In this study, Fei et al. describe the identification of NDF, a nucleosome-destabilizing factor that facilitates RNA polymerase II transcription in chromatin. Their findings demonstrate that NDF is recruited to gene bodies during transcriptional activation and facilitates Pol II transcription through nucleosomes.
Keywords: nucleosome, transcription, gene expression, RNA polymerase II, chromatin dynamics
Abstract
Our understanding of transcription by RNA polymerase II (Pol II) is limited by our knowledge of the factors that mediate this critically important process. Here we describe the identification of NDF, a nucleosome-destabilizing factor that facilitates Pol II transcription in chromatin. NDF has a PWWP motif, interacts with nucleosomes near the dyad, destabilizes nucleosomes in an ATP-independent manner, and facilitates transcription by Pol II through nucleosomes in a purified and defined transcription system as well as in cell nuclei. Upon transcriptional induction, NDF is recruited to the transcribed regions of thousands of genes and colocalizes with a subset of H3K36me3-enriched regions. Notably, the recruitment of NDF to gene bodies is accompanied by an increase in the transcript levels of many of the NDF-enriched genes. In addition, the global loss of NDF results in a decrease in the RNA levels of many genes. In humans, NDF is present at high levels in all tested tissue types, is essential in stem cells, and is frequently overexpressed in breast cancer. These findings indicate that NDF is a nucleosome-destabilizing factor that is recruited to gene bodies during transcriptional activation and facilitates Pol II transcription through nucleosomes.
In the eukaryotic nucleus, transcription of protein-coding genes is carried out by RNA polymerase II (Pol II). Our understanding of Pol II transcription is limited, however, by our knowledge of the factors that mediate this essential process. In spite of considerable progress, many important elements of Pol II transcription remain unsolved.
One particularly intriguing aspect of transcription by Pol II is the ability of the enzyme to traverse the nucleosomal template. A key protein that promotes transcription and DNA replication in chromatin is FACT (facilitates chromatin transcription/transactions) (for example, see Reinberg and Sims 2006; Formosa 2008). FACT binds to nucleosomes and can remove an H2A–H2B dimer or reorganize the nucleosome into an altered structure. In humans, however, the SSRP1 subunit of FACT is not detectable in many tissue types (Supplemental Fig. S1A; Garcia et al. 2011; Uhlén et al. 2015). It thus appears that critical components of the human transcription elongation machinery have yet to be identified.
In our studies of chromatin dynamics, we became drawn to the study of Pol II elongation in chromatin through our analysis of the prenucleosome, a conformational isomer of the nucleosome that associates with ∼80-base-pair (bp) DNA rather than ∼147-bp DNA as in a canonical nucleosome (Torigoe et al. 2011, 2013; Fei et al. 2015; Khuong et al. 2015). Notably, H3K56, which is located at the DNA entry and exit points of the nucleosome, can be acetylated by p300 in prenucleosomes but not in canonical nucleosomes (Fei et al. 2015). This observation is consistent with the partial unwrapping of the DNA in a prenucleosome relative to a nucleosome.
The properties of the prenucleosome led us to consider whether there might be factors that could partially disassemble a nucleosome and facilitate the acetylation of nucleosomal H3K56. These studies resulted in the identification of a protein that we term NDF (nucleosome-destabilizing factor), which destabilizes nucleosomes, is enriched in the transcribed regions of many active genes, facilitates transcription by Pol II through nucleosomes, and is present in all tested human tissue types. The identification of NDF reveals a new Pol II elongation factor that is structurally and functionally distinct from FACT.
Results
Identification and purification of NDF, a novel nucleosome-destabilizing factor
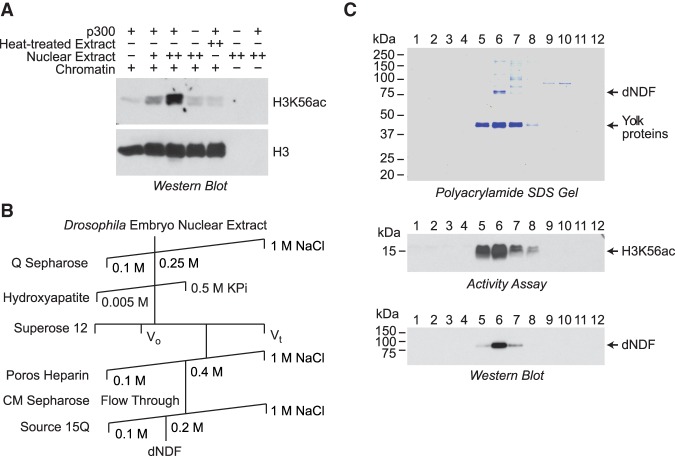
To identify factors that destabilize or disassemble nucleosomes, we used an assay that was based on our observations of H3K56 accessibility in prenucleosomes versus nucleosomes (Fei et al. 2015). Specifically, we tested for activities that would enable nucleosomal H3K56 to be acetylated by purified p300. In principle, this assay would reveal factors that either destabilize nucleosomes or unwrap DNA from the ends of nucleosomes. As shown in Figure 1A, we identified a heat-sensitive activity in a nuclear extract from Drosophila embryos that stimulates the ability of p300 to acetylate H3K56 with a chromatin substrate. This activity was purified by six column chromatography steps (Fig. 1B). The final Source 15Q column yielded a major ∼75-kDa polypeptide as well as Drosophila yolk proteins (Fig. 1C), which are among the most abundant proteins in embryos. Mass spectrometry revealed that the ∼75-kDa polypeptide is the CG4747 protein. To test whether the presence of CG4747 correlates with the biochemical activity, we synthesized full-length Drosophila CG4747 in bacteria and generated polyclonal antisera in rabbits. Western blot analysis with the antibodies revealed that CG4747 mostly correlates with the stimulation of nucleosomal H3K56 acetylation (Fig. 1C).
Figure 1.
Purification of Drosophila NDF (dNDF). (A) Identification of NDF as an activity in a Drosophila nuclear extract that facilitates nucleosomal H3K56 acetylation by purified p300. The absence (−) or presence of lower (+) or higher (++) concentrations of the indicated components is shown. Chromatin was reconstituted with plasmid DNA and purified core histones by the salt dialysis method. H3K56 acetylation was monitored by Western blot analysis. Both bands in the doublets are derived from histone H3. The heat-treated extract was subjected to incubation for 5 min at 75°C. (B) Scheme for the purification of dNDF. (C) Purification of dNDF by Source 15Q chromatography. The Source 15Q column fractions were analyzed by 10% polyacrylamide–SDS gel electrophoresis and staining by Coomassie brilliant blue R-250 (top panel), the H3K56 acetylation activity assay as in A (middle panel), and Western blot with antibodies that were raised against recombinant dNDF (bottom panel).
CG4747 was identified previously by ChIP-mass spectrometry as a factor that associates with the Drosophila MSL (male-specific lethal) complex and was found to facilitate the targeting of MSL to active genes in a manner that is dependent on H3K36 trimethylation (H3K36me3) by Set2 (Wang et al. 2013). The human homolog of the CG4747 gene is GLYR1 (glyoxylate reductase 1 homolog; also known as N-PAC and NP60). The GLYR1 protein was identified in a screen for factors that preferentially bind to H3K36me3 relative to unmethylated H3K36 (Vermeulen et al. 2010) and is present at high levels in the nuclei of all tested tissue types (Supplemental Fig. S1A). The GLYR1 gene appears to be essential in human HUES62 stem cells but not in human A375 melanoma cells (Shalem et al. 2014). GLYR1 was observed to stimulate H3K4me1 and H3K4me2 demethylation by LSD2 (KDM1b) (Fang et al. 2013); however, the region of human GLYR1 that interacts with LSD2 is not present in Drosophila CG4747 (Supplemental Fig. S1B). Both CG4747 and GLYR1 proteins contain a PWWP domain that binds to methylated lysines (Qin and Min 2014) and a dehydrogenase domain (Fig. 2A; Supplemental Fig. S1B) that does not appear to be enzymatically active in human GLYR1 (Fang et al. 2013). In addition, human GLYR1 has a putative DNA-binding AT-hook motif, but Drosophila CG4747 does not (Supplemental Fig. S1B). Based on the shared biochemical activities of CG4747 and GLYR1 (as described in this work), we designated these factors as NDF.
Figure 2.
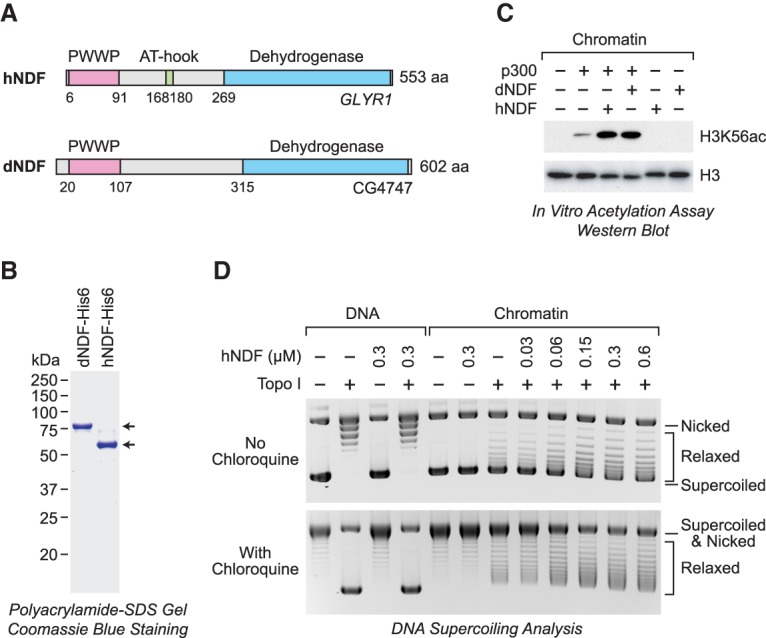
Destabilization of nucleosomes by purified recombinant dNDF and human NDF (hNDF). (A) Schematic diagram of hNDF and dNDF. The drawing is roughly to scale. Specific features are described in the text. (B) Purification of bacterially synthesized N-terminally His6-tagged dNDF and hNDF. (C) Recombinant dNDF and hNDF facilitate the acetylation of nucleosomal H3K56 by p300. This assay was performed as in Figure 1A. (D) Loss of DNA supercoiling in chromatin reveals destabilization of nucleosomes by hNDF. Chromatin was reconstituted with plasmid DNA and purified core histones by the salt dialysis method and then incubated (at 0.3 µM nucleosome concentration) with purified recombinant hNDF and topoisomerase I (Topo I) as indicated. Samples were deproteinized and subjected to 0.8% agarose gel electrophoresis in the absence or presence of 10 ng/mL chloroquine. DNA was visualized by staining with ethidium bromide. As a control, reactions were carried out in parallel with plasmid DNA.
NDF destabilizes nucleosomes in an ATP-independent manner
To test the biochemical activities of Drosophila NDF (dNDF) and human NDF (hNDF), we synthesized and purified the full-length recombinant proteins (Fig. 2B). We observed that dNDF and hNDF are able to stimulate the acetylation of nucleosomal H3K56 by p300 in a dose-dependent and ATP-independent manner (Fig. 2C; Supplemental Fig. S2A,B). Purified dNDF and hNDF did not exhibit intrinsic histone acetyltransferase activity (Fig. 2C) and also did not stimulate the acetylation of free histones by p300 (Supplemental Fig. S2C). Thus, purified recombinant dNDF and hNDF can mediate the stimulation of nucleosomal H3K56 acetylation, as seen in our original assay with the native Drosophila factor (Fig. 1).
We next examined whether NDF has the ability to disassemble nucleosomes. To this end, we investigated the ability of NDF to disrupt nucleosome-mediated supercoiling of covalently closed circular plasmid DNA. The wrapping of DNA around the core histone octamer in a nucleosome causes a change in the linking number of approximately −1 (Germond et al. 1975). Therefore, nucleosome disassembly can be monitored by the loss of negative supercoiling in chromatin in circular plasmid DNA. We reconstituted nucleosomes onto plasmid DNA by using salt dialysis methodology and then treated the chromatin with NDF in the presence of topoisomerase I. The resulting DNA species were analyzed by agarose gel electrophoresis in the absence or presence of chloroquine, which introduces positive supercoils into the DNA and enables the differentiation between relaxed circular DNA and nicked circular DNA (Fig. 2D; Supplemental Fig. S2D,E). These experiments revealed that hNDF and dNDF are able to mediate the partial disassembly of nucleosomes (as seen by the loss of negative DNA supercoiling) in a dose- and time-dependent manner. Hence, the results from the nucleosomal H3K56 acetylation and the chromatin disruption experiments indicate that hNDF and dNDF can partially disassemble nucleosomes. In addition, because NDF functions in an ATP-independent manner, it appears that the binding of NDF to chromatin provides the energy for the destabilization of nucleosomes.
The recruitment of NDF to gene bodies upon transcriptional induction is accompanied by an increase in transcript levels
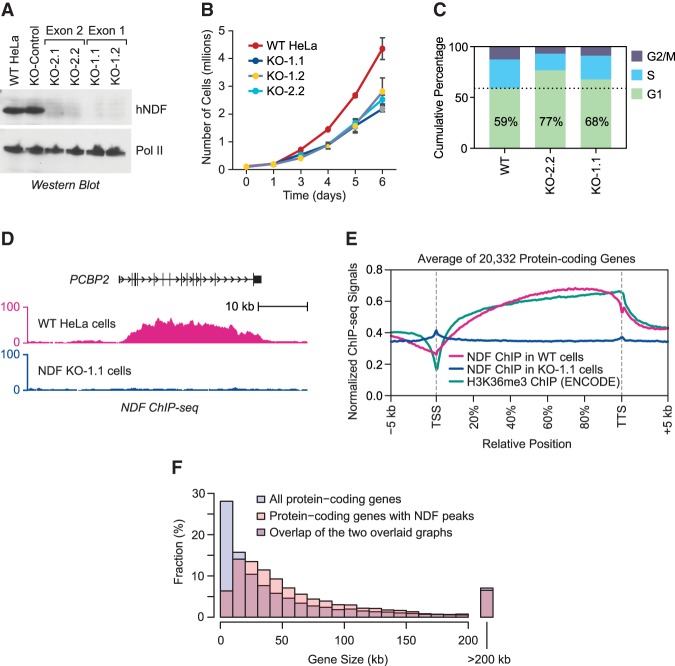
To analyze the properties of hNDF, we generated four hNDF knockout lines in HeLa cells as well as rabbit polyclonal antisera against the full-length protein (Fig. 3A; Supplemental Fig. S2F). hNDF appears to be essential in human HUES62 stem cells but not in human A375 melanoma cells (Shalem et al. 2014). The hNDF knockout HeLa cells are viable but exhibit reduced growth rates relative to wild-type cells (Fig. 3B). The hNDF knockout cells were found to have a higher percentage of cells in G1 phase than wild-type cells (Fig. 3C). Single-cell live imaging experiments showed that the hNDF knockout cells have a longer duration of the cell cycle than wild-type cells and no apparent mitotic alterations (Supplemental Fig. S3).
Figure 3.
NDF is enriched in transcribed regions of protein-coding genes, with a preference for longer genes relative to shorter genes. (A) Generation of human cell lines lacking NDF. By using CRISPR–Cas9 methodology, four knockout cell lines were generated in HeLa cells by disruption of either exon 1 (KO-1.1 and KO-1.2) or exon 2 (KO-2.1 and KO-2.2) of the gene encoding NDF. The loss of NDF was shown by Western blot analysis. The largest subunit of RNA Pol II was used as a loading control. (B) The loss of NDF reduces the growth rate of cells. On day 0, 1 × 104 cells were seeded in triplicate in 6-cm culture dishes, and attached cells were counted on the indicated days. (C) The loss of NDF increases the fraction of cells in G1, as seen by FACS analysis. (D) NDF ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data at the PCBP2 gene. (E) NDF colocalizes with H3K36me3 over transcribed regions. Metagene analysis of NDF and H3K36me3 occupancy at 20,232 protein-coding genes. (F) NDF preferentially localizes to longer protein-coding genes relative to shorter protein-coding genes. By ChIP-seq analysis, ∼4500 protein-coding genes were found to be enriched in NDF in HeLa cells. Overlaid bar graphs of the size distributions of all protein-coding genes (blue) versus NDF-enriched protein-coding genes (pink) are shown. Purple indicates regions of overlap of the two bar graphs.
To determine the localization of NDF, we carried out ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analyses in wild-type and knockout (as background control) cells. We observed that endogenous hNDF is often enriched over the transcribed regions of active genes and colocalizes with H3K36me3 over those genes (Fig. 3E,F; Supplemental Fig. S4A). This distribution of hNDF is similar to that seen with epitope-tagged dNDF in transgenic Drosophila larvae (Wang et al. 2013) and is likely to be due, at least in part, to the binding of the PWWP domain to H3K36me3 (Wang et al. 2013; Qin and Min 2014). Consistent with the presence of NDF over transcribed genes, hNDF is found predominantly in introns and exons (Supplemental Fig. S4B). Strikingly, hNDF exhibits a distinct preference for longer genes relative to shorter genes (Fig. 3F).
To investigate the relationship between hNDF (ChIP), H3K36me3 (ChIP), and gene activity (RNA sequencing [RNA-seq]), we subjected these three variables in 15,407 genes to k-means clustering and generated four clusters of genes (Fig. 4A,B; Supplemental Fig. S4C). Cluster 1 contained active genes that are enriched in NDF and H3K36me3 along the gene body. Cluster 2 had active genes with lower levels of NDF and intermediate to high levels of H3K36me3. Clusters 3 and 4 contained less active or inactive genes with low levels of NDF and H3K36me3. This analysis shows that active genes can be subdivided into a cluster that is enriched in NDF (cluster 1) and a cluster that is not enriched in NDF (cluster 2). It can also be seen that the length distribution of genes in cluster 1 was distinct from that of genes in cluster 2 (Supplemental Fig. S4D).
Figure 4.
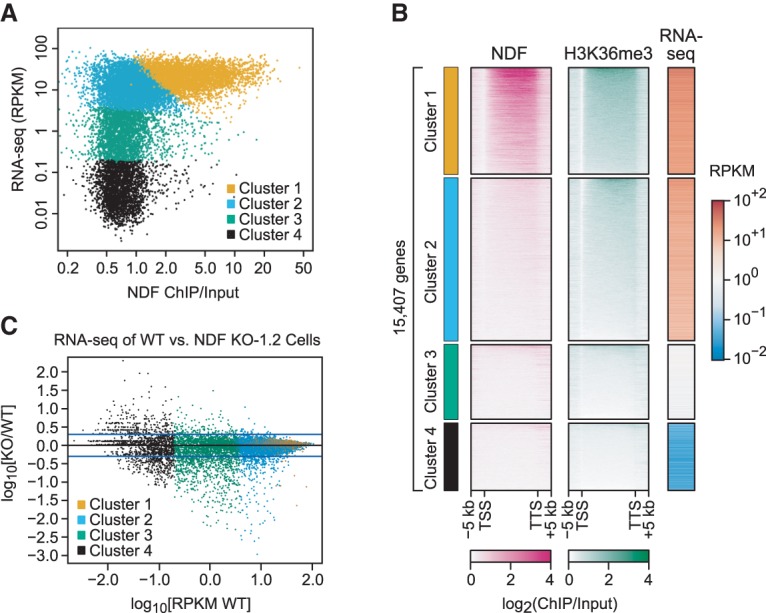
NDF contributes to the expression of many genes. Four clusters of genes were generated by k-means clustering with three variables: log10[RNA-seq signal of genes in RPKM] (reads per kilobase per million mapped reads), log10[(NDF ChIP-seq signal over gene body)/(input signal over gene body)], and log10[(H3K36me3 ChIP-seq signal over gene body)/(input signal over gene body)]. (A) NDF is enriched in active gene cluster 1 but not in active gene cluster 2. RNA-seq signal (RPKM) versus NDF ChIP/input signal for gene clusters 1–4. (B) Heat maps of NDF and H3K36me3 occupancies and gene expression in wild-type HeLa cells. Genes within each cluster were ranked by the (NDF ChIP-seq signal)/(input signal) ratio. (C) RNA-seq analysis of wild-type versus hNDF KO-1.2 cells. The plot shows log10[(RPKM knockout)/(RPKM wild type)] versus log10[RPKM wild type] for genes in clusters 1–4. Genes with RPKM = 0, such as those in cluster 4, were not included in this analysis. The top and bottom blue lines correspond to (RPKM knockout)/(RPKM wild type) ratios of 2 and 0.5, respectively.
To determine whether NDF affects gene activity, we performed RNA-seq analyses with wild-type and hNDF knockout cells. These experiments revealed that the loss of hNDF results in a decrease in the steady-state RNA levels of many genes (Fig. 4C; Supplemental Fig. S4E). This effect was observed in genes with higher hNDF levels (cluster 1) as well as genes with lower NDF levels (clusters 2–4). It is possible, for instance, that NDF is important for transcription of genes with low as well as high NDF occupancy. In addition, some of the effects could be indirect, such as through the down-regulation of transcriptional activators or repressors. Nevertheless, it appears that NDF contributes to the expression of a broad range of genes.
We also carried out global run-on sequencing (GRO-seq) analysis of wild-type and hNDF knockout cells. These experiments revealed differences in the enrichment of transcriptionally engaged polymerases over gene bodies in wild-type relative to knockout cells (Supplemental Fig. S5). These findings further support a role of NDF in the transcription process.
We then examined the dynamics of mouse NDF (mNDF) in primary bone marrow-derived macrophages (BMDMs). Because mNDF is nearly identical to hNDF (Supplemental Fig. S1B), our hNDF antibodies recognize mNDF. In these studies, we used Kdo2-lipid A (KLA), a highly specific TLR4 agonist (Raetz et al. 2006), to activate TLR4-inducible genes in BMDMs (Kaikkonen et al. 2013; Oishi et al. 2017). The gene encoding mNDF is not induced by KLA (Supplemental Fig. S6A). In these experiments, we observed that mNDF is recruited to the gene bodies of >2000 genes after 6 h of KLA activation (Fig. 5A,B; Supplemental Fig. S6B). This increase in the occupancy of NDF correlates with the increase in H3K36me3 as well as with transcript levels (Fig. 5C,D). These results thus reveal a dynamic role of NDF during gene activation.
Figure 5.
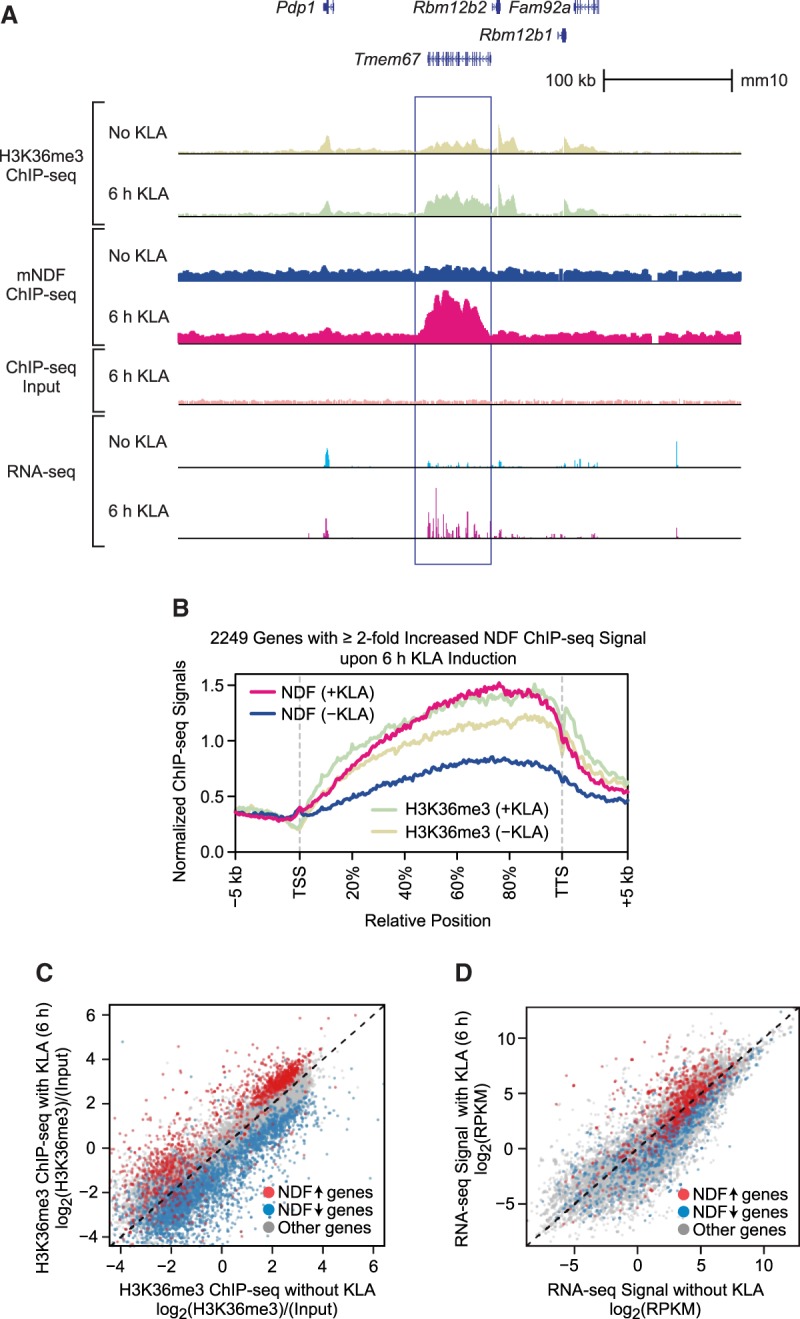
NDF is recruited to the transcribed regions of thousands of genes in mouse primary BMDMs upon transcriptional induction by KLA. (A) Genome browser view of H3K36me3 ChIP-seq, mNDF ChIP-seq, and RNA-seq with or without induction by KLA for 6 h. The region in the vicinity of the Tmem67 gene is indicated by the box. (B) Recruitment of mNDF to gene bodies upon activation of BMDMs by KLA. Metagene analysis of NDF and H3K36me3 ChIP-seq signals at 2249 genes in which NDF occupancy increases by at least twofold upon 6 h of induction with KLA. (C) The KLA-induced increase in NDF occupancy correlates with an increase in H3K36me3. The scatter plot (35,933 total genes) shows 2249 genes in which NDF occupancy increases (up arrow) by at least twofold upon KLA induction for 6 h and 4097 genes in which NDF occupancy decreases (down arrow) by at least twofold upon KLA induction for 6 h. (D) The KLA-induced increase in NDF occupancy generally correlates with an increase in transcript levels. The genes at which NDF levels change upon KLA induction are as in C.
These experiments show that NDF is recruited to the transcribed regions of >2000 genes upon transcriptional induction (Fig. 5A,B). Moreover, this recruitment of NDF is accompanied by an increase in the transcript levels of many NDF-enriched genes (Fig. 5D). In addition, the global loss of hNDF results in a decrease in the RNA levels of many genes (Fig. 4C; Supplemental Fig. S4E). These findings, combined with the ability of NDF to destabilize nucleosomes, suggested that NDF may affect transcriptional elongation in chromatin.
NDF facilitates Pol II elongation in chromatin in vitro and in cell nuclei
To test whether NDF affects transcription through nucleosomes, we performed elongation assays with purified yeast Pol II on templates with a positioned downstream nucleosome. To this end, we used a strategy that is similar to the method of Kireeva et al. (2002). As depicted in Figure 6A, these experiments involved the assembly of a Pol II elongation complex with a 5′-labeled RNA primer and the subsequent ligation of a downstream nucleosome. Transcript elongation was initiated by the addition of ribonucleoside 5′-triphosphates, and the progression of Pol II was followed by analyzing the length of the 5′ end-labeled transcripts.
Figure 6.
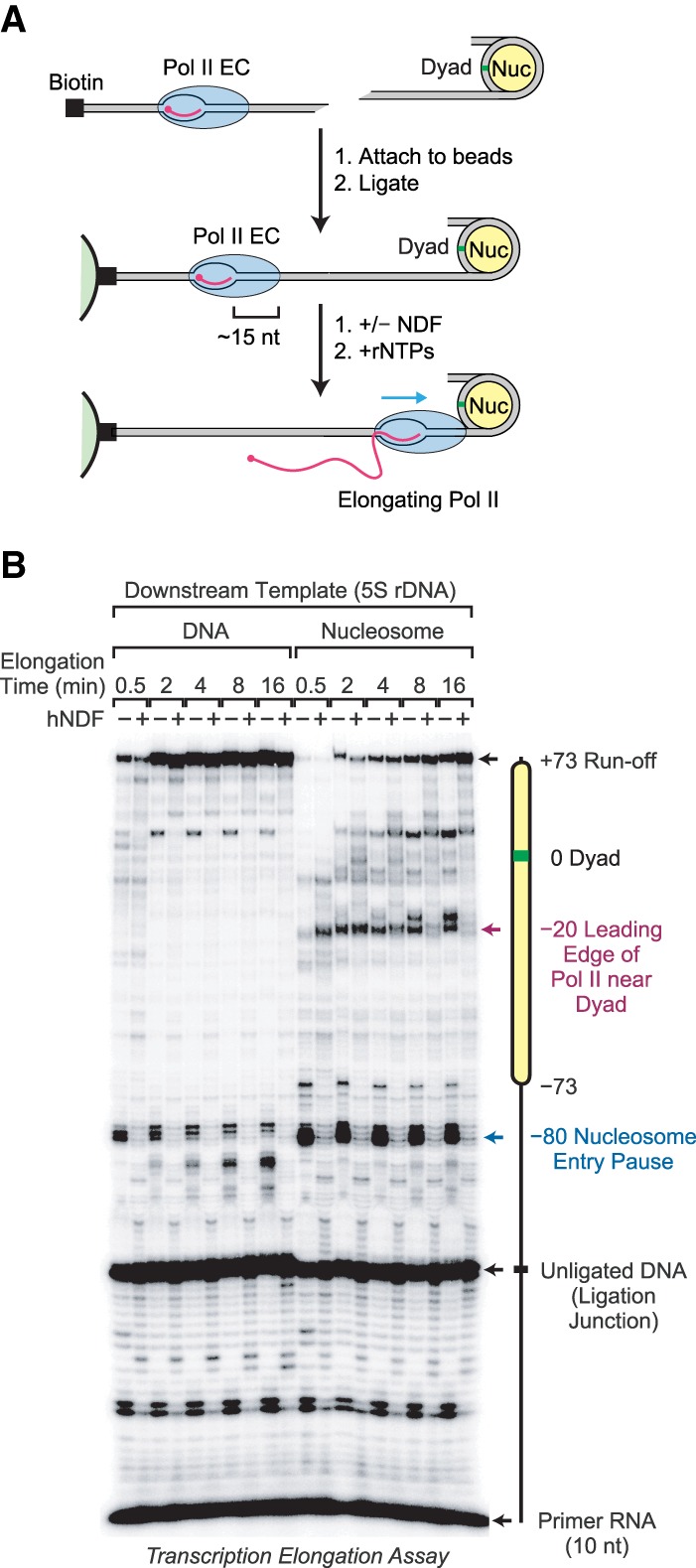
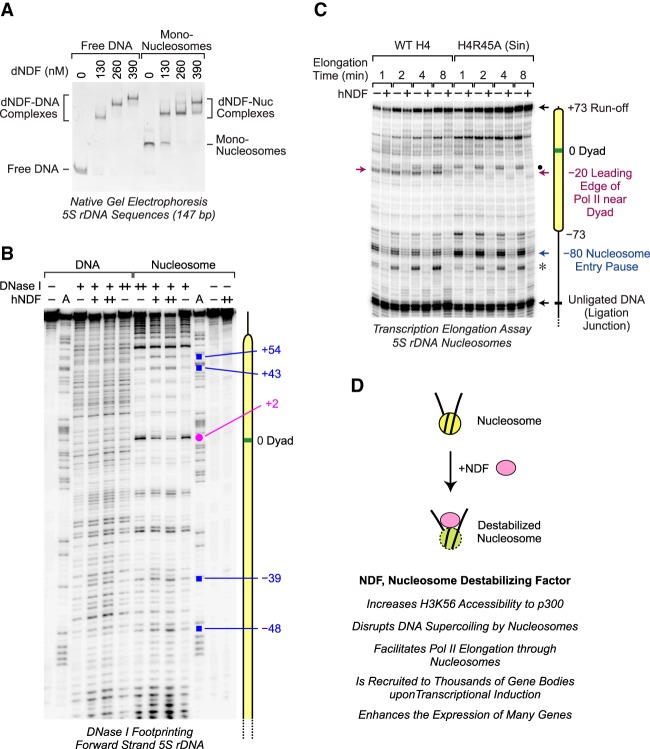
NDF facilitates transcription elongation through a nucleosome. (A) Schematic diagram of Pol II transcription through a positioned nucleosome. A transcriptionally engaged elongation complex (Pol II EC) was assembled with purified yeast Pol II and a 5′ end-labeled 10-nucleotide (nt) RNA primer. This complex was attached to streptavidin beads and then ligated to a downstream mononucleosome positioned on the 5S rDNA sequence from Xenopus borealis. Transcription elongation was initiated by the addition of ribonucleoside 5′-triphosphates (rNTPs). Where indicated, NDF was added after the ligation step but before the addition of the rNTPs. In parallel experiments, naked DNA (same 5S rDNA sequence) was ligated downstream from the elongation complex instead of a mononucleosome. The distance (∼15 nt) from the leading edge of Pol II to the 3′ end of the transcript is also shown. (B) NDF reduces the inhibition of Pol II elongation by a nucleosome. Transcription elongation reactions were performed as described in A in the presence or absence of purified hNDF for the indicated times. Experiments with nucleosomal templates included a 75-fold molar excess (0.3 µM) of free unligated mononucleosomes, which stabilize the low concentration of immobilized nucleosomes. Where indicated, hNDF was included at a concentration of 1.5 µM. The reaction products were resolved by 8% polyacrylamide–urea gel electrophoresis. The diagram shows the locations of the positioned nucleosomes, the sites of Pol II pausing, the runoff product, the ligation junction, and the 10-nt primer RNA. The sizes of the RNA species were estimated by comparison with a 25-nt radiolabeled DNA ladder.
We reconstituted mononucleosomes by salt dialysis methodology with purified bacterially synthesized core histones and DNA containing the nucleosome positioning sequence from a 5S rRNA gene of Xenopus borealis (Rhodes 1985; Hayes et al. 1990). To analyze the positioning of the nucleosomes, we used a modified version of the histone-directed DNA cleavage method of Flaus et al. (1996). These experiments showed that the reconstituted mononucleosomes were located at a single predominant position in the 5S rDNA (Supplemental Fig. S7).
We then purified yeast Pol II (Supplemental Fig. S8A) and performed the Pol II elongation experiments with the downstream 5S rDNA sequence as a mononucleosome or naked DNA (Fig. 6B). Consistent with well-established data (for example, see Izban and Luse 1991; Kireeva et al. 2002), the downstream nucleosome was observed to inhibit Pol II elongation, and, as a control, the nucleosome-mediated inhibition was lost upon dissociation of the histones at 1000 mM KCl (Supplemental Fig. S8B). Nucleosome-specific pause sites were seen at −80, −73, and −20 nucleotides (nt) relative to the dyad at position 0. When it is considered that the distance from the leading edge of Pol II to the active site is ∼15 nt (Samkurashvili and Luse 1996; Kettenberger et al. 2004), the −80 pause site roughly corresponds to the site of Pol II entry into the nucleosome, and the −20 pause site corresponds to the approach of Pol II to the dyad. These pause sites have also been observed in Drosophila cells (Weber et al. 2014).
Upon addition of hNDF, there was a significant reduction in the pausing of Pol II in the nucleosome. The effects were particularly strong at the nucleosome entry (−80) and dyad (−20) pause sites and could be seen at different reaction times (Fig. 6B) and hNDF concentrations (Supplemental Fig. S8C) as well as in the scanning profiles of the elongation products (Supplemental Fig. S8D). Moreover, in the presence of hNDF, there is a substantial amount of transcription through the nucleosome at the 8- and 16-min reaction times (Fig. 6B; Supplemental Fig. S8D). In addition, NDF reduces Pol II pausing through the naked DNA template (Fig. 6B); hence, it is possible that hNDF has a functional interaction with Pol II that is distinct from its ability to destabilize nucleosomes. We also tested dNDF in the Pol II elongation assay and observed that dNDF, like hNDF, is able to facilitate Pol II transcription through a nucleosome (Supplemental Fig. S8E). These data reveal that both hNDF and dNDF can facilitate Pol II elongation through a nucleosome.
We next examined whether NDF affects Pol II elongation in cells. To this end, we carried out nuclear run-on experiments with wild-type and NDF knockout cells. These experiments showed that the loss of NDF results in a decrease in Pol II elongation in nuclei (Supplemental Fig. S9A). Importantly, when the histones were stripped from the DNA with 1% (w/v) sarkosyl (Green et al. 1975; Scheer 1978), we observed roughly the same amount of Pol II elongation in the wild-type cell nuclei as in the knockout cell nuclei (Supplemental Fig. S9B). Hence, the reduced levels of transcription in the NDF knockout cell nuclei appear to be due to blockage by nucleosomes. In addition, the transcription was sensitive to 4 µg/mL α-amanitin and was thus mediated by Pol II (Supplemental Fig. S9). These findings therefore indicate that NDF facilitates Pol II transcription through nucleosomes in vitro as well as in cell nuclei.
NDF interacts with nucleosomes in the vicinity of the dyad
To gain a better understanding of the function of NDF in nucleosome destabilization and transcription elongation in chromatin, we examined the interaction of NDF with naked DNA and nucleosomes. First, we performed gel mobility shift analyses. (In these studies, we used dNDF instead of hNDF because the hNDF-DNA and hNDF-mononucleosome species were not stable under the gel electrophoresis conditions.) These experiments revealed that NDF binds to 147-bp 5S rDNA sequences as either naked DNA or mononucleosomes (Fig. 7A). To test for any potential sequence specificity in binding, we carried out identical experiments with a sequence from a Drosophila hsp70 promoter and obtained essentially the same results (Supplemental Fig. S10A). Thus, NDF binds to naked DNA as well as to nucleosomes. In addition, the binding of NDF to 147-bp DNA-containing mononucleosomes suggests that the nucleosomal binding does not require linker DNA.
Figure 7.
NDF interacts with nucleosomes near the dyad. (A) NDF binds to mononucleosomes as well as to free DNA. Gel mobility shift assays were performed with the indicated concentrations of dNDF and 50 nM 5S rDNA (147 bp) as either naked DNA or mononucleosomes. The resulting species were separated by 4% nondenaturing polyacrylamide gel electrophoresis and visualized by staining with ethidium bromide. The positions of the shifted complexes are indicated. (B) DNase I footprinting analysis reveals an interaction of NDF near the nucleosome dyad. DNase I footprinting reactions were carried out with the forward strand of the 5S rDNA sequence (183 bp) as either naked DNA or mononucleosomes. Purified hNDF (73 or 150 nM concentration) was included where indicated. The Maxam-Gilbert A sequencing ladder was used to map the locations of the DNase I digestion products. The pink dot shows an NDF-protected nucleotide at +2 relative to the nucleosomal dyad at position 0. The blue squares designate bands that are enhanced in the presence of hNDF. The forward strand has the same sense as the template strand of the 5S rRNA gene. (C) Comparison of the effects of NDF and the H4R45A Sin mutation on Pol II elongation through a nucleosome. Transcription elongation experiments were performed as in Figure 6 with either wild-type histone H4 or mutant histone H4R45A. The red arrow denotes the −20 Pol II pause site that is lost upon mutation of H4R45. The black dot shows the location of a pause site downstream from the −20 position that accumulates with H4R45A nucleosomes. The asterisk corresponds to a pause site that likely derives from the presence of some downstream naked DNA, as in the left panel of Figure 6B. (D) Model for the function of NDF.
Next, to investigate the binding of NDF to DNA and nucleosomes at higher resolution, we performed DNase I footprinting experiments. With naked DNA, neither hNDF nor dNDF exhibited sequence-specific interactions (Fig. 7B; Supplemental Figs. S10B, S11A,B). With mononucleosomes, DNase I digestion yielded the characteristic 10-bp digestion ladder (Noll 1974), and both hNDF and dNDF were observed to protect DNA in the vicinity of the dyad (Fig. 7B; Supplemental Figs. S10B, S11). Moreover, in the forward strand (i.e., the nontemplate strand in the transcription experiments), we observed DNase I hypersensitivity in the flanking ends of the nucleosomal DNA (Fig. 7B; Supplemental Fig. S11A,C). This hypersensitivity may reflect the destabilization of the nucleosomes by NDF and is consistent with the ability of NDF to enhance p300-mediated acetylation of H3K56, which is normally protected by nucleosomal DNA. Thus, the DNase I footprinting experiments revealed an interaction of NDF at the dyad and an alteration in histone–DNA contacts at the flanking nucleosomal DNA.
The interaction of NDF at the nucleosome dyad led us to ask whether the effects of NDF on nucleosomal transcription are similar to those of Sin (SWI-independent) mutations in histone H4. The SIN genes were identified in yeast on the basis of their ability to provide partial relief of the requirement of SWI genes for HO transcription (Sternberg et al. 1987). It was later found that some mutant alleles of the histone H4 gene have a Sin− phenotype (Kruger et al. 1995). We were specifically interested in the histone H4R45A Sin mutation. H4R45 is located near the nucleosome dyad. The H4R45A mutation results in a Sin− phenotype (Hodges et al. 2015) but has only a minimal effect on nucleosome structure (Muthurajan et al. 2004). H4R45A and the related H4R45C mutation reduce Pol II pausing on nucleosomal templates in vitro (Hsieh et al. 2010; Bintu et al. 2012). In addition, the H4R45H Sin mutation causes an increase in nuclease sensitivity as well as a reduction in nucleosome-mediated DNA supercoiling (Wechser et al. 1997). Somewhat strikingly, the effects of the H4R45 Sin mutations (i.e., reduction of Pol II pausing on nucleosomes, loss of DNA supercoiling, and increased nuclease sensitivity) are similar to the effects of NDF on chromatin.
We therefore generated chromatin with H4R45A and tested the effect of the R45A mutation on Pol II elongation (Fig. 7C). In this manner, we could compare the effects of NDF-mediated nucleosome destabilization with the effects of the H4R45A mutation. These experiments revealed that the H4R45A mutation resulted in the loss of the −20 dyad pause site and an accumulation of a downstream pause site but did not affect the −80 (nucleosome entry) or −73 pause sites. The addition of NDF facilitated Pol II transcription through the −80, −73, and other pause sites. These findings indicate that the H4R45A mutation acts specifically at the nucleosomal dyad, whereas NDF functions throughout the entire nucleosome.
Discussion
In this study, we describe the properties of NDF, a nucleosome-destabilizing factor that facilitates Pol II transcription through nucleosomes. We initially identified and purified NDF on the basis of its ability to facilitate the acetylation of nucleosomal histone H3K56 by the p300 acetyltransferase (Fig. 1). Both hNDF and dNDF can mediate the partial disassembly of nucleosomes, as detected by the loss of DNA supercoiling in circular chromatin templates (Fig. 2D; Supplemental Fig. S2D,E). hNDF is localized over gene bodies (Figs. 3D,E, 4A,B; Supplemental Fig. S4A). These results are consistent with the properties of epitope-tagged dNDF (CG4747) (Wang et al. 2013). Clustering analysis revealed that active genes can be subdivided into a cluster that is enriched in NDF and a cluster that is not enriched in NDF (Fig. 4A,B; Supplemental Fig. S4C). Thus, NDF is a nucleosome-destabilizing factor that is present in the transcribed regions of a subset of active genes.
Importantly, upon induction of transcription by KLA in mouse primary BMDMs, we found that NDF is recruited to gene bodies and that this recruitment is accompanied by an increase in the transcript levels of many of the NDF-enriched genes (Fig. 5). We also observed that the global loss of hNDF results in a decrease in the RNA levels of many genes (Fig. 4C; Supplemental Fig. S4E). NDF may thus have a direct role in gene activation. Intriguingly, NDF is preferentially associated with longer genes rather than shorter genes (Fig. 3F; Supplemental Fig. S4D). In this regard, it may be more advantageous for NDF to be located at longer genes than at shorter genes because more nucleosomes would have to be disassembled during transcription of longer genes than during transcription of shorter genes. Moreover, if there is some three-dimensional clustering of actively transcribed genes, then there may be a higher local concentration of NDF at longer transcription units than at shorter transcription units.
We directly tested whether NDF affects Pol II elongation through nucleosomes and found that hNDF and dNDF can relieve nucleosome-mediated Pol II pausing, such as at the nucleosome entry site and at the dyad (Fig. 6A,B; Supplemental Fig. S8). Moreover, in the presence of NDF, there is efficient Pol II elongation through the nucleosome (Fig. 6B; Supplemental Fig. S8D). We then performed nuclear run-on experiments to test whether NDF contributes to Pol II elongation in cells and found that NDF is important for Pol II elongation through chromatin in cell nuclei (Supplemental Fig. S9).
To gain further insight into this activity, we examined the interaction of NDF with chromatin and found that both hNDF and dNDF interact with nucleosomes in the vicinity of the dyad (Fig. 7A,B; Supplemental Figs. S10, S11). Comparison of the effects of NDF with those of the H4R45A Sin mutation revealed that H4R45A affects transcription at the dyad, whereas NDF affects transcription throughout the nucleosome (Fig. 7C). Altogether, these data indicate that NDF is a nucleosome-binding and -destabilizing factor that is recruited to gene bodies upon induction of transcription and facilitates Pol II elongation in chromatin (Fig. 7D).
NDF is present in most animals and perhaps in plants but does not appear to have a homolog in Saccharomyces cerevisiae. NDF contains a conserved N-terminal PWWP motif and a conserved C-terminal dehydrogenase domain (Fig. 2A; Supplemental Fig. S1B), but hNDF does not exhibit dehydrogenase activity (Vermeulen et al. 2010). The PWWP methyl lysine-binding domain (Qin and Min 2014) is likely to be involved in the binding of NDF to H3K36me3. However, it appears that H3K36me3 is necessary but not sufficient for the recruitment of NDF to chromatin. The proper localization of dNDF in chromatin is dependent on the Set2 histone H3K36 methyltransferase (Wang et al. 2013); however, many genes with moderate to high levels of H3K36me3 are not enriched in NDF (Fig. 4A,B; Supplemental Fig. S4C), and the sharp increase in NDF occupancy at gene bodies upon transcriptional induction is not mirrored by a similarly sharp increase in H3K36me3 (Fig. 5A,B). Thus, H3K36me3 does not appear to be sufficient for the recruitment of NDF. It is also notable that the biochemical activities of NDF, such as the destabilization of nucleosomes and relief of nucleosome-mediated Pol II pausing, do not absolutely require H3K36 methylation, as our experiments were performed with nucleosomes that were reconstituted with unmodified bacterially synthesized histones. These findings suggest that, once recruited to its site of action, NDF can destabilize nucleosomes and facilitate transcription by a mechanism that is not strictly dependent on H3K36me3.
Because NDF relieves nucleosome-mediated inhibition of Pol II elongation, it is essential to compare NDF with FACT, the well-studied factor that facilitates transcription and DNA replication in chromatin (for example, see Reinberg and Sims 2006; Formosa 2008). FACT is present in many eukaryotes, including S. cerevisiae, Drosophila, and humans. FACT and NDF are not related in terms of their primary amino acid sequences or protein domains. Moreover, FACT binds to the histones and exhibits core histone chaperone activity, whereas NDF does not appear to be a core histone chaperone (data not shown). In yeast, the Spt16 subunit of FACT is enriched over gene bodies and depleted over promoter regions of active genes (True et al. 2016). In contrast, both subunits of mouse FACT were found to be strongly enriched in the vicinity of transcription start sites but also present over gene bodies (Mylonas and Tessarz 2018). These findings suggest that there are substantial differences in the biological functions of FACT in yeast versus mice. The localization of mNDF and hNDF (Figs. 3E, 5B) is more similar to that of yeast FACT than mouse FACT. It is also notable that hNDF was observed to be present at high levels in the nuclei of 44 out of 44 tissue types tested, whereas the levels of the SSRP1 subunit of FACT were found to be low (16 out of 43) or not detectable (nine out of 43) in the same tissue types (Supplemental Fig. S1A; Garcia et al. 2011; Uhlén et al. 2015). In addition, it appears that NDF is essential in human HUES62 stem cells, whereas subunits of FACT are not (Shalem et al. 2014). Thus, in mammals, both NDF and FACT are able to destabilize nucleosomes and facilitate transcription in chromatin, but they function by distinct mechanisms and in different tissues.
It is possible and perhaps likely that there is at least one other factor that is related to NDF. The existence of an NDF-related factor is suggested by the observation that NDF is not essential for the viability of HeLa cells or A375 cells. In addition, the absence of NDF does not result in a general loss of transcription of genes that are enriched in NDF (Fig. 4). These findings could be explained by the presence of an NDF-like factor that is recruited to mostly the same genes as NDF (cluster 1 genes) (Fig. 4), such as via a PWWP motif. This NDF-like factor would then largely compensate for the absence of NDF, especially at genes that are enriched for both factors. The identification and characterization of an NDF-related factor would lead to significant new insights into the transcription process.
Based on the effects of NDF on transcription and gene expression, it is also important to consider whether alterations in the protein sequence or levels of NDF might be relevant to human diseases. In this regard, the NDF/GLYR1 gene is overexpressed in ∼21% of breast cancers (Supplemental Fig. S12A,B; Forbes et al. 2017) and mutated in ∼51% of microsatellite unstable colorectal cancers (Alhopuro et al. 2012). The up-regulation of NDF/GLYR1 also correlates with a reduced survival probability in breast cancer patients (Supplemental Fig. S12C; Uhlen et al. 2017). The knowledge of the biochemical activities of NDF would be useful in assessing the potential roles that this factor may have in breast cancer or other diseases.
In conclusion, the properties of NDF indicate that it has a key role in the transcription of chromatin by Pol II in animals. The further analysis of NDF should reveal additional factors and mechanisms that orchestrate the procession of Pol II through active genes.
Materials and methods
To ensure the reproducibility of the results, each experimental condition was performed independently at least twice.
Antibodies
Rabbit polyclonal antisera against dNDF and hNDF were generated with purified full-length Escherichia coli synthesized proteins at Pocono Rabbit Farm and Laboratory. The following commercial antibodies were used in this study: anti-H3K56ac (1:450 dilution; Millipore Sigma, 07-677I), anti-H3K56ac (1:1000 dilution; Active Motif, 39281), anti-H3K56ac (1:1000 dilution; Cell Signaling Technology, 4243), anti-H3 (1:4000 dilution; Cell Signaling Technology, 4499), anti-H3K9ac (1:1000 dilution; Abcam, ab8898), anti-H3K36me3 (1:1000 dilution; Abcam, ab9050), anti-his (1:1000 dilution; Santa Cruz Biotechnology, sc-803), and anti-Pol II (1:1000 dilution; Santa Cruz Biotechnology, sc-9001).
Purification of native NDF from Drosophila embryos
Native dNDF was purified from the 0.1 M KCl version of the soluble nuclear fraction (Kamakaka et al. 1991) from Drosophila embryos by using the scheme shown in Figure 1B. Buffer N (20 mM HEPES K+ at pH 7.5, 10% [v/v] glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 1.5 mM MgCl2, 0.1 M NaCl, 10 mM ß-glycerophosphate, 1 mM DTT, 0.2 mM PMSF) was used throughout the purification. Proteins in the peak activity fractions of the final Source 15Q chromatography step were identified by mass spectroscopy (Biomolecular/Proteomics Mass Spectrometry Facility, University of California at San Diego).
Purification of recombinant proteins
The cDNAs for dNDF (CG4747; Drosophila Genomics Resource Center) and hNDF (GLYR1; Dharmacon) (note that three mismatches in the cDNA were corrected) were subcloned into pET21b with N-terminal His6 tags to give pET21b-His6-dNDF and pET21b-His6-hNDF, and the integrity of the sequences was confirmed by DNA sequence analysis. The N-terminally His6-tagged dNDF and hNDF proteins were purified by the following method. Freshly transformed E. coli BL21(DE3) was grown in 2 L of LB medium containing 40 µg/mL ampicillin at 37°C to an A600 nm of ∼0.6, and the synthesis of the recombinant NDF was induced by the addition of IPTG to 0.4 mM final concentration. The culture was then incubated for an additional 16–18 h in an 18°C shaking water bath, and the bacteria were collected by centrifugation at 6000 rpm for 10 min at 22°C (Fiberlite F9-4x1000y rotor). Unless stated otherwise, all subsequent operations were performed at 4°C. The bacterial pellets were resuspended in 30 mL of lysis buffer (10 mM Tris-HCl at pH 7.5, 0.2% [v/v] nonidet P-40, 1 M NaCl, 10% [v/v] glycerol, 15 mM imidazole, 5 mM 2-mercaptoethanol, 0.2 mM PMSF, 1× protease inhibitor cocktail [Millipore Sigma, P9599]). The suspension was subjected to sonication on ice four times for 30 sec (Branson Sonifier 450 with a 0.25-in microtip at 20% output). Insoluble material was removed by centrifugation at 13,000 rpm for 30 min (Fiberlite F21S-8x50y rotor), and the supernatant was incubated with 3 mL (pre-equilibrated in lysis buffer) of Ni-NTA Superflow (Qiagen) for 3 h on a nutator. The mixture was transferred into a Bio-Rad Econo-Pac chromatography column, and the flowthrough, which contained ∼80% of the His6-NDF proteins, was collected and incubated with 2 mL (pre-equilibrated in lysis buffer) of Ni-NTA agarose (Qiagen) for 2 h on a nutator. The mixture was transferred into a Bio-Rad Econo-Pac chromatography column, and the resin was washed twice with 10 mL of buffer A (10 mM Tris-HCl at pH 7.5, 0.2% [v/v] nonidet P-40, 10% [v/v] glycerol, 15 mM imidazole, 5 mM 2-mercaptoethanol) containing 1 M NaCl followed by twice with 10 mL of buffer A containing 0.2 M NaCl. The His6-NDF was eluted four times with 0.5 mL of elution buffer (10 mM Tris-HCl at pH 7.5, 0.2% [v/v] nonidet P-40, 0.2 M NaCl, 10% [v/v] glycerol, 250 mM imidazole, 5 mM 2-mercaptoethanol). With His6-dNDF, it was necessary to remove a contaminating nuclease by dialysis into buffer A containing 0.1 M NaCl followed by chromatography on a 1-mL Source 15Q (GE Life Sciences) anion exchange column with linear gradient elution from 0.1 M to 1.0 M NaCl. The His6-dNDF eluted at ∼0.16 M NaCl. The purified NDF proteins were frozen in liquid nitrogen and stored at −80°C.
Human p300 protein was synthesized in Sf9 cells and purified as described in Kraus and Kadonaga (1998). Recombinant Drosophila melanogaster core histones were synthesized in E. coli and purified by the method of Khuong et al. (2017). The catalytic domain of Drosophila topoisomerase I was synthesized and purified as described by Fyodorov and Kadonaga (2003). The S. cerevisiae strain that expresses protein A-tagged Rbp3 subunit of Pol II was the generous gift of Dr. Craig Kaplan (Texas A&M University), and the Pol II was purified as described (Kaplan et al. 2008).
Nucleosome reconstitution
The X. borealis 5S rDNA and Drosophila hsp70 (87A) nucleosome positioning sequences were amplified by PCR and purified by 1.5% agarose gel electrophoresis and gel extraction (QiaQuick gel extraction kit, Qiagen). The resulting fragments were then reconstituted into mononucleosomes by salt dialysis (Stein 1989). In the transcription elongation studies, the PCR-amplified DNA was digested with TspRI restriction enzyme (New England Biolabs) for 2 h at 65°C prior to agarose gel electrophoresis, gel extraction, and mononucleosome reconstitution. Nucleosomes were reconstituted onto CsCl-purified pGIE-0 plasmid DNA (Pazin et al. 1994) by salt dialysis (Stein 1989; Fei et al. 2015) at a core histone to DNA mass ratio of 0.8:1.
H3K56 acetylation assay
The H3K56 acetylation assays were performed with either nucleosomes reconstituted onto plasmid DNA by salt dialysis (1.4 µM concentration of nucleosomes) or free core histones (1.4 µM concentration of core histone octamers) along with 1 µg of purified human p300, 30 µM acetyl-CoA, and 1.4 µg of bovine serum albumin in buffer K (25 mM HEPES K+ at pH 7.6, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% [v/v] glycerol) in a total volume of 50 µL. The mixture was incubated for 1 h at 30°C. The histones were precipitated with 25% (w/v) trichloroacetic acid, subjected to 15% polyacrylamide–SDS gel electrophoresis, and detected by Western blot analysis.
Transcription elongation assay
The transcription elongation assay is depicted in Figure 6A and is a modified version of the method of Kireeva et al. (2002). In our experiments, the 10-nt 5′ 32P-labeled primer RNA (1.6 pmol = 8 µL of a 0.2 µM stock) was annealed to an equimolar amount of template strand DNA. Purified Pol II (4 pmol) was then added to 1.6 pmol of the RNA–template strand complex, and the components were incubated for 10 min at 30°C. Next, 1.6 pmol of the 5′ biotin-labeled nontemplate strand was added and incubated for 15 min at 30°C to form the elongation complex, which was then immobilized onto 10 µL of Dynabeads MyOne Streptavidin C1 magnetic beads (Invitrogen). The beads were washed twice with 50 µL of EB1000 buffer (20 mM Tris-HCl at pH 7.5, 1 M KCl, 5 mM MgCl2, 10 mM DTT) followed by twice with 50 µL of EB40 buffer (20 mM Tris-HCl at pH 7.5, 40 mM KCl, 5 mM MgCl2, 10 mM DTT) and then resuspended in 16 µL of EB40 buffer. The immobilization efficiency of the elongation complex was estimated with a Geiger counter. Typically, ∼60% of the elongation complex was immobilized onto the Dynabeads. Next, 3 pmol of either mononucleosomes or the corresponding naked DNA fragments (both with complementary TspRI overhangs) was ligated to the immobilized elongation complex by using 2000 U of T4 DNA ligase (New England Biolabs) in 0.2× (final concentration) T4 DNA ligase reaction buffer (New England Biolabs) in a total volume of 50 µL for 25 min at 22°C. After ligation, the beads were collected and resuspended in 50 µL of EB40 buffer containing 20 µg of bovine serum albumin (Thermo Fisher Pierce), 100 U of SUPERase•In RNase inhibitor (Thermo Fisher Invitrogen), and 30 pmol of unlabeled free mononucleosomes on 147-bp DNA. The unlabeled free mononucleosomes were added only to the elongation complex templates that were ligated to a downstream nucleosome and served to stabilize the template-associated nucleosomes by increasing the overall concentration of nucleosomes in the reaction medium (for example, see Godde and Wolffe 1995). The beads were then aliquoted into tubes for the individual reactions. Each reaction contained 5 µL of the bead-containing suspension. If the ligation efficiency were 100%, then each tube would contain ∼0.08 pmol of elongation complex template. For nucleosome-containing templates, each tube additionally contained 3 pmol of unlabeled free mononucleosomes to stabilize the template-associated nucleosomes. Next, NDF or the corresponding buffer (as a negative control) was added to each tube, EB40 buffer was added to a final volume of 9 µL, and the samples were incubated for 20 min at 30°C. At this stage, the KCl concentration was 100 mM unless stated otherwise. Next, transcription elongation was initiated by the addition of 1 µL of 2.5 mM rNTPs, and the reactions were allowed to proceed for the indicated times at 22°C. The reactions were terminated with 10 µL of gel loading buffer (1 mL of gel loading buffer = 0.9 mL of deionized formamide, 0.1 mL of 0.5 M EDTA, 2 µL of 4% [w/v] bromphenol blue). The samples were analyzed by denaturing 8% polyacrylamide–urea gel electrophoresis, and the gel images were collected on a GE Typhoon imager (GE Healthcare).
Analysis of mNDF in primary BMDMs
Bone marrow was isolated from 8- to-10-wk-old C57Bl/6 (Harlan Laboratories) female mice. BMDMs were generated in DMEM supplemented with M-CSF for 7 d and activated with KLA for 6 h as described (Kaikkonen et al. 2013). RNA-seq libraries were prepared from poly(A)-enriched mRNA as described previously (Kaikkonen et al. 2013). For ChIP-seq, 5 million BMDMs were cross-linked in 1% formaldehyde for 10 min. ChIP-seq was performed as described previously (Kaikkonen et al. 2013) with 2 µL of hNDF antisera 7 or 2 µL of H3K36me3 antibodies (Abcam, ab9050). (We also performed ChIP-seq experiments with hNDF antisera 6 and obtained results essentially identical to those with hNDF antisera 7 [data not shown].) ChIP-seq libraries were prepared from ChIP DNA and inputs by blunting, A-tailing, and adapter ligation as described previously (Heinz et al. 2010) by using NextFlex barcode adapters. Libraries were amplified, size-selected by gel extraction, and subsequently sequenced on the Illumina Hi-Seq 4000 sequencer by using single-end 75-bp reads. Sequencing data were aligned to the mm10 genome by using Bowtie2, and all data were subsequently analyzed with HOMER and visualized in the University of California at Santa Cruz genome browser. Pathway analysis was performed by using Metascape (http://metascape.org).
Additional Materials and Methods are included in the Supplemental Material.
The genome-wide data have been deposited at the Gene Expression Omnibus (accession no. GSE109690).
Supplementary Material
Acknowledgments
We thank Peter Geiduschek, Grisel Cruz, and Long Vo ngoc for critical reading of the manuscript. We are grateful to Craig Kaplan for the tagged yeast Pol II strain, Gaoyang Liang for CRISPR–Cas9 vectors, and Dong Wang and Jun Xu for their invaluable advice on the elongation assay and Pol II purification. We also thank Yun-Ching Chen for help in the analysis of the RNA-seq data. J.T.K. is the Amylin Chair in the Life Sciences. F.M. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (ME 4713/1-1) and a National Institutes of Health (NIH) grant to Karen Oegema (GM074207). This work was supported by funding from the Ludwig Institute for Cancer Research to B.R., the NIH/National Institute of Diabetes and Digestive and Kidney Diseases (DK091183) and Foundation Le Ducq to C.K.G, and the NIH/National Institute of General Medical Sciences (R35 GM118060) to J.T.K.
Author contributions: This work was carried out in an interactive manner in which all authors made important intellectual contributions to the development of the project. J.F. identified and purified NDF, performed essentially all of the biochemical analyses of the protein, and generated the knockout cell lines. H.I. and B.R. contributed the genome-wide analyses of NDF in human cells. M.A.H. and C.K.G. contributed the analyses of NDF in mouse cells, including the KLA induction experiments. H.I. had a lead role in the analysis of the genome-wide data. F.M. performed the cell cycle studies. G.A.K. contributed to the design and analysis of the transcription and footprinting experiments. J.F., G.A.K., and J.T.K. oversaw the overall execution of this work. J.F. and J.T.K. were primarily responsible for writing the manuscript and received substantial contributions from all other authors in the preparation of figures and the writing and editing of the text.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.313973.118.
References
- Alhopuro P, Sammalkorpi H, Niittymäki I, Biström M, Raitila A, Saharinen J, Nousiainen K, Lehtonen HJ, Heliövaara E, Puhakka J, et al. 2012. Candidate driver genes in microsatellite-unstable colorectal cancer. Int J Cancer 130: 1558–1566. [DOI] [PubMed] [Google Scholar]
- Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. 2012. Nucleosomal elements that control the topography of the barrier to transcription. Cell 151: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Chen F, Dong Z, Hu D, Barbera AJ, Clark EA, Fang J, Yang Y, Mei P, Rutenberg M, et al. 2013. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol Cell 49: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Torigoe SE, Brown CR, Khuong MT, Kassavetis GA, Boeger H, Kadonaga JT. 2015. The prenucleosome, a stable conformational isomer of the nucleosome. Genes Dev 29: 2563–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Luger K, Tan S, Richmond TJ. 1996. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci 93: 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. 2008. FACT and the reorganized nucleosome. Mol Biosyst 4: 1085–1093. [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT. 2003. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol 371: 499–515. [DOI] [PubMed] [Google Scholar]
- Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, Omelian A, Morrison C, Gurova K. 2011. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond JE, Hirt B, Oudet P, Gross-Bellard M, Chambon P. 1975. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci 72: 1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde JS, Wolffe AP. 1995. Disruption of reconstituted nucleosomes. The effect of particle concentration, MgCl2 and KCl concentration, the histone tails, and temperature. J Biol Chem 270: 27399–27402. [DOI] [PubMed] [Google Scholar]
- Green MH, Buss J, Gariglio P. 1975. Activation of nuclear RNA polymerase by Sarkosyl. Eur J Biochem 53: 217–225. [Google Scholar]
- Hayes JJ, Tullius TD, Wolffe AP. 1990. The structure of DNA in a nucleosome. Proc Natl Acad Sci 87: 7405–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, YC Lin, Lasko P, JX Cheng, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges AJ, Gallegos IJ, Laughery MF, Meas R, Tran L, Wyrick JJ. 2015. Histone sprocket arginine residues are important for gene expression, DNA repair, and cell viability in Saccharomyces cerevisiae. Genetics 200: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. 2010. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep 11: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. 1991. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 5: 683–696. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Tyree CM, Kadonaga JT. 1991. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc Natl Acad Sci 88: 1024–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Larsson KM, Kornberg RD. 2008. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol Cell 30: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16: 955–965. [DOI] [PubMed] [Google Scholar]
- Khuong MT, Fei J, Ishii H, Kadonaga JT. 2015. Prenucleosomes and active chromatin. Cold Spring Harbor Symp Quant Biol 80: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong MT, Fei J, Cruz-Becerra G, Kadonaga JT. 2017. A simple and versatile system for the ATP-dependent assembly of chromatin. J Biol Chem 292: 19478–19490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9: 541–552. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev 12: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 9: 2770–2779. [DOI] [PubMed] [Google Scholar]
- Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. 2004. Crystal structures of histone Sin mutant nucleosomes reveal altered protein–DNA interactions. EMBO J 23: 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas C, Tessarz P. 2018. Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells. bioRxiv 10.1101/251611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. 1974. Internal structure of the chromatin subunit. Nucleic Acids Res 1: 1573–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, Kolar MJ, Matsuzaka T, Hayakawa S, Tao J, et al. 2017. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab 25: 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT. 1994. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266: 2007–2011. [DOI] [PubMed] [Google Scholar]
- Qin S, Min J. 2014. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci 39: 536–547. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, et al. 2006. Kdo2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res 47: 1097–1111. [DOI] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ III. 2006. deFACTo nucleosome dynamics. J Biol Chem 281: 23297–23301. [DOI] [PubMed] [Google Scholar]
- Rhodes D. 1985. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J 4: 3473–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samkurashvili I, Luse DS. 1996. Translocation and transcriptional arrest during transcript elongation by RNA polymerase II. J Biol Chem 271: 23495–23505. [DOI] [PubMed] [Google Scholar]
- Scheer U. 1978. Changes of nucleosome frequency in nucleolar and non-nucleolar chromatin as a function of transcription: an electron microscopic study. Cell 13: 535–549. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A. 1989. Reconstitution of chromatin from purified components. Methods Enzymol 170: 585–603. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Stern MJ, Clark I, Herskowitz I. 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48: 567–577. [DOI] [PubMed] [Google Scholar]
- Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. 2011. Identification of a rapidly formed non-nucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol Cell 43: 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigoe SE, Patel A, Khuong MT, Bowman GD, Kadonaga JT. 2013. ATP-dependent chromatin assembly is functionally distinct from chromatin remodeling. eLife 2: e00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JD, Muldoon JJ, Carver MN, Poorey K, Shetty SJ, Bekiranov S, Auble DT. 2016. The modifier of transcription 1 (Mot1) ATPase and Spt16 histone chaperone co-regulate transcription through preinitiation complex assembly and nucleosome organization. J Biol Chem 291: 15307–15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. 2017. A pathology atlas of the human cancer transcriptome. Science 357: eaan2507. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980. [DOI] [PubMed] [Google Scholar]
- Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton LM, Elledge SJ, Kharchenko PV, Garcia BA, Kuroda MI. 2013. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol 20: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. 2014. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell 53: 819–830. [DOI] [PubMed] [Google Scholar]
- Wechser MA, Kladde MP, Alfieri JA, Peterson CL. 1997. Effects of Sin– versions of histone H4 on yeast chromatin structure and function. EMBO J 16: 2086–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.