but the exact mechanism of vRNP attachment to these membranes is unknown37. Recent work also highlights the importance of viral association with vesicles as an efficient means of egress38. These studies reveal a strong propensity for diverse viruses to usurp the host membrane machinery to support various stages of the viral lifecycle. This work underscores the importance of elucidating how viral RNA genomes are recruited and anchored to membrane sites as a means to modulate infectivity and transmission.
Finally, RNA localization to endosomal membranes has also been demonstrated in the pathogenic fungus Ustilago maydis, which undergoes a morphologic switch to hyphal growth as a prerequisite to allow infection of corn39. During hyphal development, the RBP Rrm4 functions to transport ribosomes and more than 900 mRNAs to hyphal tips on endosomes40,41. These data lead to the model that the hyphal transport of endosomes provides a platform for the efficient delivery of large numbers of mRNAs and ribosomes to support the localized translation of proteins required for fungal development and infectivity.
Together, these examples demonstrate that RNA localization to intracellular membranes regulates crucial cellular functions, including mitochondrial respiration, control of translational responses to stress, host-pathogen interactions, and development. Future work will undoubtedly uncover new roles for RNA localization in the composition and function of membrane-bound organelles.
Localized RNA at the centrosome
Intriguingly, non-membrane bound organelles, such as the centrosome, are another site of RNA localization. The centrosome is the major microtubule organizing center of most animal cells and directs the formation of the mitotic spindle in dividing cells42. While the centrosome is often discussed as a proteinaceous organelle, there are longstanding hints that RNA may contribute to its biology (see Marshall and Rosenbaum43 and Alliegro44). There are multiple, non-exclusive hypotheses for the function of RNA at centrosomes (Fig. 2). RNA may regulate centrosome biology, such as assembly or biogenesis, through localized regulation of translation or through a structural role. In addition, the asymmetric distribution of RNAs at centrosomes may contribute to asymmetric cell division or cellular specializations42. Future studies are required to test these hypotheses.
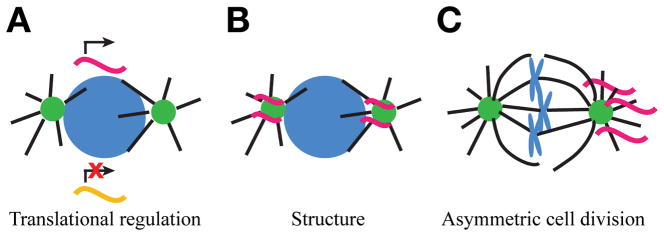
Figure 2. Proposed roles of mRNA localized to centrosomes.
While the exact function of mRNA localized to centrosomes remains unknown, several models have been proposed. Note that these models of RNA function at centrosomes are non-exclusive. (A) RNA localization to centrosomes may regulate centrosome biology through translational regulation. Centrosome localization may represent a translationally permissive or inhibitory environment. Alternatively, RNA localization may sequester transcripts away from the cytosolic translational machinery. (B) RNA localization may contribute to centrosome structure, such as through the formation of a phase separated condensate. (C) The asymmetric distribution of centrosome-localized RNAs may regulate asymmetric cell divisions.
In the mollusk Ilyanassa obsoleta, the asymmetric segregation of mRNAs on centrosomes results in the nonrandom inheritance of patterning mRNAs, such as Toll, raising the possibility that these biased patterns of inheritance contribute to embryonic patterning45. Consistent with this model, centrosomes are known to partition mRNA as cargo during cell division and contribute, for example, to the segregation of the germline determinants in Drosophila46.
Work from the last twenty years offers the first clues supporting a role for localized RNA in directly modulating centrosome function. In Xenopus oocytes, the cytoplasmic polyadenylation element binding protein (CPEB), a translation regulator, localizes Cyclin B mRNA to mitotic spindle poles 47. Neutralization of CPEB activity or mutation of the CPEB consensus site within the Cyclin B 3′UTR impairs cell division and leads to spindle formation defects, a common phenotype of centrosome dysfunction47. These mutants also disrupt enrichment of Cyclin B protein at spindle poles, raising the possibility that CPEB contributes to the local synthesis of this critical cell cycle regulator. The finding that Cyclin B is enriched at the spindle poles of diverse cell types, including Drosophila embryos and cultured mammalian cells, suggests that the regulation of cell cycle progression via RNA localization may be a conserved process7,48,49. This model is supported by proteomic analysis identifying several RBPs and processing factors in association with centrosomes50 and spindle poles49. More recently, work in cultured mammalian cells indicates the RBP Gle1 localizes to mammalian centrosomes, where it is required for efficient microtubule nucleation51. These examples support a model in which RBPs regulate the enrichment of specific RNAs to the centrosome. This model predicts that such RBPs likely interact with molecular motors to mediate RNA localization to the centrosome. At present, the motors involved in RNA enrichment at centrosomes have yet to be defined. However, we speculate that the polarized microtubule arrays nucleated by the centrosome could serve as cytoskeletal tracks for active transport. Given the role of the centrosome in nucleating the growth of microtubules with their minus-ends embedded within the pericentriolar material and their plus-ends extending into the cytosol, we predict that cytoplasmic dynein or a minus-end directed kinesin, such as HSET/Ncd, may direct RNA to the centrosome. Whether BicD, Egl, or other dynein associated factors contribute to RNA localization to centrosomes remains unknown. Collectively, these findings suggest that we are only beginning to understand the function of localized RNA at microtubule-organizing centers.
One possibility is that RNA may play a structural role in centrosomes. The emerging model that the centrosome forms by phase separation suggests that localized RNA may support the structural integrity of centrosomes, as RNA is typically a crucial component of phase separated condensates52. The phase separation model for centrosome formation is supported by the recent finding that purified C. elegans centrosomal proteins coalesce into gel-like assemblies that are capable of nucleating microtubules when mixed in the presence of a crowding agent53. RNA may be a missing component of these in vitro recombinant assemblies that would promote phase separation at physiologic concentrations without a crowding agent. Taken together, these studies indicate that while the localization of RNA to centrosomes has been established for several decades, we are only now beginning to understand the physiological significance of this association. For a few defined transcripts, most notably Cyclin B, the localization of RNA to the spindle pole appears to be important for efficient cell cycle progression and cell division.
Summary and Perspective
In this review, we highlight several examples of subcellular RNA localization from diverse cellular systems to underscore the contribution of RNA localization to cellular function beyond neuronal specialization and embryonic patterning. Examination of RNA localization within oocytes, embryos, and neurons remains an important and rich avenue of active research. Such studies often inform fundamental and disease-relevant developmental and neurological processes. Moreover, RNA localization studies within embryos and neurons typically reveal common principles that govern RNA localization control. As we emphasize in this review, such foundational studies often inform subsequent work in other cellular contexts.
We contend that the importance of RNA localization beyond embryonic and neuronal models is underappreciated. In addition to discussing some well-studied models, we present several recent studies from diverse cellular models to illuminate the role of RNA at adherens junctions, viral pathogenesis, and centrosome structure, which have only emerged within the last five years. With the advent of technologies to study RNA localization at the subcellular level, such as high throughput RNA sequencing, cross-linking immunoprecipitation, ribosome profiling, smFISH, and other advanced imaging approaches, we expect that RNA localization will increasingly be recognized as participating in multiple new and unexpected pathways. The diversity of the examples highlighted here leads us to conclude that localized RNA should be considered as a contributor to numerous biological processes, especially those that require rapid responses to changing cellular conditions. As these hypotheses are tested, we expect that the contribution of localized RNA to cellular function will be recognized as the rule rather than the exception. Comparative analyses of RNA and RBP localization patterns in response to pathological disease states will be an important future direction of research with broad implications for human health.
Synopsis.
RNA localization is a fundamental paradigm used to compartmentalize gene activity within diverse cellular contexts. When coupled to translational control, the localization of messenger RNA (mRNA) generates localized protein enrichments within cells. Long appreciated for its role in neurogenesis and embryo development, RNA localization is evolutionarily conserved across taxa and contributes to a myriad of cellular responses, including polarization, adhesion, migration, proliferation, host-pathogen interactions, the stress response, and more. Here, we summarize the importance of mRNA localization to diverse cellular responses referencing several well-studied models. We conclude that RNA localization is a widespread and conserved mechanism that generates localized protein enrichments with far-reaching consequences for cellular activity and function.
Acknowledgments
This work was support by NIH grant 1K22HL126922-01A1 to DAL. PVR was also supported by NIH grant 5K12GM000680-18.
References
- 1.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81(2):161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 2.Eliscovich C, Singer RH. RNP transport in cell biology: the long and winding road. Current Opinion in Cell Biology. 2017;45:38–46. doi: 10.1016/j.ceb.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gagnon JA, Mowry KL. Molecular motors: directing traffic during RNA localization. Critical Reviews in Biochemistry and Molecular Biology. 2011;46(3):229–239. doi: 10.3109/10409238.2011.572861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock SL. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans. 2011;39(5):1161–1165. doi: 10.1042/BST0391161. [DOI] [PubMed] [Google Scholar]
- 5.Glock C, Heumüller M, Schuman EM. mRNA transport & local translation in neurons. Curr Opin Neurobiol. 2017;45:169–177. doi: 10.1016/j.conb.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23(13):1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lécuyer E, Yoshida H, Parthasarathy N, et al. Global Analysis of mRNA Localization Reveals a Prominent Role in Organizing Cellular Architecture and Function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 9.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Meth. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffery WR, Tomlinson CR, Brodeur RD. Localization of actin messenger RNA during early ascidian development. Developmental Biology. 1983;99(2):408–417. doi: 10.1016/0012-1606(83)90290-7. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45(3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 12.Katz ZB, Wells AL, Park HY, Wu B, Shenoy SM, Singer RH. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26(17):1885–1890. doi: 10.1101/gad.190413.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz ZB, English BP, Lionnet T, et al. Mapping translation “hot-spots” in live cells by tracking single molecules of mRNA and ribosomes. Elife. 2016;5:e10415. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez N, Eromobor I, Petrie RJ, Vedula P, Cruz L, Rodriguez AJ. The β-actin mRNA zipcode regulates epithelial adherens junction assembly but not maintenance. RNA. 2014;20(5):689–701. doi: 10.1261/rna.043208.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Katz Z, Wu B, et al. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. Journal of Cell Science. 2012;125(Pt 1):81–91. doi: 10.1242/jcs.086132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kourtidis A, Necela B, Lin W-H, et al. Cadherin complexes recruit mRNAs and RISC to regulate epithelial cell signaling. J Cell Biol. 2017;216(10):3073–3085. doi: 10.1083/jcb.201612125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagaoka K, Udagawa T, Richter JD. CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nature Communications. 2012;3:675–10. doi: 10.1038/ncomms1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backes S, Herrmann JM. Protein Translocation into the Intermembrane Space and Matrix of Mitochondria: Mechanisms and Driving Forces. Front Mol Biosci. 2017;4:83. doi: 10.3389/fmolb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egea G, Izquierdo JM, Ricart J, San Martín C, Cuezva JM. mRNA encoding the beta-subunit of the mitochondrial F1-ATPase complex is a localized mRNA in rat hepatocytes. Biochem J. 1997;322:557–565. doi: 10.1042/bj3220557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol. 2000;20(21):7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margeot A, Garcia M, Wang W, Tetaud E, di Rago JP, Jacq C. Why are many mRNAs translated to the vicinity of mitochondria: a role in protein complex assembly? Gene. 2005;354:64–71. doi: 10.1016/j.gene.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 22.Liu JM, Liu DR. Discovery of a mRNA mitochondrial localization element in Saccharomyces cerevisiae by nonhomologous random recombination and in vivo selection. Nucleic Acids Res. 2007;35(20):6750–6761. doi: 10.1093/nar/gkm777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO rep. 2002;3(2):159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold VA, Chroscicki P, Bragoszewski P, Chacinska A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO rep. 2017;18(10):1786–1800. doi: 10.15252/embr.201744261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesnik C, Golani-Armon A, Arava Y. Localized translation near the mitochondrial outer membrane: An update. RNA Biology. 2015;12(8):801–809. doi: 10.1080/15476286.2015.1058686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui XA, Palazzo AF. Localization of mRNAs to the endoplasmic reticulum. Wiley Interdiscip Rev RNA. 2014;5(4):481–492. doi: 10.1002/wrna.1225. [DOI] [PubMed] [Google Scholar]
- 27.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287(8):5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10(5):e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker F, Block-Alper L, Nakamura G, Harada J, Wittrup KD, Meyer DI. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J Cell Biol. 1999;146(2):273–284. doi: 10.1083/jcb.146.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid M, Jaedicke A, Du T-G, Jansen R-P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Current Biology. 2006;16(15):1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20(1):127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yagüe E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24(9):3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- 33.Altan-Bonnet N. Lipid Tales of Viral Replication and Transmission. Trends Cell Biol. 2017;27(3):201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell NM, Lever AML. HIV Gag polyprotein: processing and early viral particle assembly. Trends in Microbiology. 2013;21(3):136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Molle D, Segura-Morales C, Camus G, et al. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J Biol Chem. 2009;284(29):19727–19743. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vale-Costa S, Amorim M. Recycling Endosomes and Viral Infection. Viruses. 2016;8(3):64–29. doi: 10.3390/v8030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce EA, Digard P, Stuart AD. The Rab11 Pathway Is Required for Influenza A Virus Budding and Filament Formation. J Virol. 2010;84(12):5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y-H, Du W, Hagemeijer MC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160(4):619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jansen R-P, Niessing D, Baumann S, Feldbrügge M. mRNA transport meets membrane traffic. Trends in Genetics. 2014;30(9):408–417. doi: 10.1016/j.tig.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Higuchi Y, Ashwin P, Roger Y, Steinberg G. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204(3):343–357. doi: 10.1083/jcb.201307164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.König J, Baumann S, Koepke J, Pohlmann T, Zarnack K, Feldbrügge M. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 2009;28(13):1855–1866. doi: 10.1038/emboj.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lerit DA, Smyth JT, Rusan NM. Organelle asymmetry for proper fitness, function, and fate. Chromosome Res. 2013;21(3):271–286. doi: 10.1007/s10577-013-9350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall WF, Rosenbaum JL. Are there nucleic acids in the centrosome? Curr Top Dev Biol. 2000;49:187–205. doi: 10.1016/s0070-2153(99)49009-x. [DOI] [PubMed] [Google Scholar]
- 44.Alliegro MC. The centrosome and spindle as a ribonucleoprotein complex. Chromosome Res. 2011;19(3):367–376. doi: 10.1007/s10577-011-9186-7. [DOI] [PubMed] [Google Scholar]
- 45.Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420(6916):682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 46.Lerit DA, Gavis ER. Transport of Germ Plasm on Astral Microtubules Directs Germ Cell Development in Drosophila. Current Biology. 2011;21(6):439–448. doi: 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103(3):435–447. doi: 10.1038/35043029. [DOI] [PubMed] [Google Scholar]
- 48.Raff JW, Whitfield WG, Glover DM. Two distinct mechanisms localise cyclin B transcripts in syncytial Drosophila embryos. Development. 1990;110(4):1249–1261. doi: 10.1242/dev.110.4.1249. [DOI] [PubMed] [Google Scholar]
- 49.Blower MD, Feric E, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179(7):1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller H, Schmidt D, Steinbrink S, et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29(19):3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jao L-E, Akef A, Wente SR. A role for Gle1, a regulator of DEAD-box RNA helicases, at centrosomes and basal bodies. Mol Biol Cell. 2017;28(1):120–127. doi: 10.1091/mbc.E16-09-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodruff JB, Hyman AA, Boke E. Organization and Function of Non-dynamic Biomolecular Condensates. Trends in Biochemical Sciences. 2017 Dec;:1–14. doi: 10.1016/j.tibs.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Woodruff JB, Gomes BF, Widlund PO, Mahamid J, Honigmann A, Hyman AA. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 2017;169(6):1066–1071e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]