Abstract
Malassezia sympodialis is a dominant commensal fungi in the human skin mycobiome but is also associated with common skin disorders including atopic eczema (AE). M. sympodialis releases extracellular vesicles, designated MalaEx, which are carriers of small RNAs and allergens, and they can induce inflammatory cytokine responses. Here we explored how MalaEx are involved in host-microbe interactions by comparing protein content of MalaEx with that of the parental yeast cells, and by investigating interactions of MalaEx with cells in the skin. Cryo-electron tomography revealed a heterogeneous population of MalaEx. iTRAQ based quantitative proteomics identified in total 2439 proteins in all replicates of which 110 were enriched in MalaEx compared to the yeast cells. Among the MalaEx enriched proteins were two of the M. sympodialis allergens, Mala s 1 and s 7. Functional experiments indicated an active binding and internalization of MalaEx into human keratinocytes and monocytes, and MalaEx were found in close proximity of the nuclei using super-resolution fluorescence 3D-SIM imaging. Our results provides new insights into host-microbe interactions, supporting that MalaEx may have a role in the sensitization and maintenance of inflammation in AE by containing enriched amounts of allergens and with their ability to interact with skin cells.
Subject terms: Clinical microbiology, Chronic inflammation, Fungal host response, Proteomics
Introduction
Our skin is colonized by a diverse microbiota, including bacteria, viruses and fungi, and we are just at the beginning of understanding their role in influencing skin health. Malassezia is the most abundant fungal skin inhabitant of humans1, and colonizes the human skin right after birth2. The Malassezia species show lipid-dependency and lipolytic enzymes such as lipases are necessary for them to acquire fatty acids from the surroundings1. The Malassezia genus currently includes 17 species1. One of the most frequently isolated species from human skin is M. sympodialis, which besides being a commensal yeast is also associated with several common skin disorders such as atopic eczema (AE)3. AE is a complex inflammatory skin disorder, which is cyclical, with relapsing flare periods4, and affects 15 to 20% of young children and up to 3% of adults5. While the pathogenesis of the disease still remains unclear, studies suggest that a genetic predisposition in combination with environmental factors facilitate the development of AE6. A defective skin barrier in turn might assist the entry of microorganisms, such as Malassezia and their allergens. Thirteen Malassezia allergens have been sequenced so far7.
M. sympodialis may also affect AE via secreted extracellular vesicles (EVs), designated MalaEx8. EVs have been shown to be released both from different mammalian cell-types, and from microorganisms and parasites through the endosomal pathway or by budding from the plasma membrane9–14. By using vesicular transport, the content of the EVs are kept protected, and large molecules are enabled to cross the cell wall. Various types of EV ranging in size from 20 nm to 1,000 nm in diameter have been described and are classified mainly on their mechanisms of biogenesis and their physiological functions10,15. Those designated exosomes have been shown to be involved in immunoregulatory mechanisms such as immune activation and suppression, and intercellular communication15, whereas EV from microorganisms with thick cell walls, such as fungi, have been related to cytotoxicity, the invasion of host cells, and the transfer of virulence factors11. As seen with the heterogeneous secreted vesicles from the parasitic yeast stage of Histoplasma capsulatum, they can contain phospholipids and proteins associated to stress responses, pathogenesis, cell wall architecture and virulence16. Previously, we demonstrated that MalaEx are carriers of allergens8, like human dendritic or B cell-derived exosomes17,18, and can induce inflammatory cytokine responses with a significantly higher IL-4 production in peripheral blood mononuclear cells (PBMC) from patients with AE compared to healthy controls8, supporting the link between AE and M. sympodialis. Recently, we also identified the presence of small RNAs in MalaEx, which appeared to have an RNAi-independent route for biogenesis19.
We have previously performed mass spectrometry (MS) based proteomics on M. sympodialis cells for gene annotation7,20. In this study, we used large-scale quantitative proteomics characterization of M. sympodialis whole yeast cells (WC) and their released MalaEx to compare their content. Cryo-electron tomography was used to visualize MalaEx’s morphology, and possible interactions between MalaEx and cells in the skin were investigated to further explore MalaEx’s role in host-microbe interactions.
Results
Characterization of harvested MalaEx
MalaEx released from M. sympodialis cultured in RPMI medium for 48 h had a vesicle size of 171 ± 12 nm (mean ± SD; n = 5 different cultures) as measured with NanoSight. For proteomics we used MalaEx produced by M. sympodialis cultured for 72 h in mDixon broth to avoid contamination from fetal calf serum within the RPMI medium, which otherwise overrides the signals of less abundant proteins. These MalaEx had a mean size of 245 ± 10.9 nm (n = 4 different cultures) in line with Ryner et al.19.
To further characterize the MalaEx we performed cryo-electron tomography. This technology revealed the presence of a heterogeneous population of vesicles with different sizes and internal vesicles, but also with varying electron-dense material suggesting different amount of internal content (Fig. 1A–C and Movie 1 Supplementary information). Some of the vesicles did also appear to have coatings, as seen on the surface of the filled vesicles in Fig. 1A,B.
Figure 1.

Cryo-electron tomography of MalaEx. (A) Central section of a tomographic reconstruction of MalaEx harvested from M. sympodialis cultured for 48 h in RPMI medium. The full tomogram is shown in Movie 1 (see Supplementary information). (B) Zoom into the green box of A showing a low electron density vesicle (red arrow) and high electron density vesicles with coated surfaces (yellow arrows). (C) Isosurface representation of the vesicles enclosed by the blue box in A. The surface representation shows two small vesicles with low electron density (in red) encircled by a larger one (in grey).
Proteomics analysis on MalaEx compared to parental cells
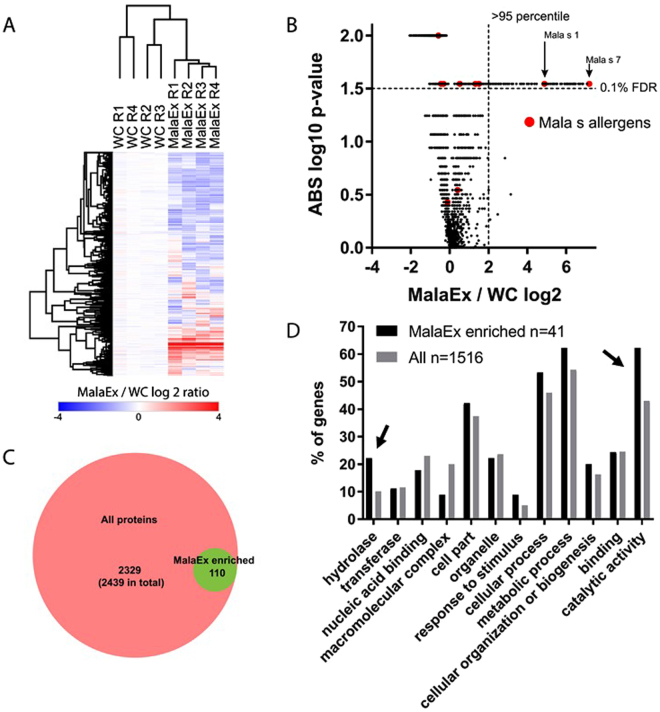
To define proteins that are enriched in MalaEx compared to WC we performed iTRAQ based quantitative proteomics comparing 4 replicates of WC cultured for 72 h, and MalaEx isolated from the their supernatants. We identified in total 3186 proteins from WC and MalaEx of which 2439 were quantified in all replicates (mainly due to missing values in the MalaEx quantification). A subset of proteins displayed higher levels in MalaEx compared to WC (Fig. 2A, Tables S1A and B). Using a combined fold change cutoff (>95 percentile) and p-value cutoff (t-test, permutation based, q-value <0.1), 110 proteins were considered enriched in MalaEx compared to WC (Fig. 2B,C, Tables S1A and B). Among these enriched proteins are 2 of the previously identified M. sympodialis allergens, Mala s 1 and Mala s 7 (Fig. 2B). In addition, the Mala s allergens 5, 6, 8–13 were identified but not considered enriched in MalaEx.
Figure 2.
Characterizing MalaEx proteins by proteomics. (A) Quantitative overview of iTRAQ based proteomics experiments to define MalaEx enriched proteins by comparing MalaEx to M. sympodialis whole cells (WC). R 1–4 denotes the 4 biological replicates cultured for 72 h in mDixon broth, and 2439 proteins with quantification across all 8 samples are shown. (B) Volcano plot to define proteins enriched in MalaEx compared to WC. Proteomics data as in (A). Vertical and horizontal dashed lines indicate 95 percentile of ratios and q-value < 0.1, respectively. Mala s allergens are indicated in red. (C) Venn diagram depicting overlap between proteins considered enriched in MalaEx compared to WC. (D) Gene ontology (GO) analysis of MalaEx. GO term distribution in proteins defined as enriched in MalaEx (from A–C) compared to all identified proteins. Numbers indicate the number of M. sympodialis proteins that could be mapped to yeast homologs for the analysis. Arrows highlight proteins most enriched in MalaEx.
To obtain an understanding of the protein content of the MalaEx enriched proteins we used gene ontology (GO) terms based on M. sympodialis yeast homologs. The protein list of MalaEx enriched proteins (Table S1B) after conversion to yeast homologs was too short to obtain significant enrichment. However, displaying GO terms percentage wise for MalaEx and WC, there were more proteins with the GO terms hydrolase and catalytic activity in the enriched MalaEx proteins than in the whole list of proteins (Fig. 2D, Table 1).
Table 1.
Gene Ontology (GO) terms and Panther classes for MalaEx enriched proteins and all identified proteins.
| Category | Category name (Accession) | MalaEx enriched | All proteins in iTRAQ dataset | ||
|---|---|---|---|---|---|
| Genes (n) | Genes %a | Genes (n) | Genes %a | ||
| Panther class | hydrolase (PC00121) | 10 | 22 | 177 | 10 |
| oxidoreductase (PC00176) | 3 | 7 | 132 | 8 | |
| enzyme modulator (PC00095) | 2 | 4 | 104 | 6 | |
| lyase (PC00144) | 1 | 2 | 53 | 3 | |
| transferase (PC00220) | 5 | 11 | 203 | 12 | |
| ligase (PC00142) | 2 | 4 | 74 | 4 | |
| nucleic acid binding (PC00171) | 8 | 18 | 404 | 23 | |
| transcription factor (PC00218) | 1 | 2 | 57 | 3 | |
| cytoskeletal protein (PC00085) | 1 | 2 | 46 | 3 | |
| Cellular component | membrane (GO:0016020) | 3 | 7 | 131 | 7 |
| macromolecular complex (GO:0032991) | 4 | 9 | 352 | 20 | |
| cell part (GO:0044464) | 19 | 42 | 659 | 37 | |
| organelle (GO:0043226) | 10 | 22 | 415 | 24 | |
| Biological process | reproduction (GO:0000003) | 1 | 2 | 6 | 0 |
| response to stimulus (GO:0050896) | 4 | 9 | 89 | 5 | |
| cellular process (GO:0009987) | 24 | 53 | 809 | 46 | |
| metabolic process (GO:0008152) | 28 | 62 | 955 | 54 | |
| biological regulation (GO:0065007) | 3 | 7 | 85 | 5 | |
| cellular component organization or biogenesis (GO:0071840) | 9 | 20 | 286 | 16 | |
| Molecular function | binding (GO:0005488) | 11 | 24 | 431 | 25 |
| structural molecule activity (GO:0005198) | 2 | 4 | 174 | 10 | |
| catalytic activity (GO:0003824) | 28 | 62 | 756 | 43 | |
aPercent of gene hit against total number of genes.
Interactions of MalaEx with human keratinocytes and monocytes
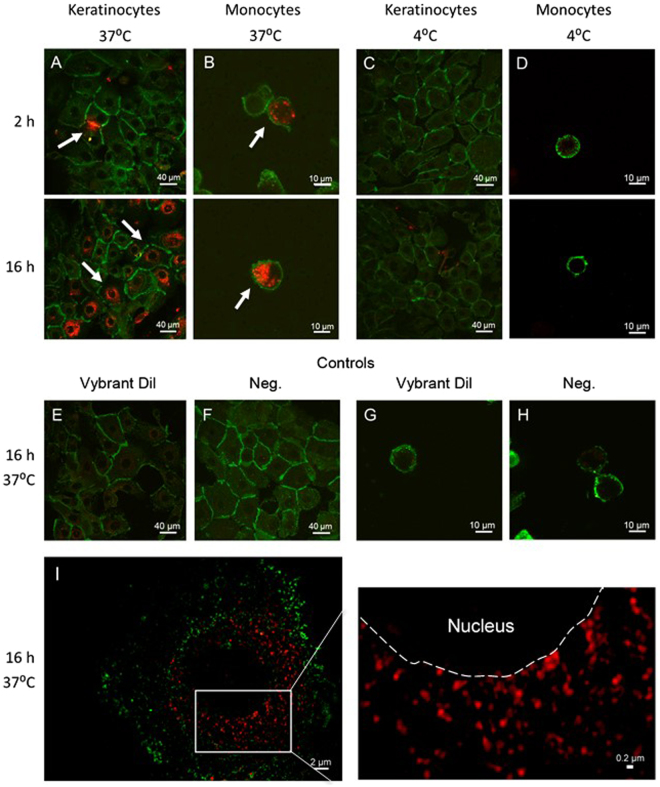
To understand how MalaEx may be involved in host-microbe interactions, we studied MalaEx interactions with keratinocytes and monocytes. Keratinocytes are the main cell type of epidermis actively participating and orchestrating the innate immune response of the skin21 and monocytes are antigen-presenting cells, which are actively recruited at sites of chronic inflammation, such as damaged and inflamed skin as seen in AE22. MalaEx labeled with Vybrant Dil were incubated with keratinocytes or monocytes for 2 h or 16 h at 4 °C and 37 °C. Confocal laser scanning microscopy (CLSM) images demonstrated interactions between MalaEx and a few keratinocytes and an uptake of MalaEx by monocytes after 2 h at 37 °C (Fig. 3A and B). After 16 h both keratinocytes and monocytes had internalized increased amounts of MalaEx (Fig. 3A and B). Internalization of MalaEx was not observed at 4 °C, neither by keratinocyte (Fig. 3C), nor by monocytes (Fig. 3D) suggesting that an active mechanism is involved in the uptake seen at 37 °C. No background was seen when the control for potentially pelleted unbound dye was added to the cells for 16 h at 37 °C (Fig. 3E and G) or when the cells were cultured alone (Fig. 3F and H). Further analysis was performed with higher resolution using 3D-SIM on the 16 h co-cultures of MalaEx with keratinocytes at 37 °C, which demonstrated MalaEx mainly to be localized in the surrounding of the nuclei (Fig. 3I).
Figure 3.
Cellular uptake of MalaEx in human primary keratinocytes and monocytes. (A–H) Confocal microscopy images following incubation of MalaEx, harvested from M. sympodialis cultured for 48 h in RPMI medium and Vybrant Dil labeled (red; 10 µg/ml), with (A,C) keratinocytes stained for E-cadherin (green) and (B,D) monocytes stained for CD14 (green) at 37 °C and 4 °C for 2 h and 16 h. Arrows highlight cells interacting with MalaEx. (E) Keratinocytes treated with Vybrant Dil control for 16 h at 37 °C and (F) cultured alone. (G) Monocytes treated with Vybrant Dil control for 16 h at 37 °C and (H) cultured alone. (I; left image) Structured Illumination Microscopy (3D-SIM) of keratinocytes incubated with MalaEx for 16 h at 37 °C from a single axial slice (depth 116 nm). (I; right image). Zoomed in 3D-SIM image of selected area (white square in left image) showing perinuclear localized MalaEx. Dashed line indicates the borderline between the nuclei and the cytoplasm of the keratinocyte.
Discussion
We have previously found that the commensal fungi M. sympodialis secretes extracellular vesicles (MalaEx), which are carriers of allergens8 and small RNAs19 and that they can induce inflammatory cytokine responses in human blood cells8. To further understand their role in host-microbe interactions, we present in this study new insights into the morphology of MalaEx, we performed the first proteome characterization of MalaEx and compared that with the whole M. sympodialis cells, and demonstrated interactions of MalaEx with skin cells.
In previous EM studies, which demonstrated a typical exosome-like morphology of MalaEx, sucrose gradient separation was used to isolate MalaEx at a density similar to that characterized for exosomes8,19. Here we decided to investigate the whole population of harvested vesicles prepared by serial ultracentrifugation and used cryo-electron tomography to be able to visualize 3D structures in more physiological conditions. Our results revealed a heterogeneous population of vesicles with different densities (Fig. 1). This is in agreement with reports on EVs from other fungal cells16,23,24, and from mammalian cell types such as platelets and mast cells25,26. The diversity of MalaEx suggests the existence of subpopulations of EVs’, which may have distinct mechanisms of biogenesis and release, and possess specific contents and thereby functions11,23,24,27. Further studies are needed to elucidate these aspects of different vesicles, which also require the development of relevant technologies28.
Quantitative proteomics identified in total 3186 proteins of which 2439 were quantified in all replicates and 110 proteins were enriched in MalaEx compared to the WC (Fig. 2). Proteomics of EV from other fungal species have identified between 100–400 proteins with large differences in protein identification between species29. A range of different GO terms were represented in both WC and MalaEx enriched proteins, as transferase, nucleic acid binding, cellular and metabolic processes, and cellular organization or biogenesis. Interestingly, our results indicate that proteins with the GO terms hydrolase and catalytic activity are enriched in MalaEx compared to WC (Fig. 2D). M. sympodialis is a lipid-dependent yeast, and therefore the ability to hydrolyze lipids from the environment by using lipolytic enzymes and integrate the fatty acids into the fungal cell are essential for their survival and are thought to play an important role in the pathogenesis of Malassezia30,31. By spreading hydrolases via MalaEx, such as lysophospholipases found in this study, in the surroundings of M. sympodialis may be beneficial for the survival of the yeast. Packaging of enzymes in EVs may e.g. bring the advantage of a higher digestive enzyme concentration for a more efficient acquisition of nutrients. However, possible consequences could be the degradation of the skin barrier. It is believed that lipase-mediated breakdown of sebaceous lipids and release of irritating unsaturated free fatty acids are important factors in the mechanism behind several skin disorders associated with Malassezia32,33.
It is still unknown how fungal vesicles traverse the cell wall. Here the EVs may also play an important role by carrying enzymes with the ability to hydrolyze cell wall components, which have the potential to promote cell wall reassembly for vesicle passage34,35. The yeast protein homologs with described functions as; aspartic proteinase yapsin, lysophospholipase, serine carboxypeptidase, acid phosphatase and glucan-glucosidase among the MalaEx proteins may be involved in degradation and passage of cell walls. Intriguingly, catalytic enzymes, such as DNA repair, ligase and helicases, histone deacetylase, ribonuclease and tRNA synthetase were identified among the MalaEx enriched proteins. They are essential for the survival of microorganisms, but the benefits of enriching them in MalaEx still need to be explored.
Furthermore, two of the M. sympodialis allergens, Mala s 1 and s 7, were found to be enriched in MalaEx (Fig. 2B). Both allergens encode proteins of unknown function without sequence homology to known allergens or to other known proteins and are predicted to be secreted3,7. Mala s 1 is mainly expressed on the cell surface of Malassezia36. A hypothesis, which merits further investigation, is that Mala s 1 via binding to phospoinositides is involved in membrane trafficking37 or may be involved in cell wall or post-secretory modifications of an as-yet-unidentified secondary metabolite, which may contribute to pathogenesis7. Interestingly, the characterized ortholog of Mala s 1 from the wheat pathogen Fusarium graminearum (Tri 14) has been proposed to be functionally associated with an adjacent gene cluster involved in biosynthesis of a mycotoxin38. Mala s 7 bears signal peptides and does not have transmembrane domains7. Genome sequencing has revealed a gene family amplification for the Mala s 7 allergen in M. sympodialis7. Notably, gene duplication is a major force driving evolution of new traits, including virulence. Thus, by spreading Mala s 1 and s 7, MalaEx may enhance host microbe interactions and may therefore have an in important role in the pathogenesis of AE.
EVs from mammalian cells have been found in diverse body fluids, such as blood39 and breast milk40, and represent important vehicles of intercellular communication in-between cells locally, but also at a distance41. Therefore, we hypothesize that MalaEx due to their small size compared to the whole yeast may be more efficient in disseminate biological signals to the host. Even though not shown yet, they might be able to reach through the outermost layer of skin, especially when the barrier is impaired such as in AE, and thereby being able to interact with host cells. Previously, it has been shown that monocyte-derived dendritic cells are able to engulf M. sympodialis and allergenic components from the yeast42. In this study, we were able to demonstrate for the first time that MalaEx interact with and are actively internalized by human primary monocytes and by keratinocytes. Since the uptake of MalaEx was not seen at 4 °C, it indicates that MalaEx were actively engulfed by endocytosis. Similar results have been reported for lysosome-derived EVs from melanocytes, which were endocytosed by keratinocytes43. Whether the role of the perinuclear localized MalaEx in keratinocytes is to deliver RNAs or proteins for intervening with the host cells, and if the interactions cause inflammation that contributes to skin diseases remains to be elucidated in future studies17,30,38. It has been suggested that vesicular secretion is a general mechanism in fungi for the transport of macromolecules related to virulence16. However, Malassezia may also have beneficial effects for the host. Recently, it was reported that Malassezia globosa, by its ability to secrete an aspartyl protease, MgSAP1, could inhibit Staphylococcus aureus biofilm formation, a virulence attribute44. Whether MalaEx participate in beneficial outcomes for the host is a topic for further studies.
Conclusions
The results of the present study offer an increased understanding of the nature of MalaEx, which seem to be morphological diverse, are enriched with certain proteins, and have the ability to interact with skin cells. These results gives us further clues for understanding more about host-microbe interactions in the sensitization and maintenance of the inflammation in AE, which is important for identifying prospective prevention and new potential therapeutic targets.
Methods
Malassezia sympodialis culture conditions
M sympodialis (ATCC 42132) was cultured on Dixon agar plates45 modified to contain 1% (vol/vol) Tween 60, 1% (wt/vol) agar, and no oleic acid (mDixon) at 32 °C. After 4 days cells were harvested, and re-suspended in PBS and counted in a Bürker chamber using trypan blue exclusion. 2 × 106 yeast cells/ml was cultured in RPMI 1640 supplemented with penicillin 100 units/ml, streptomycin 100 µg/ml, 2 mM L-glutamine, and 10% heat inactivated fetal calf serum (all from Gibco BRL, Life Technologies Ltd, Paisley, UK) incubated for 48 h in 6% CO2 at 37 °C, as previously described8.
For proteomic analysis 2 × 106 yeast cells/ml was cultured in mDixon broth supplemented with 100 units/ml penicillin and 100 µg/ml streptomycin (Gibco BRL) to avoid extensive protein contaminations derived from fetal calf serum used in the RPMI medium, and cultured for 72 h at 32 °C at 200 rpm.
Prior usage, mDixon broth and fetal calf serum had been ultracentrifuged overnight at 100 000 × g followed by filtration through a 0.22 µm filter (Nordic Biolabs, Täby, Sweden) to remove possible EV contaminants. At each culture step blood and Sabourand agar plates were inoculated in parallel to exclude bacterial and Candida contaminations, respectively.
Collection of M. sympodialis cells for proteomics
At the end of the 72 h culture period in mDixon broth, the numbers of M. sympodialis cells were counted using trypan blue exclusion. Thereafter, the culture was pelleted at 1.200 × g for 5 min, washed twice in PBS and frozen at −20 °C for future preparation of cell lysate. The culture supernatant was taken care of for further MalaEx preparations.
MalaEx preparations
M. sympodialis-derived nanovesicles (MalaEx) were prepared by serial ultracentrifugation. The culture supernatants were spun at 1.200 × g for 5 min followed by 3.000 × g for 30 min, and thereafter for the RPMI cultures filtered through a 0.22 µm filter (Nordic Biolabs) and frozen to −80 °C until further preparation. After thawing at RT, supernatants were centrifuged at 10.000 × g for 30 min. Thereafter, MalaEx were pelleted from the supernatants at 100.000 × g for 90 min, re-suspended in PBS and pelleted again at 100.000 × g for 90 min. The resulting pellet was carefully re-suspended in 100 µl PBS. Protein content was measured using a detergent compatible (DC) protein assay according to the manufacturer’s instructions (BioRad, Hercules, CA, USA). The MalaEx preparations were stored frozen at −80 °C.
NanoSight analysis
The particle size of the MalaEx preparations was measured using a LM10 platform with sCMOS camera from NanoSight Ltd, Amesbury, UK. The system is equipped with a 405 nm laser running NTA 2.3 analytical software package. The samples were analyzed at 500 × dilution in PBS with camera level 14 and detection threshold 6. Three consecutive videos were recorded for each sample using batch control to automate the procedure. The temperature was set at 20 °C before acquisition.
Cryo-electron tomography
Vitrification of MalaEx isolated from the RPMI cultures was performed with a Vitrobot (FEI Company; OR, USA). The samples were blotted for 3 sec and plunge frozen in liquid ethane. Tilt series were collected with SerialEM46 at 3° increments over a range of ±60°, with a defocus of 4 to 6 µm. Images were recorded on a 200 kV Talos Arctica (FEI Company) using a Falcon 2 direct electron detector (FEI) at a nominal magnification of 22 000× (corresponding to a pixel size of 6.61 Å). Fiducial-free alignment of tilt images and tomogram reconstructions was carried out in IMOD using the patch-track procedure47. Four tomograms were reconstructed and non-linear anisotropic diffusion (NAD) filtered48. Two tomogram were collected with the Volta phase plate using the Tomo software (FEI company) and reconstructed with Inspect3D (FEI company). Movies were generated using Fiji-ImageJ (imagej.net/Fiji open source image processing package).
Preparation of M. sympodialis and MalaEx lysates for proteomics
Cell lysate
M. sympodialis cells stored at −20 °C were thawed, dissolved in 40 ml PBS and pelleted at 3000 × g for 5 min. This was repeated once to wash the cells. The final pellets were dissolved in 200 µl PBS and added to appr. 200 µl 0.4–0.6 mm silica beads (washed with acidified water to remove fine particles, dust, and other impurities typically found in untreated beads) and homogenized in Precellyse 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France), 5 cycles (6000 rpm, 3 × 30 s). The beads were allowed to settle and 100 µl cell lysate from every sample was transferred to a new tube. 100 µl 2x lysis buffer was added (yielding final concentration of 4% SDS, 0.1 mM DTT, 25 mM Hepes pH 7.6) followed by heating for 5 min at 95 °C and sonication 3 × 20 s. Protein concentration was measured with the DC protein assay (BioRad). 100 µl of lysate from M. sympodialis cells were stored at −80 °C before proteomic analysis.
MalaEx lysate
A 1:1 volume of 2x lysis buffer (yielding final concentration of 4% SDS, 0.1 mM DTT, 25 mM Hepes pH 7.6) was added to the MalaEx preparations followed by heating for 5 min at 95 °C and sonication 3 × 20 s. Protein concentration was measured with the DC protein assay (BioRad) and the lysates were further analyzed with proteomic techniques.
Sample preparation for mass spectrometry
Biological fourplicates of either WC or MalaEx with equal amounts of protein were mixed with 1 mM DTT, 8 M urea, 25 mM HEPES, pH 7.6 in a centrifugation filtering unit, 10 kDa cutoff (Nanosep® Centrifugal Devices with Omega™ Membrane, Pall, New York, USA), and centrifuged for 15 min, 14.000 g, followed by another addition of 8 M urea buffer and centrifugation. Proteins were alkylated by 50 mM IAA, in 8 M urea, 25 mM HEPES, pH 7.6 for 10 min, centrifuged, followed by 2 more additions and centrifugations with 4 M urea, 25 mM HEPES pH 7.6. Trypsin (Promega). 1:50, trypsin:protein, was added to the samples in 0.250 M urea, 25 mM HEPES and digested overnight at 37 °C. The filter units were centrifuged for 15 min, 14.000 g, followed by another centrifugation with MQ and the flow-through was collected. iTRAQ 8plex labeling of the peptides were done according to the manufacturer’s protocol (Applied Biosystems) and cleaned by a strata-X-C-cartridge (Phenomenex, Værløse, Denmark).
IPG-IEF of peptides
The iTRAQ labelled peptides, were separated by immobilized pH gradient - isoelectric focusing (IPG-IEF) on a pH 3–10 strip as described by Branca et al.49. Peptides were extracted from the strips by a prototype liquid handling robot, supplied by GE Healthcare Bio-Sciences AB. A plastic device with 72 wells were put onto each strip and 50 µl of MQ was added to each well. After 30 min incubation, the liquid was transferred to a 96 well plate and the extraction was repeated 2 more times. The extracted peptides were dried in speed vac and dissolved in 3% acetronitrile (ACN), 0.1% formic acid.
LC-ESI-LTQ-Orbitrap analysis
Before analysis on the LTQ Orbitrap Velos (Thermo Fisher Scientific, San Jose, CA, USA), peptides were separated using an Agilent 1200 nano-LC system. Samples were trapped on a Zorbax 300SB-C18, and separated on a NTCC-360/100-5-153 (Nikkyo Technos., Ltd, Tokyo, Japan) column using a gradient of A (5% DMSO, 0.1% FA) and B (90% ACN, 5% DMSO, 0.1% FA), ranging from 3% to 40% B in 50 min with a flow of 0.4 µl/min. The LTQ Orbitrap Velos was operated in a data dependent manner, selecting 5 precursors for sequential fragmentation by CID and HCD, and analyzed by the linear iontrap and orbitrap, respectively. The survey scan was performed in the Orbitrap at 30.000 resolution (profile mode) from 300–2000 m/z, with AGC set to 1 × 106 ions. For generation of HCD fragmentation spectra, a max ion injection time of 500 ms and AGC of 5 × 104 were used before fragmentation at 37.5% normalized collision energy. For FTMS MS2 spectra, normal mass range was used, centroiding the data at 7500 resolution. Peptides for CID were accumulated for a max ion injection time of 200 ms and AGC of 3 × 104, fragmented with 35% collision energy, wideband activation on, activation q 0.25, activation time 10 ms before analysis at normal scan rate and mass range in the linear iontrap. Precursors were isolated with a width of 2 m/z and put on the exclusion list for 90 s. Single and unassigned charge states were rejected from precursor selection.
Peptide and protein identification
Orbitrap raw MS/MS files were converted to mzML format using msConvert from the ProteoWizard tool suite50. Spectra were then searched using MSGF+ (v10072)51 and Percolator (v2.08)52, where search results from 8 subsequent fractions were grouped for Percolator target/decoy analysis. All searches were done against the combined database of M. sympodialis20 and Bos Taurus (Uniprot 170217) in the Galaxy platform53. MSGF+ settings included precursor mass tolerance of 10 ppm, fully-tryptic peptides, maximum peptide length of 50 amino acids and a maximum charge of 6. Fixed modifications were iTRAQ, on lysines and peptide N-termini, and carbamidomethylation on cysteine residues, a variable modification was used for oxidation on methionine residues. Quantification of iTRAQ reporter ions was done using OpenMS project’s IsobaricAnalyzer (v2.0)54. PSMs found at 1% FDR (false discovery rate) were used to infer gene identities.
Protein quantification by iTRAQ reporter ions was calculated using ratios to the whole cell pool and normalized to the sample median. The median PSM reporter ratio from peptides unique to a protein group was used for quantification. Protein false discovery rates were calculated using the picked-FDR method using gene symbols as protein groups and limited to 1% FDR55. Identifications from Bos taurus were considered contaminants from culturing and removed.
GO enrichment analysis
M. sympodialis yeast homologs was used to map GO terms to M. sympodialis proteins20. Panther was used to assign GO terms for the distribution of GO terms in MalaEx enriched proteins56.
Co-cultures of MalaEx with human primary keratinocytes and monocytes
Epidermal keratinocytes were isolated from human abdomen ex vivo skin received from local hospitals according to manufacturer’s instructions (Gibco Invitrogen Corporation, Paisley, UK). In short, thin skin tissues were prepared by using a dermatome, which thereafter were incubated in Dispase solution (25 caseinolytic units/ml; Gibco Invitrogen Corporation) for 18 h at 4 °C. Epidermis was separated from dermis, and placed into 0.05% Trypsin-EDTA (Gibco Invitrogen Corporation) for 15 min at 37 °C for cell dissociation. After addition of Soybean Trypsin Inhibitor (Gibco Invitrogen Corporation) at a concentration of 10 mg/ml, cells were pelleted, washed and suspended in complete serum-free Keratinocyte-SFM medium supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin and 60 µg/ml Amphotericin B (Gibco Invitrogen Corporation). Approximately 3 × 106 cells where seeded in 15 ml culture medium in each T-75 culture flask and cultured at 37 °C, 6% CO2. When cell confluence reached approximately 75%, the cells were sub-cultured in new culture flasks or seeded directly onto chamber culture slides (Falcon, Corning Incorporated, NY, USA) and used for experiments.
Monocytes were achieved by first separating peripheral blood mononuclear cells (PBMC) from a buffy coat derived from one healthy blood donor (Karolinska University Hospital Blood Bank, Stockholm, Sweden) by standard gradient centrifugation with Ficoll Paque (Amersham Pharmacia Biotech AB, Uppsala, Sweden), and thereafter by separating monocytes from PBMC by using anti-CD14 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Isolated cells were analyzed by flow cytometry (FACS Canto II; BD Biosciences, CA, USA) to check for CD14+ cell purity (FITC anti-human CD14 Ab, BioLegend, San Diego, CA, USA), which was 99% and the viability was 98% as analyzed by trypan blue exclusion. Monocytes were cultured in RPMI 1640 medium (Gibco Invitrogen Corporation), supplemented with 2 mM L-glutamine (Gibco Invitrogen Corporation), 100 IU/ml penicillin, 100 µg/ml streptomycin, and 10% heat inactivated fetal calf serum (Gibco) from which exosomes had been deleted by centrifugation at 100 000 g for 16 h and filtration trough a 0.22 µm filter.
MalaEx harvested from the RPMI cultures were stained with the red fluorescent membrane dye Vybrant Dil cell-labelling solution (Life Technologies, OR, USA) according to the manufacturer’s instructions, and thereafter washed twice by centrifugation at 100,000 × g (NVT90 rotor, Beckman Coulter, Indianapolis, IN, USA). As a control the same concentration of Vybrant Dil cell-labelling solution was centrifuged in parallel to create a background control for potentially pelleted unbound dye. Ten µg MalaEx/ml were added to monocytes (0.75 cells/ml) in RPMI medium, and to human primary keratinocytes which had been pre-seeded at a concentration of 0.15 × 106 cells/ml and had reached 75% confluence onto chamber culture slides (Falcon, Corning Incorporated) in serum-free Keratinocyte-SFM (Gibco Invitrogen Corporation). Co-cultures were performed for 2 and 16 h at 37 °C or 4 °C. The study was approved by the regional ethical review board in Stockholm (2015/2082-31/1), and written informed consent was obtained from all subjects donating skin. All experiments were performed in accordance with the Helsinki Declaration ethical principles for medical research.
Confocal laser-scanning microscopy (CLSM)
Co-cultures on the chamber culture slides of MalaEx with keratinocytes were fixed in cold acetone for 5 min and with monocytes in formaldehyde for 15 min. Keratinocytes were stained by a mouse anti-36/E-Cadherin (BD Biosciences), followed by a secondary anti-mouse IgG antibody conjugated with FITC (BD Biosciences). Monocytes co-cultured with MalaEx were stained with anti-CD14 conjugated with Alexa fluor 488 (Bioss Antibodies Inc., MA, USA). Glass slides were mounted with Vectashield mounting medium (Vector Laboratories, CA, USA) and fluorescent images as z-scans were acquired on a CLSM (TCS SP2; Leica Microsystems, Mannheim, Germany).
Structured illumination microscopy (3D-SIM)
Super-resolution fluorescence 3D-SIM imaging57 was performed on a Zeiss Elyra PS.1 (Carl Zeiss, Jena, Germany) of the glass slides with MalaEx and keratinocytes co-cultures prepared for CLSM. Images were captured with an Andor iXon DU 855 EMCCD camera (Andor, Belfast, UK) with a Plan-Apochromate 100×/1.46 NA oil immersion objective. Excitation laser wavelengths used were 488 and 561 nm and fluorescence emission was collected through appropriate dichroic mirrors and single color bandpass filters (E-Cadherin 495–550 nm and MalaEx 570–620 nm). An EMCCD-gain of 10 to 20 and 5 grid rotations were applied with camera integration times of 100 to 150 ms. Image stacks with 50 nm lateral pixel size and 116 nm axial step size were sequentially acquired. Calibration measurements on 40 nm green fluorescent beads delivering a maximal focal resolution of 100 nm laterally and 250 nm axially. Raw SIM datasets were processed with the integrated ELYRA PS.1 system’s analysis software (Zen 2012 SP5 Black) with selection of automatic settings for SIM evaluations (i.e. theoretical PSF, noise filter setting, frequency weighting, baseline handling, etc.). After processing, rendered SIM images were checked for possible artefacts (e.g., amplified honeycomb noise patterns) to confirm suitable automatic analysis settings58.
Statistical analysis
Gene-E (https://software.broadinstitute.org/GENE-E/) was used for making heatmaps and defining enriched proteins in MalaEx vs. WC comparison, using permutation based t-test. Proteins were considered enriched if the ratio between MalaEx and WC was above the 95 percentile and below 0.1 false discovery rate (FDR). Graphpad Prism 7 was used to make figures.
Data access
MS proteomics data is deposited to jPOSTrepo (a repository that is in the ProteomeXchange consortium) with the dataset identifier JPST000288 & PXD009113. MS data can be previewed before publication at: https://repository.jpostdb.org/preview/13203449215a9d422a6ffc5 Using the code: 2889
Electronic supplementary material
Movie 1 Tomogram of purified MalaEx vesicles.
Acknowledgements
We thank Casper Wahlund for assistance with the NanoSight analyses and Strandkliniken, Stockholm and their patients for supplying us with skin. We also thank Felix de Haas for help with Volta phase plate tomography. AS was supported by grants from the Swedish Research Council, the Cancer and Allergy Association, and through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and the Karolinska Institutet. HV was supported by grants from the Swedish Asthma and Allergy Association’s Research Foundation. HB acknowledges support from the Swedish Foundation for Strategic Research (RIF14-0091). The EM data was collected at the Cryo-EM Swedish National Facility funded by the Knut and Alice Wallenberg Foundation, Family Erling Persson and Kempe Foundations, SciLifeLab, Stockholm University and Umeå University.
Author Contributions
H.J., H.V., T.H., U.G., J.L. and A.S. designed the study; A.A., H.V., U.G. and C.J. cultured the cells and isolated MalaEx; H.V. and T.H. characterized MalaEx and prepared cell- and MalaEx lysates; H.J. and J.L. performed proteomics; H.J. performed G.O. analyses. H.V. and C.J. performed in vitro experiments and H.V. the CLSM. analyses; M.C. performed the cryo-EM and tomography and H.B. the 3D-SIM analyses. H.J., H.V., J.L., M.C., H.B. and A.S. analyzed and interpreted the data and prepared the manuscript. All authors participated in the writing and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Henrik J. Johansson and Helen Vallhov contributed equally to this work.
Change history
10/15/2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has not been fixed in the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27451-9.
References
- 1.Sparber F, LeibundGut-Landmann S. Host Responses to Malassezia spp. in the Mammalian Skin. Front Immunol. 2017;8:1614. doi: 10.3389/fimmu.2017.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagata R, et al. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr Int. 2012;54:350–355. doi: 10.1111/j.1442-200X.2012.03563.x. [DOI] [PubMed] [Google Scholar]
- 3.Saunders CW, Scheynius A, Heitman J. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 2012;8:e1002701. doi: 10.1371/journal.ppat.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garmhausen D, et al. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy. 2013;68:498–506. doi: 10.1111/all.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 6.Kim Jung, Kim Jong, Cho Dae, Park Hyun. Molecular Mechanisms of Cutaneous Inflammatory Disorder: Atopic Dermatitis. International Journal of Molecular Sciences. 2016;17(8):1234. doi: 10.3390/ijms17081234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gioti A, et al. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. MBio. 2013;4:e00572–00512. doi: 10.1128/mBio.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehrmann U, et al. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses–novel mechanisms for host-microbe interactions in atopic eczema. PLoS One. 2011;6:e21480. doi: 10.1371/journal.pone.0021480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyer F, Baur A. Biogenesis and functions of exosomes and extracellular vesicles. Methods Mol Biol. 2016;1448:201–216. doi: 10.1007/978-1-4939-3753-0_15. [DOI] [PubMed] [Google Scholar]
- 11.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf JM, Casadevall A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr Opin Microbiol. 2014;22:73–78. doi: 10.1016/j.mib.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coakley G, Maizels RM, Buck AH. Exosomes and other extracellular vesicles: The new communicators in parasite infections. Trends Parasitol. 2015;31:477–489. doi: 10.1016/j.pt.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belting M, Christianson HC. Role of exosomes and microvesicles in hypoxia-associated tumour development and cardiovascular disease. J Intern Med. 2015;278:251–263. doi: 10.1111/joim.12393. [DOI] [PubMed] [Google Scholar]
- 15.Yanez-Mo M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albuquerque PC, et al. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cellular Microbiology. 2008;10:1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallhov H, et al. Dendritic cell-derived exosomes carry the major cat allergen Fel d 1 and induce an allergic immune response. Allergy. 2015;70:1651–1655. doi: 10.1111/all.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Admyre C, et al. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J Allergy Clin Immunol. 2007;120:1418–1424. doi: 10.1016/j.jaci.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Rayner S, et al. Identification of small RNAs in extracellular vesicles from the commensal yeast Malassezia sympodialis. Scientific reports. 2017;7:39742. doi: 10.1038/srep39742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu YF, et al. Proteogenomics produces comprehensive and highly accurate protein-coding gene annotation in a complete genome assembly of Malassezia sympodialis. Nucleic Acids Research. 2017;45:2629–2643. doi: 10.1093/nar/gkx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bitschar K, Wolz C, Krismer B, Peschel A, Schittek B. Keratinocytes as sensors and central players in the immune defense against Staphylococcus aureus in the skin. J Dermatol Sci. 2017;87:215–220. doi: 10.1016/j.jdermsci.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- 23.Casadevall A, Nosanchuk JD, Williamson P, Rodrigues ML. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009;17:158–162. doi: 10.1016/j.tim.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vargas G, et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 2015;17:389–407. doi: 10.1111/cmi.12374. [DOI] [PubMed] [Google Scholar]
- 25.Brisson AR, Tan S, Linares R, Gounou C, Arraud N. Extracellular vesicles from activated platelets: a semiquantitative cryo-electron microscopy and immuno-gold labeling study. Platelets. 2017;28:263–271. doi: 10.1080/09537104.2016.1268255. [DOI] [PubMed] [Google Scholar]
- 26.Zabeo D, et al. Exosomes purified from a single cell type have diverse morphology. J Extracell Vesicles. 2017;6:1329476. doi: 10.1080/20013078.2017.1329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira DL, et al. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One. 2010;5:e11113. doi: 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamith-Miranda D., Nimrichter L., Rodrigues M.L., Nosanchuk J.D. Fungal extracellular vesicles: modulating host–pathogen interactions by both the fungus and the host. Microbes and Infection. 2018;20(9-10):501–504. doi: 10.1016/j.micinf.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues ML, Nakayasu ES, Almeida IC, Nimrichter L. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. Journal of Proteomics. 2014;97:177–186. doi: 10.1016/j.jprot.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juntachai W, Oura T, Murayama SY, Kajiwara S. The lipolytic enzymes activities of Malassezia species. Med Mycol. 2009;47:477–484. doi: 10.1080/13693780802314825. [DOI] [PubMed] [Google Scholar]
- 31.Wu G, et al. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet. 2015;11:e1005614. doi: 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeAngelis YM, et al. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J Invest Dermatol. 2007;127:2138–2146. doi: 10.1038/sj.jid.5700844. [DOI] [PubMed] [Google Scholar]
- 33.Pini G, Faggi E. Extracellular phospholipase activity of Malassezia strains isolated from individuals with and without dermatological disease. Rev Iberoam Micol. 2011;28:179–182. doi: 10.1016/j.riam.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Joffe, L. S., Nimrichter, L., Rodrigues, M. L. & Del Poeta, M. Potential Roles of Fungal Extracellular Vesicles during Infection. mSphere1, 10.1128/mSphere.00099-16 (2016). [DOI] [PMC free article] [PubMed]
- 35.Rodrigues ML, Godinho RM, Zamith-Miranda D, Nimrichter L. Traveling into Outer Space: Unanswered Questions about Fungal Extracellular Vesicles. PLoS Pathog. 2015;11:e1005240. doi: 10.1371/journal.ppat.1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zargari A, Emilson A, Hallden G, Johansson S, Scheynius A. Cell surface expression of two major yeast allergens in the Pityrosporum genus. Clin Exp Allergy. 1997;27:584–592. doi: 10.1111/j.1365-2222.1997.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 37.Vilhelmsson M, et al. Crystal structure of the major Malassezia sympodialis allergen Mala s 1 reveals a beta-propeller fold: a novel fold among allergens. J Mol Biol. 2007;369:1079–1086. doi: 10.1016/j.jmb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Dyer RB, Plattner RD, Kendra DF, Brown DW. Fusarium graminearum TRI14 is required for high virulence and DON production on wheat but not for DON synthesis in vitro. J Agric Food Chem. 2005;53:9281–9287. doi: 10.1021/jf051441a. [DOI] [PubMed] [Google Scholar]
- 39.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 40.Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 41.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buentke E, et al. Uptake of the yeast Malassezia furfur and its allergenic components by human immature CD1a+ dendritic cells. Clin Exp Allergy. 2000;30:1759–1770. doi: 10.1046/j.1365-2222.2000.00937.x. [DOI] [PubMed] [Google Scholar]
- 43.Waster P, Eriksson I, Vainikka L, Rosdahl I, Ollinger K. Extracellular vesicles are transferred from melanocytes to keratinocytes after UVA irradiation. Sci Rep. 2016;6:27890. doi: 10.1038/srep27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Hao, Goh Bee Na, Teh Wooi Keong, Jiang Zhenze, Goh Joleen Pei Zhen, Goh Amelia, Wu Guangxi, Hoon Shawn S., Raida Manfred, Camattari Andrea, Yang Liang, O’Donoghue Anthony J., Dawson Thomas L. Skin Commensal Malassezia globosa Secreted Protease Attenuates Staphylococcus aureus Biofilm Formation. Journal of Investigative Dermatology. 2018;138(5):1137–1145. doi: 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 45.Gueho E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996;69:337–355. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 46.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 48.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 49.Branca RM, et al. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat Methods. 2014;11:59–62. doi: 10.1038/nmeth.2732. [DOI] [PubMed] [Google Scholar]
- 50.Holman JD, Tabb DL, Mallick P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr Protoc Bioinformatics. 2014;46(13):24 11–19. doi: 10.1002/0471250953.bi1324s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Granholm V, et al. Fast and accurate database searches with MS-GF+ Percolator. J Proteome Res. 2014;13:890–897. doi: 10.1021/pr400937n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boekel J, et al. Multi-omic data analysis using Galaxy. Nat Biotechnol. 2015;33:137–139. doi: 10.1038/nbt.3134. [DOI] [PubMed] [Google Scholar]
- 54.Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol Cell Proteomics. 2015;14:2394–2404. doi: 10.1074/mcp.M114.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komis G, et al. Superresolution live imaging of plant cells using structured illumination microscopy. Nature protocols. 2015;10:1248–1263. doi: 10.1038/nprot.2015.083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1 Tomogram of purified MalaEx vesicles.