Abstract
Large efforts have been devoted to the genetic code engineering in the past decade, aiming for unnatural amino acid mutagenesis. Recently, an increasing number of studies were reported to employ quadruplet codons to encode unnatural amino acids. We and others have demonstrated that the quadruplet decoding efficiency could be significantly enhanced by an extensive engineering of tRNAs bearing an extra nucleotide in their anticodon loops. In this work, we report the identification of tRNA mutants derived from directed evolution to efficiently decode a UAGA quadruplet codon in mammalian cells. Intriguingly, the trend of quadruplet codon decoding efficiency among the tested tRNA variants in mammalian cells was largely the same as that in E. coli. We subsequently demonstrate the utility of quadruplet codon decoding by the construction of the first HIV-1 mutant that lacks any in-frame amber nonsense codons and can be precisely activated by the decoding of a genomically embedded UAGA codon with an unnatural amino acid. Such conditionally activatable HIV- 1 mutant can likely facilitate both fundamental investigations of HIV-1 as well as vaccine developments. The use of quadruplet codon, instead of an amber nonsense codon, to control HIV-1 replication has the advantage in that the correction of a frameshift caused by a quadruplet codon is much less likely than the reversion of an amber codon back into a sense codon in HIV-1.
Keywords: Quadruplet codon, genetic code, unnatural amino acid, HIV, virus engineering
Graphical abstract

Triplet codons are the predominant form of genetic code in nature with rare exceptions of programmed frameshift, e.g., +1 frameshift (an apparent quadruplet codon decoding).1 Recent developments in genetic code engineering showed that quadruplet codons could be decoded under experimental conditions with modified translation machinery.2–13 We11, 12 and others13 also demonstrated that the decoding efficiency for quadruplet codons (e.g., AGGA, UAGA, UAGC, etc.) could be significantly enhanced by engineering the anticodon stem and loop of tRNAPyl, a tRNA from Methanosarcina maize, bearing an extra nucleotide in the anticodon loop. Unlike the naturally existing +1 frameshift, decoding quadruplet codons by the engineered tRNA variants does not require any recoding signals embedded in the mRNA. The generation of a collection of quadruplet codon decoding tRNAs have significant values in both fundamental studies and practical applications. On one hand, these tRNA variants can be used to investigate how mutations in tRNA mediate the switch from a triplet to quadruplet reading frame within a universal translational machinery that prefers reading triplet codons. On the other hand, quadruplet codon decoding tRNAs can be employed to further expand cellular genetic code. This will augment the current efforts in unnatural amino acid (unAA) mutagenesis so that more unAAs with defined chemical properties can be simultaneously incorporated into a single protein in living cells.14–20
The directed evolutions of quadruplet codon decoding tRNAPyl variants were carried out in E. coli.11–13 Such evolution aims to optimize the interactions between a quadruplet codon decoding tRNAPyl bearing an expanded anticodon loop and the host’s translation machinery, especially the ribosome. In a previous report, we have shown for the first time that an evolved tRNAPyl mutant from bacteria can be directly utilized to decode an AGGA codon in mammalian cells.11 In this work, we investigated the decoding of UAGA codon and were able to identify two tRNAPyl variants that could efficiently decode a UAGA codon in mammalian cells. Our approach to use engineered tRNAPyl mutants from E. coli host to decode quadruplet codon in mammalian system can potentially be generalized to other tRNAPyl variants for the decoding of additional quadruplet codons, which will greatly facilitate the genetic code expansion in mammalian cells.
To further demonstrate the utility of quadruplet codon decoding in mammalian cells, we engineered a genomically recoded HIV-1 and showed that the replication of this variant can be precisely controlled by an unnatural amino acid. Such conditionally activatable HIV-1 mutant can likely be used to facilitate fundamental studies of HIV-1. Furthermore, the use of quadruplet codon, instead of the amber nonsense codon, as a cue to control HIV-1 replication is expected to augment our novel strategy to construct HIV-1 vaccine.21, 22 Historically, vaccines have been our best choice against the world’s deadliest infectious diseases. The development of an HIV-1 vaccine would undoubtedly be the best solution for the ultimate control of HIV-1 pandemic, but remains elusive.23 While live-attenuated virus vaccines confer good protections,24 the potential pathogenic consequences associated with the uncontrolled virus replication preclude such vaccines from clinical applications.25–27 A strategy that can be used to tightly control HIV-1 replication would be useful to overcome the safety concerns. In our previous reports,21, 22 we developed an approach to control HIV-1 replication through an unAA- mediated suppression of amber nonsense codons. With relatively high mutation rate, HIV-1 can potentially regain functional replication by mutating an installed amber codon back to a sense codon. One approach to mitigate this problem is to introduce multiple amber codons (e.g., three or more) into several essential genes of HIV-1.21 As an alternate approach, a quadruplet codon, instead of a triplet amber codon, can be sued as part of the control element for HIV-1 replication. The +1 frameshift caused by the insertion of a quadruplet codon is more difficult to be corrected (needs a much less common deletion event in HIV-1 genome) than the reversion of the amber codon into a sense codon.28
RESULTS AND DISCUSSION
Decoding a UAGA codon in mammalian cells
In a previous report, we conducted directed evolution of a tRNAPyl variant, tRNAUCCU (contain an apparent UCCU anticodon), in E. coli.11 We observed that a tRNAUCCU mutant that could efficiently decode AGGA codon in E. coli also functioned well in mammalian cells. We hypothesized that the observation can be generalized to other quadruplet decoding tRNAPyl variants evolved from a bacterial host. It is likely that the ribosome decoding centers from different kingdoms have certain conservations although prokaryotes have 70S ribosomes (30S small subunit, 50S large subunit) whereas eukaryotes have 80S ribosomes (40S small subunit, 60S large subunit). To further examine the above hypothesis, we studied a different set of tRNAPyl (tRNAUCUA) variants, which were obtained through directed evolution in E. coli,12 for their ability to decode the UAGA codon in mammalian cells.
From a collection of previously evolved tRNAUCUA, we selected three mutants (referred hereafter as tRNAUCUA-1, tRNAUCUA-2, and tRNAUCUA-3; Table S2) with varied UAGA decoding efficiency in E. coli.12 Among them, tRNAUCUA-1 and tRNAUCUA-3 displayed the highest and the lowest UAGA decoding efficiency, respectively (Figure S1). To examine their ability to decode UAGA in mammalian cells (293T), we employed a fluorescence-based assay, where the decoding efficiency was directly linked to the expression level of an EGFP mutant (EGFP-Tyr40UAGA) of which the codon at position Tyr40 was changed to UAGA. A two-plasmid system was used for protein expression. One plasmid (pEGFP-Tyr40UAGA) harbors the gene of EGFP-Tyr40UAGA. The other plasmid (pBocLysRS-tRNAUCUA) encodes a tRNAUCUA mutant of interest under the control of a human U6 promoter and a copy of a BocLysRS under the control of a non-regulated CMV promoter. BocLysRS specifically aminoacylates the tRNA with N£-(tert-butyloxy-carbonyl)-L-lysine (BocLys). Following transfection with the two plasmids, 293T cells were cultivated in the presence (5 mM; Figure 1) or the absence (Figure S2) of Boc-Lys for an additional 24 h before visualization under a confocal fluorescence microscope. As shown in Figure 1, strong fluorescence signals were observed in the presence of BocLys when tRNAUCUA-1 or tRNAUCUA-2 was used to decode the UAGA codon in EGFP-Tyr40UAGA. On the other hand, cells expressing tRNAUCUA-3 only displayed weak fluorescence signal, which was similar to that of the cells expressing the wild-type tRNAUCUA that was derived from the wild-type tRNAPyl 29, 30 by changing its anticodon sequence into quadruplet UCUA. In control experiments, no detectable fluorescence signal was observed when transfected cells were cultivated in the media without BocLys (Figure S2).
Figure 1. Decoding a UAGA codon in the EGFP-Tyr40UAGA-encoding gene in 293T cells.

All media contained BocLys (5 mM). The left panel shows fluorescent images of 293T cells in the EGFP channel (488 nm excitation and 530/25 nm emission), the middle panel shows bright-field images of the same 293T cells, and the right panel shows composite images of bright-field and fluorescent images. Scale bars, 20 μm. These are representative images from triplicate experiments.
To obtain a more quantitative view of UAGA decoding efficiency and specificity, we conducted flow cytometry analysis. After transfection and cultivation, cells were detached from culture plates with trypsin digestion and re-suspended in PBS buffer for fluorescence measurements (Figure S3). To compare decoding efficiency, we used the normalized fluorescence intensity, which was the product of the mean fluorescence intensity of the fluorescent cell population multiplied by the percentage of fluorescent cells (100,000 cells sorted/sample). As shown in Figure 2, tRNAUCUA-1 displayed the highest UAGA decoding efficiency in 293T cells followed by tRNAUCUA-2 and tRNAUCUA-3. The UAGA decoding efficiency of these three tRNA variants are 9.5-fold, 6.1-fold and 2.3-fold higher than that of the parental wild-type tRNAUCUA. The flow cytometry analyses also confirmed our observation from the cell imaging experiments that tRNAUCUA-3 had only slightly higher decoding efficiency than that of the wild-type tRNAUCUA. The expression level of the EGFP-Tyr40UAGA mutant when tRNAUCUA-1 was used was about 2.8% of that of the wild-type EGFP. Intriguingly, the trend of quadruplet codon decoding efficiency among the four tRNA variants in mammalian cells (Figure 2) was the same as that in E. coli (Figure S1). Although a larger data set will be needed in order to perform more meaningful statistical analysis, current observations largely supported our hypothesis that tRNAPyl variants derived from directed evolution in E. coli can be potentially transferred into mammalian system with similar trends in activities.
Figure 2. The efficiency and specificity of UAGA decoding by tRNAUCUA variants in mammalian cells.
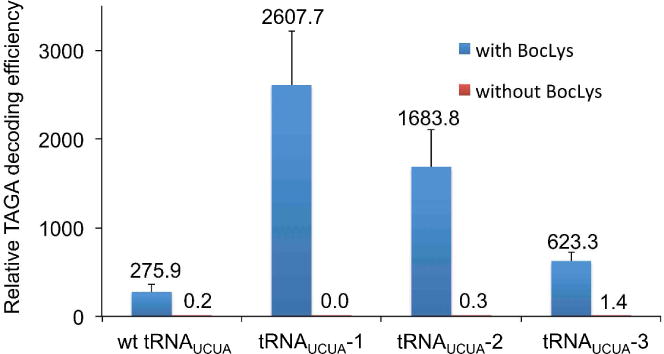
The relative efficiencies of UAGA decoding were calculated by multiplying the mean fluorescence intensity by the percentage of fluorescent cells in flow cytometry analyses. Each data point is the average of triplet measurements with standard deviation. The raw data of flow cytometry experiments are shown in Figure S3.
Construction of a genomically recoded HIV-1 mutant
Our goal was to demonstrate the utility of quadruplet codon decoding as a control element to regulate the replication of HIV-1. Since HIV-1 uses amber codon (UAG) as a stop signal in protein translation, we sought to eliminate all functional amber codons so that the genome is free of in-frame UAGA sequence. This would prevent any undesirable read-through once the quadruplet decoding mechanism is installed. The HIV-1 variant, NL4-3, contains five amber codons that function as translational stops for Vif, Vpu, Vpr, Tat, and Rev proteins. Due to the highly efficient coding nature of HIV-1 genome,31 all five amber codons are also part of the reading frames of other proteins. Therefore, the replacement of amber codons with one of the other two nonsense codons will lead to amino acid changes. To minimize this effect, we chose to substitute the amber codon with ochre codon (UAA), which resulted in one nucleotide change per replacement.
We first constructed four NL4-3 mutants. Each of them had a single amber to ochre mutation in Vif, Vpu, Vpr, or Rev. These four HIV-1 mutants, NL4-3-vif-ochre, NL4-3-vpu-ochre, NL4-3-vpr-ochre, and NL4-3-rev-ochre, were individually assembled by transfecting their encoding plasmids into 293T cells. Following cultivation of the cells for 48 h, viruses were harvested and the infectivities were examined by infection assays of the TZM-bl reporter cells. TZM-bl cells contain a genomic copy of a Tat-driven bacterial lacZ gene. The infection of TZM-bl cells by an active HIV-1 variant turns on the expression of lacZ-encoded β-galactosidase. As a result, TZM-bl cells are stained with blue color in the presence of X-gal. As shown in Figure S4, the infectivity of the four mutants was qualitatively similar to that of the parent NL4-3 virus, which indicated that the amino acid substitutions did not have any obvious effects on HIV-1 function. Encouraged by the results, we proceeded to generate an amber-codon-free NL4-3 mutant, NL4-3-all-ochre, by replacing all five amber codons with ochre codons. This led to mutations in four proteins at six positions, including Vpr (Glu21Lys), Tat (Arg7Lys), Rev (Arg41Lys), and Env (Ser29Asn, Glu732Lys, and Ser808Asn). Functional test using above infectivity assay showed that the NL4-3-all-ochre mutant retained a similar level of infectivity relative to the parental NL4-3 virus (Figure 3). The data suggested that a complete amber to ochre mutation was well tolerated by HIV-1 virus.
Figure 3. Infectivity assay of NL4-3, NL4-3-all-ochre, and NL4-3-control.
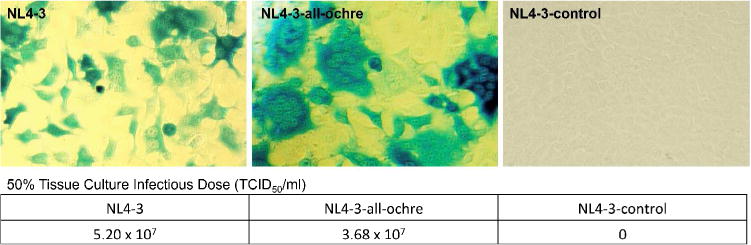
The three HIV-1 variants, NL4-3, NL4-3-all-ochre, and NL4-3-control were assembled in 293T cells, then used to infect TZM-bl cells. TZM-bl cells contain a genomic copy of a Tat-driven bacterial lacZ gene. Functional infection of TZM-bl cells by HIV-1 turns on the expression of lacZ-encoded p-galactosidase. As a result, TZM-bl cells are stained with blue color in the presence of X-gal. These are representative images from triplicate experiments. The molecular clone, NL4-3-control, was directly derived from NL4-3 with an amber mutation at position Trp36 of p17 matrix protein and cannot be assembled in 293T cells.
Controlling HIV-1 replication through quadruplet codon decoding
In previous reports,21, 22 we demonstrated that HIV-1 replication could be controlled by decoding an amber codon with an unnatural amino acid. Since HIV-1 has relatively high genetic variability, it is still possible that HIV-1 amber mutant can regain functional replication by mutating the installed amber codon back to a sense codon. Although we did not observe any such instance in our in vitro experiments, this possibility might become greater in a longer time span when the mutant replicates in vivo. The reversion mutation will cause safety concerns if HIV-1 amber mutant would be used as a vaccine. One approach to solve this problem is to introduce multiple amber codons (e.g., more than three) into the essential genes of HIV- 1.21 The possibility for HIV-1 to recover all amber mutations is small. Another potential strategy is to use quadruplet UAGA codon, instead of triplet amber codon, as part of the control element for HIV-1 replication. In this case, simple substitution of nucleotides in the UAGA codon does not allow HIV-1 to replicate since it causes +1 frameshift and cannot lead to the expression of functional protein. Nucleotide insertion/deletion at or close to the UAGA codon may lead to nonessential amino acid changes and allow the expression of functional proteins. However, such event is less common than nucleotide substitution.28 Therefore, we envisage that the use of one or more UAGA codons will efficiently prevent HIV-1 from regaining its function in the absence of UAGA-decoding machinery, which includes an unnatural amino acid (e.g., BocLys), a UAGA decoding tRNA (e.g., tRNAUCUA-1), and an aminoacyl-tRNA synthetase (e.g., BocLysRS) that can charge the tRNA with the unnatural amino acid.
To examine if we can use UAGA decoding to control HIV-1 replication, we constructed two HIV-1 quadruplet mutants based on NL4-3-all-ochre, which lacks any functional amber codons. In the first construct (NL4-3-Trp36UAGA), a UAGA codon was introduced into the p17 matrix protein at position Trp36. In the second construct (NL4-3-Tyr59UAGA), a UAGA codon was introduced into the HIV-1 protease protein at position Tyr59. Both of the two positions are at the protein surface and should not interfere with protein function according to our previous studies.21
To examine the assembly of the virus, each HIV-1 quadruplet construct was transfected into 293T cells together with a plasmid (pBocLysRS-tRNAUCUA-1) that encodes BocLysRS and the best UAGA decoding tRNA. Cells were cultivated either in the presence or in the absence of 5 mM BocLys for 48 h. Viruses were subsequently harvested, diluted (1:5), and used to infect TZM-bl reporter cells. To gauge the infectivity, we used a Tat-induced luciferase reporter that is integrated on the chromosome of TZM-bl cells (Figure 4). The NL4-3-Trp36UAGA mutant displayed good infectivity when it was assembled in the presence of BocLys. In contrast, no functional virus was detected in the absence of BocLys. The NL4-3-Tyr59UAGA mutant only showed low infectivity. Nevertheless, again, no background production of virus was detected in the absence of BocLys. The above data suggest that the decoding of UAGA codon with unnatural amino acid can be used as a very stringent control of HIV- 1 replication.
Figure 4. Infectivity assay of NL4-3-Trp36UAGA and NL4-3-Tyr59UAGA mutants in the presence and in the absence of BocLys.
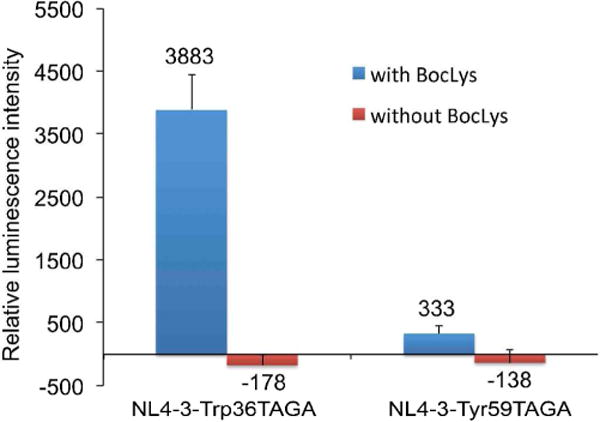
The two HIV-1 variants were assembled in 293T cells. TZM-bl cells were infected with virus (with 1:5 dilution) collected from the supernatant of 293T cell cultures. TZM-bl cells contain a genomic copy of a Tat-driven luciferase gene. Functional infection of TZM-bl cells by HIV-1 turns on the expression of luciferase. As a result, luminescence signal is directly linked to HIV-1 infectivity.
In summary, we have demonstrated for the second time that tRNAPyl mutants evolved in bacteria could be readily used to decode quadruplet codons in mammalian cells. The trends of decoding efficiency and fidelity were largely the same between bacterial and mammalian systems. A plausible explanation for this observation is that our engineering efforts focused on a region of tRNA that could lead to intrinsic structural changes to accommodate the extended anticodon loop. As a result, applicability of the obtained tRNA mutants was not affected by certain differences in translational machineries of the host. Therefore, the current approach can potentially be generalized to the engineering of additional quadruplet decoding tRNAPyl (or other tRNAs that are orthogonal both in E. coli and mammalian cells) variants for the expansion of mammalian genetic code. On the other hand, it should be noted that evolution of regions of a tRNA that exclusively engage in interactions with the translational machinery may need to be conducted directly in the targeting or a closely related host cell. In the second part of this work, we further demonstrated the utility of mammalian quadruplet codon decoding by the construction of a genomically recoded and conditionally active HIV-1 variant whose replication could be controlled by the decoding of a UAGA codon in its essential gene. While we have not examined the replication capacity of our HIV-1 mutants with HIV-1 susceptible wild-type cells (e.g., human peripheral blood mononuclear cells), our investigation with TZM-bl cells represents a good surrogate for such study. This work will augment our current efforts in the development of live attenuated HIV-1 vaccine.
METHODS
Materials and General Methods
Infectious molecular clone NL4-3 was obtained from the NIH AIDS Reagent Program. BocLys was purchased from Bachem. Primers were ordered from Sigma. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. KOD hot start DNA polymerase was purchased from EMD Millipore. Standard molecular biology techniques32 were used throughout. Site-directed mutagenesis was carried out using overlapping PCR. E. coli GeneHogs were used for routine cloning and DNA propagation for none-HIV plasmids. E. coli MAX Efficiency Stbl2 (Thermo Fisher Scientific Inc) was used for routine cloning and DNA propagation for HIV plasmids. All solutions were prepared in deionized water further treated by Barnstead Nanopure® ultrapure water purification system (Thermo Fisher Scientific Inc). Antibiotics were added where appropriate to following final concentrations: ampicillin, 100 mg L-1; kanamycin, 50 mg L-1; tetracycline, 12.5 mg L-1.
Cell transfection, fluorescence imaging and flow cytometry assay
293T cells were grown in a medium containing DMEM (Gibco), 10% FBS, and 2 mM L-glutamine at 37°C in a humidified atmosphere of 5% CO2. About 5×105 293T cells were plated into each well of 6-well plates and cultured overnight. Transfection was performed using Lipofectamine 2000 (ThermoFisher) by following the manufacture’s instruction. Six hours post transfection, BocLys was added to each sample well at a final concentration of 5 mM and an equal volume of growth medium was added to each control well. For fluorescence imaging, cells were grown for an additional 12 h before being washed with DMEM base medium, fixed with 4% paraformaldehyde (w/v), washed three times with DPBS, and visualized by an Inverted (Olympus IX 81) confocal microscope. The samples were excited at 488 nm to acquire EGFP fluorescence images at 530/25 nm. For flow cytometry assay, the cells were detached from culture plate with 0.25% trypsin and washed twice with growth medium, resuspended in 1 ml 1xPBS (containing 4% paraformaldehyde), and fixed at 4 °C. The expression of EGFP was measured using a Cytek DxP10 flow cytometer (Cytek) and 100,000 cells were recorded for each sample. The flow cytometry data were analyzed using FlowJo v10 software (Tree Star).
Reconstitution and infectivity assay of HIV-1 viruses
To reconstitute HIV-1 viruses, appropriate DNA constructs were transfected into 293T cells in 6-well plate using Lipofectamine 2000 (ThermoFisher). After 72 h of cultivation in the presence or absence of 5 M BocLys, culture supernatant was collected from each well. Suspending cells and debris were removed by passing through 0.45 μm filter. The infectivity of reconstituted HIV-1 viruses were assayed using TZM-bl cells by following previously described procedures.21, 33 For the X-Gal staining assay, diluted virion- containing samples, mixed with 10% DMEM growth medium and DEAE-dextran at a final concentration of 40 mg/mL, were added into each well of a 6-well flat bottom plate that contains TZM-bl cells. After 48 hours of cultivation at 37 °C in a humidified atmosphere of 5% CO2, TZM-bl cells were washed, fixed, and stained using X-gal solution (Genlantis) for 2 hours at 37 °C. Infectivity was evaluated by counting X-gal-positive cells under a light microscope. For the luminescence-based assay, collected virion samples were first diluted fivefold in culture media. A 100 μl of diluted virion was then mixed with 100 μL of fresh TZM-bl cells (10,000) and transferred into wells of black-wall, clear-bottom 96-well plates (Corning-Costar). After 48 h of incubation, medium was removed and 50 μl of cell lysis buffer (Promega) was added to each well. After 20 min of incubation at room temperature, 50 μl luciferase assay substrate (Promega) was added to each well and luminescence was quantified with a VICTOR3 multilabel plate reader (Perkin Elmer). The analyses were done in quadruplicate.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (grant 1553041 to J.G.) and National Institute of Health (grant 1R01AI111862 to J.G., Q.L., and W.N.).
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at DOI: Additional experimental procedures, tables, figures, and data.
ORCID
Jiantao Guo: 0000-0001-6983-9953 Qingsheng Li: 0000-0003-4101-4073 Wei Niu: 0000-0003-3826-1276
Author Contributions
Yan Chen and Yanmin Wan contributed equally to this work. Yan Chen constructed HIV-1 mutant strains and plasmids, and conducted quadruplet codon decoding studies in mammalian cells. Yanmin Wan conducted most of experiments involving live HIV-1 variants and assisted in quadruplet codon decoding studies in mammalian cells. Nanxi Wang and Zhe Yuan contributed equally to this work. Nanxi Wang constructed several HIV-1 mutant strains and plasmids, conducted quadruplet codon decoding studies in E. coli. Zhe Yuan conducted several experiments involving live HIV-1 variants. Wei Niu, Qingsheng Li, and Jiantao Guo designed the study and wrote the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Atkins JF, Bjoerk GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohsaka T, Ashizuka Y, Murakami H, Sisido M. Incorporation of nonnatural amino acids into streptavidin through In vitro frame-shift suppression. J Am Chem Soc. 1996;118:9778–9779. [Google Scholar]
- 3.Hohsaka T, Ashizuka Y, Taira H, Murakami H, Sisido M. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry. 2001;40:11060–11064. doi: 10.1021/bi0108204. [DOI] [PubMed] [Google Scholar]
- 4.Magliery TJ, Anderson JC, Schultz PG. Expanding the Genetic Code: Selection of Efficient Suppressors of Four-base Codons and Identification of "Shifty" Four-base Codons with a Library Approach in Escherichia coli. J Mol Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JC, Magliery TJ, Schultz PG. Exploring the limits of codon and anticodon size. Chem Biol. 2002;9:237–244. doi: 10.1016/s1074-5521(02)00094-7. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taki M, Matsushita J, Sisido M. Expanding the genetic code in a mammalian cell line by the introduction of four-base codon/anticodon pairs. ChemBioChem. 2006;7:425–428. doi: 10.1002/cbic.200500360. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc Natl Acad Sci U S A. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee A, Lajoie MJ, Xiao H, Church GM, Schultz PG. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem. 2014;15:1782–1786. doi: 10.1002/cbic.201402104. [DOI] [PubMed] [Google Scholar]
- 11.Niu W, Schultz PG, Guo J. An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Shang X, Cerny R, Niu W, Guo J. Systematic evolution and study of UAGN decoding tRNAs in a genomically recoded bacteria. Sci Rep. 2016;6:21898. doi: 10.1038/srep21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, Mehl RA, Chin JW. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labeling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Schultz PG. Synthesis at the interface of chemistry and biology. J Am Chem Soc. 2009;131:12497–12515. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin JW. Reprogramming the genetic code. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- 17.Ai H-w. Biochemical analysis with the expanded genetic lexicon. Anal Bioanal Chem. 2012;403:2089–2102. doi: 10.1007/s00216-012-5784-2. [DOI] [PubMed] [Google Scholar]
- 18.Liu WR, Wang YS, Wan W. Synthesis of proteins with defined posttranslational modifications using the genetic noncanonical amino acid incorporation approach. Mol BioSyst. 2011;7:38–47. doi: 10.1039/c0mb00216j. [DOI] [PubMed] [Google Scholar]
- 19.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl- tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 20.Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment - Expanding the chemistry in biology. Chem Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N, Li Y, Niu W, Sun M, Cerny R, Li Q, Guo J. Construction of a live- attenuated HIV-1 vaccine through genetic code expansion. Angew Chem, Int Ed. 2014;53:48674871. doi: 10.1002/anie.201402092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Z, Wang N, Kang G, Niu W, Li Q, Guo J. Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth Biol. 2017;6:721731. doi: 10.1021/acssynbio.6b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fauci AS. 25 years of HIV. Nature. 2008;453:289–290. doi: 10.1038/453289a. [DOI] [PubMed] [Google Scholar]
- 24.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 25.Baba TW, Jeong YS, Pennick D, Bronson R, Greene MF, Ruprecht RM. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 26.Wyand MS, Manson KH, Lackner AA, Desrosiers RC. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 27.Baba TW, Liska V, Khimani AH, Ray NB, Dailey PJ, Penninck D, Bronson R, Greene MF, McClure HM, Martin LN, Ruprecht RM. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 28.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 31.Hryckiewicz K, Bura M, Kowala-Piaskowska A, Bolewska B, Mozer-Lisewska I. HIV RNA splicing. HIV & AIDS Review. 2011;10:61–64. [Google Scholar]
- 32.Sambrook JF, Russell DW, editors. Molecular cloning: A laboratory manual. third. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 33.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C-l, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


