Figure 4. The CRL2 adaptors KLHDC2, KLHDC3 and KLHDC10 target distinct C-terminal glycine degrons.
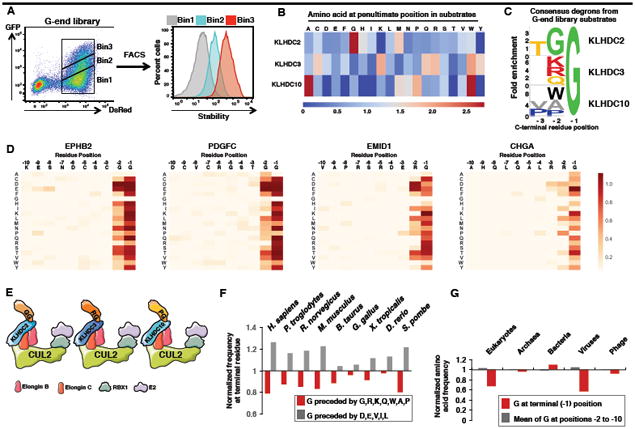
(A) Schematic representation of the G-end GPS library screen.
(B) Comparison of the amino acid frequencies observed at the -2 position preceding the C-terminal glycine residue among KLHDC2, KLHDC3 or KLHDC10 substrates.
(C) Consensus sequences for the C-terminal degrons recognised by KLHDC2, KLHDC3 or KLHDC10.
(D) Saturation mutagenesis was performed for two Cul2KLHDC2 substrates (EPHB2 and PDGFC) and for two Cul2KLHDC3 substrates (EMID1 and CHGA). In each case, darker colors represent a greater degree of stabilization conferred by the mutation.
(E) Summary of the C-terminal degrons recognized by KLHDC2, KLHDC3 and KLHDC10.
(F) Comparison of the normalized frequency at the -2 position of the indicated “favored” (G,R, K, Q, W, P, A) or “disfavored” (D, E, V, I and L) amino acids for recognition by KLHDC2, KLHDC3 and KLHDC10.
(G) Depletion of C-terminal glycine is specific to the proteomes of eukaryotes and eukaryotic viruses. See also Figure S4.
