This Outlook by Wan and Hopper discusses the study on circular RNAs (circRNAs) in this issue of Genes & Development by Huang et al. There, they show that nuclear export of circRNAs is length-dependent, and the length measurement mechanism is shown to be controlled by the four-amino-acid motifs in Hel25E and its homologs.
Keywords: circRNA, Hel25E, UAP56, URH49, DDX39A, DDX39B
Abstract
Circular RNAs (circRNAs) comprise a recently appreciated category of RNAs that are in high abundance and serve important biological functions. Although several discoveries have been made regarding the biogenesis and functions of circRNAs, their subcellular trafficking has remained largely unknown. In this issue of Genes & Development, Huang and colleagues (pp. 639–644) reported the first study of the nuclear export of circRNAs. Drosophila Hel25E and its human homologs, UAP56 and URH49, are required for nuclear export of circRNAs. Nuclear export of circRNAs is surprisingly length-dependent, and the length measurement mechanism was shown to be controlled by motifs in Hel25E and its homologs consisting of four amino acids.
Circular RNAs (circRNAs) were discovered >40 years ago, but their biogenesis, subcellular trafficking, and biological functions are still under investigation. circRNAs are generated in the nucleus from many protein-encoding genes when a splice donor is joined with an upstream splice acceptor via “backsplicing” (Ebbesen et al. 2016). circRNA levels are regulated by several factors, including RNA-binding proteins, exon-skipping events, the presence of complementary sequences of flanking introns, and the availability of splicing machinery components (Wilusz 2018). Certain circRNAs accumulate to higher levels than their associated linear mRNAs due to inherent stability (Werfel et al. 2016). Although some circRNAs may function in the nucleus, most circRNAs localize to the cytoplasm (Wilusz 2018). The biological roles of most circRNAs await discovery, but some circRNAs function as sponges for microRNAs (Hansen et al. 2013; Memczak et al. 2013), and some function in regulation of transcription and alternative splicing of the genes from which they were derived (Wilusz 2018), whereas others serve as templates for translation (Tatomer and Wilusz 2017). In addition, aberrant expression of circRNAs has been implicated in promoting cancer (Wilusz 2017), and the cell type and tissue specificity of circRNA expression suggest their involvement in development (Barrett and Salzman 2016).
Many circRNAs are expressed in nondividing cells (e.g., neurons); thus, nuclear export of circRNAs must occur by an active process rather than diffusion after nuclear envelope breakdown during mitosis. In this issue of Genes & Development, Huang et al. (2018) explored the unknown process for nuclear export of circRNAs.
To discover factors involved in nuclear export of circRNAs, Huang et al. (2018) screened 26 candidate proteins with known functions in RNA nuclear export using RNAi knockdowns in Drosophila DL1 cells. They discovered that the DExH/D-box helicase Hel25E, known to be required for nuclear export of mRNAs, is necessary and sufficient for nuclear export of some, but not all, tested circRNAs. Depletion of Hel25E resulted in nuclear accumulation of both nascent and steady-state long circRNA circdati (1120 nucleotides [nt]), but not short circRNA circlaccase2 (490 nt), both of which predominantly localize to the cytoplasm in wild-type Drosophila DL1 cells (Fig. 1). They also showed that nuclear accumulation is not due to overall changes in the total circRNA levels.
Figure 1.
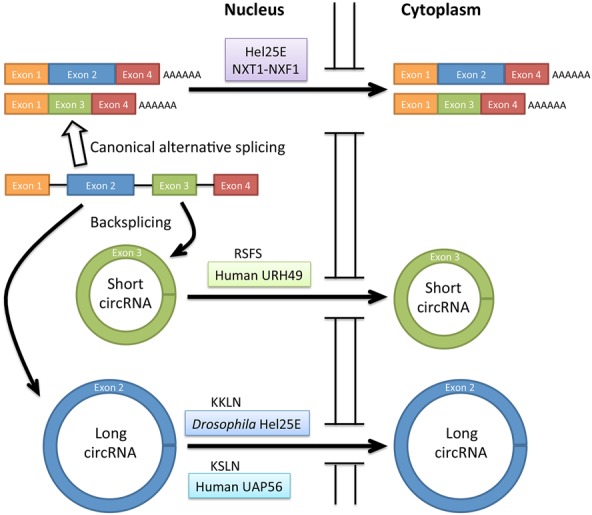
Nuclear export of short and long circRNAs. Short and long circRNAs are generated by backsplicing of pre-mRNA. In human cells, the nuclear export of short circRNA (<400 nt) requires URH49 with the four-amino-acid motif RSFS. The nuclear export of long circRNA requires Drosophila Hel25E with the four-amino-acid motif KKLN for circRNA >800 nt or human UAP56 with four-amino-acid motif KSLN for circRNA >1200 nt.
Testing the possibility that Hel25E may have a size preference for circRNAs, Huang et al. (2018) studied 12 additional endogenous circRNAs of different lengths and exon numbers. Depletion of Hel25E caused nuclear retention of long (>811-nt) but not short (<702-nt) circRNAs. The investigators further explored this putative size-dependent nuclear export mechanism for circRNAs by generating plasmid-encoded mature circRNAs derived from the firefly luciferase gene (circfirefly). Interestingly, nuclear export dependency of the circfirefly RNAs on Hel25E increased concomitantly with the lengths of the circfirefly (from 900 nt to 1100 nt to 1677 nt). The results mirror the observation of Hel25E depletion on the nuclear accumulation of endogenous circRNAs, suggesting that a gradient effect of length may exist for the nuclear export of circRNAs.
Huang et al. (2018) extended their studies to human cells. The investigators showed that the two human homologs of Hel25E (UAP56 [DDX39B] and URH49 [DDX39A]), which are involved in nuclear export of human mRNAs, also function in nuclear export of circRNAs. The results are consistent with a previous study showing that UAP56 and URH49 bind circRNAs in human cells (Chen et al. 2017). However, UAP56 functions in nuclear export of long circRNAs (>1298 nt), whereas URH49 functions in nuclear export of short circRNAs (<356 nt). The nuclear export of circRNAs between 411 and 1099 nt seems to be complicated, as nuclear accumulation of these circRNAs does not correlate with the depletion of UAP56 or URH49, perhaps due to secondary structures of the circRNAs.
To study how Hel25E and its human homologs selectively regulate nuclear export of circRNAs, Huang et al. (2018) tested whether Hel25E mutant constructs lacking ATP binding or helicase activity could rescue circRNA nuclear export defects in DL1 cells depleted of endogenous Hel25E. Surprisingly, these activities are dispensable. To identify amino acids important for circRNA length-dependent nuclear export, the investigators reasoned that the proteins regulating nuclear export of long circRNAs (Hel25E and UAP56) might share a sequence that is absent from the protein (URH49) regulating nuclear export of short circRNAs. The investigators identified a four-amino-acid motif that is conserved between Hel25E and UAP56 but not conserved in URH49. In an elegant experiment using chimeric constructs, they demonstrated that the four-amino-acid motif conserved between Hel25E and UAP56 is both necessary and sufficient to control circRNA length preference for nuclear export of long circRNA.
In summary, the study from Huang et al. (2018) showed that nuclear export of circRNAs is an active process, that proteins conserved from Drosophila (Hel25E) to human (UAP56 and URH49) cells function in nuclear export of circRNAs, and that, unexpectedly, the nuclear export process has a length preference that maps to a four-amino-acid sequence (Fig. 1). The findings are timely, novel, and important and certainly invite more questions for future studies: (1) Are there additional factors for nuclear export of circRNAs? As depletion of Hel25E and homologs does not result in complete nuclear retention of circRNAs, it is likely that additional export factors exist. Nuclear export of circRNAs could be achieved by direct binding of the identified proteins, similar to Exportin-t binding to transfer RNAs (tRNAs) or Exportin-5 binding to microRNAs, or, instead, circRNA nuclear export might be mediated by adapter proteins, similar to the mechanisms for nuclear export of mRNAs. (2) What is the mechanism for circRNA length measurement? It will be interesting to learn whether different factors copurify with the endogenous and chimeric proteins specifying nuclear export of long versus short circRNAs. (3) Are the circRNAs with known functions (microRNA sponges and templates for translation) regulated by similar nuclear export mechanisms? Answers to these questions regarding circRNA subcellular trafficking will also provide insights into the biogenesis and functions of circRNAs.
Acknowledgments
This work was supported by funding from National Institute of Health grant GM122884 to A.K.H., and The Ohio State University Comprehensive Cancer Center Pelotonia Graduate Fellowship to Y.W.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.316216.118.
References
- Barrett SP, Salzman J. 2016. Circular RNAs: analysis, expression and potential functions. Development 143: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. 2017. Sensing self and foreign circular RNAs by intron identity. Mol Cell 67: 228–238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen KK, Kjems J, Hansen TB. 2016. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta - Gene Regul Mech 1859: 163–168. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, Wilusz JE. 2018. A length-dependent, evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev (this issue) 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Tatomer DC, Wilusz JE. 2017. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol Cell 66: 1–2. [DOI] [PubMed] [Google Scholar]
- Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. 2016. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 98: 103–107. [DOI] [PubMed] [Google Scholar]
- Wilusz JE. 2017. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol 14: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2018. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]


