Abstract
Comparing the abundance of one RNA molecule to another is crucial for understanding cellular functions but most sequencing techniques can target only specific subsets of RNA. In this study, we used a new fragmented ribodepleted TGIRT sequencing method that uses a thermostable group II intron reverse transcriptase (TGIRT) to generate a portrait of the human transcriptome depicting the quantitative relationship of all classes of nonribosomal RNA longer than 60 nt. Comparison between different sequencing methods indicated that FRT is more accurate in ranking both mRNA and noncoding RNA than viral reverse transcriptase-based sequencing methods, even those that specifically target these species. Measurements of RNA abundance in different cell lines using this method correlate with biochemical estimates, confirming tRNA as the most abundant nonribosomal RNA biotype. However, the single most abundant transcript is 7SL RNA, a component of the signal recognition particle. Structured noncoding RNAs (sncRNAs) associated with the same biological process are expressed at similar levels, with the exception of RNAs with multiple functions like U1 snRNA. In general, sncRNAs forming RNPs are hundreds to thousands of times more abundant than their mRNA counterparts. Surprisingly, only 50 sncRNA genes produce half of the non-rRNA transcripts detected in two different cell lines. Together the results indicate that the human transcriptome is dominated by a small number of highly expressed sncRNAs specializing in functions related to translation and splicing.
Keywords: high-throughput sequencing, noncoding RNA, snoRNA, RNA detection, thermostable group II intron reverse transcriptase, transcriptome analysis
INTRODUCTION
The study of gene expression is a rapidly growing area of genomic research (Ozsolak and Milos 2011; Jiang et al. 2015). The amount of RNA sequencing is expected to surge as the need for understanding gene function and the identification of new biomarkers increases (Rabbani et al. 2016). However, despite the increase in RNA sequencing in the last five years, established methods for the detection of midsize sncRNAs remain elusive (Veneziano et al. 2016). Indeed, RNA ranging in size between 50 and 300 nucleotides (nt) was termed the “black hole” of RNA biology due to the lack of sequencing information (Steitz 2015). Most standard sequencing methods are focused on the detection of polyadenylated messenger RNAs with sizes typically larger than 1 kb (Costa et al. 2010; Liang and Zeng 2016). As such, these methods are not useful for the detection of nonpolyadenylated transcripts, many of which are shorter than 500 nt (e.g., small nuclear RNA [snRNA], small nucleolar RNA [snoRNA], transfer RNA [tRNA], and many long noncoding RNA [lncRNA]) (Veneziano et al. 2016). In addition, selection of polyadenylated RNA prevents the detection of RNA processing and maturation intermediates. Current approaches for the sequencing of sncRNAs depend on selection techniques that enrich RNAs based on their size or localization in the cell (Deschamps-Francoeur et al. 2014; Bai and Laiho 2016). Recent studies suggest that most of these techniques introduce bias in the relative representation of noncoding RNAs even for those with similar sizes (Deschamps-Francoeur et al. 2014; Nottingham et al. 2016).
Classical estimates of RNA abundance are usually generated by using targeted in vivo labeling experiments, as in the case of rRNA, tRNA, snRNA, and snoRNA, or by using microarrays as in the case of mRNA and miRNA (Waldron and Lacroute 1975; Wolf and Schlessinger 1977; Bissels et al. 2009). These class-specific estimates of RNA abundance indicate that ∼90% of the human transcriptome by mass is composed of rRNA, while the highest number of molecules per cell (MPC) is attributed to tRNA (Waldron and Lacroute 1975; Wolf and Schlessinger 1977). Other methods such as reverse transcription quantitative PCR (RT-qPCR) (Ginzinger 2002; Shakeel et al. 2017), digital PCR (dPCR) (Whale et al. 2012; Hayden et al. 2013; Witwer et al. 2013; Morley 2014; Sager et al. 2015), and in situ hybridization (e.g., FISH) (Vera et al. 2016) are also being used for RNA quantification, but their use remains limited to a relatively low number of RNAs and they are rarely utilized for comparisons between different classes of RNA. More recently, transcriptome sequencing has become the most frequently used method for large scale profiling of the transcriptome (Casamassimi et al. 2017). However, these techniques are efficient in comparing the relative levels of transcript abundance within the same class of RNA but they cannot directly compare between different classes of RNA (e.g., between coding and sncRNA). Therefore, while the number of different RNA classes might be established, measurements of their true relative abundance remain to be verified.
Recently, a new sequencing method using a thermostable group II intron reverse transcriptase (TGIRT) was developed, which exploits the ability of this highly processive enzyme to reverse transcribe full-length, highly structured noncoding RNAs (Nottingham et al. 2016; Qin et al. 2016). This method of sequencing (TGIRT-seq) does not require ligation of adapters to the RNA but instead uses the proficient template switching activity of TGIRT to couple adapter addition to the 3′ terminal nucleotide of an RNA template. This method avoids sequence and structure biases in RNA ligation, as well as interference from the 5′ cap structure of mRNAs (Nottingham et al. 2016). Sequencing of human reference RNA samples with external RNA controls consortium (ERCC) spike-in control RNAs showed that TGIRT-seq has less bias than the widely used TruSeq method and enables sequencing of tRNAs and other sncRNAs together with mRNAs and lncRNAs (Nottingham et al. 2016; Qin et al. 2016). As such, the method provides a potentially useful tool for direct comparison of the abundance of different classes of RNA (Zheng et al. 2015; Nottingham et al. 2016).
In this study, we compare the capacity of different sequencing methods including TGIRT-based methods to faithfully rank the abundance of different classes of RNA and depict the overall landscape of the human transcriptome. The results indicate that sequencing of fragmented ribodepleted cellular RNA using TGIRT not only ranks transcripts of the same class of RNA more accurately than targeted sequencing approaches but also provides the most complete and experimentally supported portrait of the human transcriptome. Using this method, we were able to confirm the overall conclusions of previous estimates of RNA abundance showing that tRNA are the most abundant RNA species in terms of number of molecules. However, unlike previous estimates our results show that snRNAs are actually more abundant than mRNAs and snoRNAs and that in general sncRNAs are at least 1000 times more abundant than mRNAs encoding proteins functioning in the same biological complex. Interestingly, direct comparisons between the coding and noncoding RNAs participating in the assembly of ribonucleoprotein complexes permitted the identification of specific components with either regulatory or multiple functions. Together our results indicate that simultaneous detection of both coding and noncoding RNA by TGIRT-seq not only increases the number of transcript types analyzed but also improves the precision of RNA ranking and estimates of abundance within each class of RNA.
RESULTS
Comparison between the capacities of different sequencing methods to quantify different components of the human transcriptome
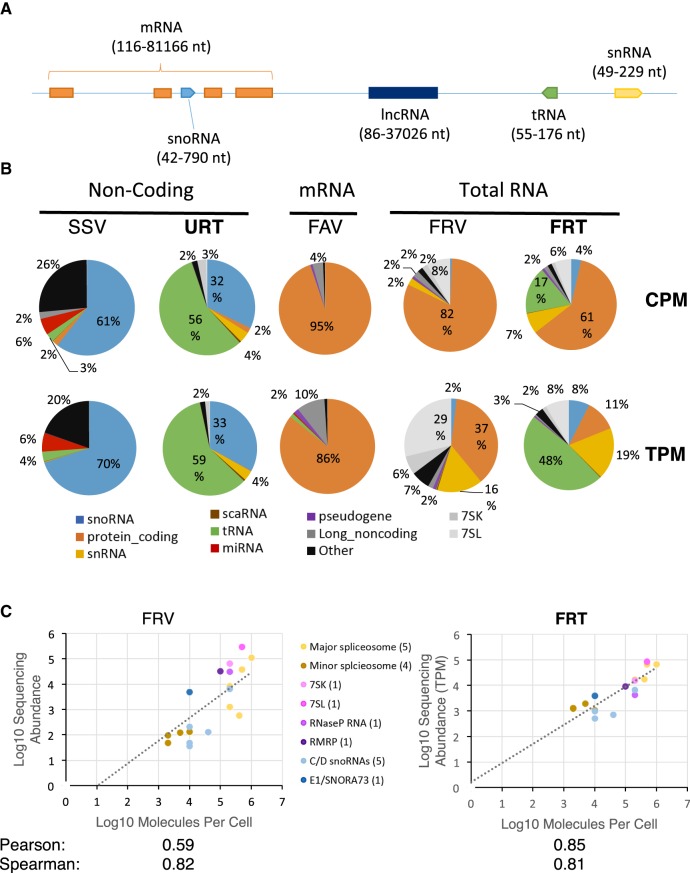
Most sequencing methods deal with coding and noncoding RNAs separately (Fig. 1A), providing little information about the overall landscape of the human transcriptome. To identify the best approach for an integrated analysis of the human transcriptome, we evaluated the capacity of different sequencing methods to quantify transcripts both within the same class and between different classes of RNAs. We chose five sequencing protocols, three that target a specific class of RNA as reference for comparison between RNA within the same class, and two general methods targeting all RNA species other than rRNA. The class-specific methods include (i) size-selected viral reverse transcriptase sequencing (abbreviated SSV), (ii) TGIRT-seq of unfragmented, ribodepleted whole-cell RNA (abbreviated URT), and (iii) fragmented poly(A)-selected viral reverse transcriptase sequencing (abbreviated FAV), while the general methods include (i) fragmented ribodepleted viral reverse transcriptase sequencing (abbreviated FRV), and (ii) fragmented ribodepleted TGIRT-seq (abbreviated FRT). These five different approaches cover the most commonly used methods and test two newly developed techniques (URT and FRT) that use the thermostable group II intron reverse transcriptase, TGIRT-III. TGIRT has high processivity and strand displacement activity, which makes it possible to generate cDNA from short highly structured RNA without size selection (Mohr et al. 2013; Nottingham et al. 2016; Qin et al. 2016). Two of the methods targeting specific RNA species used RNA selection steps like size selection (SSV) or poly(A) tail selection (FAV), while in the remaining methods (URT, FRT, and FRV), RNAs were ribodepleted and sequenced without size selection. The RNA was extracted from the model ovarian cancer cell-line SKOV3ip1 and two biological replicates for each protocol were sequenced to read depths that vary between 15 and 300 million reads (Supplemental Table S1). The correlation between the biological replicates for each method is indicated in Supplemental Table S2. In general, replicates for each method were highly correlated. All comparisons between methods were performed using counts per million (CPM) and transcripts per million (TPM) to eliminate read depth biases. TPM is preferred when comparing RNAs of different lengths since longer RNAs generate more fragments, hence more reads. CPM reflects the mass of molecules whereas TPM depicts the number of molecules when using a fragmented library.
FIGURE 1.
Sequencing methods permitting simultaneous detection of transcripts with different sizes and structures reveal a human transcriptome dominated by noncoding RNA. (A) Schematic of the human genome illustrating the predicted size distribution of different classes of RNA (size range based on Ensembl annotations shown in parentheses). (B) Distribution of RNA in the human transcriptome as detected by different sequencing methods. The RNA was extracted from the ovarian cancer model cell line SKOV3ip1 and subjected to different sequencing protocols using different RNA selection methods and reverse transcriptases including size-selected viral reverse transcriptase sequencing (SSV), unfragmented, ribodepleted RNA TGIRT-seq (URT), fragmented poly(A) selected viral reverse transcriptase sequencing (FAV), fragmented ribodepleted viral reverse transcriptase sequencing (FRV), fragmented ribodepleted RNA TGIRT-seq (FRT). The intended target of the different methods is indicated above the method names. The results are shown in the form of pie charts illustrating the distribution of RNA abundance in counts per million (CPM) or transcripts per million (TPM). The results are the average of two biological replicates. The percentage of the main classes (≥2%) is indicated. The color legend for the different RNA classes is shown at the bottom. (C) Comparison between the capacity of viral and group II intron-encoded RTs to predict the abundance of noncoding RNA. The noncoding RNA abundance obtained by the viral RT- or TGIRT-based sequencing methods FRV or FRT was plotted against established estimates of the number of molecules per cell for each biotype (Tycowski et al. 2006). Pearson and Spearman coefficients are indicated at bottom. A legend of the different classes of noncoding RNA and the number of genes considered from each type tested is shown in the middle.
RNA used for preparing the SSV library was extracted using a mirVana kit but without a gel purification step, enabling the consideration of all RNA <200 nt in length (Deschamps-Francoeur et al. 2014). This method is expected to accurately detect uncapped RNA shorter than 200 nt. As expected, SSV data sets possessed few reads generated from protein-coding genes and instead were enriched in reads from noncoding RNAs (Fig. 1B, left panel). However, the reads generated from noncoding RNA were not appropriately distributed among the different classes of noncoding RNA nor within the same class of RNA. For example, while it is well established that tRNA is the most abundant RNA family in the cell, most reads (61% CPM and 70% TPM) corresponded to snoRNAs, the vast majority of which were mapped to box C/D snoRNA, while 6% of the reads originated from miRNAs, and only 3% originated from tRNAs, which are difficult for retroviral RTs to reverse transcribe. Bias in the detection of different RNAs using this method could also be explained by adapter ligation bias to RNA ends, especially the 5′ end in the case of capped RNA and by the difficulty of viral RTs in reverse transcribing sncRNAs. Thus, while size selection may enrich certain RNA species, its application to total untreated RNA is not sufficient to accurately detect all noncoding RNAs shorter than 200 nt.
In contrast, sequencing of total unfragmented ribodepleted RNA using TGIRT (URT) succeeded in detecting all five classes of the main noncoding RNA with lengths between 60 and 700 nt: tRNA, snoRNA, snRNA, 7SL, and 7SK. The majority of the reads (56% CPM and 59% TPM) corresponds to tRNA, followed by snoRNA (32% CPM and 33% TPM), while both long noncoding RNA and miRNA were detected at low levels (Fig. 1B, second panel). This suggests that unfragmented RNA sequencing using TGIRT provides a better representation of sncRNAs than size selection but performs equally poorly in the detection of long polyadenylated RNAs and very short RNAs.
Sequencing libraries produced by viral reverse transcriptases after poly(A) selection (FAV) resulted in marked enrichment (95% CPM and 86% TPM) of reads corresponding to protein-coding RNA and a quasi-absence of noncoding RNA with the exception of long noncoding RNA, which represent 10% of the TPMs (Fig. 1B, middle panel). The relatively large proportion of TPM attributed to long noncoding RNA suggests that a considerable number of long noncoding RNAs are polyadenylated.
In contrast, sequencing of fragmented, ribodepleted reverse transcribed RNA using standard viral reverse transcriptases (FRV) sampled a much larger number of RNA classes (Fig. 1B, fourth panel). Most reads were still generated from coding RNAs (82% CPM and 37% TPM) but 7SL RNA followed close behind, generating 29% of the TPMs. Most other noncoding RNAs (e.g., snRNA, snoRNA, and 7SK) were detected, with the exception of tRNA and miRNA. Therefore, while sequencing of fragmented RNAs using viral reverse transcriptases permits the detection of both protein-coding and many classes of noncoding RNA, it misses the most highly expressed class of nonribosomal RNA in the cell (i.e., tRNA) and biases the transcriptome composition in favor of protein-coding RNA.
Strikingly, reverse transcription of fragmented, ribodepleted RNA using TGIRT (FRT) permitted the detection of all classes of coding and noncoding RNA but poorly detects miRNA, likely due to fragmentation and bead purification (Nottingham et al. 2016). The distribution of the read counts generated by FRT indicates that while the majority of reads (CPM) are produced from protein-coding RNA, tRNA is the most abundant RNA class (48%) as measured in TPM (Fig. 1B, right panel). Surprisingly, the second most abundant transcripts are the snRNAs, accounting for 19% of the TPM, while snoRNAs and 7SL each represent 8% of the TPM (Fig. 1B, right panel). Since FRT efficiently detects tRNA while poorly detecting miRNA, we conclude that it is best suited for detection of RNA larger than 60 nt. Most importantly, FRT was able to correctly identify all ectopically added (spiked in) RNA species from the ERCC (Supplemental Fig. S1). The spike-in, which was added to the RNA extracted from different cell lines, consists of a set of polyadenylated transcripts covering a wide range of transcript lengths (250 to 2000 nt) and several orders of magnitude in concentration. The Pearson correlation between the sequencing estimated abundance values and the actual concentration of the spike-in RNAs added to cellular RNA during library preparation was 0.99, confirming the accuracy of the FRT-seq estimates for polyadenylated RNAs (Supplemental Fig. S1). Therefore, FRT appears to be a good tool for the simultaneous detection of different classes of coding and noncoding RNA longer than 60 nt in a single RNA sample.
Accurate RNA quantification using fragmented ribodepleted thermostable group II intron reverse transcriptase sequencing (FRT)
Out of the five sequencing methods, only two, FRV and FRT, were able to significantly detect both coding and noncoding RNAs in a single RNA sample. FRV generated a higher proportion of reads from 7SL and 7SK than FRT, while FRT was the only method to detect both tRNA and protein-coding RNA in a single RNA sample. To address the question of which method most accurately represents the true hierarchy of RNA abundance in human cells, we first compared the sequencing estimates of different species of noncoding RNA to the number of MPC previously established using immunoprecipitation of in vivo labeled RNA (Mimori et al. 1984; Tycowski et al. 2006; Palazzo and Lee 2015). The group of RNAs considered covers the main classes of sncRNAs, including major and minor spliceosome components, 7SK, 7SL, RNase P, RNase MRP, and snoRNA. As most of these RNAs are encoded by multiple genes we grouped all reads generated from these repeated genes together to permit comparisons with previous biochemical estimates that consider all transcripts regardless of their origin (Mimori et al. 1984; Tycowski et al. 2006). Most sequencing methods using a selection step, even those that specifically enrich for short RNA (<200 nt) like SSV and URT correlated very poorly with MPC measurements (Supplemental Fig. S2). In contrast, methods sequencing fragmented, ribodepleted total RNA like FRV and FRT correlated better with MPC values (Fig. 1C). In general, FRT was better at detecting transcripts of all sizes while FRV overestimated longer RNA species and underestimated shorter RNA species (Supplemental Fig. S3). Indeed, most abundance values produced by FRT strongly correlated with MPC values with much less variation than FRV.
Strikingly, FRT showed similar amounts of many sncRNAs of the same class. For example, U4, U5, and U6, which compose the major spliceosomal tri-snRNP, have almost identical abundance values using FRT (between 2% and 9% pairwise difference in abundance), as do minor spliceosome components U4atac and U6atac (2% difference in abundance), as indicated in Supplemental Table S3. Consistently, the minor spliceosome snRNAs, which have fewer splicing targets, were less abundant than their major spliceosome counterpart, as would be expected (Fig. 1C, right panel).
To better understand the differences between bacterial (FRT) and viral (FRV) reverse transcriptase sequencing methods, we visually compared these two methods using splatterplots (Mayorga and Gleicher 2013). As indicated in Supplemental Figure S4A, in general FRV tended to underestimate sncRNAs in comparison with FRT. This FRV bias was most pronounced in the case of snoRNA and tRNA (Supplemental Fig. S4B). The bias in the case of snoRNA appeared to include all classes of snoRNA but was most pronounced in the case of box C/D snoRNA (Supplemental Fig. S5). Together these comparisons clearly indicate that sequencing using FRT most accurately predicts the hierarchy of sncRNA abundances within total nonfractionated RNA samples.
We used PCR as an independent test to determine which sequencing method performed best for the quantification of the different classes of RNAs. For sncRNAs, we used dPCR, which accurately counts the number of RNA molecules in a given volume and thus supports the comparison of the abundance of different molecules from the same sample. As indicated in Supplemental Figure S6, the selection-based sequencing methods performed poorly when compared to dPCR, while the ribodepleted sequencing methods FRV and FRT correlated much better for sncRNAs.
The abundance of different protein-coding RNAs obtained by sequencing was also compared to RT-qPCR, which generally showed good correlation for all methods (Supplemental Fig. S7). However, methods selecting for noncoding RNAs like SSV and URT did not detect several coding RNAs resulting in lower correlation with RT-qPCR than the FAV, FRV, and FRT methods. Overall, FRV and FRT correlated slightly better with RT-qPCR than FAV, even though the latter is specifically enriched in protein-coding genes.
To evaluate the capacity of the different sequencing methods to detect the ratio of splice variants, we compared the percent splicing index (PSI) estimated by each method and quantified by rMATS (replicate multivariate analysis of transcript splicing) (Shen et al. 2014) to that generated by the well-established splice sensitive endpoint PCR technique (Klinck et al. 2012). As indicated in Supplemental Figure S8, the best correlation with the endpoint PCR value was obtained by FAV, which enriches for protein-coding RNAs, while methods selecting for noncoding RNA either failed to produce PSI values (SSV) or did not correlate well (URT) due to poor detection of the target mRNA. The total RNA sequencing methods FRV and FRT were close seconds after FAV, producing similarly good correlation values with PCR. The slightly better correlation (0.04 difference in both Spearman and Pearson coefficients) of FAV is due mostly to FRV and FRT producing lower estimates of a few PSI values, possibly due to the lack of mRNA selection in the case of FRV and FRT. Overall, while selection for protein-coding genes through poly(A) enrichment may slightly improve estimation of the ratio of splice variants, FRT provides the best option to evaluate splicing ratio within a sample without losing the ability to detect other classes of RNA.
The transcriptome of model cell lines is defined by a small number of highly expressed noncoding genes and a large number of moderately expressed protein-coding genes
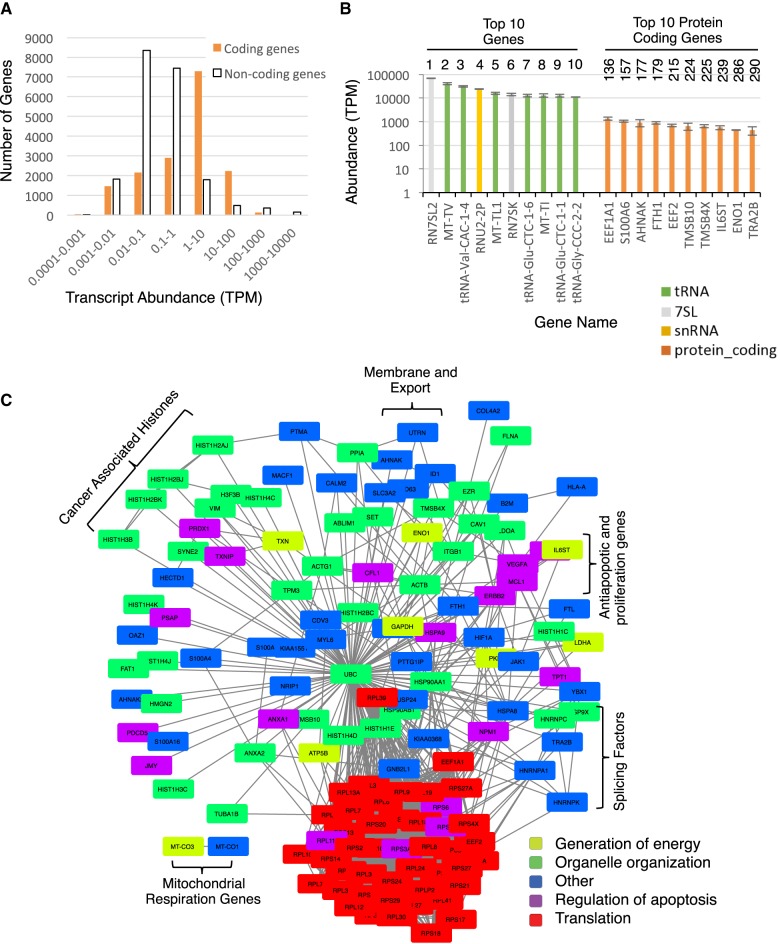
The capacity to accurately rank the abundance of one RNA molecule relative to another provides a unique opportunity to probe the composition of the human transcriptome. Accordingly, we used the sequencing reads generated by FRT to study the distribution of different coding and noncoding RNAs in the human transcriptome. To characterize the origin of the noncoding RNA dominance of the transcriptome, we first classified coding and noncoding RNAs according to their different levels of RNA abundance. The distribution of abundance per gene indicates that most noncoding genes are poorly expressed (<1 TPM), while most protein-coding genes are expressed at levels varying from 1 to 10 TPM (Fig. 2A). In contrast, 519 noncoding RNAs have abundances over 100 TPM, 164 of which are above 1000 TPM. Therefore, it appears that the human ribodepleted transcriptome is dominated by transcripts generated from a small subset of noncoding RNA genes. Indeed, 50% of the human transcriptome is produced from only 50 short noncoding RNA genes (Supplemental Fig. S9). To ensure that these results are not specific to cancer cell lines, we compared the results obtained from the ovarian cancer model cell line SKOV3ip1 to that generated from the immortalized normal ovarian cell line INOF. As indicated in Supplemental Figures S10 and S11, the overall hierarchy of RNA abundance did not change, confirming that most of the transcriptome is produced by a small number of noncoding RNA. Indeed, the main differences between the two cell lines are in the amount of tRNA, which is higher in the INOF and in the order of abundance of certain mRNAs that reflects the different nature of these two cell lines.
FIGURE 2.
The composition of the human transcriptome is dominated by a small subset of highly expressed noncoding RNA genes and a large number of moderately expressed protein-coding genes reflecting cellular phenotypes. (A) The abundance of both coding and noncoding gene transcripts was determined using FRT, separated into bins based on transcript abundance, and the number of genes per bin illustrated in the form of a bar graph. (B) The genes producing the top 10 overall most abundant RNAs and the top 10 most abundant protein-coding RNAs are shown as a bar graph. The rank of each transcript based on abundance in transcript per million (TPM) is indicated on top. (C) Interaction map of the most expressed protein-coding genes in the model ovarian cancer cell line SKOV3ip1. Genes producing RNAs with more than 100 TPM were identified, and their functional, genetic, and physical interactions obtained from STRING (Szklarczyk et al. 2015) and illustrated as an interaction network. The main gene ontology annotations for the genes are indicated at bottom right (also see Supplemental Table S6). Open brackets indicate examples of complexes associated with cancer phenotypes and other established phenotypes of SKOV3ip1 cells.
Examination of the top 10 most expressed genes in the model ovarian cancer cell line SKOV3ip1 indicated that after removal of rRNA, the most abundant transcripts are RN7SL2 (>68,000 TPM), which encodes the noncoding RNA component (7SL) of the signal recognition particle followed by tRNA genes, a gene encoding the U2 snRNA (RNU2-2P) and the RN7SK gene encoding the noncoding RNA 7SK (Fig. 2B). As expected, seven out of the top 10 most abundant RNAs are tRNAs, but surprisingly three of these are mitochondrial tRNAs, with MT-TV, encoding a valine mitochondrial tRNA, being the most abundant cellular tRNA. This could reflect the large number of mitochondria per cell or that mitochondrial tRNAs are more stable than their cytoplasmic counterparts. Interestingly, no single U1 gene figures among the 10 most expressed non-rRNA genes. Indeed, over a hundred copies of U1 are annotated in the human genome, and the transcripts of 10 of these copies produced 3000 and 9000 TPM (Supplemental Fig. S12), resulting in an extremely high overall abundance for U1. Greater than 96% of U1 transcripts are produced by 10 highly expressed copies, while in contrast other highly expressed noncoding RNAs such as 7SL and 7SK, although encoded by multiple genes in humans, are mostly expressed from a single locus in the SKOV3ip1 cell line.
Overall, the top 10 most abundant RNAs are noncoding RNAs and have relative transcript counts between 15,000 and 85,000 TPM, while the top 10 most abundant protein-coding RNAs only reach between 400 and 1500 TPM (Supplemental Table S4). This suggests that the maximum steady state output of coding and noncoding RNA genes differs by one to two orders of magnitude and likely reflects a combination of higher transcription rate and increased RNA stability. However, it is important to note that the most abundant protein-coding RNA in this cell line, EEF1A1, ranks 136th overall and is followed by other noncoding RNAs (Fig. 2B; Supplemental Table S4), suggesting that at least some protein-coding genes are more expressed than most noncoding RNAs. Nevertheless, in general the most abundant RNAs are generated by noncoding RNA genes.
Examination of the function of the genes encoding the top 10 most abundant protein-coding RNAs suggested that the most highly expressed genes are implicated in nontissue-specific functions like translation (e.g., EEF1A1 and EEF2), cytoskeleton and organelle organization (e.g., TMSB10, TMSB4X, and AHNAK) (Smart et al. 2010; Davis et al. 2014; Abbas et al. 2015; Zhang et al. 2017). Consistently, analysis of the functional relationship between the most abundant (>100 TPM) protein-coding RNAs revealed a tight functional network involved in organelle organization, regulation of apoptosis, and translation (Fig. 2C; Supplemental Tables S5, S6). Notably, almost all these genes are ubiquitously expressed in all human tissues, and many produce some of the most abundant proteins in the human proteome (Ramsköld et al. 2009; Beck et al. 2011). Comparison between the transcriptome of SKOV3ip1, an invasive epithelial cell line, and INOF, a mesenchymal normal immortalized cell line, revealed a similar distribution of the RNA biotypes (Supplemental Fig. S10). The distribution of coding versus noncoding RNAs was also similar for INOF and SKOV3ip1, with the most abundant noncoding RNAs being 10 to 100 times more abundant than the most abundant coding RNAs (cf. Supplemental Fig. 10C,D to Fig. 2A,B). Once again, the top 10 most abundant RNAs in INOF are dominated by tRNAs and 7SL. Additionally, the 50 most abundant RNAs in INOF (all noncoding) represent over 50% of all ribodepleted TGIRT-seq detected transcripts, as found for SKOV3ip1. However, while the mRNAs coding for the housekeeping proteins like the translation factors EEF1A1 and EEF2 were of similar abundance in both cell lines, major differences were found in the order of the mRNA coding for cancer associated proteins like ENO1, SOD2, and S100A4. For example, the mRNA of the enolase 1 gene (ENO1) known to be overexpressed in multiple cancers (Tsai et al. 2010; Zhang et al. 2010; Yu et al. 2012, 2014; Principe et al. 2017) was ranked 9 among protein-coding genes for its expression in SKOV3ip1 and 83 in INOF. Consistently the mRNA coding for the cancer-associated calcium binding protein S100A4 (Kikuchi et al. 2006; Maelandsmo et al. 2009) was ranked 21 in SKOV3ip1 and 10953 in INOF (Supplemental Tables S4, S5). Therefore, while FRT may detect cell type–specific differences in the mRNA of protein-coding genes, the overall hierarchy of RNA abundance remains similar in both normal and ovarian cancer cell lines. These results support the overall reproducibility of the FRT-based transcriptome profiling and suggest that the detected proportions of RNA biotypes could be generalized to different cell types.
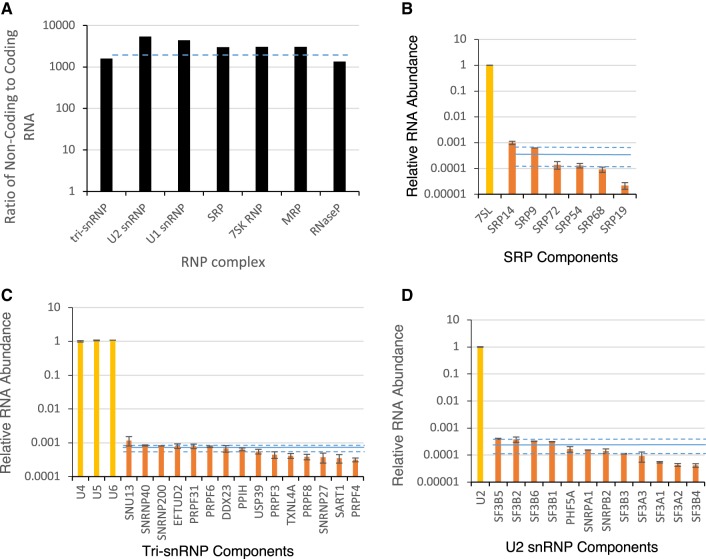
Ribonucleoprotein particles are generated from highly abundant noncoding RNA and proteins produced by uniformly less abundant protein-coding RNA
After establishing the accuracy of TGIRT-seq for the comparison among and between different RNA families, we further interrogated our SKOV3ip1 data sets to characterize the relationships between different RNAs. RNAs forming stable ribonucleoprotein complexes are among the most studied RNA in cells, yet we know very little about the relative abundance of the noncoding and protein-coding RNAs used in the biogenesis of the same RNP complex. Examination of the ratio of noncoding and coding RNAs of the main RNP classes in SKOV3ip1 indicated that on average noncoding RNAs were 3000-fold more abundant than transcripts encoding proteins associated with the same complex (Fig. 3A). The lowest ratio between noncoding and coding RNAs was found in the tri-snRNP and RNase P, while the highest was found in the U2 and U1 snRNPs. This suggests that the noncoding RNA component of RNPs may be more highly transcribed and more stable than their protein-coding counterparts to match the translational output of a mostly uniform population of mRNAs.
FIGURE 3.
Major ribonucleoprotein complexes are generated from mostly uniformly abundant populations of protein-coding transcripts and highly abundant noncoding RNAs. (A) The ratio of the noncoding and coding RNAs associated with seven established ribonucleoprotein complexes as determined using FRT are illustrated in the form of a bar chart. The dashed line indicates the average ratio of noncoding to coding RNA, which is approximately 3000:1. The abundance of mRNAs coding for key protein components of SRP (B), tri-snRNP (C), and U2 snRNP (D) complexes are plotted as a fraction of their respective noncoding RNA. The solid line indicates the average abundance level of the protein-coding RNA of the complex, and the dashed lines indicate 5% and 95% confidence intervals. The standard deviation of two biological replicates is indicated in the form of error bars.
Examination of the relative abundance of different protein-coding RNAs within each RNP complex indicated that the mRNAs coding for the protein components of each complex often accumulated at similar levels, except those encoding certain proteins like SRP19 (Fig. 3B–D; Supplemental Fig. S13). In the case of SRP, the SRP19 mRNA was much less abundant than the other five protein-coding RNA, suggesting that this RNA, if translated with the same efficiency as those for the other proteins, may function as a limiting factor for this complex (Fig. 3B). Indeed, SRP19 is a key regulator of 7SL RNA folding and assembly (Maity et al. 2008). Differences in the abundance of the protein-coding RNA associated with the same complex could also be offset by differences in translation levels or in protein stability. However, genes associated with the same complex generally appeared to produce similar amounts of protein coding RNA (Supplemental Fig. S13). Overall the results indicate that while the mRNA abundance of certain RNP components might be limiting, most are similarly expressed and dominated by the noncoding RNA component.
snoRNA abundance depends on the type of snoRNA and the function of the host gene
While tRNAs and snRNAs dominate the nonribosomal RNA noncoding transcriptome, snoRNAs compose the largest class by number of genes with distinct transcripts. It is generally assumed that both methylation and pseudouridylation snoRNAs are expressed at the same level, given their common function in ribosome biogenesis (Dieci et al. 2009). However, this notion has never been directly investigated. Examination of box H/ACA and box C/D snoRNA abundance indicated that ∼30% of box C/D snoRNA genes do not produce detectable amounts of RNA versus 20% of box H/ACA genes in SKOV3ip1 cells (Supplemental Fig. S14A). Most of the nonexpressed or poorly expressed snoRNA genes are recently added annotations that exist in Ensembl but not in the manually curated snoRNAbase database (Supplemental Fig. S15). Overall, there was a higher proportion of H/ACA genes that generated a detectable quantity of transcripts than C/D genes. However, for the snoRNAs that are detected, the abundance of RNAs produced by H/ACA and C/D snoRNA genes was similar (Supplemental Fig. S14A).
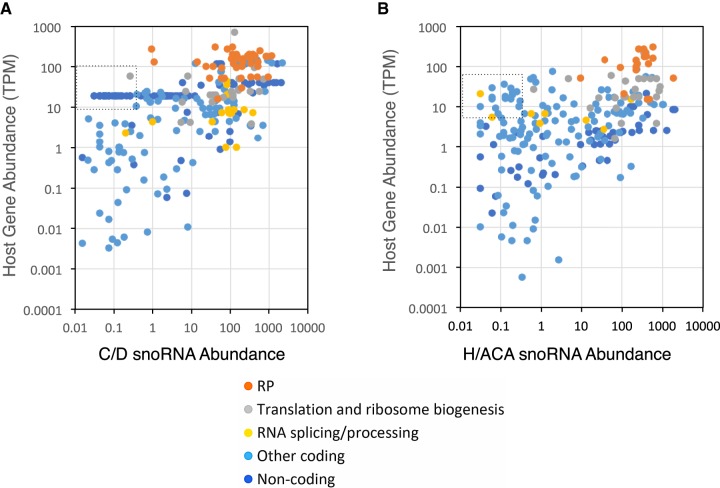
Since many snoRNAs in the human transcriptome are produced from introns of protein-coding genes, we examined the abundance of these snoRNAs relative to the function and abundance of their host gene mRNAs. Interestingly, the abundance of snoRNAs was found to vary in a subtype-specific fashion based on the function of the host genes. For example, H/ACA snoRNAs encoded in the introns of genes coding for ribosomal proteins, and noncoding RNAs were significantly more abundant than C/D snoRNAs encoded in introns of the same type of genes (Supplemental Fig. S14B). In contrast, C/D snoRNAs were significantly more abundant than H/ACA snoRNAs when they are found in genes implicated in RNA processing and splicing (Supplemental Fig. S14B). In some cases, snoRNA abundance exceeded that of the host gene (Supplemental Fig. S14C) and in general, more H/ACA than C/D snoRNA were at least 10-fold more abundant than their host gene RNAs (35% versus 19%).
To better understand the relationship between the impact of host gene expression and its influence on resident snoRNA abundance, we compared the host and snoRNA abundances for each snoRNA and categorized them based on host gene function. As indicated in Figure 4A,B and Supplemental Figure S16, there were more C/D than H/ACA snoRNAs encoded in the introns of ribosomal protein genes (41 C/D versus 18 H/ACA), and the abundance of the 41 C/D snoRNA varied greatly from 100 times less abundant to 50 times more abundant than the host gene mRNA (Fig. 4; Supplemental Fig. S16). In contrast, only 18 H/ACA snoRNAs were found encoded within introns of ribosomal protein genes and most were expressed at a level equal to or greater than their host genes (Fig. 4B; Supplemental Fig. S16). Overall, comparison between the abundance of snoRNA and their host genes indicated that in general there is little correlation between the abundances of the host and passenger snoRNA (Supplemental Fig. S14D). We conclude that the abundance of snoRNA is not obligatorily linked to the expression level of its host RNA (i.e., a highly abundant snoRNA could be produced from the same gene as a scarce mRNA).
FIGURE 4.
The abundance of snoRNAs relative to the host mRNA in which they are encoded depends on the type of snoRNA and the function of the host genes. (A,B) Scatter plots illustrating the relationship between the abundance of box C/D (A) and H/ACA (B) snoRNAs and the protein-coding RNA produced from their host genes, as determined by FRT. The function of the different host genes is indicated in the legend at the bottom. RP indicates ribosomal protein. The dashed boxes indicate area with the most visible difference between C/D and H/ACA snoRNA.
The abundance of snoRNA depends at least partially on the nature of the targeted modification sites
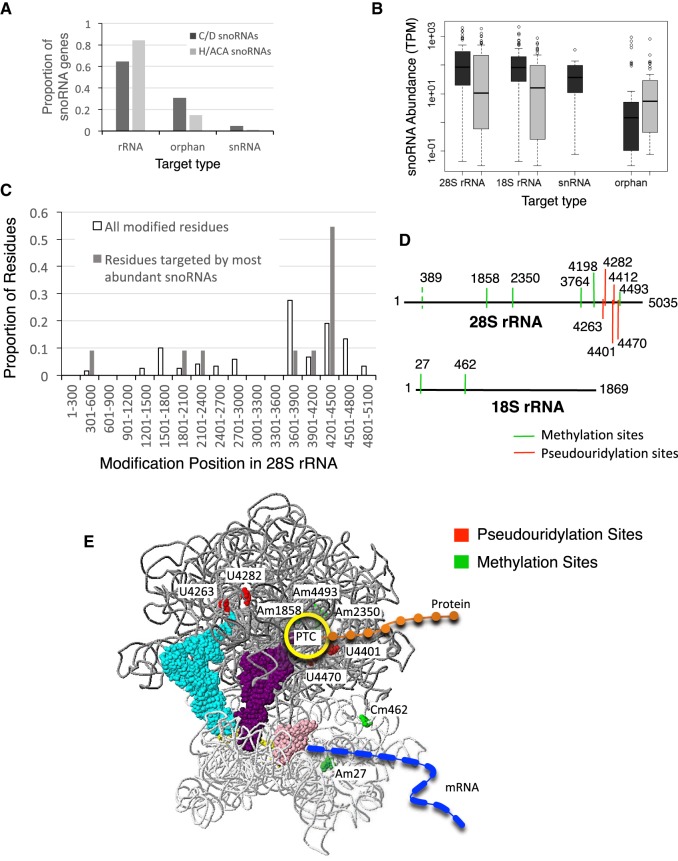
Since the most well-characterized function of snoRNA is guiding RNA modifications, we next examined the relationship between the abundance of each snoRNA and the location of their target modification site. We compared the abundance of different snoRNAs modifying rRNA to each other and to those targeting snRNA including scaRNA (Fig. 5; Supplemental Fig. S17). In general, the proportion of H/ACA snoRNAs targeting a site in the rRNA modification site was more than that of C/D snoRNAs, while more C/D snoRNAs targeted snRNA or have no known targets (orphan) than H/ACA snoRNAs (Fig. 5A). The abundance of orphan snoRNAs was on average lower than the abundance of snoRNA with known rRNA targets (Fig. 5B). Indeed, the most abundant snoRNAs (>1000 TPM) are either involved in the processing of pre-rRNA (U3/SNORD3 and E1/SNORA73) or target four specific regions in 28S rRNA and two regions in 18S rRNA (Fig. 5C–E). The snoRNA targeting snRNA in most cases had similar transcript levels and were about 100 times less abundant than their target RNAs (Supplemental Fig. S17A–E). In contrast, almost all of the highly abundant snoRNAs (>1000 TPM) were found to target modifications in the 28S and 18S rRNA structure surrounding the peptidyl transferase center (PTC), the site immediately adjacent to tRNA binding sites, and the mRNA and protein tunnels (Fig. 5E).
FIGURE 5.
The abundance of snoRNAs correlates with their function. (A) Distribution of snoRNA by target type. The proportion of expressed box C/D snoRNAs (dark gray) and box H/ACA (light gray) targeting rRNA, snRNA, or no known target (orphan) is indicated in the form of a bar graph. (B) Box plot displaying the distribution of abundance of both box C/D (dark gray) and H/ACA (light gray) snoRNAs as a function of their target type. The abundance of snoRNAs targeting the 28S rRNA, 18S rRNA, snRNA, and those with no known target (orphan) were identified using FRT and the average value of two biological replicates plotted, with the solid line indicating the median value. (C) Position of the 28S rRNA modification sites targeted by the most abundant snoRNA. The 28S methylated or pseudouridylated residues were binned according to their position in the molecule, counted, and then their proportion plotted as a bar graph. The white bars indicate the proportion of all known modified residues found at the indicated position, while the gray bars indicate the proportion of those residues modified by the most abundant snoRNA (>1000 TPM) as determined by FRT. (D) Position of the 28S (top) and 18S (bottom) rRNA modification sites targeted by the most abundant snoRNA. (E) Three-dimensional model of the ribosome featuring the modification sites targeted by the most abundant snoRNA. The model was generated by the 3D rRNA modification maps database tool kit (Piekna-Przybylska et al. 2008). The rRNA is shown in dark gray for the 28S large subunit rRNA and light gray for the 18S small subunit rRNA. A tRNA is shown in the A (light blue), P (purple), and E (pink) sites and the approximate position of the mRNA and nascent peptide are indicated in blue and orange, respectively. The pseudouridylation and methylation sites targeted by the most abundant snoRNAs are shown in red and green, respectively. The position of the peptidyl transferase center (PTC) is indicated by the yellow circle.
DISCUSSION
Most approaches for transcriptome analysis and RNA quantification compare RNA levels under different conditions and in most cases focus on a specific class of RNA (Ozsolak and Milos 2011). In this study, we show that most current sequencing techniques are not suitable for comparing the abundance of RNA from different classes of coding and noncoding RNA. Current techniques that use ribodepleted total RNA and standard viral RTs tend to underrepresent sncRNAs and in particular tRNA, while techniques enriching for short noncoding RNAs do not detect protein-coding RNA. Surprisingly, RNA class-specific sequencing techniques like size or poly(A) selection-based techniques either do not provide a particular advantage or simply fail to properly detect the relative abundance of RNA within the targeted class. In contrast, the ribodepleted TGIRT-seq technique (FRT) used in this study shows a more faithful representation of the distribution of all classes of coding and noncoding RNA longer than 60 nt in two different human cell lines. Indeed, FRT accurately predicts the hierarchy of both noncoding and coding RNA abundance relative to themselves and each other as compared to both biochemical and RT-qPCR estimates (Fig. 1; Supplemental Fig. S2–S8). It is now possible to detect protein-coding RNAs and their regulatory, nested or associated noncoding RNAs in the same sequencing reaction.
Using this newly developed technique, we were able to show that the nonribosomal RNA human transcriptome is composed mainly of tRNA and snRNA. This is consistent with previous studies showing that tRNA genes produce the highest number of transcripts in the cell (Palazzo and Lee 2015). Surprisingly, we found that RNAs representing the RNA component (7SL) of the signal recognition particle, mainly produced from two loci (ENSG00000274012 and ENSG00000265735), constitute 4%–8% of the nonribosomal RNA transcriptome (Figs. 1, 2; Supplemental Fig. S10). Consistently, examining the number of sequencing reads generated from different genes indicates that a large number of noncoding transcripts are generated mainly from a few highly expressed genes (Fig. 2; Supplemental Figs. S9, S10; Supplemental Table S4), while protein-coding transcripts are generated from a large number of modestly expressed genes (Fig. 2; Supplemental Fig. S10; Supplemental Tables S4, S5).
The capacity of a sequencing method to faithfully reproduce the natural diversity of a transcriptome depends on its ability to detect the largest number of RNA classes and transcripts within each class. Based on this feature, FRT appears to generate the most comprehensive picture as it detects the largest number of RNA classes and RNA transcripts within a class (Fig. 1). However, both URT and FRT seem to underrepresent RNAs shorter than 60 nt due to either fragmentation or difficulty in separating cDNAs of short RNAs from similarly sized primer-dimers by bead purification (Nottingham et al. 2016). Unfortunately, enriching for miRNA using size selection based methods, while increasing the number of miRNA detected, would not necessarily provide the correct ranking between different miRNA and related (e.g., precursor) noncoding RNA. This is evident from the results of the sequencing methods using selection steps (e.g., SSV and FAV), which do not enhance their capacity to correctly rank the RNAs within the selected, presumably due to bias within the selection process (Fig. 1; Supplemental Figs. S2, S6). The endpoint PCR determined splicing indexes correlated well with those determined by FAV and FRT (Supplemental Fig. S8), suggesting that enrichment of polyadenylated mRNA is not essential for the ratio of alternative splicing. Therefore, it appears that the best way forward for miRNA would likely be a modified TGIRT-seq method that mitigates the formation of primer dimers enabling better recovery of RNAs smaller than 60 nt.
Another way in which to evaluate different methods is how well these methods rank RNA transcripts relative to one another. This is complicated as it is difficult to ascertain the absolute correct rank of RNA. Spike-in RNAs have been used previously to compare the abilities of different methods to discriminate between the abundance of distinct transcripts (Jiang et al. 2011; Nottingham et al. 2016). According to this standard of quality the FRT sequencing method performs very well with an almost perfect detection of ERCC spike-ins pattern (Supplemental Fig. S1). We used three additional criteria to evaluate the capacity of a given method to rank RNA abundance relative to each other. The first comparison was between the sequencing results and established ranks of transcripts determined using quasi-linear detection techniques like in vivo labeling techniques (Mimori et al. 1984; Tycowski et al. 2006; Ryan et al. 2008). The second comparison was the correlations with dPCR, RT-qPCR, and endpoint RT-PCR that compares the abundance of two splice variants using a single primer pair. In the case of end-point RT-PCR, we did not compare one amplicon to another but rather between each amplicon and the value obtained from sequencing data. Thirdly, we evaluated the capacity of each technique to identify the abundance of different classes of RNA. The results indicated that the highest correlation between methods was found between FRT and the MPC data obtained using in vivo labeling (Fig. 1C). Furthermore, there is a good general correlation between dPCR, RT-qPCR and certain sequencing techniques like FRT (Supplemental Figs. S6E, S7E). The endpoint RT-PCR estimates of the ratio of splice variants correlated only slightly better with FAV, which enriches for polyadenylated RNA. Most importantly the data obtained from FRT matched the expected tendency of protein-coding genes with similar functions to be expressed at similar levels (Figs. 1, 3, 4).
Direct comparison between different classes of RNA indicates that the majority of nonribosomal RNA transcripts (89%–96% according to FRT) are noncoding. This high abundance of noncoding RNA was previously predicted by class-specific analyses and would be expected for RNA families with stoichiometric functions like tRNA (Palazzo and Lee 2015). Indeed, a very high number of tRNAs is required to deliver the amino acids needed to supply the translation of all protein-coding genes in cells (Wilusz 2015). However, it was surprising to detect almost 1.8 times more spliceosomal snRNAs than protein-coding gene RNAs (Fig. 1B). Given the catalytic nature of the splicing reaction one might expect a much lower number of the subunits of the catalytic core of the spliceosome compared to its protein-coding RNA substrates (Wachtel and Manley 2009). Explanations for the high abundance of snRNA, could include (i) the previously suggested cotranscriptional assembly of the splicing complex prior to synthesis of the 3′ splice site, which may slow the turnover rate of the spliceosome, (ii) the large number of splice sites per mRNA that need recognition by matching number of snRNA, or (iii) some snRNAs serve other nonspliceosomal functions (Blázquez and Fortes 2013; Naftelberg et al. 2015). Indeed, one of the most highly expressed snRNAs, U1, was shown to form a stable complex with protein-coding RNAs to protect it from premature cleavage and polyadenylation (Kaida et al. 2010; Blázquez and Fortes 2013). Similarly, one might hypothesize that U2 snRNA may also have an extra-spliceosomal function given its abundance level that almost matches that of U1 (Supplemental Table S4). Regardless of this possibility, it is clear that the spliceosome is in high demand since all the spliceosomal snRNAs are among the most abundant RNAs in the cell (Fig. 1B; Supplemental Fig. S10; Supplemental Table S4).
The high-abundance of many noncoding RNAs could be partially explained by the high transcription rate of RNA polymerase III (Arimbasseri et al. 2014). Indeed, RNA Pol III plays a central role in shaping the transcriptome landscape, as it is responsible for the transcription of two of the most abundant gene families, tRNA and 7SL RNA (White 2004). However, it is now clear that this cannot be the only explanation for the strong abundance of sncRNAs given the relatively high abundance of snRNA transcribed by RNA polymerase II (Egloff et al. 2008). The mechanisms controlling RNA abundance are likely a combination of gene multiplication and rates of transcription and RNA turnover. In the case of snRNAs, each RNA species is encoded by multiple genes but not all are expressed. However, those that are expressed produce many more transcripts than most mRNA genes (Figs. 2, 3; Supplemental Table S3; Supplemental Figs. S12, S13; Egloff et al. 2008). Overall, it is clear that most of the human transcriptome is populated by transcripts originating from a limited set of highly productive genes.
One obvious advantage of total RNA quantification is the capacity to study the expression and biogenesis of stable ribonucleoprotein complexes. The data shown in Figure 3 and Supplemental Figure S13 indicate that while components of the same RNP complex may have similar transcript abundance there are few exceptions that deviate from this rule. In some cases, this deviation from the consensus (e.g., Fig. 3B–D) might signal key components that may regulate or ensure the overall quality of the RNP biogenesis. For example, the study of the different components of SRP indicates that the RNAs coding for the protein components of this RNP are divided into three subclasses based on their abundance (Maity et al. 2008). The first includes two highly abundant mRNAs (SRP14 and SRP9), the second includes moderately abundant mRNAs (SRP72, SRP54, and SR68) and the third consists of a single mRNA accumulating at a much lower level than the others (SRP19) (Akopian et al. 2013). The most highly abundant mRNAs, SRP14, and SRP9, encode structural proteins of SRP that are constitutively bound to the RNA (Fig. 3B; Leung and Brown 2010). In contrast, the protein encoded by the least abundant mRNA, SRP19, functions as an activation signal that restructures the RNA to signal its nuclear export and the completion of RNP assembly in the cytoplasm (Fig. 3B; Maity et al. 2008). Therefore, the RNA abundance in this case appears to be consistent with the biological function of its encoded protein. In general, the structural noncoding RNA components of the tri-snRNP, the U1 and U2 snRNPs, the SRP, the 7SK RNP, the MRP and the RNaseP complexes are one thousand times more abundant than the mRNAs that encode their protein partners (Fig. 3A). This suggests that on average each single mRNA molecule needs to produce a thousand proteins to meet the demand of its noncoding RNA counterparts.
To date, one of the most difficult RNA classes to detect using sequencing techniques has been snoRNA (Veneziano et al. 2016). In general, snoRNAs are much longer than miRNAs but shorter than most protein-coding genes, making their abundances either not detectable or highly variable using standard sequencing library preparation techniques. Indeed, we have found that the use of size selection techniques favors the detection of the shorter and less structured box C/D snoRNA over the longer H/ACA snoRNA (Deschamps-Francoeur et al. 2014). However, the use of FRT now permits comprehensive inter-and intra-class comparison of snoRNA abundance (Fig. 4; Supplemental Figs. S14, S16). The results of this comparison presented in Supplemental Figure S14 suggest that in general the abundance of H/ACA and C/D snoRNA is similar as would be expected from RNA with similar functions in the modification of rRNA (Dieci et al. 2009; Watkins and Bohnsack 2012). However, we noticed that there are many more undetectable C/D snoRNA than H/ACA snoRNA in the SKOV3ip1 transcriptome (Supplemental Figs. S14A, S15). This could be due to the larger number of orphan C/D snoRNA (those that do not have an annotated target site), which are expected to be less expressed than those targeting rRNA (Dupuis-Sandoval et al. 2015). The fact that most of the unexpressed genes come from annotations in the Ensembl database but are not included in the carefully curated snoRNAbase suggests that the differences could come from mis-annotation or a high number of pseudogenes (Supplemental Fig. S15; Hubbard et al. 2007; Xie et al. 2007). Sequencing of a large number of tissues and cell lines may better differentiate between these possibilities. In any case, it is clear that the differences between snoRNA abundance are not necessarily a broad class-specific feature. Our study indicates that most snoRNAs are tightly linked to the expression of their host gene, which would be expected. However, we found that many snoRNAs have marked differences in abundance with their host genes and H/ACA and C/D snoRNA exhibit different dependencies on their host genes (Fig. 4; Supplemental Fig. S14D). For example, H/ACA snoRNA abundance correlated better with noncoding host genes than C/D snoRNA. Similarly, the abundance of C/D snoRNAs nested in genes encoding proteins involved in translation did not correlate as well as that of H/ACA snoRNAs and their host translation protein genes (Fig. 4; Supplemental Fig. S14D). The origin of this variation is not readily clear but could be explained by differences in the biogenesis or stability of both H/ACA and C/D snoRNA. With the advent of improved sequencing methods like TGIRT-based FRT that enable direct comparison of most classes of coding and noncoding RNA components, we can study the mechanisms of snoRNA biogenesis and generate a comprehensive model of the interplay between the coding and noncoding components of the transcriptome.
MATERIALS AND METHODS
Cell culture
The ovarian adenocarcinoma SKOV3ip1 and the ovarian immortalized INOF cell line were grown in DMEM/F12 (50/50) medium and OSE medium (Wisent), respectively. The medium in both cases was supplemented with 10% fetal bovine serum and 2 mM L-glutamine. Cell propagation and passaging were as recommended by ATCC (American Type Culture Collection). Cells were trypsinized and collected in 5 × 106 pellets, resuspended in 700 µL TRIzol (Ambion) and kept at −80°C until RNA extraction.
RNA extraction and conventional sequencing library preparation
The RNA used for SSV sequencing was extracted using a low molecular weight RNA extraction kit (mirVana, Invitrogen) as previously described (Deschamps-Francoeur et al. 2014), and from these samples, cDNA libraries were prepared using the TruSeq Small RNA Sample Prep Kit (Illumina), which includes adapter ligation, reverse transcription, and PCR amplification. The RNA used for FAV and FRV sequencing was isolated and purified from 5 µg DNA-free total RNA extracted using either a NEBNext Poly(A) mRNA Isolation Module (New England Biolabs) in the case of FAV sequencing or Ribo-Zero Gold (Illumina) in the case of FRV, according to the manufacturers’ protocol. Library preparations were performed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) in order to generate an RNA-seq library from 100 ng of purified RNA. In the cases of URT and FRT, the RNA was extracted using RNeasy Kit from Qiagen.
Construction and sequencing of TGIRT-seq libraries
TGIRT-seq libraries were constructed as previously described (Nottingham et al. 2016; Qin et al. 2016). ERCC spike-ins (Kralj and Salit 2013; Tong et al. 2016) were added to selected SKOV3ip1 library and used as control for detection uniformity. Further details are provided in the Supplemental Methods section.
RNA-seq analysis
All data sets were passed through a quantification pipeline to obtain CPM and TPM values. Fastq files were checked for quality using FastQC and trimmed using Cutadapt (Martin 2011) and Trimmomatic (Bolger et al. 2014) (with TRAILING:30) to remove adapters and portions of reads of low quality, respectively. Further details are provided in the Supplemental Methods section.
Annotation modification
An annotation file in gene transfer format (.gtf) was obtained from Ensembl (Yates et al. 2016) (hg38, v87). The annotation file was supplemented with tRNA genes from GtRNAdb (Chan and Lowe 2016) and with snoRNA genes from Refseq (O'Leary et al. 2016) that were missing in Ensembl annotations (Supplemental Table S7). Further details are provided in the Supplemental Methods section.
Gene biotype pooling
Gene biotypes as given by the Ensembl annotation files were pooled for simplicity. The groups “Protein_coding,” “Pseudogene,” and “Long_noncoding” were obtained by pooling biotypes as recommended by Ensembl (http://useast.ensembl.org/Help/Faq?id=468). The group “Other” corresponds to any other biotype not listed.
RT-qPCR, end-point RT-PCR, and dPCR analysis
RT-qPCR and dPCR primer design and validation were performed by the Université de Sherbrooke RNomics Platform (http://rnomics.med.usherbrooke.ca/) as previously described (Brosseau et al. 2010). (Primers used are listed in Supplemental Tables S8–S10.) Further details are provided in the Supplemental Methods section.
DATA DEPOSITION
Additional data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). The SSV samples are available under the accession number GSE55946 (sample names SKOV3ip1_WT_1 and SKOV3ip1_ WT_2). The remaining samples were deposited under accession number GSE99065.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
Thermostable group II intron reverse transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas and East Tennessee State University to InGex, LLC. A.M.L. and the University of Texas are minority equity holders in InGex, LLC, and A.M.L. and some present and former Lambowitz laboratory members receive royalty payments from the sale of TGIRT enzymes and kits and from the licensing of intellectual property. S.A. and M.S.S. and affiliated laboratory members declare no conflict of interest.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the excellent help and input of Mr. Fabien Dupuis-Sandoval for establishing parts of the bioinformatics pipeline used in this work. Endpoint, digital, and quantitative RT-PCR were performed by the RNomics Platform of the Université de Sherbrooke. Libraries were sequenced either at the Genome Sequencing and Analysis Facility (GSAF) at The University of Texas at Austin or at the Génome Québec Innovation Centre. This work was supported by Canadian Institutes of Health Research (CIHR) grant PJT 153171 (S.A. and M.S.S.), National Institutes of Health grant R01 GM37949 and Welch Foundation grant F1706 (A.M.L.), and Research Chair in RNA Biology and Cancer Genomics (S.A.). G.D.F. was supported by a Natural Sciences and Engineering Research Council (NSERC) Masters scholarship and a scholarship from the Faculty of Medicine and Health Sciences of the Université de Sherboroke. V.B. was supported by a Masters scholarship from the Fonds de Recherche du Québec - Santé (FRQS). M.S.S. holds a FRQS Research Scholar Junior 2 Career Award.
Author contributions: V.B., G.D.F., S.C., and R.M.N. conducted experiments and analyzed data. P.B.B. helped analyze data. A.M.L., M.S.S., and S.A. conceived the project, designed the experiments, analyzed data, and wrote the manuscript with help from the other authors.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064493.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Abbas W, Kumar A, Herbein G. 2015. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front Oncol 5: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan SO. 2013. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82: 693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ. 2014. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription 5: e27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Laiho M. 2016. Deep sequencing analysis of nucleolar small RNAs: RNA isolation and library preparation. Methods Mol Biol 1455: 231–241. [DOI] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. 2011. The quantitative proteome of a human cell line. Mol Syst Biol 7: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A. 2009. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez L, Fortes P. 2013. U1 snRNP control of 3′-end processing and the therapeutic application of U1 inhibition combined with RNA interference. Curr Mol Med 13: 1203–1216. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, Gervais-Bird J, Tremblay K, Perreault JP, Elela SA. 2010. High-throughput quantification of splicing isoforms. RNA 16: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A. 2017. Transcriptome profiling in human diseases: new advances and perspectives. Int J Mol Sci 18: E1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V, Angelini C, De Feis I, Ciccodicola A. 2010. Uncovering the complexity of transcriptomes with RNA-seq. J Biomed Biotechnol 2010: 853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TA, Loos B, Engelbrecht AM. 2014. AHNAK: the giant jack of all trades. Cell Signal 26: 2683–2693. [DOI] [PubMed] [Google Scholar]
- Deschamps-Francoeur G, Garneau D, Dupuis-Sandoval F, Roy A, Frappier M, Catala M, Couture S, Barbe-Marcoux M, Abou-Elela S, Scott MS. 2014. Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Res 42: 10073–10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Preti M, Montanini B. 2009. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 94: 83–88. [DOI] [PubMed] [Google Scholar]
- Dupuis-Sandoval F, Poirier M, Scott MS. 2015. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6: 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594. [DOI] [PubMed] [Google Scholar]
- Ginzinger DG. 2002. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30: 503–512. [DOI] [PubMed] [Google Scholar]
- Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM. 2013. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J Clin Microbiol 51: 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. 2007. Ensembl 2007. Nucleic Acids Res 35: D610–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, Gingeras TR, Oliver B. 2011. Synthetic spike-in standards for RNA-seq experiments. Genome Res 21: 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Zhou X, Li R, Michal JJ, Zhang S, Dodson MV, Zhang Z, Harland RM. 2015. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci 72: 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi N, Horiuchi A, Osada R, Imai T, Wang C, Chen X, Konishi I. 2006. Nuclear expression of S100A4 is associated with aggressive behavior of epithelial ovarian carcinoma: an important autocrine/paracrine factor in tumor progression. Cancer Sci 97: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinck R, Chabot B, Abou Elela S. 2012. High-throughput analysis of alternative splicing by RT-PCR. Wiley, NY. [Google Scholar]
- Kralj JG, Salit ML. 2013. Characterization of in vitro transcription amplification linearity and variability in the low copy number regime using external RNA control consortium (ERCC) spike-ins. Anal Bioanal Chem 405: 315–320. [DOI] [PubMed] [Google Scholar]
- Leung E, Brown JD. 2010. Biogenesis of the signal recognition particle. Biochem Soc Trans 38: 1093–1098. [DOI] [PubMed] [Google Scholar]
- Liang H, Zeng E. 2016. RNA-seq experiment and data analysis. Methods Mol Biol 1366: 99–114. [DOI] [PubMed] [Google Scholar]
- Maelandsmo GM, Flørenes VA, Nguyen MT, Flatmark K, Davidson B. 2009. Different expression and clinical role of S100A4 in serous ovarian carcinoma at different anatomic sites. Tumour Biol 30: 15–25. [DOI] [PubMed] [Google Scholar]
- Maity TS, Fried HM, Weeks KM. 2008. Anti-cooperative assembly of the SRP19 and SRP68/72 components of the signal recognition particle. Biochem J 415: 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Mayorga A, Gleicher M. 2013. Splatterplots: overcoming overdraw in scatter plots. IEEE Trans Vis Comput Graph 19: 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori T, Hinterberger M, Pettersson I, Steitz JA. 1984. Autoantibodies to the U2 small nuclear ribonucleoprotein in a patient with scleroderma-polymyositis overlap syndrome. J Biol Chem 259: 560–565. [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. 2013. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19: 958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley AA. 2014. Digital PCR: a brief history. Biomol Detect Quantif 1: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, Kornblihtt AR. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84: 165–198. [DOI] [PubMed] [Google Scholar]
- Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, Lambowitz AM. 2016. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22: 597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. 2011. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. 2008. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res 36: D178–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe M, Borgoni S, Cascione M, Chattaragada MS, Ferri-Borgogno S, Capello M, Bulfamante S, Chapelle J, Di Modugno F, Defilippi P, et al. 2017. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J Hematol Oncol 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, Lambowitz AM. 2016. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22: 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani B, Nakaoka H, Akhondzadeh S, Tekin M, Mahdieh N. 2016. Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol Biosyst 12: 1818–1830. [DOI] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Zeeberg BR, Caplen NJ, Cleland JA, Kahn AB, Liu H, Weinstein JN. 2008. SpliceCenter: a suite of web-based bioinformatic applications for evaluating the impact of alternative splicing on RT-PCR, RNAi, microarray, and peptide-based studies. BMC Bioinformatics 9: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager M, Yeat NC, Pajaro-Van der Stadt S, Lin C, Ren Q, Lin J. 2015. Transcriptomics in cancer diagnostics: developments in technology, clinical research and commercialization. Expert Rev Mol Diagn 15: 1589–1603. [DOI] [PubMed] [Google Scholar]
- Shakeel M, Rodriguez A, Tahir UB, Jin F. 2017. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol Lett 40: 227–236. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci 111: E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Dubé KN, Riley PR. 2010. Identification of Thymosin β4 as an effector of Hand1-mediated vascular development. Nat Commun 1: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. 2015. RNA–RNA base-pairing: theme and variations. RNA 21: 476–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Yang C, Wu PY, Wang MD. 2016. Evaluating the impact of sequencing error correction for RNA-seq data with ERCC RNA spike-in controls. IEEE EMBS Int Conf Biomed Health Inform 2016: 74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai ST, Chien IH, Shen WH, Kuo YZ, Jin YT, Wong TY, Hsiao JR, Wang HP, Shih NY, Wu LW. 2010. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur J Cancer 46: 1712–1723. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. 2006. The ever-growing world of small nuclear ribonucleoproteins. In The RNA world, 3rd ed (ed. Gesteland RF et al. ), pp. 327–368. Cold Spring Harbor Laboratory Press, NY. [Google Scholar]
- Veneziano D, Di Bella S, Nigita G, Laganà A, Ferro A, Croce CM. 2016. Noncoding RNA: current deep sequencing data analysis approaches and challenges. Hum Mutat 37: 1283–1298. [DOI] [PubMed] [Google Scholar]
- Vera M, Biswas J, Senecal A, Singer RH, Park HY. 2016. Single-cell and single-molecule analysis of gene expression regulation. Annu Rev Genet 50: 267–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtel C, Manley JL. 2009. Splicing of mRNA precursors: the role of RNAs and proteins in catalysis. Mol Biosyst 5: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C, Lacroute F. 1975. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol 122: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT. 2012. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414. [DOI] [PubMed] [Google Scholar]
- Whale AS, Huggett JF, Cowen S, Speirs V, Shaw J, Ellison S, Foy CA, Scott DJ. 2012. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res 40: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. 2004. RNA polymerase III transcription and cancer. Oncogene 23: 3208–3216. [DOI] [PubMed] [Google Scholar]
- Wilusz JE. 2015. Controlling translation via modulation of tRNA levels. Wiley Interdiscip Rev RNA 6: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, McAlexander MA, Queen SE, Adams RJ. 2013. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol 10: 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SF, Schlessinger D. 1977. Nuclear metabolism of ribosomal RNA in growing, methionine-limited, and ethionine-treated HeLa cells. Biochemistry 16: 2783–2791. [DOI] [PubMed] [Google Scholar]
- Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. 2007. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res 35: D183–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. 2016. Ensembl 2016. Nucleic Acids Res 44: D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi J, Cheng S, Zhu Y, Zhao X, Yang K, Du X, Klocker H, Yang X, Zhang J. 2012. Estrogen promotes prostate cancer cell migration via paracrine release of ENO1 from stromal cells. Mol Endocrinol 26: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shen J, Mannoor K, Guarnera M, Jiang F. 2014. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin Lung Cancer 15: 372–378.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li M, Liu Y, Han N, Zhang K, Xiao T, Cheng S, Gao Y. 2010. [ENO1 protein levels in the tumor tissues and circulating plasma samples of non-small cell lung cancer patients]. Zhongguo Fei Ai Za Zhi 13: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren D, Guo L, Wang L, Wu S, Lin C, Ye L, Zhu J, Li J, Song L, et al. 2017. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res 19: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.