Abstract
The Microprocessor complex, consisting of an RNase III DROSHA and the DGCR8 dimer, cleaves primary microRNA transcripts (pri-miRNAs) to initiate microRNA (miRNA) maturation. Pri-miRNAs are stem–loop RNAs, and ∼79% of them contain at least one of the three major and conserved RNA motifs, UG, UGU, and CNNC. We recently demonstrated that the basal UG and apical UGU motifs of pri-miRNAs interact with DROSHA and DGCR8, respectively. They help orient Microprocessor on pri-miRNA in a proper direction in which DROSHA and DGCR8 localize to the basal and apical pri-miRNA junctions, respectively. In addition, CNNC, located at ∼17 nucleotides (nt) from the Microprocessor cleavage site, interacts with SRSF3 (SRp20) to stimulate Microprocessor to process pri-miRNAs. The mechanism underlying this stimulation, however, is unknown. In this study, we discovered that SRSF3 recruits DROSHA to the basal junction in a CNNC-dependent manner, thereby enhancing Microprocessor activity. Furthermore, by generating various pri-miRNA substrates containing CNNC at different locations, we demonstrated that such stimulation only occurs when CNNC is located at ∼17 nt from the Microprocessor cleavage site. Our findings reveal the molecular mechanism of SRSF3 in pri-miRNA processing and support the previously proposed explanation for the highly conserved position of CNNC in SRSF3-enhanced pri-miRNA processing.
Keywords: SRSF3, microprocessor, microRNA, DROSHA, DGCR8
INTRODUCTION
MicroRNAs (miRNAs), short and single-stranded RNAs of ∼22 nucleotides (nt), are mostly produced via the canonical microRNA biogenesis pathway (Ha and Kim 2014). This process involves two consecutive cleavages by RNase III-type enzymes. The primary miRNA transcript (pri-miRNA) synthesized in the nucleus is initially cleaved by the Microprocessor complex composed of nuclear RNase III DROSHA and its cofactor DGCR8 (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004, 2006; Landthaler et al. 2004; Auyeung et al. 2013; Nguyen et al. 2015; Fareh et al. 2016; Herbert et al. 2016; Kwon et al. 2016). The resulting product, precursor miRNA (pre-miRNA) is exported to the cytoplasm and is further cleaved by another RNase III enzyme, Dicer, that produces a miRNA duplex of ∼22 nt (Ma et al. 2004; MacRae et al. 2006a,b; Park et al. 2011; Gu et al. 2012; Tian et al. 2014). Only one strand of each duplex is loaded into an Argonaute (Ago) protein, and the other strand is eliminated (Kawamata and Tomari 2010). The Ago-miRNA interaction forms the core of an RNA-induced silencing complex that plays a critical role in posttranscriptional gene regulation in various essential biological processes (Bartel 2009; Ameres and Zamore 2013; Ha and Kim 2014).
Pri-miRNA (Fig. 1A), a substrate of Microprocessor, has a local stem–loop, consisting of a three-helix stem flanked by the basal and apical junctions at two ends (Han et al. 2006; Ha and Kim 2014). DROSHA, a catalytic subunit of the Microprocessor complex, must localize at the pri-miRNA basal junction to cleave pri-miRNA at the productive sites (∼13 nt from the basal junction) and generates pre-miRNA (Fig. 1A). However, the somewhat symmetrical nature of pri-miRNA secondary structure often leads Microprocessor to interact with the apical junction, thereby cleaving pri-miRNAs at the unproductive sites ∼13 nt from the apical junction (Han et al. 2006; Nguyen et al. 2015). Multiple primary sequence motifs that have been recently identified (Auyeung et al. 2013; Fang and Bartel 2015) create an asymmetry for pri-miRNAs and facilitate association of DROSHA with the basal junction (Nguyen et al. 2015; Kwon et al. 2016). The U and G at positions -14 and -13 (Fig. 1A) whose frequencies in human pri-miRNAs conserved to mice, so-called representative pri-miRNAs, are ∼40% and 48%, respectively, independently contribute to pri-miRNA processing (Auyeung et al. 2013). The basal UG motif (Fig. 1A) at the basal junction (Auyeung et al. 2013), which is present in ∼24% of the representative pri-miRNAs, directly recruits DROSHA to the basal junction, thereby stimulating the productive cleavage by DROSHA (Nguyen et al. 2015). The apical UGU motif at the apical loop (Fig. 1A), which appears in ∼29% of the representative pri-miRNAs (Auyeung et al. 2013), interacts with the DGCR8 dimer, indirectly favoring the basal junction-DROSHA interaction by preventing DROSHA from binding to the apical junction (Nguyen et al. 2015). The GHG motif (Fig. 1A), found in the stem of 25% of the representative pri-miRNAs, enhances the enzymatic activity by an unknown mechanism (Fang and Bartel 2015). The most abundant motif identified in pri-miRNAs is CNNC (Fig. 1A), which is present in ∼60% of the representative pri-miRNAs and is located at the 3′-RNA segment ∼16–18 nt from the cleavage sites (Auyeung et al. 2013). The CNNC motif associates with SRSF3 (SRp20), a splicing factor that stimulates the Microprocessor activity in vitro (Auyeung et al. 2013). In addition, SRSF3 enhances Microprocessor to cleave a pri-mir-30c variant in which the CNNC motif is exposed in a single-stranded RNA region, thereby affecting miR-30c expression in vivo (Fernandez et al. 2017). In contrast, SRSF3 fails to stimulate the Microprocessor activity for pri-mir-30c wild-type (WT), where the CNNC motif is present in the duplex RNA region (Fernandez et al. 2017). This emphasizes the physiological significance of the SRSF3–CNNC interaction in the appropriate cellular context. However, a molecular mechanism explaining how SRSF3 stimulates pri-miRNA processing and why the position of CNNC is restricted to an area within 16–18 nt from the DROSHA cleavage sites remains elusive.
FIGURE 1.
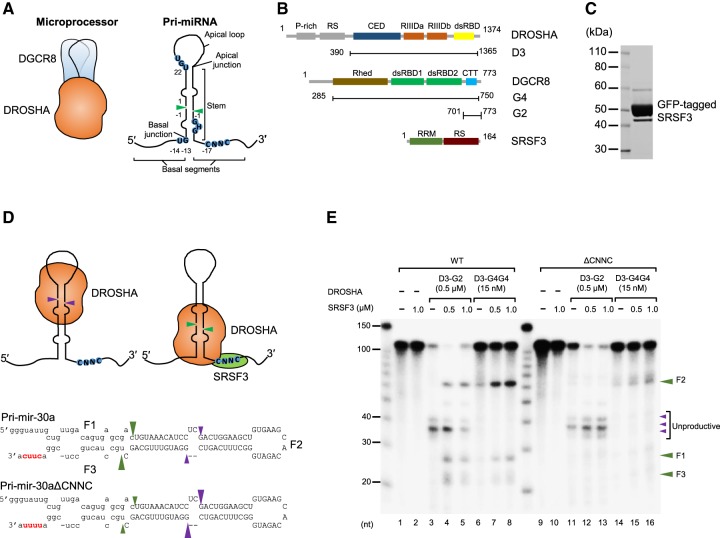
SRSF3 recruits DROSHA to the CNNC-containing basal junction. (A) Microprocessor is a trimeric complex consisting of an RNase III DROSHA and a DGCR8 dimer. The typical pri-miRNA, a substrate of the Microprocessor complex, has a stem–loop structure. The green arrowheads indicate the DROSHA cleavage sites. The basal junction and the basal UG motif located at 14 nt from the cleavage site are recognized by DROSHA. The apical UGU motif of the apical loop that appeared at 22 nt from the cleavage site is recognized by DGCR8. The CNNC motif of the basal segment that resides at 17 nt from the cleavage site interacts with SRSF3. (B) Protein constructs used in this study. The first and end residue numbers for each construct are shown. (P-rich) Proline-rich domain, (RS) arginine/serine-rich domain, (CED) central domain, (RIIIDa and RIIIDb) RNase III domains, (dsRBD) dsRNA-binding domain, (Rhed) RNA-binding heme domain, (CTT) C-terminal tail region, (RRM) RNA recognition motif. SRSF3 is composed of 164 amino acids and its molecular weight is about 19 kDa. It has one RRM at the N terminus and one RS domain at the C terminus with extensive phosphorylation on serine residues. (C) The purity of the GFP-tagged SRSF3 on SDS–PAGE. (D) The pri-mir-30a substrates. The uppercase letters represent the pre-miRNA region. The CNNC and ΔCNNC motifs are highlighted in red. The green and purple arrowheads indicate the productive and unproductive cleavage sites of DROSHA, respectively. (E) Processing of the pri-mir-30a substrates by D3–G2 or D3–G4G4 and SRSF3. The internally labeled pri-mir-30a substrates were incubated with the indicated proteins for 1 h under the conditions described in Materials and Methods. The three products, a 5′-fragment, pre-miRNA, and a 3′-fragment were named as F1, F2, and F3, respectively.
In this study, we explored the mechanism of SRSF3 action using purified recombinant proteins from human cells (Fig. 1B). We discover that SRSF3 stimulates Microprocessor by recruiting DROSHA to the pri-miRNA basal junction. This recruitment is dependent on the CNNC motif of pri-miRNA at the precise position.
RESULTS
SRSF3 recruits DROSHA to the basal junction of pri-miRNA in a CNNC-dependent manner
To determine how SRSF3 affects the pri-miRNA processing, we purified the SRSF3 proteins tagged with GFP from suspension HEK293E cells (Fig. 1C) and selected pri-mir-30a (Fig. 1D) harboring a CNNC motif (CUUC) 17 nt from the DROSHA cleavage sites. The D3–G2 complex contains all necessary domains for DROSHA activity (D3, Fig. 1B) and a stabilizing fragment of DGCR8 (G2, Fig. 1B). The D3–G4G4 complex consists of the D3 fragment of DROSHA and the G4G4 dimer of DGCR8 (G4, Fig. 1B). These two complexes were purified as described previously (Nguyen et al. 2015). First, we tested the effect of SRSF3 on the D3–G2 complex. Consistent with previous studies (Nguyen et al. 2015; Kwon et al. 2016), D3–G2 alone primarily cleaved pri-mir-30a at the unproductive sites (Fig. 1E, lane 3). Interestingly, however, the addition of SRSF3 caused D3–G2 to successfully cleave pri-mir-30a at the productive sites (Fig. 1E, lanes 4,5). As a result, SRSF3 strongly stimulated the trimeric D3–G4G4 complex to cleave at the productive sites (Fig. 1E, lanes 6–8; Supplemental Fig. S1A, lanes 3–5). In contrast, when the CUUC motif was replaced with the UUUU motif, SRSF3 did not enhance the productive activity of the D3–G2 and D3–G4G4 complexes (Fig. 1E, lanes 11–16; Supplemental Fig. S1A, lanes 8–10). These data suggest that by interacting with the CNNC motif at the 3′-segment of pri-miRNAs, SRSF3 might recruit DROSHA to the basal junction, thereby stimulating the occurrence of productive cleavages. The other proteins, SRSF7 (9G8), DDX5, and DDX17 that were proposed to interact with the 3′-segment of pri-miRNA were also tested on the pri-miRNA processing of DROSHA (Wang et al. 2012a; Auyeung et al. 2013; Dardenne et al. 2014; Mori et al. 2014; Moy et al. 2014). Unlike SRSF3, they did not enhance the DROSHA activity (Supplemental Fig. S1B,C), suggesting that they might use different mechanisms in the pri-miRNA processing.
SRSF3 and DROSHA form a complex with the CNNC-containing pri-mRNAs
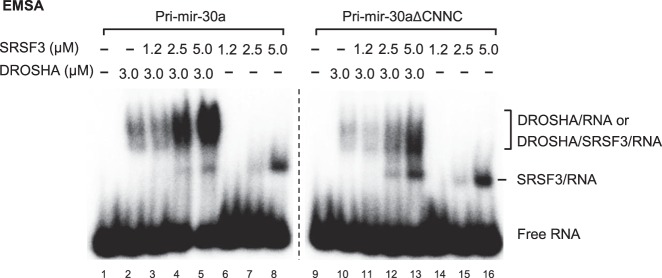
To confirm that SRSF3 recruits DROSHA to the basal junction, we examined if SRSF3 and DROSHA can physically interact with each other. We used purified SRSF3 and DROSHA proteins in immunoprecipitation assays but did not identify a direct protein–protein interaction (data not shown). Thus, their interaction is likely to be either transient or RNA-dependent. Therefore, we used an electrophoretic mobility shift assay (EMSA) to test whether SRSF3 and DROSHA can form a stable complex on RNA. The resulting EMSA data showed that individual DROSHA and SRSF3 proteins interacted with pri-mir-30a and distinctly shifted the electrophoretic mobility of RNA bands (Fig. 2, lane 2 for DROSHA and lane 8 for SRSF3; Supplemental Fig. S2A). DROSHA seemed to have a slightly higher affinity to the pri-mir-30a than pri-mir-30aΔCNNC (Fig. 2, cf. lane 2 with 10; Supplemental Fig. S2B,C). When two proteins were simultaneously incubated with pri-mir-30a, the shifted RNA bands were denser than those observed using any one of the proteins and RNA (Fig. 2, cf. lane 5 with 2 or 8; Supplemental Fig. S2A). This indicates that SRSF3 and DROSHA coordinately interact with RNA. Furthermore, the SRSF3-bound RNA band was further shifted by the addition of DROSHA (Fig. 2, cf. lane 8 with 5), suggesting that SRSF3 and DROSHA associate together on the same pri-miRNA molecule. In contrast with CUUC-containing pri-mir-30a, SRSF3 inefficiently stimulated the complex formation of DROSHA and UUUU-containing pri-mir-30a (Fig. 2, cf. lanes 12–13 with 4–5). This declined stimulatory effect of SRSF3 could be due to its weak interaction with the UUUU motif (Schaal and Maniatis 1999; Hargous et al. 2006; Änkö et al. 2012). In addition, the band of UUUU-containing pri-mir-30a bound with SRSF3 was not efficiently shifted by the addition of DROSHA (Fig. 2, cf. lane 16 with 13). The data from EMSA experiments support a model that SRSF3 interacts with CNNC and recruits DROSHA to the basal junction.
FIGURE 2.
SRSF3 and DROSHA (D3–G2) form a complex with pri-miRNA. The electrophoretic mobility shift assay (EMSA) experiments were carried out with the pri-mir-30a substrates and the indicated proteins in the figure. The increasing amounts of the indicated proteins were incubated with the radiolabeled pri-mir-30a WT or ΔCNNC substrate in 20 μL of the buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 0.2 mg/mL BSA, 1 mM DTT, and 2 mM EDTA. The reaction mixtures were incubated on ice for 1 h and supplemented with 4 μL of the 6× sample buffer composed of 0.01% (w/v) bromophenol blue, 60% (v/v) glycerol. Finally, 10 μL of each sample was loaded on 6% native PAGE and run at 4°C for 1 h.
Proper position of CNNC is critical for SRSF3 to stimulate productive processing by DROSHA
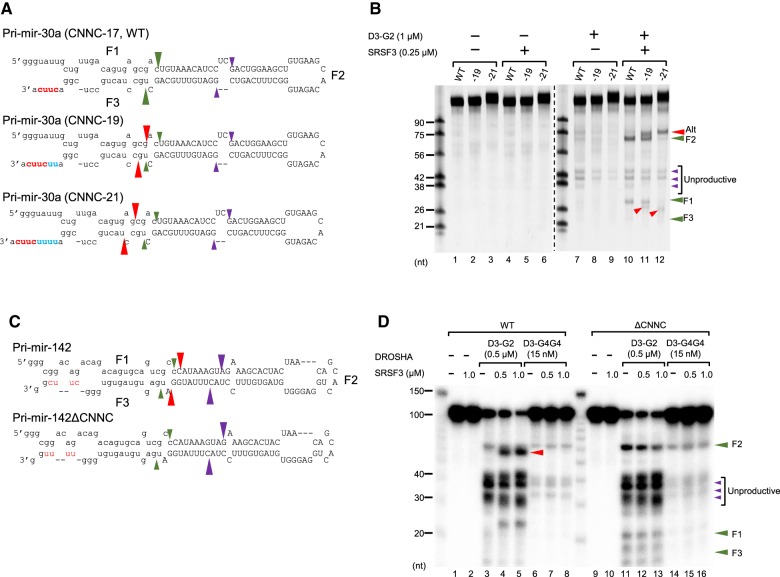
Since the CNNC motif is highly conserved at the 3′-segment of pri-miRNAs ∼16 to 18 nt from the DROSHA cleavage sites and this position is important for miRNA expression in vivo (Auyeung et al. 2013), we examined the positional effect of the CNNC motif on the pri-miRNA processing. We generated two pri-mir-30a variants with the CUUC motif at either 19 or 21 nt from the DROSHA cleavage sites (Fig. 3A) and used them in the processing assays with D3–G2 and SRSF3 (Fig. 3B). Here, we found that SRSF3 stimulated DROSHA (D3–G2) to cleave these variants more at the basal sites (Fig. 3B, cf. lanes 11–12 with 8–9; Supplemental Fig. S3A,B), suggesting that SRSF3 could also recruit DROSHA to the basal junction of these variants. Unlike pri-mir-30a WT, however, SRSF3 enhanced the DROSHA cleavage of the variants more at alternative sites 1 or 2 nt closer to the basal junction (Fig. 3B, cf. lanes 11 and 12 with 10; Supplemental Fig. S3B). Consistently, SRSF3 facilitated D3–G4G4 to cleave pri-mir-30a WT and its variants more efficiently at the basal junction (Supplemental Fig. S3C). However, compared with D3–G2, D3–G4G4 was less stimulated by SRSF3 to cleave these variants at the alternative sites (Supplemental Fig. S3C), implying that DGCR8 (G4G4) might hold DROSHA at the productive cleavage sites (Nguyen et al. 2015). This result confirms the role of SRSF3 in DROSHA recruitment and suggests that SRSF3 stimulates the productive processing by DROSHA as the CNNC motif is present at a position ∼17 nt downstream from the cleavage sites.
FIGURE 3.
SRSF3 stimulates the productive processing by DROSHA in a CNNC position-dependent manner. (A,C) The pri-mir-30a and pri-mir-142 substrates. The uppercase letters represent the pre-miRNA region. The CNNC motif and the added nucleotides are highlighted in red and blue, respectively. The green, purple, and red arrowheads indicate the productive, unproductive, and alternative cleavage sites of DROSHA, respectively. (B,D) Processing of the pri-mir-30a and pri-mir-142 substrates by D3–G2 or D3–G4G4 and SRSF3. The unlabeled pri-mir-30a substrate (0.5 μM) or the internally labeled pri-mir-142 substrate was incubated with the proteins that are shown in the figure for 1 h under the conditions described in Materials and Methods. F1, F2, and F3 are the productive products. The unproductive and alternative products are indicated with the green and red arrowheads, respectively.
To further confirm the importance of the CNNC position, we investigated the effect of SRSF3 on the Microprocessor processing of pri-mir-142 in which the CNNC motif is located at 15 nt downstream from the cleavage sites (Fig. 3C). SRSF3 persistently induced DROSHA (D3–G2) to cleave at the basal sites on pri-mir-142 WT (Fig. 3D, cf. lanes 4–5 with 3) but not pri-mir-142ΔCNNC (Fig. 3D, cf. lanes 12–13 with 11). However, SRSF3 stimulated DROSHA to cleave at a position 1 or 2 nt away from the productive sites cut by DROSHA alone (Fig. 3D, cf. lanes 4–5 with 3). In addition, SRSF3 failed to stimulate D3–G4G4 to cleave pri-mir-142 (Fig. 3D, cf. lanes 7–8 with 6). This further indicates that SRSF3 functions in a CNNC position-dependent manner to facilitate the pri-miRNA processing.
DISCUSSION
Previous studies demonstrated that human Microprocessor interacts with multiple cis-acting RNA elements to accurately and efficiently cleave pri-miRNAs (Han et al. 2006; Auyeung et al. 2013; Nguyen et al. 2015; Kwon et al. 2016). The basal junction and its UG motif are recognized by DROSHA (Han et al. 2006; Nguyen et al. 2015). The apical loop and the UGU motif interact with the DGCR8 dimer assisted by hemin (Nguyen et al. 2015, 2018; Partin et al. 2017). The CNNC motif at the 3′ flanking region of pri-miRNA interacts with SRSF3, a trans-acting factor that stimulates Microprocessor (Auyeung et al. 2013). In this study, we reveal a molecular mechanism in which SRSF3 binds to the CNNC motif, thereby recruiting DROSHA to the basal junction of pri-miRNAs. Consequently, SRSF3 influences the orientation of Microprocessor on the CNNC-containing pri-miRNAs (Fig. 4 for the model).
FIGURE 4.
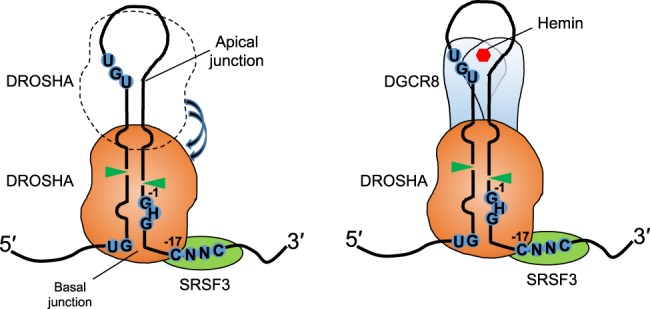
Model for the SRSF3's molecular mechanism in pri-miRNA processing. The basal and apical junctions are two single-stranded RNA/double-stranded RNA junctions of pri-miRNA. DROSHA can recognize and cleave at either junction. By interacting with the CNNC motif optimally located at the 3′-segment of pri-miRNA ∼17 nt from the DROSHA cleavage sites, SRSF3 strengthens interactions between DROSHA and the basal junction, thereby stimulating DROSHA to cleave at the productive sites. As a result, SRSF3 enhances pri-miRNA processing of the Microprocessor complex.
This study demonstrates that SRSF3 can recruit DROSHA to the basal junction by interacting with the CNNC motif at the various positions (15–21 nt from the DROSHA cleavage sites). Interestingly, SRSF3 can alter the DROSHA cleavage sites at the stem of pri-miRNAs. For example, SRSF3 can shift DROSHA from the productive cleavage sites to the alternative sites toward the basal junction when the CNNC motif is farther from the basal junction (19 or 21 nt from the cleavage sites) (Fig. 3B). In contrast, SRSF3 can move DROSHA to the alternative sites away from the basal junction as the CNNC motif resides closer to the basal junction (15 nt from the cleavage sites) (Fig. 3D). These indicate that SRSF3 and DROSHA might make a protein–protein contact upon interaction with pri-miRNA and that the two proteins are likely positioned at a certain fixed distance. In addition, the EMSA results (Fig. 2; Supplemental Fig. S2) support that SRSF3 and DROSHA might form a stable complex on the CNNC-containing pri-miRNA. Of note, SRSF3 does not cause the DROSHA/RNA complex to obviously further shift in the gel, suggesting that either the DROSHA/RNA/SRSF3 complex has a similar size as the DROSHA/RNA complex or SRSF3 might dissociate from pri-miRNA after facilitating DROSHA binding. Future structural studies are expected to reveal the atomic mechanisms underlying the DROSHA/RNA/SRSF3 interaction.
Unlike DROSHA, the cleavage sites by the Microprocessor complex are less shifted by SRSF3 (pri-mir-30a, Supplemental Fig. S3C; pri-mir-142, Fig. 3D) as the CNNC motif is not located at the optimal position (16–18 nt from the productive cleavage sites). This could be explained by a role of DGCR8 in the Microprocessor complex, that strongly retains DROSHA at the productive cleavage sites (Nguyen et al. 2015) despite the effect of SRSF3. Therefore, the proper position of CNNC (16–18 nt from the DROSHA cleavage sites) is crucial for SRSF3 to coordinate with DROSHA and DGCR8 in effective stimulation of the pri-miRNA processing in vitro. However, a broader window (16–21 nt) of CNNC might be functional in vivo when a large group of pri-miRNAs was investigated (Conrad et al. 2014). It is possible that the secondary structure surrounding the CNNC motif or other cofactors may contribute to the CNNC-dependent processing of pri-miRNAs by Microprocessor.
SRSF3, the smallest member of the highly conserved SR-rich splicing factor family, regulates the splicing of numerous genes (Jumaa and Nielsen 1997, 2000; Jumaa et al. 1997; Galiana-Arnoux et al. 2003; Yu et al. 2004; de la Mata and Kornblihtt 2006; Gonçalves et al. 2008, 2009; Sen et al. 2009; Manley and Krainer 2010; Wang et al. 2012b; Wong et al. 2012; Ajiro et al. 2016). Furthermore, SRSF3 interacts with various CNNC-containing mRNAs (Hargous et al. 2006; Änkö et al. 2012). Therefore, it will also be interesting to investigate if the DROSHA/RNA/SRSF3 interaction may be involved in mRNA metabolism.
MATERIALS AND METHODS
Processing assay
The processing assay was carried out at 37°C in 20 μL assay buffer containing 50 mM Tris-hydrochloride (Tris-HCl [pH 7.5]), 100 mM sodium chloride (NaCl), 10% glycerol, 0.2 μg/μL bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), and 2 mM magnesium chloride (MgCl2). Approximately 10,000 cpm of the RNA substrates was used, and the enzyme concentrations and incubation time are indicated in the figures. The reaction was stopped by adding 20 μL TBE-urea sample buffer (Bio-Rad). Finally, the mixture was heated at 95°C for 10 min and quickly chilled on ice before loading onto a 10% Urea-polyacrylamide gel electrophoresis (Urea-PAGE) gel with the RNA size markers (Decade marker, Ambion). The RNA substrates were prepared as described previously (Nguyen et al. 2015).
Purification of recombinant SRSF3 from human cells
The SRSF3 fused to the C-terminal GFP and 10× His-tag was cloned into the pXG vector as used in the previous studies (Nguyen et al. 2015; Kwon et al. 2016). The plasmids were transfected into 3.6 L HEK293E suspension cell culture and the cells were harvested after 2 d. The cell pellets were resuspended in 200 mL T500 buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, and 4 mM β-mercaptoethanol) supplemented with 2 μg/mL RNase A and protease inhibitor cocktail. The clear lysate obtained by sonication and centrifugation was loaded on a Ni-NTA column. The column was washed with 200 mL T500 supplemented 20 mM imidazole and eluted with 50 mL T500 supplemented 200 mM imidazole. The eluted proteins were diluted to 100 mM NaCl with T0 (20 mM Tris-HCl [pH 7.5] and 4 mM β-mercaptoethanol) and bound with Q-sepharose. The Q-sepharose was washed with 150 mM NaCl-containing buffer, and the proteins were eluted with T500. The proteins were further purified using gel filtration with a Superdex 200 10/300 GL column and were finally frozen with LN2 and stored at −80°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of our laboratories for discussion and technical assistance. We thank Dr. Narry Kim for her enormous support and invaluable discussion. K.K. was supported by the Institute for Basic Science from the Ministry of Science, ICT and Future Planning of Korea (IBS-R008-D1). T.D.N. and S.H.L. were supported by Hong Kong University of Science and Technology PhD Scholarship. This work was supported by National Natural Science Foundation of China (31770884). T.A.N. is an investigator of Hong Kong University of Science and Technology.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065862.118.
REFERENCES
- Ajiro M, Jia R, Yang Y, Zhu J, Zheng ZM. 2016. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res 44: 1854–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14: 475–488. [DOI] [PubMed] [Google Scholar]
- Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. 2012. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. 2013. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152: 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Marsico A, Gehre M, Orom UA. 2014. Microprocessor activity controls differential miRNA biogenesis in vivo. Cell Rep 9: 542–554. [DOI] [PubMed] [Google Scholar]
- Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, et al. 2014. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep 7: 1900–1913. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR. 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRSF3. Nat Struct Mol Biol 13: 973–980. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Fang W, Bartel DP. 2015. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol Cell 60: 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareh M, Loeff L, Szczepaniak M, Haagsma AC, Yeom KH, Joo C. 2016. Single-molecule pull-down for investigating protein-nucleic acid interactions. Methods 105: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N, Cordiner RA, Young RS, Hug N, Macias S, Cáceres JF. 2017. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun 8: 15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Lejeune F, Gesnel MC, Stevenin J, Breathnach R, Del Gatto-Konczak F. 2003. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRSF3. J Biol Chem 278: 32943–32953. [DOI] [PubMed] [Google Scholar]
- Gonçalves V, Matos P, Jordan P. 2008. The β-catenin/TCF4 pathway modifies alternative splicing through modulation of SRSF3 expression. RNA 14: 2538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves V, Matos P, Jordan P. 2009. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 18: 3696–3707. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. 2012. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 151: 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The DROSHA-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the DROSHA-DGCR8 complex. Cell 125: 887–901. [DOI] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. 2006. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Sarkar SK, Mills M, Delgado De la Herran HC, Neuman KC, Steitz JA. 2016. A heterotrimer model of the complete Microprocessor complex revealed by single-molecule subunit counting. RNA 22: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. 1997. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J 16: 5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. 2000. Regulation of SRp20 exon 4 splicing. Biochim Biophys Acta 1494: 137–143. [DOI] [PubMed] [Google Scholar]
- Jumaa H, Guénet JL, Nielsen PJ. 1997. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol 17: 3116–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. 2010. Making RISC. Trends Biochem Sci 35: 368–376. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng S, Kim VN, Woo JS. 2016. Structure of Human DROSHA. Cell 164: 81–90. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. 2006a. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Li F, Zhou K, Cande WZ, Doudna JA. 2006b. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harbor Symp Quant Biol 71: 73–80. [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR. 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24: 1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. 2014. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 156: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy RH, Cole BS, Yasunaga A, Gold B, Shankarling G, Varble A, Molleston JM, tenOever BR, Lynch KW, Cherry S. 2014. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell 158: 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. 2015. Functional anatomy of the human Microprocessor. Cell 161: 1374–1387. [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Park J, Dang TL, Choi YG, Kim VN. 2018. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res 10.1093/nar/gky248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. 2011. DICER recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin AC, Ngo TD, Herrell E, Jeong BC, Hon G, Nam Y. 2017. Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs. Nat Commun 8: 1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. 1999. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol 19: 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Talukdar I, Webster NJ. 2009. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol 29: 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ. 2014. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human DICER. Mol Cell 53: 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Guo J, Zhou J. 2004. A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J Neurochem 90: 164–172. [DOI] [PubMed] [Google Scholar]
- Wang D, Huang J, Hu Z. 2012a. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol Cell Proteomics 11: M111.011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chatterjee D, Jeon HY, Akerman M, Vander Heiden MG, Cantley LC, Krainer AR. 2012b. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J Mol Cell Biol 4: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Garner B, Halliday GM, Kwok JB. 2012. Srp20 regulates TrkB pre-mRNA splicing to generate TrkB-Shc transcripts with implications for Alzheimer's disease. J Neurochem 123: 159–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.