SUMMARY
NusG/RfaH/Spt5 transcription elongation factors are the only transcription regulators conserved across all life. Bacterial NusG regulates RNA polymerase (RNAP) elongation complexes (ECs) across most genes, enhancing elongation by suppressing RNAP backtracking and coordinating ρ-dependent termination and translation. The NusG paralog RfaH engages the EC only at operon polarity suppressor (ops) sites and suppresses both backtrack and hairpin-stabilized pausing. We used single-particle cryo-EM to determine structures of ECs at ops with NusG or RfaH. Both factors chaperone base pairing of the upstream duplex DNA to suppress backtracking, explaining stimulation of elongation genome-wide. The RfaH-opsEC structure reveals how RfaH confers operon specificity through specific recognition of an ops hairpin in the single-stranded nontemplate DNA and tighter binding to the EC to exclude NusG. Tight EC binding by RfaH sterically blocks the swiveled RNAP conformation necessary for hairpin-stabilized pausing. The universal conservation of NusG/RfaH/Spt5 suggests that the molecular mechanisms uncovered here are widespread.
Graphical Abstract
INTRODUCTION
Multi-subunit RNA polymerases (RNAPs) interact with a wide array of accessory proteins that modulate every step of RNA synthesis. Among them, NusG/Spt5 is the only regulator that is conserved in all domains of life (Werner, 2012). NusG/Spt5 co-localizes with elongating RNAP across most genes (Mayer et al., 2010; Mooney et al., 2009a), typically enhancing transcript elongation by reducing RNAP pausing (Herbert et al., 2010; Hirtreiter et al., 2010) but also connecting transcription to diverse cellular processes through contacts with other regulators of RNA biogenesis (Werner, 2012). For example, Escherichia coli (Eco) NusG contacts ribosomes (Saxena et al., 2018), termination factor ρ (Li et al., 1993), or NusA (Said et al., 2017); metazoan Spt5 interacts with the negative regulator NELF to stimulate promoter-proximal pausing (Yamaguchi et al., 1999), as well as many other interaction partners (Werner, 2012; Hartzog and Fu, 2013). Specialized NusG/Spt5 paralogs, generated during evolutionary diversification, have been identified in bacteria (Goodson et al., 2017) and eukaryotes (Bies-Etheve et al., 2009).
Bacterial NusG is a two-domain monomer. Spt5 forms a heterodimer with Spt4 in archaea and eukaryotes (Werner, 2012). The NusG N-terminal domain (NGN), present in all NusG/Spt5-family proteins, contacts RNAP and is followed by a single KOW (Mooney et al., 2009b) domain in bacteria and archaea, five KOWs and a C-terminal phosphorylated repeat region in eukaryotes, and two more KOWs in metazoans and some plants. The NGN contacts the β′/RPB1 clamp helices (CH) and β/RPB2 on opposite sides of the active site cleft in RNAPs from all domains of life (Bernecky et al., 2017; Ehara et al., 2017; Hirtreiter et al., 2010; Klein et al., 2011; Martinez-Rucobo et al., 2011; Sevostyanova et al., 2011; 2008), although an alternative location has been proposed based on a NusG-RNAP co-crystal structure lacking nucleic acids (Liu and Steitz, 2017). By bridging the cleft, the NGN has been proposed to function as a processivity clamp that ensures uninterrupted RNA synthesis.
The NGN alone modulates RNAP pausing (Belogurov et al., 2007; Hirtreiter et al., 2010; Mooney et al., 2009b). The KOWs serve as contact sites for interacting proteins, and the Spt5 KOWs also contact RNA or DNA (Bernecky et al., 2017; Ehara et al., 2017) to aid elongation or stabilize binding to the EC (Crickard et al., 2016).
Life’s only universal transcription factor plays a central role in pausing, underscoring the importance of pausing in gene regulation. Pausing is proposed to aid timely recruitment of transcription regulators, guide nascent RNA folding, oppose chromatin silencing and genome instability, permit termination, and match the rate of transcription to those of other coupled processes, such as translation and splicing (Mayer et al., 2017). However, excessive pausing may lead to arrest, particularly in protein-coated DNA such as eukaryotic chromatin. Indeed, yeast Spt5 assists RNAP progression through the nucleosome (Crickard et al., 2017). Although recent results have suggested biophysical mechanisms for pausing via uncoupling of RNA and DNA translocation and for pause stabilization via rotation of an RNAP “swivel module” (Kang et al., 2018), the mechanism by which NusG/Spt5 suppresses pausing is unclear.
Eco RfaH is a NusG/Spt5 paralog that does not stimulate ρ, but exhibits strong anti-backtracking activity (like NusG/Spt5), can recruit ribosomes to nascent RNAs via its KOW, and unlike NusG, can counteract the pause-stabilizing effects of nascent RNA hairpins (pause hairpins; PHs) (Kolb et al., 2014). RfaH is recruited early in transcription units containing the ops sequence, which is exposed in the non-template strand DNA (nt-DNA) of paused elongation complexes (PECs) (Artsimovitch and Landick, 2002).
To investigate how interactions of NusG/Spt5/RfaH suppress backtrack and PH-stabilized pausing, and to visualize sequence-specific interaction of RfaH with the nt-DNA of ECs, we determined single-particle cryo-electron microscopy (cryo-EM) structures of Eco NusG or RfaH bound to an ops-containing EC (opsEC). We used the structures to design and interpret biochemical experiments that probe the interactions of RfaH and NusG with ECs and the mechanism by which they modulate pausing. Together, our results suggest a molecular model for the effects of NusG/Spt5-family proteins on transcription elongation.
RESULTS
Cryo-EM structures of a NusG-opsEC and an RfaH-opsEC
For cryo-EM structure determination of the NusG-opsEC and RfaH-opsEC, we designed an RNA-DNA scaffold based on the A20 (20mer RNA transcript with A at the 3′-end) scaffold used previously for cryo-EM structure determination of an Eco EC (Kang et al., 2017) except containing the ops sequence in the nt-DNA (Figure 1A). NusG suppresses pausing at and downstream from ops, whereas RfaH induces a strong pause 1–2 nucleotides after the ops pause (A20 on the opsEC scaffold; Figure 1A) (Artsimovitch and Landick, 2000; 2002). To ascertain that the opsEC obtained by direct reconstitution on the ops-scaffold supports NusG and RfaH function, we monitored RNA extension of A20 RNA in the presence of NusG or RfaH. In agreement with data obtained on standard templates (Artsimovitch and Landick, 2002, Mooney et al., 2009b), NusG reduced RNAP pausing downstream of ops whereas RfaH inhibited escape due to specific ops/RfaH interactions (Figure S1). We conclude that NusG or RfaH bind the directly reconstituted opsEC and modulate its function as expected.
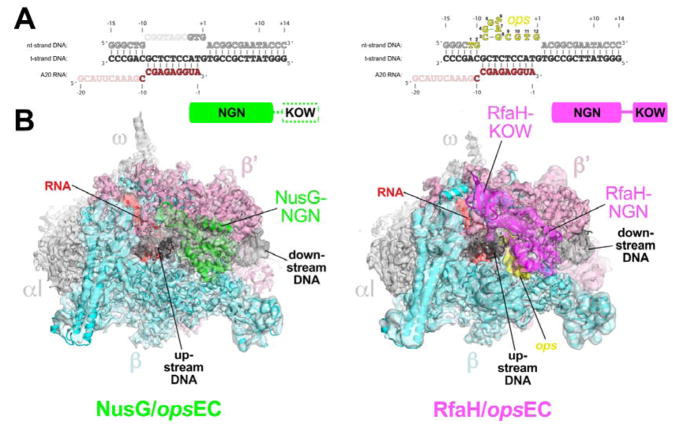
Figure 1. Structures of the NusG-opsEC and RfaH-opsEC.
A. Nucleic acid scaffold sequence used for cryo-EM. The same sequences were used in the NusG-opsEC (left) and RfaH-opsEC (right) structures. Disordered segments in each structure are faded. The nt-DNA ops sequence (nt-DNA −11 to +1, colored yellow for the RfaH-opsEC and numbered according to the ops position) forms a short hairpin that interacts specifically with RfaH but was disordered in the NusG-opsEC (left).
B. The cryo-EM density maps for the NusG-opsEC (left, 3.7 Å nominal resolution, low-pass filtered to the local resolution) and RfaH-opsEC (right, 3.5 Å nominal resolution, but shown is the 3.7 Å nominal resolution map with full-length RfaH, low-pass filtered to the local resolution) are rendered as transparent surfaces and colored as labeled. Superimposed are the final models; proteins are shown as backbone ribbons, the nucleic acids as sticks. The domain organization of NusG (green) and RfaH (magenta) are indicated above. The disordered NusG-KOW is shown in white with a dashed green outline.
See also Tables S1, S2, and Figures S1 – S7.
We used cryo-EM to determine the Eco NusG-opsEC and RfaH-opsEC structures (Figure 1B). The NusG-opsEC structure was determined to a nominal resolution of 3.7 Å (Figures 1B, S2A, S3, S4, Table S1). Local resolution calculations indicate that the central core of the structure, including much of NusG, was determined to 3.0 – 3.5 Å resolution (Figure S3H, S4A, B). Although full-length NusG was used, cryo-EM density for only the NusG-NGN was observed; the flexibly tethered NusG-KOW was disordered (Figure 1B). Density corresponding to the NusG-KOW could not be recovered by focused classification approaches (Scheres, 2012). The NusG-NGN alone is necessary and sufficient for NusG-mediated modulation of RNAP pausing (Belogurov et al., 2007; Hirtreiter et al., 2010; Mooney et al., 2009b) whereas the NusG-KOW links the EC to ribosomes (Saxena et al., 2018) and ρ (Li et al., 1993; Mooney et al., 2009b).
The RfaH-opsEC structure was determined to a nominal resolution of 3.5 Å (Figures S2B, S5, S6, Table S1). Local resolution calculations indicate that the central core of the structure, including much of RfaH, was determined to 2.9 – 3.5 Å resolution (Figure S5H, S6). Although full-length RfaH was used, cryo-EM density for only the RfaH-NGN was observed in the 3.5 Å resolution map. A particle classification focused on the flap tip, RNA exit channel, and upstream duplex DNA gave rise to a second RfaH-opsEC reconstruction from a sub-population of the particles (3.7 Å nominal resolution) that revealed cryo-EM density corresponding to the RfaH-KOW (Figures 1B, S2B, Table S1).
The RNAPs of the NusG- and RfaH-opsEC structures were very similar to previously reported Eco EC cryo-EM structures (Kang et al., 2017), with rmsds of 0.64 Å (2,666 Cα′s aligned) and 0.59 Å (2,732 Cα′s aligned), respectively. The NusG- and RfaH-opsEC structures were nearly as similar to an EC structure as two independently determined EC structures were to each other (Table S2).
Initial examination of the structures revealed several key observations. First, both NusG and RfaH-NGN bound to the same location on the upstream face of the EC cleft, covering the single-stranded nt-DNA and upstream fork junction of the transcription bubble (Figure 1B). The location and orientation of the NGN domains is consistent with biochemical analyses of NusG and RfaH interactions with the EC (Belogurov et al., 2007; 2010; Mooney et al., 2009b) as well as structural analyses of archaeal and metazoan Spt4/5 complexes (Bernecky et al., 2017; Ehara et al., 2017; Martinez-Rucobo et al., 2011) (Figures S7A, B). Our structures are not consistent with a crystal structure of an Eco NusG/core RNAP complex (Liu and Steitz, 2017) that lacks nucleic acids so may not be relevant to NusG function; further, the binding mode of the NusG-NGN seen in our NusG-EC structure (Figure 1B) is precluded by neighboring symmetry-related RNAP molecules in this crystal lattice, explaining the discrepancy (Figure S7C).
Second, binding of the NusG- and RfaH-NGNs constrain the path of the upstream duplex DNA and the upstream segment of the single-stranded nt-DNA, facilitating rewinding of the upstream duplex DNA and possibly explaining the suppression of backtrack pausing (Figure 2). In the RfaH-opsEC, the ops sequence in the single-stranded nt-DNA forms a short hairpin-like structure that interacts sequence-specifically with RfaH (Figures 3, S6A). Space exists in the NusG-opsEC to accommodate the nt-DNA but NusG lacks the capacity to interact sequence-specifically with the DNA and much of the ops sequence in this context is disordered (Figure 1A).
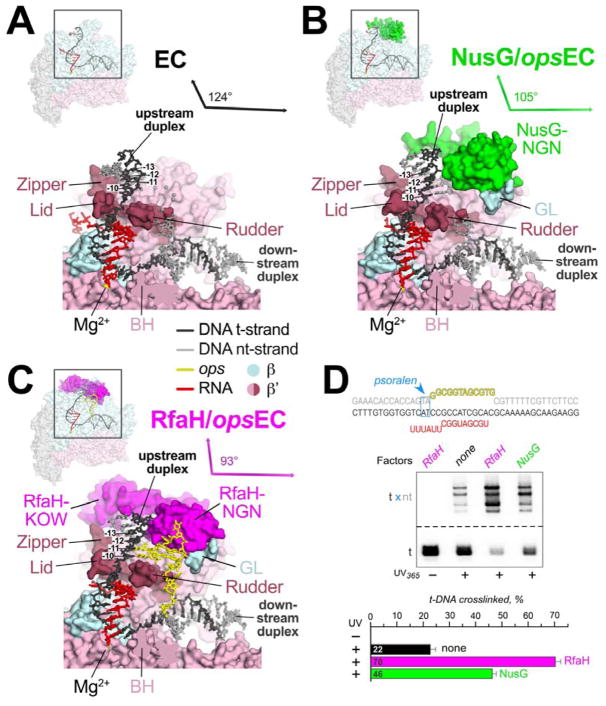
Figure 2. NusG and RfaH remodel and stabilize the upstream duplex DNA.
A. (top left) Overall view of the EC structure (Kang et al., 2017). The RNAP is shown as a transparent molecular surface, revealing the nucleic acid scaffold inside (shown in cartoon format and colored according to the legend). The boxed region is magnified below. (bottom) Magnified view. Most of the β subunit (light cyan) has been removed to reveal the inside of the RNAP active site cleft. The β′ subunit is light pink but with the zipper, lid, and rudder highlighted in brown. The Bridge-Helix (BH) is also labeled. The nucleic acids are shown as sticks (with the first four base pairs of the upstream duplex, - 10 through −13, labeled). The RNAP active site Mg2+-ion is shown as a yellow sphere. The thin black arrows are drawn parallel to the downstream duplex DNA axis (nearly horizontal) and the upstream duplex, which subtends an angle of 124°.
B. As in (A) but showing the NusG-opsEC structure. NusG is green.
C. As in (A) but showing the RfaH-opsEC structure. RfaH is magenta.
D. Probing the upstream fork junction by psoralen crosslinking. The opsECs were assembled on the scaffold shown on top, with the TA intercalation motif (blue) positioned immediately upstream from the ops element; the t-DNA was labeled with [γ32P]-ATP. Following incubation with RfaH or NusG, the ECs were illuminated with 365 nm UV light. The crosslinked products were analyzed on 12 % gels and the fraction of t-DNA crosslinked to the nt-DNA was quantified (bottom). Error bars indicate the s.d. of triplicate measurements.
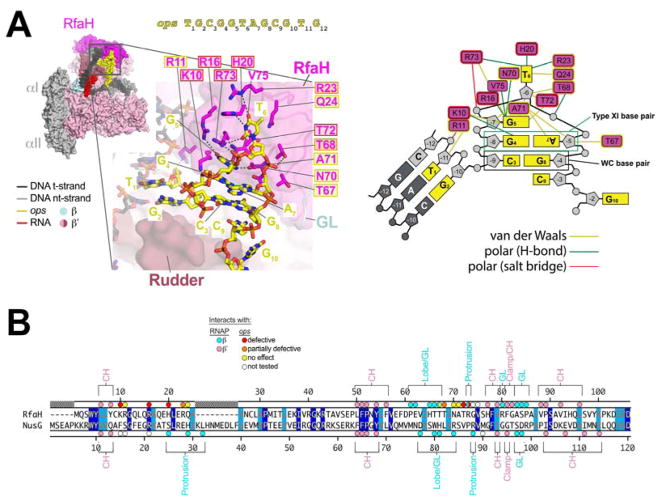
Figure 3. RfaH/ops interactions.
A. (left panel, top) View of the RfaH-opsEC (similar to Figure 2C); the nt-DNA sequence shown on top is numbered according to ops position. Proteins are shown as molecular surfaces. The nucleic acids are shown in CPK format and colored according to the legend. The boxed region is magnified with RfaH rendered transparent, revealing the α-carbon backbone (in cartoon format) and amino acid side chains that interact with ops. Polar RfaH/ops interactions, H-bonds (≤ 3.5 Å) or salt bridges (≤ 4.5 Å) are denoted by gray dashed lines. The ops sequence (yellow) and RfaH (magenta) residues are numbered. The shaded boxes denote the effect of substitutions on RfaH recruitment to ops (red shaded box, defective; orange, partially defective; yellow, no effect; no box, not tested; (Belogurov et al., 2010).
(right panel) Schematic of RfaH/ops interactions. The DNA is color-coded as in (A). The magenta rectangles denote RfaH residues contacting the DNA. Colored lines denote interactions: yellow, van der Waals (≤ 4.5 Å); green, H-bond (≤ 3.5 Å); red, salt bridge (≤ 4.5 Å).
B. Structure-based sequence alignment of the Eco RfaH- and NusG-NGN, numbered above and below, respectively. Identical residues are shaded dark blue, homologous residues light blue. The colored dots on top (RfaH) and bottom (NusG) denote RNAP and ops contacts (color-coded as shown in the legend). The RNAP structural elements that the RfaH (top) and NusG (bottom) residues interact with are noted.
Third, the NusG- and RfaH-NGNs make contacts with the RNAP that bridge across the upstream face of the active site cleft (Figures 4A, B), stabilizing the overall active conformation of the EC and disfavoring the swiveled conformation associated with PH-stabilized pausing (Figure 4C) (Kang et al., 2018). The stabilization of the active EC RNAP conformation by RfaH is stronger than NusG (Figure 5A–F), and RfaH is more effective at inhibiting hairpin-stabilized pauses than NusG (Figure 5C) (Belogurov et al., 2009). We will describe these structural features in the context of the roles of NusG and RfaH in transcription elongation, and present biochemical evidence for their roles.
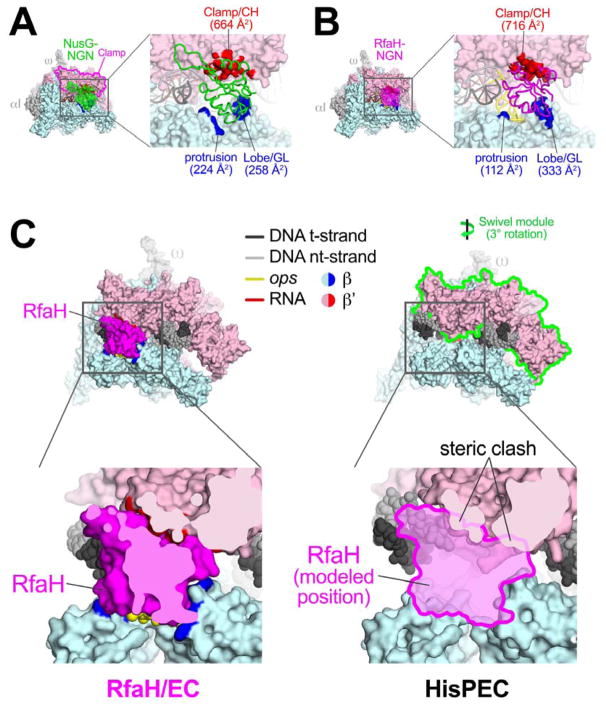
Figure 4. NusG-NGN and RfaH-NGN interactions with RNAP bridge the active site cleft and are incompatible with PH-induced RNAP swiveling.
A. (left) Overall view of the NusG-opsEC structure. Proteins are shown as molecular surfaces, nucleic acids in cartoon format (color-coded as shown in the key or as labeled).
(right) Magnified view of the boxed region on the left. The NusG-NGN is shown as an α-carbon backbone worm. Surfaces of RNAP that contact NusG (≤ 4.5 Å) are colored red (β′ subunit) or blue (β subunit) and labeled along with the buried surface area of the protein/protein interaction.
B. Same as (A) but for the RfaH-opsEC.
C. (left, top) Overall view of the RfaH-opsEC. The boxed region is magnified below.
(left, bottom) Magnified view, sliced at the level of RfaH to reveal the close fit between RfaH and elements of the RNAP β (light cyan) and β′ (light pink) subunits.
(right, top) Overall view of the his PH-stabilized PEC (Kang et al., 2018). The formation of the PH in the RNAP RNA exit channel induces an ~3° rotation (as shown) of the ‘swivel module’ (outlined in green). The boxed region is magnified below.
(right, bottom) Magnified view, but also showing the modeled position of RfaH. The swiveled conformation of the hisPEC is not compatible with RfaH binding due to steric clashes (noted).
Figure 5. RfaH inhibits PH stimulation of pausing and RNAP swiveling more effectively and binds ECs more tightly than NusG.
A. Scaffold used to test asRNA stimulation of pausing. The sequence is identical to that used to determine the hisPEC structure (Kang et al., 2018), except that the PH is replaced with an asRNA binding target (Kolb et al., 2014).
B. Experimental scheme to measure PH stimulation of pausing using asRNA.
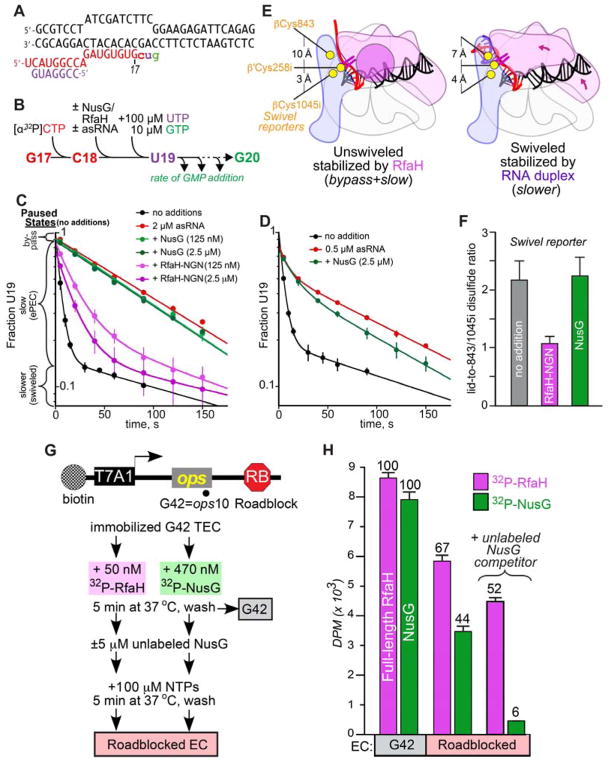
C. Effect of RfaH-NGN and NusG on stimulation of pausing by an RNA duplex. Following UTP and GTP addition to the radiolabeled C18 EC, the fraction of U19 present as a function of time was determined in the presence of indicated ligands. EC fractions corresponding to bypass, elemental pause, and swiveled states are indicated on the y-axis. Data shown are means and s.d. from at least three replicates.
D. Effect of NusG on stimulation of pausing by 0.5 μM asRNA. NusG decreased the slower, apparently swiveled fraction of U19 only modestly, from 0.57 ± 0.02 to 0.49 ± 0.02. Data shown are means and s.d. from at least three replicates.
E. Depiction of RNAP swiveling and location of CTR Cys residues. PH-induced swiveling changes distance from β′C258i to its potential disulfide partners, βC1045i or β′C843 from 3 Å and 10 Å, respectively, to 4 Å and 7 Å (Kang et al., 2018).
F. RfaH-NGN but not NusG decreased the ratio of CTR reporter disulfides on the hisPEC scaffold (containing PH; Kang et al., 2018), indicating that RfaH-NGN but not NusG shifts the equilibrium from the swiveled to the unswiveled RNAP conformation. Data shown are means and s.d. from at least three replicates.
G. RfaH and NusG retention on the EC. A linear DNA template with a T7A1 promoter, the ops element, and an EcoRI site 128 nt downstream from the opsP site was immobilized on streptavidin beads via a biotin on the nt-DNA (top). A cleavage-deficient EcoRIQ111 protein (RB) was bound to roadblock the transcribing RNAP. Halted ops10 (G42) ECs were formed by step-wise transcription with NTP subsets and incubated with radiolabeled RfaH or NusG. After washing away the unbound RfaH/NusG, transcription was resumed by addition of all NTPs with or without an excess of unlabeled NusG, which is expected to bind to RNAP upon dissociation of the pre-bound factor. G42 and roadblocked ECs were washed to remove unbound RfaH/NusG and analyzed by scintillation counting.
H. RfaH (magenta bars) and NusG (green bars) binding to the immobilized ECs. The residual factor binding to the roadblocked ECs is expressed relative to that observed with the G42 EC, which is defined as 100%. Data shown are means and s.d. from three biological replicates.
NusG and RfaH remodel the EC nucleic acids, chaperoning upstream duplex DNA reannealing and explaining the suppression of backtrack pausing
After separating from the RNA-DNA hybrid, the template strand DNA (t-DNA) in the EC is directed out of the RNAP active site cleft through a channel between the β′lid and the β′rudder where it immediately anneals with the nt-DNA (−10 position, Figures 1A, 2A) (Kang et al., 2017). The upstream DNA duplex in the EC is relatively unconstrained and mobile, making few interactions with the RNAP (Kang et al., 2017; Korzheva et al., 2000).
Other than RfaH-R11, which interacts with the nt-DNA phosphate backbone at the −11 position (ops T1; Figure 3A), stable interactions between NusG or RfaH and upstream duplex DNA are not observed and consequently the DNA segment is mobile. Nevertheless, both NusG and RfaH alter the path of the upstream duplex DNA, decreasing the subtended angle with the downstream duplex DNA (Figures 2B, C, S4C, D).
Psoralen intercalates into double-stranded DNA at TA steps and forms a T-T interstrand crosslink when activated by UV light. Psoralen crosslinking efficiency serves as a probe of DNA structure since psoralen intercalates most efficiently in stable, B-form duplex DNA. Based on results of psoralen crosslinking and fluorescence quenching, Turtola and Belogurov (2016) proposed that the −10 base pair at the upstream fork-junction of the EC transcription bubble was distorted, as later observed in the EC structure (Kang et al., 2017). We found that NusG increased the efficiency of psoralen crosslinking at the upstream fork-junction more than two-fold over the EC (Figure 2D), as observed previously (Turtola and Belogurov, 2016). RfaH increased the crosslinking efficiency more than 3.5-fold over the EC. Taken together, these results suggest that NusG and RfaH chaperone and stabilize the formation of the −10 bp.
RfaH recognizes ops as an nt-DNA hairpin
In the EC, the ten nucleotides of single-stranded nt-DNA (−9 to +1) in the transcription bubble span from the −10 to +2 nucleotides, which are base-paired in the upstream and downstream DNA duplexes, respectively (Figure 1A). Repositioning of the upstream duplex DNA by NusG/RfaH reduces the distance separating the −10 and +2 nt-DNA phosphates from 41 Å in the EC to 33 Å in the NusG-opsEC and 30 Å in the RfaH-opsEC. In the RfaH-opsEC, a short hairpin with a two base-pair stem forms in the single-stranded nt-DNA and this DNA structure is specifically recognized by RfaH (Figures 3A, S6A).
Using the ops sequence numbering (Figure 3A) rather than the scaffold numbering, ops T1 and G2 are base-paired as part of the upstream DNA duplex (Figure 3A). C3 forms a Watson-Crick base pair with G8, while G4 and A7 participate in a Saenger type XI base pair (Saenger, 1984) to form the ops hairpin stem. G5 stacks on the upstream face of G4 while T6 is flipped out of the base stack. C9 stacks with the downstream face of G8. The rest of the ops sequence (G10T11G12) is single-stranded and does not interact with RfaH but interacts with RNAP as in the EC structure.
Other than RfaH-K10, which hydrogen-bonds (H-bonds) with G4(O6), the base pairs C3:G8 and G4:A7 do not make extensive base-specific interactions with RfaH – these bases are conserved in the ops sequence because of their role in forming the ops hairpin stem, the geometry of the which sets up extensive base-specific interactions of RfaH with G5 and T6 (Figures 3A, S6A). The three H-bonding atoms of the Watson-Crick edge of G5 (N2, N1, O6) H-bond with the backbone carbonyls of RfaH-N70 and V75, and with the side chain of R16, respectively. The three H-bonding atoms of the T6 base (O2, N3, O4) participate in H-bonds with the side chains of R73, H20, and R23, respectively. The aliphatic chain of RfaH-Q24 makes van der Waals contact with the T6 exocyclic methyl. All of the RfaH side chains that make base-specific H-bonds (K10, R16, H20, R23, R73) are important for RfaH function. Single Ala substitutions of these residues interfere with RfaH recruitment at ops (Figure 3) (Belogurov et al., 2010).
In the structure-based alignment with NusG, RfaH-K10, H20, R23, and R73 correspond to NusG-F15, S25, E28, and P87, each unable to participate in the equivalent interactions with ops (Figure 3B). The disposition of the DNA in the NusG-opsEC seems compatible with ops hairpin formation (Figure 2B) but cryo-EM density for most of the ops sequence (C3 to C9) is completely absent and the DNA is presumed to be disordered.
NusG and RfaH contacts bridge the RNAP active-site cleft
The RNAP is like a crab claw, with one pincer comprising primarily the β′ subunit, and the other primarily the β subunit (Figure 4) (Zhang et al., 1999). A large cleft between the pincers contains the active site and accommodates nucleic acids in the EC (Gnatt et al., 2001; Kang et al., 2017; Korzheva et al., 2000; Vassylyev et al., 2007). The clamp (Figure 4A), a mobile structural module that makes up much of the β′ pincer (Gnatt et al., 2001), undergoes swinging motions that open the channel to allow entry of nucleic acids during initiation, or that close the channel around the DNA and RNA-DNA hybrid to enable processive elongation (Chakraborty et al., 2012; Gnatt et al., 2001).
The NusG and RfaH-NGN bind the upstream face of the EC, bridging the active-site cleft by contacting the clamp of the β′ pincer and the protrusion and lobe of the β pincer (Figures 4A, B, S4A, B, S6B, C). NusG and RfaH interactions with RNAP are analogous (Figure 3B) with one exception – the first α-helix of NusG (residues 18–34) interacts with the protrusion while the same region of RfaH (residues 13–24) interacts with the ops hairpin, which inserts between RfaH and the protrusion (Figure 4B). Despite these additional NusG/RNAP interactions, overall the NusG/RNAP and RfaH/RNAP interface buries a similar total surface area (NusG, 1,150 Å2; RfaH, 1,160 Å2). After RNAP escape from ops, the specific RfaH-ops contacts are lost and RfaH may establish interactions with the protrusion, significantly increasing the RfaH/RNAP interaction interface and affinity relative to NusG. Consistently, RfaH outcompetes NusG for EC binding in vitro even when NusG-NGN is at a 10-fold excess over RfaH, and excludes NusG from RNAP transcribing ops-operons in the cell (Belogurov et al., 2009) despite NusG being present in large excess (50 to 100-fold) over RfaH in vivo (Schmidt et al., 2016).
The primary interaction determinant for both NusG and RfaH is the clamp helices (CH; Figures 4A, B, S4B, S6C), consistent with previous analyses (Belogurov et al., 2007; 2010; Mooney et al., 2009b; Sevostyanova et al., 2008). Both factors interact with the GL (Figures S4A, S6B). However, interactions of the RfaH HTT motif (residues 65–67; Figures 3B, S6B) with the GL are required for RfaH function (Belogurov et al., 2010; Sevostyanova et al., 2011), whereas deletion of the GL supports normal NusG activity (Nandymazumdar et al., 2016; Turtola and Belogurov, 2016).
Both NusG and RfaH contacts are incompatible with RNA pause hairpin-induced EC swiveling
RfaH efficiently suppresses both backtrack- and PH-stabilized pausing (Artsimovitch and Landick, 2002; Kolb et al., 2014). Nascent PHs can increase pause lifetimes tenfold or more (Toulokhonov et al., 2001). Recent cryo-EM studies revealed that formation of the hisPH in the RNAP RNA exit channel induced a previously unseen global conformational change in the RNAP termed ‘swiveling’ (Kang et al., 2018). In the swiveled RNAP, the clamp, shelf, and other structural features of the β′ pincer (called the swivel module; Figure 4C) undergo a concerted rotation of about 3° about an axis roughly parallel with the BH (perpendicular to the RNA-DNA hybrid). A previously described conformational state of the RNAP termed ‘ratcheting’ has in common with swiveling the rotation of the shelf and other structural features of the β′ pincer, but RNAPs in the ratcheted conformation have an open clamp (Tagami et al., 2010; Weixlbaumer et al., 2013; Zhang et al., 1999). PH-induced swiveling is unique in combining rotation of the shelf and other structural features with rotation of the closed clamp (Kang et al., 2018). Swiveling is thought to increase pause lifetimes by allosterically inhibiting trigger-loop folding (Kang et al., 2018). Swiveling alters the relative positions of the β′ and β pincers which the bound RfaH bridges (Figure 4B), and modeling reveals that RfaH binding is incompatible with the swiveled state (Figure 4C).
Modeling indicates that NusG binding is also incompatible with the swiveled conformation. The greater inhibition of PH action and of RNAP swiveling by RfaH is likely due to stronger binding of RfaH to ECs. To explore these differences, we used a scaffold resembling the ops cryo-EM scaffold but containing his pause sequences that form an RNA-duplex-stabilized pause upon addition of an antisense RNA oligonucleotide (asRNA; Figure 5A) (Kang et al., 2018). RfaH, but not NusG, can outcompete binding of an 8mer asRNA to a similar scaffold (Kolb et al., 2014); thus, we used a 7mer asRNA to maximize the ability of NusG to compete (Figure 5A).
C18 ECs formed one nucleotide upstream of the pause were reacted with UTP and GTP (Figure 5B). In the absence of ligands, the fraction of U19 present as a function of time was triphasic (Figure 5C). A small fraction of ECs failed to pause (bypass fraction). Most U19 ECs entered the elemental paused state and exhibited slow addition of G20. A small fraction of ECs added G20 even more slowly; we propose that this fraction may represent the swiveled state. Added at 2 μM, the 7mer asRNA stimulated pause dwell time at U19 ~20-fold. Whereas 125 nM RfaH could suppress pause stimulation by 2 μM asRNA almost as effectively as 2.5 μM RfaH, 2.5 μM NusG had little or no effect on asRNA action (Figure 5C). When asRNA was lowered to 0.5 μM, 2.5 μM NusG gave a minimally detectable effect (Figure 5D).
Consistent with an ability of RfaH but not NusG to suppress asRNA-induced swiveling, a cysteine-triplet reporter (CTR) that detects swiveling by a shift in disulfide bond formation by β′-lid Cys258i from βCys1045i to βCys843 (Kang et al., 2018) reported RfaH but not NusG suppression of the swiveled conformation on a hisPEC scaffold (Figures 5E, F).
To ask if this reduced effect of NusG could be explained by weaker binding of NusG vs. RfaH to ECs, we performed a NusG-RfaH competition experiment (Figures 5G, H). RfaH or NusG was bound to ECs halted at ops by step-wise walking of RNAP after initiation on a T7 A1 promoter template. Upon addition of all four NTPs, ECs moved along the template until encountering a roadblock generated by a noncleaving mutant EcoRI endonuclease bound 128 nt downstream of ops (Pavco and Steege, 1990), so ops-RfaH contacts were no longer possible. When the roadblocked ECs were washed with buffer, radiolabeled RfaH was retained to a greater extent than radiolabeled NusG (Figure 5H). Almost no radiolabled NusG was retained when unlabeled NusG competitor was present at 5 μM, whereas most radiolabeled RfaH remained bound in the presence of NusG competitor. These data establish that, once associated with an EC at ops, RfaH remains bound even in the absence of ops contacts, whereas NusG is readily lost. In support of our structural observations that, once RNAP escapes from ops, the RfaH-EC interactions likely bury a larger surface area than NusG-EC interactions, these results are consistent with much weaker NusG-EC binding thatn RfaH-EC binding.
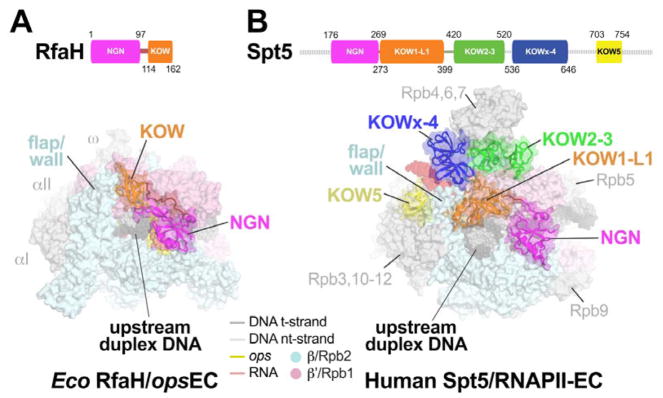
RfaH KOW domain binds RNAP and remodels the upstream duplex DNA path and RNA exit channel like Spt5
The positioning of Spt5 KOW1-5 domains on the surface of RNAPII by cryo-EM (Bernecky et al., 2017; Ehara et al., 2017) suggests that the Spt5-KOWs may contact upstream DNA, RNA, and transcription regulators from fixed locations on the EC, rather than on freely rotating tethers. The RfaH-KOW binds RNAP at a location similar to that occupied by Spt5-KOW1 (Figure 6). The side-chain contacts and orientations of Spt5-KOW1 and RfaH-KOW are not conserved, but their locations relative to RNAP and upstream duplex DNA are similar. For both structures, the open face of the large KOW domain β sheet faces away from RNAP, but the RfaH-KOW is rotated ~45° and shifted about 10 Å toward the RNA exit channel relative to Spt5-KOW1. Both KOWs appear to guide the upstream duplex DNA. Spt5-KOW1 contains an eukaryotic-specific insertion called L1 that increases the extent of upstream DNA contact. These DNA contacts are consistent with increased protection of upstream DNA against exonuclease III digestion by Spt4/5 (Crickard et al., 2016).
Figure 6. Comparison of the RfaH-opsEC with an Spt5-RNAPII-EC.
A. (top) Schematic showing RfaH domains (NGN, magenta; KOW, orange), with amino acid residues numbered.
(bottom) overall view of the RfaH-opsEC. The RNAP is shown as a molecular surface, the nucleic acids in CPK format (colored as shown in the key or as labeled). RfaH is shown as a backbone ribbon with a transparent molecular surface, colored as in the schematic above. The RfaH-KOW binds directly above the upstream DNA duplex and next to the β flap/wall.
B. (top) Schematic showing the domain organization of Spt5 (Bernecky et al., 2017), with amino acid residues numbered and disordered regions denoted as a dashed line.
(bottom) overall view of the human Spt5-RNAPII-EC (Bernecky et al., 2017). This structure also includes Spt4, which has been removed for clarity. The RNAP is shown as a molecular surface, the nucleic acids in CPK format (colored as shown in the key or as labeled). Spt5 is shown as a backbone ribbon with a transparent molecular surface, colored as in the schematic above. The Spt5-KOW1-L1 domain binds directly above the upstream DNA duplex and next to the Rbp2 flap/wall. Also see (Ehara et al., 2017).
See also Figure S8.
Both RfaH-KOW and Spt5-KOWs affect the RNA exit channel, whose mouth is formed in part by the tip of a module called the flap in bacteria and the wall in eukaryotes. The bacterial flap tip is capped with an α-helix that contacts either σ factors or NusA and is required for their function (Guo et al., 2018; Kuznedelov et al., 2002). In contrast, the flap tip appears to be dispensable for human RNAPII transcription (Palangat et al., 2011). Both the bacterial flap tip and the eukaryotic wall/flap tip are flexible and often disordered in crystal and cryo-EM structures, but strikingly both are contacted by RfaH-KOW/Spt5-KOW1. These contacts pull the bacterial flap tip away from the RNA exit channel into a novel location (Figure 6). It is possible that this change contributes to RfaH effects on PH-stabilized pausing, since deletion of the flap tip eliminates PH stimulation of pausing (Hein et al., 2014). Given the lack of documented function for the wall, its contacts with Spt5-KOW1 may not have significant effects on transcription. However, the Spt5-KOWx-4 domain binds near the path of the exiting RNA immediately upstream from a bacterial PH, and would partially clash with the location of the PH (Kang et al., 2018). Thus, Spt5 KOWx-4 could inhibit formation of nascent RNA structures in the RNAPII RNA exit channel or shift to accommodate them.
DISCUSSION
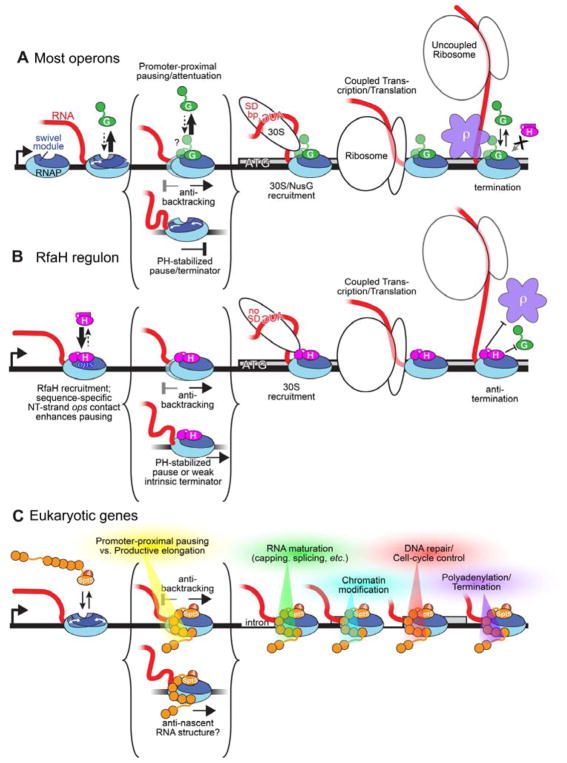
Transcription in all cellular organisms is a focal point for the regulation of gene expression. Although the central enzyme of transcription, the multisubunit cellular RNAP, is evolutionarily conserved across all life, this conservation does not extend to regulation. For instance, each RNAP faces similar mechanistic challenges during promoter-specific initiation but relies on evolutionarily unrelated basal factors (Werner, 2012). Similarly, unrelated factors control the elongation and termination phases of the transcription cycle, with one exception. NusG/Spt5 elongation factors are structurally (Figure 6) and functionally homologous, making them the only transcription regulators that are conserved in all domains of life (Werner, 2012). We report here key insights into the mechanistic basis for regulation of RNAP function by this family of universal regulators from cryo-EM structures of Eco NusG and its operon-specific paralog Eco RfaH engaged with an EC (Figure 1). We show that (i) NusG and RfaH stabilize base-pairing of the upstream duplex DNA (Figure 2), providing an explanation for their ability to suppress backtrack pausing; (ii) RfaH achieves operon-specificity in part by specific recognition of an ops DNA hairpin in the exposed nt-DNA of the EC transcription bubble (Figures 3, S6A), and (iii) RfaH suppresses PH-stabilized pausing by preventing RNAP swiveling (Figures 4, 5).
The mechanistic basis for NusG/RfaH regulation of RNAP pausing
RNAP pausing is a key mechanism for regulating gene expression in all organisms (Mayer et al., 2017). The mechanistic and structural basis for transcriptional pausing is understood in greatest detail in bacteria (Artsimovitch and Landick, 2000; Kang et al., 2018). Pauses initially arise when specific sequences prevent complete translocation of the DNA, resulting in an offline elemental paused EC (ePEC) with post-translocated RNA transcript but pre-translocated DNA (Kang et al., 2018). This hybrid state resists NTP binding, providing time for the ePEC to isomerize into other, more long-lived paused states: (i) Backtracking prolongs pausing by disengaging the RNA 3′-terminus from the RNAP active site (Artsimovitch and Landick, 2000); (ii) PH formation stabilizes the RNAP in the swiveled conformation (Figure 4C), prolonging pausing by allosterically inhibiting trigger-loop folding required for the optimal active site configuration (Kang et al., 2018). The results presented here allow us to propose mechanistic hypotheses for the complex effects of NusG and RfaH on RNAP elongation.
The primary effects of NusG-NGN and RfaH-NGN on the EC are i) stabilizing base-pairing in the upstream duplex DNA (NusG and RfaH; Figure 2), and ii) inhibiting RNAP swiveling (RfaH; Figures 4C, 5). Formation of the ePEC is associated with an incompletely translocated intermediate without major conformational changes in the RNAP (Kang et al., 2018), and accordingly NusG and RfaH are not known to have strong effects on the ePEC (Larson et al., 2014).
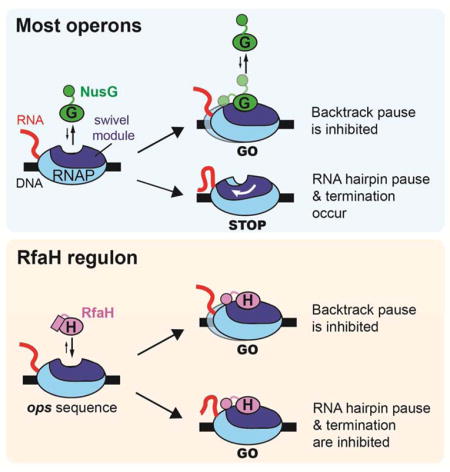
Both NusG and RfaH increase the overall transcription elongation rate by suppressing backtrack pausing (Belogurov et al., 2010; Turtola and Belogurov, 2016). Our structural and biochemical analyses (Figure 2) support the conclusion that both NusG and RfaH suppress backtracking by stabilizing the −10 bp of the upstream duplex DNA, as proposed by Turola and Belogurov (2016) for NusG.
Both NusG and RfaH binding are sterically incompatible with RNAP swiveling (Figure 4C), but only RfaH efficiently suppresses PH-stabilized pausing (Figure 5C). The results of the CTR assay (Figures 5E, F) and the NusG/RfaH retention assay (Figures 5G, H) argue that the binding energy of RfaH to the EC is sufficient to inhibit PH formation and suppress RNAP swiveling, whereas NusG binding energy is not. These results explain how RfaH can suppress PH-stabilized pausing by counteracting RNAP swiveling, whereas NusG cannot.
Adaptations in RfaH confer operon-specificity
In bacteria, the specialized NusG paralog RfaH has maintained key functions of the NGN (EC binding, suppression of backtrack pausing) and tethered KOW (ribosome interactions) but has developed an elaborate regulatory mechanism to confer operon-specificity. These include specific recognition of an ops-hairpin in the exposed nt-DNA of the transcription bubble (Figures 3A, S6A) and a molecular switch that auto-inhibits RfaH-NGN-RNAP interactions in the absence of ops recognition (Belogurov et al., 2007) (Figures 7A, B).
Figure 7. Comparison of NusG, RfaH, and Spt5 action as elongation regulators.
A. On most bacterial operons, NusG (G, green) associates with RNAP weakly until RNAP enters a protein-coding gene and NusG-KOW interaction with 30S is possible. Weak NusG interaction is sufficient to inhibit backtracking but not to inhibit PH-induced RNAP swiveling. When transcription and translation become significantly uncoupled, NusG activates Rho-dependent termination via NusG-KOW-Rho interaction.
B. On ops-containing operons, RfaH (H, magenta) is recruited by contacts to the ops hairpin in the exposed nt-DNA (yellow; Figure 3A). The refolded RfaH-KOW domain interacts with RNAP and the tightly bound RfaH excludes NusG, inhibits backtracking, and inhibits PH stimulation of pausing by preventing RNAP swiveling (Figure 4C). RfaH associates with 30S similarly to NusG, but inhibits Rho indirectly upon significant transcription-translation uncoupling by excluding NusG.
C. Spt4/5 (4 and 5, orange) associates with RNAPII in the promoter-proximal region though NusG homologous contacts of its NGN and of 5 its 7 KOWs. Spt5 inhibits backtracking, may inhibit swiveling and formation of nascent RNA structures, and mediates the regulatory switch between promoter-proximal pausing/attenuation and productive elongation in part through the actions of NELF and P-TEFb. The full set of Spt5 interactions involved in this switch as well as in downstream roles in mediating RNA maturation, chromatin modifications, recruiting of DNA repair factors, cell-cycle control, polyadenylation, and termination remains incompletely defined.
The cryo-EM structure of the RfaH-opsEC represents the RfaH ‘loading’ complex, revealing the structural adaptations that allow specific ops-RfaH recognition (Figure 3). The auto-inhibitory RfaH C-terminal helical hairpin sterically blocks the protein-protein interactions between RfaH-NGN and the RNAP β′ CH (Figure 4B) but would not interfere with ops-RfaH interactions. Presumably, pausing of the EC precisely at ops displays the ops hairpin in the nt-DNA to allow for RfaH recognition and establishment of RfaH/RNAP β subunit interactions (Figure 4B). Thermal fluctuations of the RfaH auto-inhibitory helical hairpin allow RfaH-RNAP β′ CH interactions, stabilizing the RfaH-NGN-EC interactions and freeing the RfaH-C-terminal domain (CTD) to refold into the KOW (Figure 1B). Unlike the NusG-KOW, which is disordered in our cryo-EM maps, the RfaH-KOW was pinned down through interactions with the EC in a sub-population of the particles (Figures 1B, 6A, S2B), similar to the Spt5-KOW1-L1 domain (Figure 6B). Weak RfaH-KOW-EC interactions could help prevent the RfaH-CTD from competing with the RNAP β′ CHs for RfaH-NGN interactions in the absence of other RfaH-KOW interactors, such as the ribosome (Figure 7B).
The RfaH-NGN binds to the EC with a higher affinity than the NusG-NGN (Belogurov et al., 2009) (Figure 5H), a second adaptation that allows RfaH to function in an operon-specfic manner by excluding NusG from the RfaH-EC. Moreover, the tighter binding of the RfaH-NGN confers its ability to counteract RNAP swiveling, providing a mechanistic basis for the suppression of PH-stabilized pauses (Figures 5C, 7B).
Complex roles of NusG/Spt5 factors in vivo
The in vivo roles of both NusG and Spt5 are complex. They both stimulate transcript elongation by RNAP over much of the genome, but more importantly for the cell they serve as recruitment platforms for accessory factors to coordinate transcription elongation with other cellular functions (Figure 7). For example, NusG plays a crucial role in ρ-dependent termination through direct NusG-KOW-ρ interactions (Li et al., 1993) and functional/scaffolding roles in multiprotein assemblies that effect antitermination (Said et al., 2017).
RfaH function is confined to transcription units containing an ops sequence in an upstream segment but is no less complex. RfaH uses its ability to suppress backtrack and RNA hairpin-stabilized pausing and to coordinate with the ribosome (but not ρ) through its KOW to ensure the efficient transcription of long operons.
The control and release of promoter-proximal pausing by RNAPII is a major checkpoint for regulating gene expression in metazoans, and Spt4/5 (called DSIF in metazoa) plays a key role therein (Kwak and Lis, 2013). DSIF cooperates with the metazoan-specific NELF to enable promoter-proximal pausing (Yamaguchi et al., 1999), and is modified by P-TEFb to transition into an elongation activator that remains bound to the EC with incompletely understood roles during genic transcription (Kwak and Lis, 2013). A key question now is if DSIF and its regulatory partners control RNAPII pausing using similar mechanisms as those revealed here for NusG and RfaH.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
S. A. Darst, darst@rockefeller.edu
METHODS DETAILS
RNAP expression and purification for Cryo-EM
Eco RNAP lacking the αCTDs was prepared as described previously (Twist et al., 2011). The αCTD has not been implicated in transcription control by NusG nor RfaH. Glycerol was added to the purified RNAP to 15% (v/v), and the sample was aliquoted and flash-frozen in liquid nitrogen. The aliquots were stored at −80 °C until use.
NusG expression and purification
NusG was purified from pRM1160. pRM1160 was generated by Gibson assembly of the wild-type nusG gene PCR amplified from Eco chromosomal DNA using primers 10224 and 10225 with a vector fragment amplified from pRM756 using primers 10226 and 10027. pRM1160 is kanamycin-resistant, contains the T7 promoter upstream of nusG, and encodes nusG followed by a precision protease cleavage site and ten histidine residues.
To prepare full-length Eco NusG, plasmids encoding NusG with a C-terminal (His)6-tag were grown in Eco BL21 (DE3) in LB with 50 μg/ml kanamycin at 37 °C to an OD600 of 0.5, induced for protein expression by addition of IPTG to final concentration of 0.5 mM, grown for an additional 3 hours, then harvested by centrifugation at 17,600 g for 30 minutes at 4°C. Harvested cells were resuspe nded in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 0.1 mM PMSF, and lysed in a continuous flow French Press (Avestin, Ottawa, Ontario, Canada). The lysate was centrifuged (17,600 g, 30 minutes, 4°C) and the sup ernatant was loaded onto a HiTrap IMAC column (GE healthcare Life Sciences, Pittsburgh, PA) charged with Ni2+. The column was washed with IMAC buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, β-mercaptoethanol) with 30 mM imidazole, then eluted with an imidazole gradient to 250 mM. The protein eluted at about 70 mM imidazole and was inclubated with human rhinovirus (HRV) 3C protease to cleave the C-terminal (His)6-tag and dialyzed for overnight at 4 °C against 20 mM Tris-HCl, pH 8.0, 1 00 mM NaCl, 1 mM DTT. The dialyzed and tag-cleaved protein was loaded onto a Hitrap Q (GE Healthcare Life Sciences) and eluted with a NaCl gradient to 1 M. The protein eluted at about 200 mM NaCl and was subsequently purified by gel filtration chromatography on a HiLoad Superdex75 column (GE Healthcare Life Sciences) in 10 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% (v/v) glycerol, 0.5 mM EDTA, 1 mM DTT. Additional glycerol was added to the purified NusG to 15% (v/v), and the sample was aliquoted and flash-frozen in liquid nitrogen. The aliquots were stored at −80 °C until use.
RfaH expression and purification
Wild-type Eco RfaH was expressed and purified as described previously (Vassylyeva et al., 2006), flash-frozen in liquid nitrogen and stored at −80 °C until use.
Purification and labeling of HMK-tagged RfaH and NusG
Plasmids encoding NusG and RfaH with an N-terminal (His)6-tag and a protein kinase A recognition site (RRASV) were grown in Eco XJb (DE3) in LB with 40 μg/ml kanamycin at 37 °C to an OD 600 of 0.4, induced by addition of IPTG to final concentration of 0.2 mM, and grown at 18 °C overnight; L-arabinose w as added to 0.07% 2 hours before harvesting. The cells were harvested by centrifugation (18,000 g, 30 minutes, 4°C), resuspended in 50 mM Tris-HCl, pH 6.9, 500 mM NaCl, 5% Glycerol and 1 mM β-mercaptoethanol supplemented with complete, EDTA-free Protease Inhibitor Cocktail (Roche) and incubated with 0.25 mg/ml of Lysozyme on ice for 30–40 minutes. The cells were then lysed by sonication. The lysate was centrifuged (18,000 g, 30 minutes, 4°C) and the supernatant was loaded onto a gravity column with Ni Sepharose 6 Fast Flow resin (GE Healthcare Life Sciences, Pittsburgh, PA) equilibrated with 50 mM Tris-HCl, pH 6.9, 500 mM NaCl, 5% Glycerol and 1 mM β-mercaptoethanol. The column was washed 20 mM imidazole. The proteins were eluted with an imidazole gradient to 250 mM in 50 mM Tris-HCl, pH 6.9, 80 mM NaCl, 5% (v/v) Glycerol and 1 mM β-mercaptoethanol, directly loaded onto a 5-ml HiTrap Heparin column, and eluted with an NaCl gradient to 1 M. The peak fractions were dialyzed against 10 mM Tris-HCl, pH 7.9, 250 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 50% (v/v) glycerol and flash-frozen for storage at −80 °C.
RfaH-opsEC and NusG-opsEC preparation for Cryo-EM
Synthetic DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA), RNA oligonucleotides from GE Healthcare Dharmacon (Lafayette, CO). The nucleic acids for the ops-scaffold (Figure 1A) were dissolved in RNase-free water (Ambion/ThermoFisher Scientific, Waltham, MA) at 0.2–1 mM. Template DNA and RNA were annealed at a 1:1 ratio in a thermocycler (95 °C for 2 min, 75 °C for 2 min, 45 °C for 5 min, followed by steady cooling to 25 °C at 1 °C/min). The annealed RNA-DNA hybrid was stored at −80 °C until use. Purified Eco RNAP was buffer-exchanged over a Superose 6 INCREASE (GE Healthcare Life Sciences) column into 20 mM Tris-HCl, pH 8.0, 120 mM potassium acetate, 5 mM MgCl2, 5 mM DTT. The eluted protein was mixed with the pre-annealed RNA-DNA hybrid at a molar ratio of 1:1.3 and incubated for 15 min at room temperature. Nt-DNA and additional 5 mM MgCl2 was added and incubated for 10 min. RfaH or NusG (buffer exchanged over the Superose 6 INCREASE column in the same buffer as RNAP) was added to a molar ratio of 1:3 (RNAP:RfaH or RNAP:NusG). The complex was concentrated by centrifugal filtration (EMD Millipore, Billerica, MA) to 4.0–5.5 mg RNAP/ml concentration before grid preparation.
Cryo-EM grid preparation
Before freezing, CHAPSO was added to the samples to 8 mM final concentration. C-flat (Protochips, Morrisville, NC) CF-1.2/1.3 400 mesh gold grids were glow-charged for 15 s prior to the application of 3.5 μl of the complex sample (4.0–5.5 mg/ml protein concentration), then plunge-frozen in liquid ethane using a Vitrobot mark IV (FEI, Hillsboro, OR) with 100% chamber humidity at 22 °C.
Cryo-EM data acquisition and processing for NusG-opsEC
The grids were imaged using a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan, Pleasanton, CA). Images were recorded with Leginon (Suloway et al., 2005) in counting mode with a pixel size of 1.07 Å and a defocus range of 0.8 to 2.5 μm (Figure S3C). Data were collected with a dose of 8 electrons/physical pixel/s. Images were recorded with a 10 s exposure and 0.2 s sub-frames (50 total frames) to give a total dose of 69.9 electrons/Å2. Structural biology software was accessed through the SBGrid consortium (Morin et al., 2013). Dose fractionated subframes were aligned and summed using Unblur (Grant and Grigorieff, 2015). The contrast transfer function was estimated for each summed image using CTFFIND4 (Rohou and Grigorieff, 2015). From the summed images, particles were automatically picked in Gautomatch (Zhang, unpublished; see Key Resource Table), manually inspected, and then individually aligned using direct-detector-align_lmbfgs software (Rubinstein and Brubaker, 2015). The aligned particles were subjected to 2D classification in RELION specifying 100 classes (Scheres, 2012), and poorly populated classes were removed, resulting in 514,900 particles (Figure S3D). These particles were 3D autorefined in RELION using a map of Eco elongation complex (EMD-8585; Kang et al., 2017), low-pass filtered to 60 Å resolution as an initial 3D template. With this initial model, 3D classification was performed without alignment with a soft mask generated in Chimera (Pettersen et al., 2004) and RELION. The soft mask excluded flexible RNAP domains (SI1, SI3, flap tip helix, and single-stranded nucleic acids) of EC. Among the classes from the 3D classification, the best-resolved and most-populated class was 3D autorefined and subjected to the second 3D classification without alignment with the soft mask that was used in the first 3D classification. From this classification, the best-resolved class containing 171,900 particles was 3D autorefined with solvent flattening, and post-processed in RELION, yielding the final reconstruction at 3.7 Å resolution (Figures S2A, S3F). Local resolution calculations (Figure S3H) were performed using blocres (Cardone et al., 2013).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Eco BL21(λDE3)T-H/pEcrpo(HX-)ABCZ | (Twist et al., 2011) | N/A |
| Eco BLR λDE3 | Novagen | |
| Eco XJb λDE3 | Zymo Research | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| [α-32P]CTP | Perkin Elmer | Cat# BLU008H |
| [γ-32P]ATP | Perkin Elmer | Cat# BLU002Z |
| Bio-Rex 70 cation exchange resin, analytical grade, 100–200 mesh | Bio-Rad | Cat# 1425842 |
| 3-([3-Cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO) | Sigma-Aldrich | Cat# C4695 |
| Dynabeads® MyOne™ Streptavidin C1 | Dynal Biotech ASA | Cat# 650.01 |
| HiLoad 16/600 Superdex 75 pg | GE Healthcare Life Sciences | Cat# 28989333 |
| HiLoad 26/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat# 28989336 |
| HIS-Select® Nickel Affinity Gel | Sigma-Aldrich | Cat# M6611 |
| HiTrap IMAC HP | GE Healthcare Life Sciences | Cat# 17092003 |
| HiTrap Q HP | GE Healthcare Life Sciences | Cat# 17115401 |
| 8-Methoxypsoralen | Sigma-Aldrich | Cat# M3501 |
| Nuclease-free water (not DEPC treated) | Ambion | Cat# 4387936 |
| Polyethyleneimine, ~M.W.60,000, 50% wt.% aqueous solution, branched, Acros Organics 178572500 | Fisher Scientific | Cat# AC178572500 |
| Protein kinase A, catalytic subunit | Sigma-Aldrich | Cat# P2645 |
| Superose 6 INCREASE 10/300 GL | GE Healthcare Life Sciences | Cat# 29091596 |
| T4 polynucleotide kinase | New England Biolabs | Cat# M0201 |
| ΔαCTD Eco RNAP polymerase (cryo-EM samples) | (Twist et al., 2011) | N/A |
| CTR RNAP: β′1045iC 258iC, β843C | Kang et al, 2018 | N/A |
| Eco NusG (cryo-EM) | This paper | |
| Eco NusG (transcription assays) | (Mooney et al., 2009b) | |
| Eco PKA-tagged NusG | This paper | |
| Eco RfaH | (Vassylyeva et al., 2006) | |
| Eco RfaH-NGN (transcription assays) | (Hein et al., 2014) | |
| Eco PKA-tagged RfaH | (Artsimovitch and Landick, 2002) | |
| EcoRIQ111; a mutant variant of EcoRI used as a roadblock | (Strobel et al., 2017) | |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Coordinates of Eco NusG-opsEC | This paper | PDB: 6C6U |
| Coordinates of Eco RfaH-NGN-opsEC | This paper | PDB: 6C6T |
| Coordinates of Eco RfaH-full-length-opsEC | This paper | PDB: 6C6S |
| Coordinates of Eco EC | (Kang et al., 2017) | PDB: 6ALF |
| Coordinates of Eco NusG-NGN | (Mooney et al., 2009b) | PDB: 2K06 |
| Coordinates of Eco RfaH-NGN | (Belogurov et al., 2007) | PDB: 2OUG |
| Coordinates of Eco RfaH-KOW | (Burmann et al., 2012) | PDB: 2LCL |
| Coordinates of E. coli hisPEC | (Kang et al., 2018) | PDB: 6ASX |
| Coordinates of Human DSIF/Pol II-EC | (Bernecky et al., 2017) | PDB: 5OIK |
| Coordinates of Pyrococcus furiosis Spt5-RNAP clamp domain | (Martinez-Rucobo et al., 2011) | PDB: 3QQC |
| Coordinates of Eco NusG/RNAP | (Liu and Steitz, 2017) | PDB: 5TBZ |
| Cryo-EM map of Eco EC | (Kang et al., 2017) | EMD-8585 |
| Cryo-EM map of Eco NusG-opsEC | This paper | EMD-7351 |
| Cryo-EM map of Eco RfaH-NGN-opsEC | This paper | EMD-7350 |
| Cryo-EM map of Eco RfaH-full-length-opsEC | This paper | EMD-7349 |
| Oligonucleotides | ||
|
hisPEC non-template DNA (6ASX) GCGTCCTATCGATCTTCGGAAGAGATTCAGAG |
IDT | Lab stock #10924 |
|
hisPEC template DNA (6ASX) CTCTGAATCTCTTCCAGCACACATCAGGACGC |
IDT | Lab stock #10919 |
|
his
ePEC G17 RNA UCAUCCGGCGAUGUGUG |
IDT | Lab stock #6593 |
|
his
7-nt antisense RNA CCGGAUG |
IDT | Lab stock #12196 |
|
NusG forward primer GTTTAACTTTAAGAAGGAGATATACATATGTCTGAAGCTCCTAAAAAG |
IDT | Lab stock #10224 |
|
NusG reverse primer GAACAGAACTTCCAACTCGAGGGCTTTTTCAACCTGGCTG |
IDT | Lab stock #10225 |
|
opsEC non-template DNA GGGCTGCGGTAGCGTGACGGCGAATACCC |
IDT | |
|
opsEC template DNA GGGTATTCGCCGTGTACCTCTCCTAGCCC |
IDT | |
|
opsEC RNA GCAUUCAAAGCGGAGAGGUA |
GE Healthcare Dharmacon | |
|
TAops
nt-DNA GAAACACCACCAGTAGGCGGTAGCGTGCGTTTTTCGTTCTTCC |
IDT | |
|
TAops
t-DNA GGAAGAACGAAAAACGCACGCTACCGCCTACTGGTGGTGTTTC |
IDT | |
|
TAops
RNA UUAUUCGGUAGCGU |
IDT | |
|
EcoRI roadblock ATAGGCAGTCATGGAATTCACCACTGGAAGATCTGAA |
Sigma-Aldrich | Lab stock #2600 |
|
T7A1 template primer Bio-GGAGAGACAACTTAAAGAGA |
Sigma-Aldrich | Lab stock #44 |
|
T7 vector forward CTTTTTAGGAGCTTCAGACATATGTATATCTCCTTCTTAAAGTTAAAC |
IDT | Lab stock #10226 |
|
T7 vector reverse primer GAACAGAACTTCCAactcgagGGCTTTTTCAACCTGGCTG |
IDT | Lab stock #10227 |
| Recombinant DNA | ||
| pACYCDuet-1_Ec_rpoZ | Twist et al., 2011 | N/A |
| pEcrpo(HX-)ABCZ | Twist et al., 2011 | N/A |
| pIA244 wild-type NusG with His6+HMK tag at the N-terminus |
This work | N/A |
| pIA270 wild-type RfaH with His6+HMK tag at the N-terminus |
(Artsimovitch and Landick, 2002) | |
|
pIA349 transcription template with T7A1 promoter followed by the ops and his pause signals |
(Artsimovitch and Landick, 2002) | |
|
pIA900 Wildtype RNAP overexpression plasmid with the TEV protease and His10-tags at rpoC-C-terminus |
(Svetlov and Artsimovitch, 2015) | |
| pRM1160 | ||
| pIA244 wild-type NusG with His6+HMK tag at the N-terminus |
This work | N/A |
| pIA270 wild-type RfaH with His6+HMK tag at the N-terminus |
(Artsimovitch and Landick, 2002) | |
|
pIA349 transcription template with T7A1 promoter followed by the ops and his pause signals |
(Artsimovitch and Landick, 2002) | |
| pVS12 Wild-type RfaH fused to the chitin-binding and intein domains |
(Vassylyeva et al., 2006) | |
|
pIA900 Wildtype RNAP overexpression plasmid with the TEV protease and His10-tags at rpoC-C-terminus |
(Svetlov and Artsimovitch, 2015) | |
|
pRM756 RNAP overexpression plasmid. His10-ppx tag at rpoC-C-terminus |
Windgassen et al., 2014 | Lab stock #2956 |
|
pRM950 RNAP overexpression plasmid. β′258iC. β1045iC 843C. HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus |
Kang et al., 2018 | Lab stock #5250 |
|
pRM1160 NusG overexpression plasmid. His10-ppx tag at NusG C-terminus |
This work | Lab stock #5460 |
| Software and Algorithms | ||
| Blocres | (Cardone et al., 2013) | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera |
| COOT | (Emsley and Cowtan, 2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| CTFFIND4 | (Rohou and Grigorieff, 2015) | http://grigoriefflab.janelia.org/ctffind4 |
| Direct-detector-align_lmbfgs | (Rubinstein and Brubaker, 2015) | https://sites.google.com/site/rubinsteingroup/direct-detector-align_lmbfgs |
| Gautomatch | Zhang, Unpublished | http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/ |
| Leginon | (Suloway et al., 2005) | http://www.leginon.org |
| MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu |
| PHENIX | (Adams et al., 2010) | https://www.phenix-online.org/documentation/index.html |
| Pymol | Schrödinger, LLC | http://www.pymol.org |
| RELION | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion |
| Serial EM | (Mastronarde, 2005) | http://bio3d.colorado.edu/SerialEM |
| Unblur & Summovie | (Grant and Grigorieff, 2015) | http://grigoriefflab.janelia.org/unblur |
| Other | ||
| C-flat CF-1.2/1.3 400 mesh gold grids | Electron Microscopy Sciences | CF413-100-Au |
Cryo-EM data acquisition and processing for RfaH-opsEC
The grids were imaged using a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan, Pleasanton, CA). Images were recorded with Serial EM (Mastronarde, 2005) in super-resolution counting mode with a super resolution pixel size of 0.650 Å and a defocus range of 0.8 to 2.4 μm (Figure S5B). Data were collected with a dose of 8 electrons/physical pixel/s (1.3 Å pixel size at the specimen). Images were recorded with a 15 s exposure and 0.3 s sub-frames (50 total frames) to give a total dose of 71.0 electrons/Å2. Dose fractionated subframes were 2 × 2 binned (giving a pixel size of 1.3 Å), aligned and summed using Unblur (Grant and Grigorieff, 2015). The contrast transfer function was estimated for each summed image using CTFFIND4 (Rohou and Grigorieff, 2015). From the summed images, particles were automatically picked in Gautomatch (Zhang, unpublished; see Key Resource Table), manually inspected, and then individually aligned using direct-detector-align_lmbfgs software (Rubinstein and Brubaker, 2015). The aligned particles were subjected to 2D classification in RELION specifying 100 classes (Scheres, 2012), and poorly populated classes were removed, resulting in 389,200 particles (Figure S5C). These particles were 3D autorefined in RELION using a map of Eco elongation complex (EMD-8585; Kang et al., 2017), low-pass filtered to 60 Å resolution as an initial 3D template. With this initial model, 3D classification was performed without alignment with a soft mask generated in Chimera and RELION. The soft mask excluded flexible RNAP domains (SI1, SI3, flap tip helix, and single-stranded nucleic acids) of EC. Among the 3D classes, the two best-resolved classes were combined, 3D autorefined, and subjected to second 3D classification without alignment with the soft mask that was used in the first 3D classification. From this classification, the best-resolved class was 3D autorefined with solvent flattening, and post-processed in RELION, yielding the final reconstruction at 3.5 Å resolution (Figures S5, S5E).
To resolve heterogeneity around RfaH the upstream duplex DNA, focused 3D classification on the unmodeled region was performed (Scheres, 2012). A soft map containing RNAP, nucleic acids, and the RfaH-NGN was generated in Chimera and RELION to make a subtracted particle stack in RELION. The subtracted particles were 3D classified into six classes without alignment, reverted to the original (unmasked) particles, and 3D autorefined. Among the classes, one class resolved the RfaH-KOW domain and flap-tip helix. The particles in this class, containing 107,500 particles (about 28% of the starting particles after 2D classification; Figure S2B), was 3D autorefined with solvent flattening and post-processed, yielding the final reconstruction at 3.7 Å resolution (Figure S2B). Local resolution calculations (Figure S5H) were performed using blocres (Cardone et al., 2013).
Model building, refinement and validation
To build initial models, The Eco EC (PDB ID 6ALF; Kang et al., 2017) was fitted into the electron density maps using Chimera. NusG-NGN (PDB ID 2K06; (Mooney et al., 2009b), RfaH-NGN (PDB ID 2OUG; (Belogurov et al., 2007), and RfaH-KOW (PDB ID 2LCL; (Burmann et al., 2012) were also fitted into NusG-opsEC, RfaH-NGN-opsEC, and focused RfaH-opsEC cryo-EM maps accordingly. These initial models were real-space refined against the working half map using Phenix real-space-refine (Adams et al., 2010). In the refinement, domains in the core and nucleic acids were rigid-body refined, then subsequently refined with secondary structure restraints. At the end of refinement, Fourier shell correlations (FSC) were calculated between the refined model and the half map used for refinement (work), the other half map (free), and the full map to assess over-fitting (Figures S3G, S5F, G).
Psoralen crosslinking
Scaffolds were assembled from synthetic TAops oligonucleotides, t-DNA was end-labeled with [32P]-ATP using T4 polynucleotide kinase (PNK; NEB). Following labeling, oligonucleotides were purified using QIAquick Nucleotide Removal Kit (Qiagen). To assemble a scaffold, RNA and t-DNA oligonucleotides were combined in PNK buffer and annealed in a PCR machine as follows: 5 min at 45 °C; 2 min each at 42, 39, 36, 33, 30, and 27 °C, 10 min at 25 °C. 12 pmoles of t-DNA/RNA hybrid were mixed with 14 pmoles of His-tagged core RNAP in 30 μl of TB [20 mM Tris-HCl, pH 7.9, 5% (v/v) Glycerol, 40 mM KCl, 5 mM MgCl2, 10 mM β-mercaptoethanol], and incubated at 37 °C for 10 min. 15 μl of His-Select® HF Nickel Affinity Gel (Sigma-Aldrich) was washed once in TB and incubated with 20 μg Bovine Serum Albumin in a 40-μl volume for 15 min at 37 °C, followed by a single wash step in TB. The t-DNA/RNA/RNAP complex was mixed with the Affinity Gel for 15 min at 37 °C on a ther momixer (Eppendorf) at 900 rpm, and washed twice with TB. 30 pmoles of the nt-DNA oligonucleotide were added, followed by incubation for 20 min at 37 °C, one 5-min incuba tion with TB supplemented with 1 M KCl in a thermomixer, and five washes with TB. The assembled ECs were eluted from beads with 90 mM imidazole in a 15-μl volume, purified through a Durapore (PVDF) 0.45 μm Centrifugal Filter Unit (Merck Millipore), and resuspended in TB. For crosslinking, the ECs were supplemented with 6.3% (v/v) DMSO and 0.92 mM 8-methoxypsoralen and incubated for 2 min at 37 °C, f ollowed by addition of 50 nM RfaH, 500 nM NusG, or storage buffer, and a 3-min incubation at 37 °C. Complexes were then exposed to 365 nm UV light (8W Model UVLMS-38; UVP, LLC) for 20 min on ice. The reactions were quenched with an equal volume of Stop buffer (8 M Urea, 20 mM EDTA, 1 x TBE, 0.5 % Brilliant Blue R, 0.5 % Xylene Cyanol FF). Samples were heated for 2 min at 95 °C and separated by electrophoresis in denaturing acrylamide (19:1) gels (7 M Urea, 0.5X TBE). The gels were dried and the products were visualized and quantified using a FLA9000 Phosphorimaging System (GE Healthcare), ImageQuant Software, and Microsoft Excel.
Cys Triplet Reporter (CTR) assays
Nucleic-acid scaffolds used to reconstitute hisPEC for Cys triplet reporter cross-linking assays (Figure 5E) were assembled on purified DNA and RNA oligonucleotides as described (Hein et al., 2014). Briefly, 10 μM RNA, 12 μM template DNA, and 15 μM nt-DNA (Key Resource Table) were annealed in reconstitution buffer (RB; 20 mM Tris-HCl, pH 7.9, 20 mM NaCl, and 0.1 mM EDTA). To assemble complexes, scaffold (2 μM) was mixed with limiting CTR RNAP (1 μM; CTR RNAP: β′1045iC 258iC, β843C) in transcription buffer (50 mM Tris-HCl, pH 7.9, 20 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% glycerol, and 2.5 μg of acetylated bovine serum albumin/ml). NusG-NGN or RfaH-NGN proteins were added to 1 μM and combined with cystamine and DTT to final concentrations of 2.5 mM and 2.8 mM, respectively, to generate a redox potential of −0.36. Reactions were incubated for 60 min at room temperature and then stopped by addition of iodoacetamide to 15 mM. The formation of cysteine cross-links was then evaluated by non-reducing SDS-PAGE (4–15% gradient Phastgel; GE Life Sciences) as described previously (Hein et al., 2014). Gels were stained with Coomassie Blue and imaged with a CCD camera. The fraction cross-linked was quantified with ImageJ software (Schneider et al., 2012). The experimental error was determined as the standard deviation of measurements from three or more independent replicates.
RNAP pause assays
The nucleic-acid scaffold (Figure 5A; Key Resources Table) used to reconstitute ECs for pause assays (Figures 5C, D) was assembled as previously described (Hein et al., 2014). Briefly, PAGE-purified G17 RNA (2 nt upstream of the pause site, 10 μM), t-DNA (15 μM), and nt-DNA (20 μM) were annealed in reconstitution buffer (RB; 10 mM Tris-HCl, pH 7.9, 40 mM KCl, and 5 mM MgCl2). Scaffolds (2 μM) were incubated with 0.5 μM RNAP for 15 min at 37 °C in Elongation Buffer (E B; 25 mM HEPES-KOH, pH 8.0, 130 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, DTT, 0.15 mM EDTA, 5% glycerol, and 25 μg of acetylated bovine serum albumin/ml) to form ECs, diluted to 0.1 μM in EB, then incubated with heparin (0.1 mg/ml final) for 3 min at 37 °C, and labeled by incorporation of 2 μM [α-32P]CMP for 1 min at 37 °C. NusG-NGN or RfaH-NGN (or EB for the ±asRNA conditions) were added to the C18 complexes and incubated at 37 °C for 10 min before adding 7-mer asRNA or equal volume TE for the minus asRNA condition, and then incubated for another 10 min at 37°C to fo rm an RNA duplex mimic of the hisPEC hairpin. ECs were then assayed for pause-escape kinetics by addition of 100 μM UTP and 10 μM GTP in EB at 37 °C. Reaction sampl es were removed at time points and quenched with an equal volume of 2X urea stop buffer (8 M urea, 50 mM EDTA, 90 mM Tris-borate buffer, pH 8.3, 0.02% each bromophenol blue and xylene cyanol). All active PECs were then chased out of the pause by addition of 1 mM GTP for 1 min at 37 °C to aid quantitation. RNAs in each quenched re action sample were separated on a 15% urea-PAGE gel. The gel was exposed to a PhosphorImager screen, and the screen was scanned using Typhoon PhosphorImager software and quantified in ImageQuant (GE Life Sciences). The fraction of RNA at pause (U19) as a function of time was fit to single- or double-exponential decay functions using KaleidaGraph to obtain the amplitudes of bypass, slow pause, and slower pause species and pause escape rates.
Retention of RfaH and NusG on the EC
A linear template was generated by PCR of pIA349 using a top biotinylated primer and a bottom primer with an EcoRI recognition site. The template (8 pmoles) was incubated with EcoRIQ111 (3 μM; to achieve complete occupancy) in 40 μl BB (20 mM Tris-HCl, pH 7.9, 6% glycerol, 50 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol) for 15 min at 37°C. To form an immobilized halted G37 EC, holo-RNAP (8 pmoles), ApU (100 μM) and 5 μM each CTP, GTP and ATP were added together with 20 μl of prewashed Streptavidin coated magnetic beads (Dynabeads® MyOne™ Streptavidin C1) and incubated for 15 min at 37 °C. The halted complexes were washed three times with 500 μl of BB using a Magnetic Separation Stand (Promega). UTP was added at 5 μM for 5 min at 37 °C, followed by three washes. Then ATP, GTP, and CTP were added at 5 μM to form G42 (ops10) EC. The sample was divided into two aliquots; to one, 32P-labeled RfaH was added to 50 nM, and to the other 32P-labeled NusG was added to 470 nM, followed by a 5-min incubation at 37°C and three washes. Each reaction was split again into three aliquots: a) no further treatment; b) 5-min chase at 37°C with 100 μM NTPs; and c) 5-min chase at 37°C with 100 μM NTPs and 5 μM unlabeled NusG. After three washes with BB, samples were measured in a LS6500 Multi-Purpose Scintillation Counter (Beckman Coulter). The experiment was done in triplicates.
Supplementary Material
NusG and RfaH effects on RNA chain elongation on the scaffold used for cryo-EM experiments (shown on top). Scaffolds were assembled with the 32P-labeled RNA strand in the cryo-EM buffer and preincubated with RfaH (100 nM) or NusG (1000 nM), in the absence or in the presence of 8 mM CHAPSO. Elongation was restarted upon addition of 0.15 mM ATP, GTP, and CTP, aliquots were withdrawn at the indicated times and analyzed on 12% denaturing urea-acrylamide gels (19:1) in 0.5x TBE. Positions of RNA products are indicated, with the region encompassing C21, A22, C23 and G24 RNAs highlighted in black. RfaH and NusG display their characteristic effects immediately downstream from the ops site (A20). Consistent with the patterns observed on standard transcription templates, RfaH promotes RNAP pausing in the 21–24 region, whereas NusG decreases pausing in this region. The bar graph shows the fraction of C21-G24 RNAs (as % of total RNA) at the 40-sec time point. The addition of CHAPSO has only a minor effect on RNAP elongation and response to NusG and RfaH.
A. Flowchart showing the image processing pipeline for the cryo-EM NusG-opsEC data starting with 6,318 dose-fractionated movies collected on a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan). Movie frames were aligned and summed using Unblur (Grant and Grigorieff, 2015). Particles were autopicked with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and manually revised from these summed images. The revised particles were polished using direct-detector-align_lmbfgs (Rubinstein and Brubaker, 2015) for subsequent 2D classification using RELION (Scheres, 2012). After 2D classification, the dataset contained 514,900 aligned particles. These particles were auto-refined in RELION using a model of the Eco EC (PDB ID 6ALF; (Kang et al., 2017) as an initial 3D template. 3D classification into six classes was performed on the particles using the refined model and alignment angles. The best class (containing the most particles and having the highest resolution) was subjected to a second 3D classification into four classes. After the second 3D classification, one class containing 33.4% of the starting particles (171,900 particles) was autorefined and post-processed in RELION, yielding the final reconstruction at 3.7 Å resolution.
B. Flowchart showing the image processing pipeline for the cryo-EM RfaH-opsEC data starting with 3,495 dose-fractionated movies collected on a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan). Movie frames were aligned and summed using Unblur (Grant and Grigorieff, 2015). Particles were autopicked with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and manually revised from these summed images. The revised particles were polished using direct-detector-align_lmbfgs (Rubinstein and Brubaker, 2015) for subsequent 2D classification using RELION (Scheres, 2012). After 2D classification, the dataset contained 389,200 aligned particles. These particles were auto-refined in RELION using a model of the Eco EC (PDB ID 6ALF; (Kang et al., 2017) as an initial 3D template. 3D classification into six classes (right branch) was performed on the particles using the refined model and alignment angles. The best two classes (containing the most particles and having the highest resolutions) was subjected to a second 3D classification into four classes. After the second 3D classification, one class containing 44.9% of the starting particles (174,600 particles) was autorefined and post-processed in RELION, yielding the final reconstruction at 3.5 Å resolution.
For the focused 3D classification (left branch of the flowchart), a soft mask that excluded the upstream duplex DNA and nearby protein regions was generated using Chimera and RELION. The mask was used to make a subtracted particle stack in RELION with the filtered map generated in the initial autorefinement. The subtracted particles were 3D classified into six classes without alignment. Among the six classes, one class containing 24% of the starting particles (107,500 particles) had density for the RfaH-KOW domain. The original (unmasked) particles in this class were autorefined and post-processed in RELION, yielding the final reconstruction at 3.7 Å resolution.
A. SDS polyacrylamide gel of purified Eco ΔαCTD-RNAP
B. SDS polyacrylamide gel of purified Eco NusG.
C. Representative micrograph of the NusG-opsEC in vitreous ice.
D. The fifteen most populated classes from 2D classification.
E. Angular distribution for NusG-opsEC particle projections.
F. Gold-standard FSC of the NusG-opsEC. The gold-standard FSC was calculated by comparing the two independently determined half-maps from RELION. The dotted line represents the 0.143 FSC cutoff, which indicates a nominal resolution of 3.7 Å.
G. FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
H. (top) The 3.7-Å resolution cryo-EM density map of the NusG-opsEC is colored as follows: αI, αII, ω subunits, gray; β, cyan; β′, pink; NusG, green; t-DNA, dark blue; nt-DNA, yellow; RNA, red. The rightmost view is sliced as indicated in the leftmost view.
(bottom) Same views as (top) but colored by local resolution (Cardone et al., 2013).
A. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein models for the NusG/RNAP GL interactions.
B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein models for the NusG/RNAP CH interactions.
C. Cryo-EM density (filtered to the local resolution) showing the RfaH-KOW and upstream duplex DNA of the RfaH-opsEC.
D. Cryo-EM density (filtered to the local resolution) showing the upstream duplex DNA of the NusG-opsEC
A. SDS polyacrylamid gel showing purified Eco RfaH.
B. Representative micrograph of the RfaH-opsE in vitreous ice.
C. The fifteen most populated classes from 2D classification.
D. Angular distribution for RfaH-opsEC particle projections.
E. Gold-standard FSC of the RfaH-opsEC. The gold-standard FSC was calculated by comparing the two independently determined half-maps from RELION. The dotted line represents the 0.143 FSC cutoff, which indicates a nominal resolution of 3.5 Å.
F. RfaH-NGN-opsEC: FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
G. RfaH-full-length-opsEC: FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
H. (top) The 3.5-Å resolution cryo-EM density map of the RfaH-opsEC is colored as follows: αI, αII, ω subunits, gray; β, cyan; β′, pink; RfaH, orange; t-DNA, dark blue; nt-DNA, yellow; RNA, red. The rightmost view is sliced as indicated in the leftmost view.
(bottom) Same views as (top) but colored by local resolution (Cardone et al., 2013).
A. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the RfaH/ops G5 and T6 interactions.
B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the NusG/RNAP GL interactions.
C. B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the NusG/RNAP CH interactions.
Previously determined structures of NusG/Spt5 factors bound to the archaeal RNAP clamp domain (A; 3QQC; (Martinez-Rucobo et al., 2011), bound in a RNAPII-EC (B; 5OIK; (Bernecky et al., 2017), or bound to Eco core RNAP (C; 5TBZ; (Liu and Steitz, 2017)) are superimposed with the NusG-opsEC via RNAP α-carbons to compare the resulting dispositions of the NusG/Spt5. The color-coding in each panel is indicated in the color keys below.
A. The archaeal (Pyrococcus furiousus) RNAP clamp domain was superimposed with the Eco RNAP from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.23 Å. The positions of the NusG-NGN (green) and Spt5 (orange) are shifted relative to each other by about 2.8 Å but the orientation of the domain is conserved.
B. The Rpb1 and Rpb2 subunits of human RNAPII from the human DSIF(Spt4/Spt5)/RNAPII-EC were superimposed with the β′ and β subunits (respectively) from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.52 Å. The positions of the NusG-NGN (green) and Spt5 (orange) are shifted relative to each other by about 7.9 Å but the orientation of the domain is conserved.
C. The Eco core RNAP from the NusG/RNAP was superimposed with the RNAP from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.94 Å. The orientation of overall fold of the NusG-NGN (orange) is not consistent with the NusG-NGN from the NusG-opsEC (green).
HIGHLIGHTS.
Cryo-EM structures of NusG and RfaH-bound transcription elongation complexes
NusG and RfaH suppress backtracking by stabilizing the upstream duplex DNA
RfaH recognizes an ops DNA hairpin in the nontemplate strand of the transcription bubble
RfaH suppresses RNA hairpin-stabilized pausing by preventing RNAP swiveling
Acknowledgments
We thank K. Uryu and D. Acehan at The Rockefeller University Electron Microscopy Resource Center for help with EM sample preparation, M. Ebrahim and J. Sotiris at The Rockefeller University Cryo-EM Resource Center and E. Eng at the New York Structural Biology Center for help with data collection, and members of our research groups for helpful comments on the manuscript. Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the NIH National Institute of General Medical Sciences (GM103310) and the Simons Foundation (349247). This work was supported by National Institute of Health grants R01 GM67153 to I.A., R01 GM38660 to R.L. and R35 GM118130 to S.A.D.
Footnotes
Supplemental Information includes 8 figures and 2 tables and can be found with this article online at…
DECLARATION OF INTERESTS
The authors declare no competing interests.
Accession numbers
The cryoEM density maps have been deposited in the EM Data Bank with accession codes EMD-7351 (NusG-opsEC), EMD-7350 (RfaH-NGN-opsEC), and EMD-7349 (RfaH-full-length-opsEC). Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6C6U (NusG-opsEC), 6C6T (RfaH-NGN-opsEC), and 6C6S (RfaH-full-length-opsEC).
AUTHOR CONTRIBUTIONS
Conceptualization, I.A., R.L., S.A.D.; Investigation, J.Y.K., R.A.M., Y.N., J.S., T.V.M.; Analysis, J.Y.., I.A., R.L., S.A.D.; Writing, J.Y.K., I.A., R.L., S.A.D.; Supervision, I.A., R.L., S.A.D.; Funding Acquisition, I.A., R.L., S.A.D.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Micro. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural Basis for Converting a General Transcription Factor into an Operon-Specific Virulence Regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernecky C, Plitzko JM, Cramer P. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nature Struct Mol Biol. 2017;24:809–815. doi: 10.1038/nsmb.3465. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, Artsimovitchm I, Rösch P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj SR, Irschik H, Jansen R, et al. Opening and Closing of the Bacterial RNA Polymerase Clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Fu J, Reese JC. Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest. J Biol Chem. 2016;291:9853–9870. doi: 10.1074/jbc.M116.716001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Lee J, Lee TH, Reese JC. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucl Acids Res. 2017;45:6362–6374. doi: 10.1093/nar/gkx220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine SI. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Goodson JR, Klupt S, Zhang C, Straight P, Winkler WC. LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nat Microbiol. 2017;2:17003. doi: 10.1038/nmicrobiol.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A. Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell. 2018;69:816–827.e4. doi: 10.1016/j.molcel.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–115. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nature Struct Mol Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Zhou J, Mooney RA, La Porta A, Landick R, Block SM. E. coli NusG Inhibits Backtracking and Accelerates Pause-Free Transcription by Promoting Forward Translocation of RNA Polymerase. J Mol Biol. 2010:1–14. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirtreiter A, Damsma GE, Cheung ACM, Klose D, Grohmann D, Vojnic E, Martin ACR, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucl Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell. 2018;69:802–815.e1. doi: 10.1016/j.molcel.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PDB, Chen J, Campbell EA, Mustaev A, Chait BT, Gottesman ME, Darst SA. Structural basis of transcription arrest by coliphage HK022 nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017;6:e25478. doi: 10.7554/eLife.25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J Biol Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM IUCr. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Liu B, Steitz TA. Structural insights into NusG regulating transcription elongation. Nucl Acids Res. 2017;45:968–974. doi: 10.1093/nar/gkw1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase–Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mayer A, Landry HM, Churchman LS. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol. 2017;46:72–80. doi: 10.1016/j.ceb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator Trafficking on Bacterial Transcription Units In Vivo. Mol Cell. 2009a;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rösch P, Gottesman ME, Landick R. Two Structurally Independent Domains of E. coli NusG Create Regulatory Plasticity via Distinct Interactions with RNA Polymerase and Regulators. J Mol Biol. 2009b;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandymazumdar M, Nedialkov Y, Svetlov D, Sevostyanova A, Belogurov GA, Artsimovitch I. RNA polymerase gate loop guides the nontemplate DNA strand in transcription complexes. Proc Natl Acad Sci USA. 2016;113:14994–14999. doi: 10.1073/pnas.1613673114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Grass JA, Langelier MF, Coulombe B, Landick R. The RPB2 flap loop of human RNA polymerase II is dispensable for transcription initiation and elongation. Mol Cell Biol. 2011;31:3312–3325. doi: 10.1128/MCB.05318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, Brubaker MA. Alignment of cryo-EM movies of individual particles by optimization of image translations. J Struct Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Saenger W. Principles of Nucleic Acid Structure. New York, NY: Springer Science & Business Media; 1984. [Google Scholar]
- Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, Lee CT, Loll B, Behrmann E, Bürger J, et al. Structural basis for λN-dependent processive transcription antitermination. Nat Microbiol. 2017;2:17062. doi: 10.1038/nmicrobiol.2017.62. [DOI] [PubMed] [Google Scholar]
- Saxena S, Myka KK, Washburn R, Costantino N, Court DL, Gottesman ME. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol Microbiol. 2018 doi: 10.1111/mmi.13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. The quantitative and condition-dependent Escherichia coli proteome. Nature Biotech. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc Natl Acad Sci USA. 2008;105:865–870. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Watters KE, Nedialkov Y, Artsimovitch I, Lucks JB. Distributed biotin-streptavidin transcription roadblocks for mapping cotranscriptional RNA folding. Nucl Acids Res. 2017;45:e109. doi: 10.1093/nar/gkx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: tools and protocols. Methods Mol Biol. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Sekine SI, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric Control of RNA Polymerase by a Site That Contacts Nascent RNA Hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Turtola M, Belogurov GA. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife. 2016;5:e18096. doi: 10.7554/eLife.18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twist KA, Husnain SI, Franke JD, Jain D, Campbell EA, Nickels BE, Thomas MS, Darst SA, Westblade LF. A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the alpha C-terminal domain. Protein Sci. 2011;20:986–995. doi: 10.1002/pro.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyeva MN, Svetlov V, Klyuyev S, Devedjiev YD, Artsimovitch I, Vassylyev DG. Crystallization and preliminary crystallographic analysis of the transcriptional regulator RfaH from Escherichia coli and its complex with ops DNA. Acta Crystallogr F Struct Biol Cryst Commun. 2006;62:1027–1030. doi: 10.1107/S174430910603658X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Leon K, Landick R, Darst SA. Structural Basis of Transcriptional Pausing in Bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen TA, Mooney RA, Nayak D, Palangat M, Zhang J, Landick R. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucl Acids Res. 2014;42:12707–12721. doi: 10.1093/nar/gku997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NusG and RfaH effects on RNA chain elongation on the scaffold used for cryo-EM experiments (shown on top). Scaffolds were assembled with the 32P-labeled RNA strand in the cryo-EM buffer and preincubated with RfaH (100 nM) or NusG (1000 nM), in the absence or in the presence of 8 mM CHAPSO. Elongation was restarted upon addition of 0.15 mM ATP, GTP, and CTP, aliquots were withdrawn at the indicated times and analyzed on 12% denaturing urea-acrylamide gels (19:1) in 0.5x TBE. Positions of RNA products are indicated, with the region encompassing C21, A22, C23 and G24 RNAs highlighted in black. RfaH and NusG display their characteristic effects immediately downstream from the ops site (A20). Consistent with the patterns observed on standard transcription templates, RfaH promotes RNAP pausing in the 21–24 region, whereas NusG decreases pausing in this region. The bar graph shows the fraction of C21-G24 RNAs (as % of total RNA) at the 40-sec time point. The addition of CHAPSO has only a minor effect on RNAP elongation and response to NusG and RfaH.
A. Flowchart showing the image processing pipeline for the cryo-EM NusG-opsEC data starting with 6,318 dose-fractionated movies collected on a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan). Movie frames were aligned and summed using Unblur (Grant and Grigorieff, 2015). Particles were autopicked with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and manually revised from these summed images. The revised particles were polished using direct-detector-align_lmbfgs (Rubinstein and Brubaker, 2015) for subsequent 2D classification using RELION (Scheres, 2012). After 2D classification, the dataset contained 514,900 aligned particles. These particles were auto-refined in RELION using a model of the Eco EC (PDB ID 6ALF; (Kang et al., 2017) as an initial 3D template. 3D classification into six classes was performed on the particles using the refined model and alignment angles. The best class (containing the most particles and having the highest resolution) was subjected to a second 3D classification into four classes. After the second 3D classification, one class containing 33.4% of the starting particles (171,900 particles) was autorefined and post-processed in RELION, yielding the final reconstruction at 3.7 Å resolution.
B. Flowchart showing the image processing pipeline for the cryo-EM RfaH-opsEC data starting with 3,495 dose-fractionated movies collected on a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan). Movie frames were aligned and summed using Unblur (Grant and Grigorieff, 2015). Particles were autopicked with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and manually revised from these summed images. The revised particles were polished using direct-detector-align_lmbfgs (Rubinstein and Brubaker, 2015) for subsequent 2D classification using RELION (Scheres, 2012). After 2D classification, the dataset contained 389,200 aligned particles. These particles were auto-refined in RELION using a model of the Eco EC (PDB ID 6ALF; (Kang et al., 2017) as an initial 3D template. 3D classification into six classes (right branch) was performed on the particles using the refined model and alignment angles. The best two classes (containing the most particles and having the highest resolutions) was subjected to a second 3D classification into four classes. After the second 3D classification, one class containing 44.9% of the starting particles (174,600 particles) was autorefined and post-processed in RELION, yielding the final reconstruction at 3.5 Å resolution.
For the focused 3D classification (left branch of the flowchart), a soft mask that excluded the upstream duplex DNA and nearby protein regions was generated using Chimera and RELION. The mask was used to make a subtracted particle stack in RELION with the filtered map generated in the initial autorefinement. The subtracted particles were 3D classified into six classes without alignment. Among the six classes, one class containing 24% of the starting particles (107,500 particles) had density for the RfaH-KOW domain. The original (unmasked) particles in this class were autorefined and post-processed in RELION, yielding the final reconstruction at 3.7 Å resolution.
A. SDS polyacrylamide gel of purified Eco ΔαCTD-RNAP
B. SDS polyacrylamide gel of purified Eco NusG.
C. Representative micrograph of the NusG-opsEC in vitreous ice.
D. The fifteen most populated classes from 2D classification.
E. Angular distribution for NusG-opsEC particle projections.
F. Gold-standard FSC of the NusG-opsEC. The gold-standard FSC was calculated by comparing the two independently determined half-maps from RELION. The dotted line represents the 0.143 FSC cutoff, which indicates a nominal resolution of 3.7 Å.
G. FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
H. (top) The 3.7-Å resolution cryo-EM density map of the NusG-opsEC is colored as follows: αI, αII, ω subunits, gray; β, cyan; β′, pink; NusG, green; t-DNA, dark blue; nt-DNA, yellow; RNA, red. The rightmost view is sliced as indicated in the leftmost view.
(bottom) Same views as (top) but colored by local resolution (Cardone et al., 2013).
A. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein models for the NusG/RNAP GL interactions.
B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein models for the NusG/RNAP CH interactions.
C. Cryo-EM density (filtered to the local resolution) showing the RfaH-KOW and upstream duplex DNA of the RfaH-opsEC.
D. Cryo-EM density (filtered to the local resolution) showing the upstream duplex DNA of the NusG-opsEC
A. SDS polyacrylamid gel showing purified Eco RfaH.
B. Representative micrograph of the RfaH-opsE in vitreous ice.
C. The fifteen most populated classes from 2D classification.
D. Angular distribution for RfaH-opsEC particle projections.
E. Gold-standard FSC of the RfaH-opsEC. The gold-standard FSC was calculated by comparing the two independently determined half-maps from RELION. The dotted line represents the 0.143 FSC cutoff, which indicates a nominal resolution of 3.5 Å.
F. RfaH-NGN-opsEC: FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
G. RfaH-full-length-opsEC: FSC calculated between the refined structure and the half map used for refinement (work), the other half map (free), and the full map.
H. (top) The 3.5-Å resolution cryo-EM density map of the RfaH-opsEC is colored as follows: αI, αII, ω subunits, gray; β, cyan; β′, pink; RfaH, orange; t-DNA, dark blue; nt-DNA, yellow; RNA, red. The rightmost view is sliced as indicated in the leftmost view.
(bottom) Same views as (top) but colored by local resolution (Cardone et al., 2013).
A. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the RfaH/ops G5 and T6 interactions.
B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the NusG/RNAP GL interactions.
C. B. Stereo view showing cryo-EM density (blue mesh) and the superimposed protein model for the NusG/RNAP CH interactions.
Previously determined structures of NusG/Spt5 factors bound to the archaeal RNAP clamp domain (A; 3QQC; (Martinez-Rucobo et al., 2011), bound in a RNAPII-EC (B; 5OIK; (Bernecky et al., 2017), or bound to Eco core RNAP (C; 5TBZ; (Liu and Steitz, 2017)) are superimposed with the NusG-opsEC via RNAP α-carbons to compare the resulting dispositions of the NusG/Spt5. The color-coding in each panel is indicated in the color keys below.
A. The archaeal (Pyrococcus furiousus) RNAP clamp domain was superimposed with the Eco RNAP from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.23 Å. The positions of the NusG-NGN (green) and Spt5 (orange) are shifted relative to each other by about 2.8 Å but the orientation of the domain is conserved.
B. The Rpb1 and Rpb2 subunits of human RNAPII from the human DSIF(Spt4/Spt5)/RNAPII-EC were superimposed with the β′ and β subunits (respectively) from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.52 Å. The positions of the NusG-NGN (green) and Spt5 (orange) are shifted relative to each other by about 7.9 Å but the orientation of the domain is conserved.
C. The Eco core RNAP from the NusG/RNAP was superimposed with the RNAP from the NusG-opsEC, resulting in an rmsd for the 40 α-carbons of the CHs of 1.94 Å. The orientation of overall fold of the NusG-NGN (orange) is not consistent with the NusG-NGN from the NusG-opsEC (green).