Datta et al. define three major classes of enhancers that are differentially sensitive to binding and transcriptional activation by Bicoid (Bcd) and Orthodenticle (Otd). The specific activities of enhancers in each class are mediated by DNA motif variants preferentially bound by Bcd or Otd and the presence or absence of sites for cofactors that interact with these proteins.
Keywords: Bicoid, Orthodenticle, enhancer, embryo, transcription factor binding, suboptimal binding
Abstract
The K50 (lysine at amino acid position 50) homeodomain (HD) protein Orthodenticle (Otd) is critical for anterior patterning and brain and eye development in most metazoans. In Drosophila melanogaster, another K50HD protein, Bicoid (Bcd), has evolved to replace Otd's ancestral function in embryo patterning. Bcd is distributed as a long-range maternal gradient and activates transcription of a large number of target genes, including otd. Otd and Bcd bind similar DNA sequences in vitro, but how their transcriptional activities are integrated to pattern anterior regions of the embryo is unknown. Here we define three major classes of enhancers that are differentially sensitive to binding and transcriptional activation by Bcd and Otd. Class 1 enhancers are initially activated by Bcd, and activation is transferred to Otd via a feed-forward relay (FFR) that involves sequential binding of the two proteins to the same DNA motif. Class 2 enhancers are activated by Bcd and maintained by an Otd-independent mechanism. Class 3 enhancers are never bound by Bcd, but Otd binds and activates them in a second wave of zygotic transcription. The specific activities of enhancers in each class are mediated by DNA motif variants preferentially bound by Bcd or Otd and the presence or absence of sites for cofactors that interact with these proteins. Our results define specific patterning roles for Bcd and Otd and provide mechanisms for coordinating the precise timing of gene expression patterns during embryonic development.
Animal body plans are established in large part by transcriptional networks that provide positional information during embryogenesis (Peter and Davidson 2015). The evolution of body plans is driven by alterations to these networks, including changes to cis-regulatory elements and neofunctionalization of transcription factors (TFs) after gene duplication (Thornton et al. 2003; Carroll 2008; Peter and Davidson 2011; Tautz and Domazet-Loso 2011; Abascal et al. 2013).
Network interactions involve direct binding of TFs to binding sites in enhancers of target genes, and binding events are integrated into spatial and temporal patterns of expression along the major axes during development. Understanding the mechanisms controlling the dynamics of gene expression remains a challenge, in part because TFs exist in families of closely related proteins and bind very similar sequence motifs in vitro (Noyes et al. 2008b). While possible mechanisms by which TF specificity is achieved (TF-preferred binding sequences, cofactor binding, etc.) have been proposed (Slattery et al. 2011), it is unclear how binding events are coordinated in vivo so that each protein can regulate specific gene targets.
The early Drosophila melanogaster embryo provides a unique system to study the spatiotemporal complexity of TF–DNA interactions. Drosophila and other cyclorrhaphan Diptera develop via a “long germ” mode, in which all segments of the body plan are specified and positioned during the syncytial blastoderm stage (Lynch and Desplan 2003; Liu and Kaufman 2005; Peel 2008) In fruit flies, the maternal TF Bicoid (Bcd) sets up the first positional instructions for anterior patterning (Driever et al. 1990). bcd mRNA is sequestered at the anterior pole of the mature oocyte via sequences in its 3′ untranslated region (UTR) (Berleth et al. 1988). After fertilization, translation and diffusion establish an anterior gradient of Bcd protein (Fig. 1A,B; Driever and Nusslein-Volhard 1988; Little et al. 2011). Bcd contains a homeodomain (HD) and is a transcriptional activator of >50 target genes (Driever and Nusslein-Volhard 1989; Struhl et al. 1989; Finkelstein and Perrimon 1990; Stanojevic et al. 1991; Chen et al. 2012), which form a network of downstream regulatory interactions that precisely patterns the embryo. Embryos lacking Bcd fail to form any anterior structures, including all cephalic and thoracic segments, and show duplications of posterior structures (e.g., Filzkoerper [Fk]) in anterior regions (Frohnhofer and Nusslein-Volhard 1986). While the majority of its target genes is expressed during and after cellularization, the Bcd protein gradient is active only during the syncytial blastoderm stage, prior to cellularization.
Figure 1.
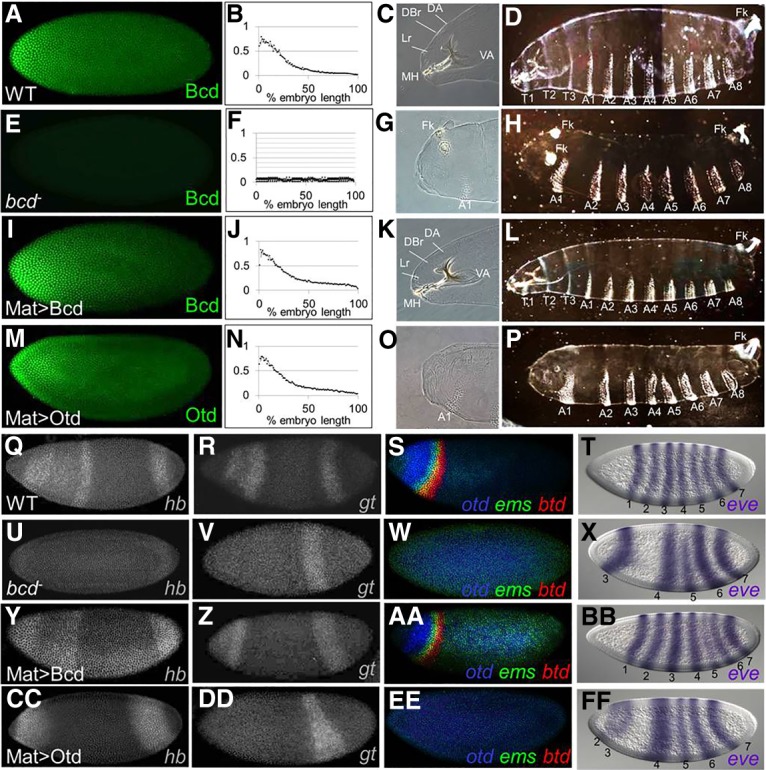
A maternal Otd gradient (Mat > Otd) cannot replace most Bcd-like functions. (A–P) Protein expression patterns (A,E,I,M), gradient quantifications (averaged from five embryos; B,F,J,N), anterior cuticle structures (C,G,K,O), and whole larva cuticles (D,H,L,P) are shown for wild type (A–D), bcd mutants (E–H), bcd mutants containing the Mat > Bcd transgene (I–L), and bcd mutants containing the Mat > Otd transgene (M–P). Labeled structures in anterior regions (C,G,K,O) include the dorsal arm (DA), dorsal bridge (DBr), labrum (Lr), mouthhooks (MH), ventral arm (VA), Filzkoerper (Fk), and first abdominal segment (A1). (D,H,L,P) Thoracic (T1–T3) and abdominal (A1–A8) segments are labeled. (Q–FF) mRNA expression patterns for six Bcd target genes (hunchback [hb], giant [gt], otd, empty spiracles [ems], btd, and even-skipped [eve]) in wild type (Q–T), bcd mutants (U–X), bcd mutants containing the Mat > Bcd transgene (Y–BB), and bcd mutants containing the Mat > Otd transgene (CC–FF). Assayed target genes are labeled in the bottom right corner of each panel. (T,X,BB,FF) Numbers correspond to eve stripes. All embryos in this study are oriented with anterior to the left.
Despite its critical functions in Drosophila, Bcd is not well conserved even within insects. Rather, Bcd arose after a recent gene duplication event and rapidly evolved to play an important role in embryonic patterning (Stauber et al. 1999; Casillas et al. 2006). In three species lacking Bcd (Tribolium castaneum, Nasonia vitripennis, and Acyrthosiphon pisum), at least some of the anterior patterning functions of Bcd are performed by Orthodenticle (Otd) (Schroder 2003; Lynch et al. 2006; Huang et al. 2010). otd is maternally expressed in these species, and its disruption causes severe bcd-like defects in anterior patterning. In Drosophila, otd has evolved to become a zygotic target gene of Bcd-dependent activation (Finkelstein and Perrimon 1990). Loss of Drosophila otd causes embryonic lethality, but otd mutants show cephalic defects that are much less dramatic than the complete loss of anterior structures observed in bcd mutants (Finkelstein and Perrimon 1990; Finkelstein et al. 1990). Later in development, Otd is critical for central nervous system and eye development (Finkelstein et al. 1990)—roles that are conserved in vertebrates, including humans (Finkelstein and Boncinelli 1994).
Both Otd and Bcd contain a lysine at amino acid position 50 (K50) of their respective HDs and bind in vitro to the consensus sequence TAATCC (Treisman et al. 1989; Noyes et al. 2008a). The K50 residue and preference for TAATCC are conserved among all Otd homologs (Finkelstein and Boncinelli 1994) and thus are ancient, but the ancestral protein that gave rise to Bcd was a Hox3 protein with a glutamine (Q) at HD position 50 (Q50) (Stauber et al. 1999). This suggests that an important step in Bcd's evolution was the conversion of Q50 in the ancestor to K50, which changed its DNA-binding preference and allowed it to usurp some of the anterior patterning roles played by Otd in ancestral insects. All known Bcd target gene enhancers contain multiple copies of TAATCC (Chen et al. 2012). We hypothesize that Otd regulated many of these enhancers in the ancestral species that gave rise to Drosophila and regulates a similar battery of enhancers in extant insect species that do not contain Bcd. If this is the case, perhaps Otd can replace many of Bcd's functions in Drosophila if it is maternally expressed and distributed in an anterior gradient.
We tested whether Otd can provide Bcd-like patterning functions through a transgenic gene replacement assay. We present here a comprehensive comparison of the in vitro binding preferences and in vivo binding profiles of Bcd and Otd. We also demonstrate that the two proteins bind sequentially to enhancers in feed-forward relays (FFRs) in which Bcd-binding initiates target gene activation, and Otd-binding maintains expression after the maternal Bcd gradient decays. Each protein also binds independently to distinct enhancers. We present evidence that Otd- and Bcd-specific binding activities in vivo are controlled by two mechanisms: subtle differences in binding preference and protein-specific interactions with cofactors. Our results define specific roles for Bcd and Otd in embryonic patterning in Drosophila and shed light on the molecular mechanisms that alter regulatory networks during evolution.
Results
A maternal gradient of Otd cannot replace Bcd in Drosophila
To assess the functional similarity between Bcd and Otd, we tested whether Otd could mediate Bcd-like activities using a gene replacement assay (Fig. 1). Coding regions for both Bcd and Otd were inserted into transgenes containing the bcd promoter (for maternal expression) and the bcd 3′ UTR (for anterior mRNA localization) and were integrated into the same genomic position (Bateman et al. 2006). These constructs (designated Mat > Bcd and Mat > Otd) were crossed into bcdE1-null mutant females, and embryos laid by those females were assayed for RNA and protein expression. mRNAs from the transgenes were maternally expressed and localized to the anterior pole region (Supplemental Fig. 1A). Antibody stains showed very similar gradients of Bcd and Otd in early embryos containing the transgenes, and the Bcd gradient from the rescue transgene was indistinguishable from the endogenous (wild-type) gradient (Fig. 1A,B,I,J,M,N).
Wild-type embryos develop cephalic structures and three thoracic segments in anterior regions during embryogenesis (Fig. 1C,D). All of these structures are missing in embryos laid by bcd mutant females, and posterior structures (Fk) are duplicated near the anterior pole (Fig. 1G,H). At the molecular level, bcd mutants fail to activate all Bcd target genes, including hunchback (hb), giant (gt), otd, buttonhead, (btd), empty spiracles (ems), and even-skipped (eve) stripes 1 and 2 (eve-1 and eve-2) (Fig. 1U–X, cf. with Q–T), and fail to repress translation of Cad in anterior regions (Supplemental Fig. 1B).
As expected, when the control construct (Mat > Bcd) was crossed into embryos lacking Bcd, it completely rescued all morphological defects (Fig. 1K,L), and 95% of the embryos developed into fertile adults. This construct also activated the expression patterns of all six tested Bcd target genes (Fig. 1Y–BB). In contrast, when the Mat > Otd construct was crossed into bcd mutants, none of the morphological structures missing in bcd mutants structures was rescued (Fig. 1O,P). However, we did detect the suppression of ectopic posterior Fk (Fig. 1O, cf. with G). Mat > Otd activated only two Bcd target genes (hb and eve-2) but failed to activate four others (gt, otd, btd, and ems) (Fig. 1CC–FF) and failed to translationally repress caudaI (Supplemental Fig. 1B). We also tested whether Mat > Bcd and Mat > Otd could activate 24 other Bcd-dependent reporter genes (Chen et al. 2012). The Mat > Bcd construct activated expression of all 24 of these reporters, while Mat > Otd activated only two (Supplemental Fig. 2). Taken together, these results show that Otd cannot rescue most Bcd functions when provided maternally despite the fact that the two proteins bind very similar DNA sequences in vitro.
HDs mediate distinct in vivo activities of Otd and Bcd
The inability of Mat > Otd to activate most Bcd target genes or repress Cad is consistent with its failure to rescue anterior segments in embryos lacking Bcd. This result is not surprising in view of the fact that the evolved Bcd- and Otd-coding regions show very little sequence conservation (38% sequence identity within their HDs) (Fig. 2A) and no detectable homology outside the HD. To map regions of Bcd and Otd that mediate their distinct functions, we generated rescue constructs with chimeric proteins in which the DNA-binding HDs were precisely swapped (Mat > Bcd:OtdHD and Mat > Otd:BcdHD) (Fig. 2B,C,F,G). If the structural differences preventing Otd from rescuing bcd mutants lie inside its HD, inserting the Bcd HD into the Otd protein (Mat > Otd:BcdHD) should cause a strong rescue of the phenotype. If those differences lie outside the HD, inserting the Otd HD into the Bcd protein (Mat > Bcd:OtdHD) should result in a strong rescue.
Figure 2.
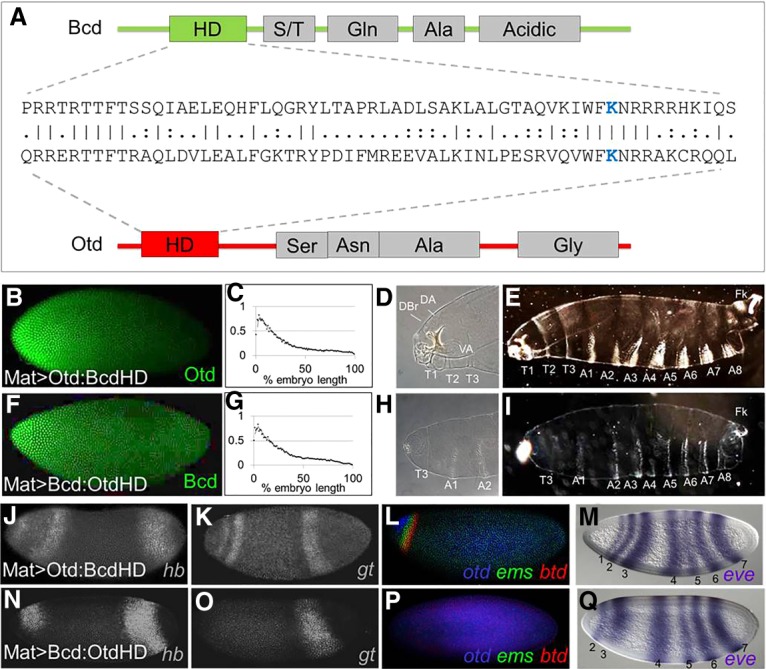
(A) A structural comparison of the Bcd and Otd proteins. Schematic representations show the positions of the HDs and mapped activation domains (gray boxes). An amino acid sequence comparison of the HDs is shown in the middle. Identical amino acids are indicated by vertical lines, and similarities are shown as two dots. The critical lysine at position 50 (K50) is shown in blue. (B–I) Testing HD swap chimeras for Bcd-like activities. (B–I) Protein expression patterns (B,F), gradient quantifications (averaged from five embryos; C,G), anterior cuticle structures (D,E,H,I), and whole larva cuticles (E,I) are shown for bcd mutants containing the Mat > Otd:BcdHD transgene (B–E) and bcd mutants containing the Mat > Bcd:OtdHD transgene (F–I). (D,H) Labeled structures in anterior regions include the dorsal arm (DA), dorsal bridge (DBr), ventral arm (VA), thoracic segments (T1–T3), and two abdominal segments (A1 and A2). (E,I) Thoracic (T1–T3) and abdominal (A1–A8) segments are labeled. (J–Q) mRNA expression patterns for six Bcd target genes (hb, gt, otd, ems, btd, and eve) in bcd mutants containing the Mat > Otd:BcdHD transgene (J–M) and bcd mutants containing the Mat > Bcd:OtdHD transgene (N–Q). (M,Q) Numbers correspond to eve stripes. Assayed target genes are labeled in the bottom right corner of each panel.
Inserting the Bcd HD into the Otd protein (Mat > Otd:BcdHD) caused a dramatic rescue of anterior structures (Fig. 2D,E). Although rescue was incomplete (embryos died before hatching), all embryos containing the Mat > Otd:BcdHD formed three thoracic segments and at least some identifiable cephalic structures (Fig. 2D,E). Mat > Otd:BcdHD also activated all six tested Bcd target genes (Fig. 2J–M) and repressed Cad translation (Supplemental Fig. 1B). In contrast, the reciprocal swap (Mat > Bcd:OtdHD) caused very little rescue of the morphological defects of bcd mutants (Fig. 2H,I) and activated only hb and eve2, similar to rescue by Mat > Otd (Fig. 2N–Q, cf. with 1EE–HH). Together, these results indicate that important functional differences between the two proteins lie within their HDs, and regions outside their HDs are largely interchangeable.
In vivo and in vitro binding activities of Bcd and Otd
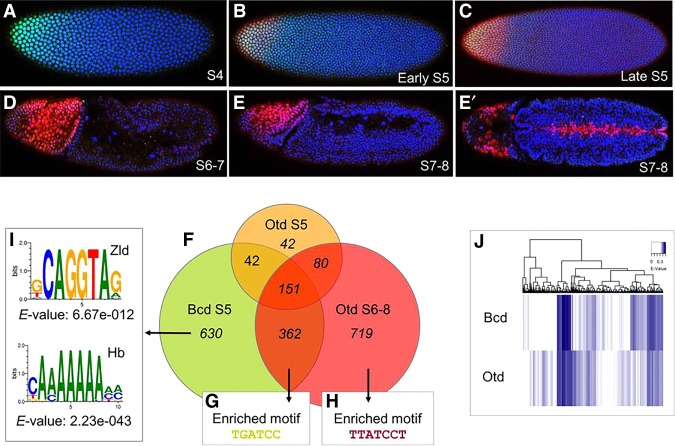
The result that the Bcd and Otd HDs are not interchangeable seems in conflict with the observation that Bcd and Otd bind the same TAATCC consensus sequence in vitro (Noyes et al. 2008b). However, it is possible that preferential binding to suboptimal sites might enable their distinct functionalities in vivo. Alternatively, the Bcd and Otd HDs might differentially interact with cofactors. To compare the binding activities of Bcd and Otd in wild-type embryos, we performed ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) experiments. To determine the best time intervals for embryo collection, we double-stained embryos at five consecutive time points after egg laying (AEL) using anti-Bcd and anti-Otd antibodies in wild-type embryos. These experiments show the Bcd gradient at stage 4 (S4) but no Otd expression at this stage (Fig. 3A). The Otd protein is first visible at early S5 (Fig. 3B), and its expression increases at mid to late S5, when both the Bcd and Otd proteins are strongly expressed (Fig. 3C). After cellularization during S6–S8, the Bcd protein becomes undetectable (Fig. 3D), while Otd expression is maintained in the head and up-regulated along the ventral midline (Fig. 3D,E′).
Figure 3.
(A–E′) A temporal comparison of Bcd and Otd expression. All embryos are stained for the Bcd protein (green), the Otd protein (red), and DAPI (blue), which marks individual nuclei (blue). The embryos shown represent a temporal series from A (youngest) to E (oldest). The ages of embryos are labeled by stage (S). (F–I) Genome-wide binding activities of Bcd and Otd. (F) A Venn diagram showing the number Bcd and Otd peaks in euchromatin (false discovery rate of <5%) in wild type embryos at the S5 and S6–S8 time points. (G–I) Motifs enriched in each data set are indicated. (J) Heat map comparisons of 9-mers bound by Bcd and Otd hierarchical clustering analysis of E-scores. All 9-mers bound by Bcd and Otd with an E-value of >0 are represented. Every position on the heat map is a single 9-mer.
Based on these experiments, we performed ChIP-seq on collections of embryos at S5 (when the two proteins are coexpressed) and at S6–S8 (when only Otd is detectable) (see the Materials and Methods). We detected 1185 Bcd-bound peaks in S5 embryos (Fig. 3F). Ninety-nine percent of these peaks mapped to euchromatic regions of the genome, and 65 of 66 previously known Bcd-dependent enhancers (Chen et al. 2012) were detected in this experiment. Otd bound to 524 peaks at S5, but only 60% (315 peaks) mapped to euchromatin. The remaining 40% (209 peaks) mapped to heterochromatic or uncharacterized regions of the genome. At the later time point, no significant Bcd-binding was detected, but Otd bound to 1312 peaks, 98% of which mapped to euchromatic regions (Fig. 3F).
Comparisons of the ChIP-seq profiles at both time points showed that Bcd and Otd bind to 630 and 719 unique peaks, respectively (Fig. 3F), supporting the observation that the two proteins are not functionally interchangeable in vivo (Fig. 1). The molecular basis for differential binding in vivo is not clear, but one possibility is that the Bcd and Otd HDs have inherent binding preferences that were not detected in previous in vitro binding studies. To test this, we used universal protein-binding microarrays (PBMs) in which purified GST-tagged Otd and Bcd HDs were tested for binding to all possible 9-mer nucleotide sequences (Materials and Methods) (Berger and Bulyk 2006). Previous work has shown that if a Drosophila HD is bound to a 9-mer sequence in a PBM with an E-score of >0.31, it is likely to be a functionally relevant site in vivo (Busser et al. 2012). A comparative heat map of Bcd and Otd binding to every possible 9-mer is shown in Figure 3J. Only 9-mers with an E-score of >0.31 are shown in color. The PBM E-score binding profiles indicate differences in binding preferences between Bcd and Otd (Fig. 3J).
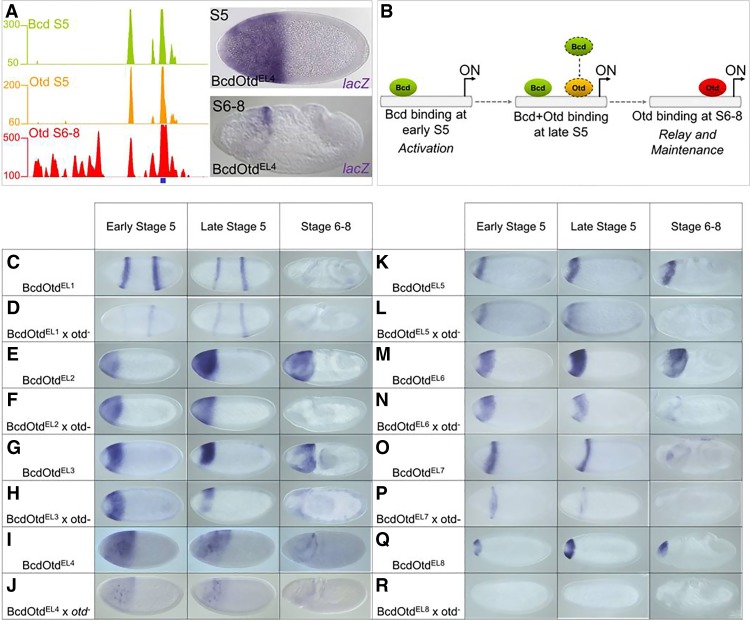
The ChIP-seq experiments also detected 513 peaks that were bound by Bcd early and then by Otd at the late time point (Fig. 3F). These overlapping peaks include 53 of 66 previously characterized Bcd-dependent enhancers (Fig. 4A; Supplemental Fig. 3; Chen et al. 2012). We hypothesize that these enhancers are regulated by a common FFR that integrates the activities of Otd and Bcd. In this model, genes controlled by these relay enhancers are initially activated by Bcd and then regulated by Otd after the Bcd gradient decays (Fig. 4B), and the transfer of control from Bcd to Otd would effectively extend the time of expression for a specific set of target genes. We classify these enhancers as BcdOtdEL (where E indicates bound early [S5], and L indicates bound late [S6–S8]) and use a consistent nomenclature here.
Figure 4.
Temporal regulation of FFR enhancer activity by Bcd and Otd. (A) ChIP-seq peaks for Bcd and Otd around the hb locus. The position of a known Bcd-dependent enhancer is shown as a blue box. This enhancer drives expression early at S5 and later at S6–S8. (B) A model for a FFR coordinated by Bcd and Otd. (C–T) Testing FFR enhancers in otd mutant embryos. Reporter gene expression patterns in wild-type embryos (C,E,G,I,K,M,O,Q) and otd mutant embryos (D,F,H,J,L,N,P,R). Embryos are shown at three time points, as indicated at the top.
A FFR regulates enhancers bound by Bcd and Otd
To test the FFR hypothesis, we examined the activities of eight candidate relay enhancers that were bound by both Bcd and Otd, expressed in anterior regions, and continuously active during the period between S5 and S6–S8 (Fig. 4C–R; Supplemental Fig. 3A). In a previous study, we showed that removing Bcd function (which also removes Otd) completely abolished expression driven by all of these enhancers (Chen et al. 2012). If Otd is involved in maintaining the expression patterns driven by these enhancers, removing its function should cause a loss or reduction of expression at S6–S8. To ablate Otd function, we used CRISPR–Cas9 mutagenesis to delete the Otd HD in the endogenous locus (Materials and Methods). This mutation caused embryonic lethality but did not affect the shape of the Bcd gradient (Supplemental Fig. 4).
Expression patterns driven by the eight candidate relay enhancers were assayed in otd mutant embryos at three time points: early S5 (high Bcd and low Otd), late S5 (high Bcd and high Otd), and S6–S8 (no Bcd and high Otd). For six of the eight tested enhancers, removing Otd caused a substantial reduction in expression at late S5 and/or a complete loss of expression at S6–S8 but had little effect on early activation (Fig. 4C–H,K–P; Supplemental Fig. 3). One of these enhancers was the EHE enhancer from the otd gene itself (BcdOtdEL6) (Supplemental Fig. 3), and the strong reduction of expression driven by that enhancer (Fig. 4M,N) suggests that maintenance of the normal otd pattern is mediated by a positive autoregulatory loop. The other two reporter genes (BcdOtdEL4 and BcdOtdEL8) tested in otd mutants showed strong reductions even in early S5 (Fig. 4I,J,Q,R; Supplemental Fig. 3), suggesting that Otd is required for the initial activation of some Bcd target genes. We define these as “relay enhancers”; they are bound and activated by Bcd prior to S5 and are then maintained by Otd in later development. Taken together, these results and the large number of shared peaks in the ChIP-seq experiments suggest that the FFR between Bcd and Otd is an important mechanism for extending the timing of expression of a subset of target genes in anterior regions of the embryo. In this relay mechanism, Otd's primary role is the maintenance of gene expression.
The Bcd–Otd FFR involves sequential binding to a suboptimal site
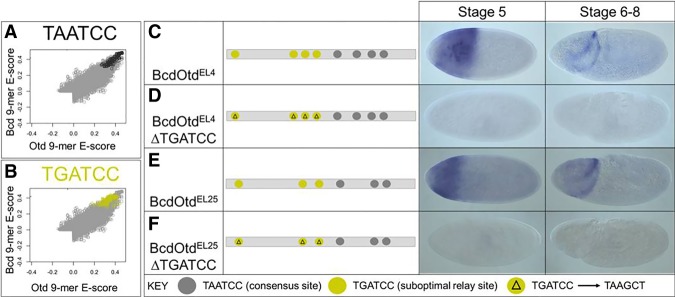
One possibility is that FFR-regulated enhancers contain specific sequence motifs that facilitate sequential binding of Bcd and Otd. Using a discriminative motif search (see the Materials and Methods), we identified a single base variant of the K50 consensus, TGATCC, which is enriched in peaks bound by both proteins compared with peaks bound by Bcd or Otd alone (Fig. 3G). This site does not contain the TAAT core recognition site preferred by most HD-containing TFs (Treisman et al. 1989; Noyes et al. 2008b). This TGATCC motif appears in 62% of the overlapping peaks compared with 23% of Bcd S5-only peaks (P = 0) and 38% of Otd S6–S8-only peaks (P = 9.05 × 10−8). We also searched the data from our PBM experiments to see how this single base change affects the binding preferences of Bcd and Otd (Fig. 5). This search showed that TGATCC-containing 8-mers are suboptimal (less strongly bound by both proteins) compared with the TAATCC consensus (Fig. 5A). It also appears that Bcd prefers TGATCC-containing 8-mers more strongly than does Otd (Fig. 5B).
Figure 5.
(A,B) PBM comparisons of DNA-binding preferences between Bcd and Otd. Scatter plots showing all 9-mers that contain the K50-binding consensus motif TAATCC (black circles in A) and all 9-mers containing the variant TGATCC that is enriched in feed-forward enhancers (yellow circles in B). (C–F) Functional tests of the TGATCC motif. lacZ expression patterns are shown at two time points for two wild-type enhancers (C,E) and those same enhancers with point mutations in TGATCC motifs (D,F). A binding site key is shown at the bottom.
To test the in vivo function of the TGATCC suboptimal site, we mutated it in two different enhancers (BcdOtdEL4 and BcdOtdEL25). Both are relay enhancers because they (1) are activated at both S5 and S6–S8 (Fig. 5C,E), (2) are bound by both Bcd and Otd, (3) lose expression in bcd mutants, and (4) lose expression late in otd mutants (Fig. 4; Chen et al. 2012). The BcdOtdEL4 and BcdOtdEL25 enhancers contain three copies each of the TGATCC site and four and three copies, respectively, of the TAATCC consensus site. We converted the TGATCC sites in these enhancers to TAAGCT—a lower-affinity site for both proteins that is equally represented in all ChIP data sets. For both enhancers, these mutations caused a complete loss of expression (Fig. 5D,F), suggesting that this motif is important for both Bcd-mediated activation and Otd-dependent maintenance of expression and that the TGATCC-binding site is critical for the Bcd/Otd relay.
Anterior patterning by the integration of three classes of enhancers
In addition to the relay enhancers identified by overlapping peaks of Bcd and Otd binding, our ChIP-seq experiments yielded large numbers of unique binding peaks for each protein (Fig. 3). The 630 peaks unique to Bcd included 13 previously known Bcd-dependent enhancers (Chen et al. 2012). The expression patterns of eight of these enhancers perdured after the Bcd gradient decays (Supplemental Fig. 5) but were not bound by Otd, so we hypothesize that other factors must be involved in the maintenance of their patterns (see the Discussion).
The ChIP experiments also identified many peaks bound only by Otd (122 peaks at S5 and 799 at S6–S8); 80 peaks were detected at both time points (Fig. 3F). We used lacZ reporter genes to test 19 of these peaks for enhancer activity. In our first experiments, we tested nine of the 719 fragments bound specifically by Otd only at the late time point (OtdL) (Fig. 6B; Supplemental Fig. 5). As expected, all nine enhancers showed anterior expression patterns in S6–S8 embryos (Fig. 6B; Supplemental Fig. 5). We also tested 10 genomic fragments bound uniquely by Otd only at S5 (OtdE; four fragments) or at both time points (OtdEL; six fragments). Surprisingly, none of the 10 tested fragments directed any reporter gene expression at either stage of development (Fig. 6A; Supplemental Fig. 5). These results are consistent with the failure of Otd to rescue bcd mutant embryos (Fig. 1) and suggest that Otd binding to euchromatic regions at S5 does not lead to enhancer activation. The reason for Otd's failure to activate expression at S5 is not clear, but one possibility is that activation of anterior genes in the early embryo requires prior binding by Bcd. Alternatively, because activation by Bcd requires interactions with cofactors (Simpson-Brose et al. 1994; Crauk and Dostatni 2005; Xu et al. 2014), Otd's inability to activate may be caused by a failure to make critical protein–protein contacts.
Figure 6.
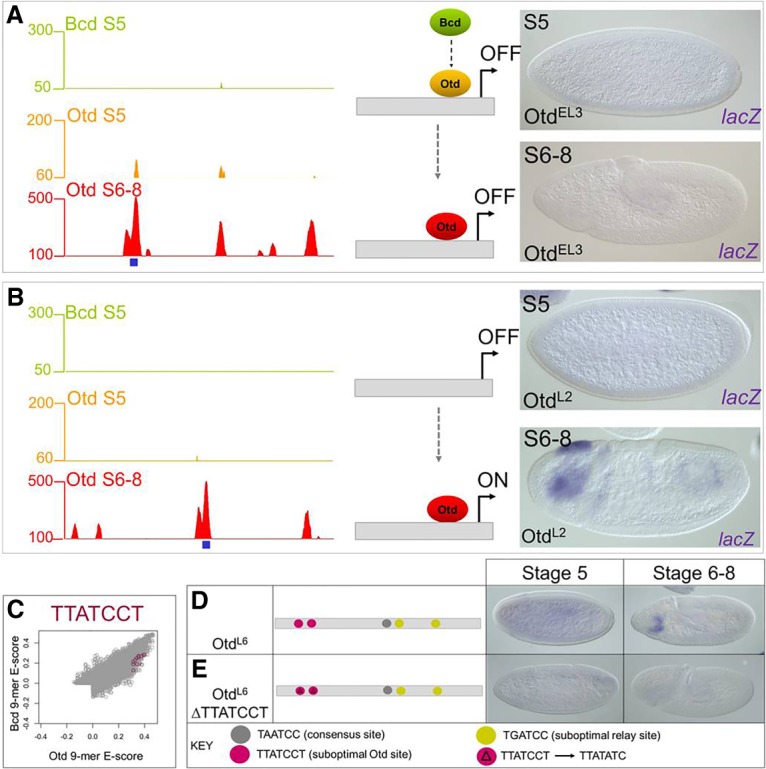
Reporter gene tests of genomic fragments bound only by Otd. (A) OtdEL3 is bound by Otd at S5 and S6–S8, but not by Bcd, and is inactive. (B) The OtdL2 enhancer is bound by Otd at S6–S8 and is active in the head. (C) Otd-preferred sequence TTATCCT that is enriched in the Otd S6–S8 ChIP data is preferred in vitro by Otd. (D,E) Mutating the Otd suboptimal site TTATCCT abolishes enhancer activity in a late acting Otd enhancer.
Taken together, our results define three classes of enhancers that mediate Bcd and/or Otd functions in vivo. Class 1 enhancers relay transcriptional initiation by Bcd to transcriptional maintenance by Otd, Class 2 enhancers are activated by Bcd and are maintained by an unknown factor. Class 3 enhancers are activated at a later time point by an Otd-dependent mechanism that is completely independent of Bcd.
Otd-dependent activation is mediated by another suboptimal site
We next searched for overrepresented motifs within the set of peaks bound specifically by Otd at S6–S8. This search identified an enriched K50 variant motif in Otd, TTATCCT, an extended variant of the canonical TAATCC motif optimally preferred by Bcd and Otd (Fig. 3H). It appears in 26% of Otd S6–S8 peaks, 10% of Bcd + Otd peaks, and only 9% of Bcd S5 peaks (P = 1.89 × 10−15). An analysis of our PBM data showed that this suboptimal site is preferentially bound by Otd compared with Bcd (Fig. 6C), consistent with its overrepresentation in Otd-bound genomic fragments.
We tested the role of the TTATCCT motif in an enhancer (OtdL6) that is bound by Otd and transcriptionally active only at S6–S8 in wild-type embryos and inactive in otd mutants (Fig. 6D; Supplemental Fig. 5). The OtdL6 enhancer contains two exact copies of the TTATCCT motif (Fig. 6D), one copy of the canonical K50HD-binding motif (TAATCC), and two copies of the TGATCC motif that is overrepresented in relay enhancers. We mutated the TTATCCT motifs, leaving the canonical and relay motifs intact, which caused a complete loss of expression (Fig. 6E). Based on our PBM data, ChIP data enrichment, and the enhancer site change data, we conclude that these suboptimal sites are necessary for activation by Otd at S6–S8.
Timing of enhancer activity is controlled by suboptimal motif preferences and cofactor interactions
Our motif searches identified suboptimal binding sites required for the activities of FFR enhancers (Fig. 5) and OtdL enhancers (Fig. 6), respectively. These results suggest that subtle binding preferences between the two proteins contribute to correctly timing enhancer activation in vivo. Alternatively, it is possible that the differential timing of enhancer activity is controlled by the presence or absence of binding sites for protein-specific cofactors. Previous studies identified two TFs, Hb and Zelda (Zld), as critical for Bcd-dependent activation (Small et al. 1992; Simpson-Brose et al. 1994; Porcher et al. 2010). Binding motifs for both of these proteins are enriched in Bcd-dependent enhancers (Schroeder et al. 2004; Ochoa-Espinosa et al. 2005; Nien et al. 2011; Xu et al. 2014). Consistent with this, a discriminative motif analysis revealed an enrichment of Hb and Zld motifs in regions bound in vivo by Bcd and Bcd + Otd compared with Otd only (Fig. 3I).
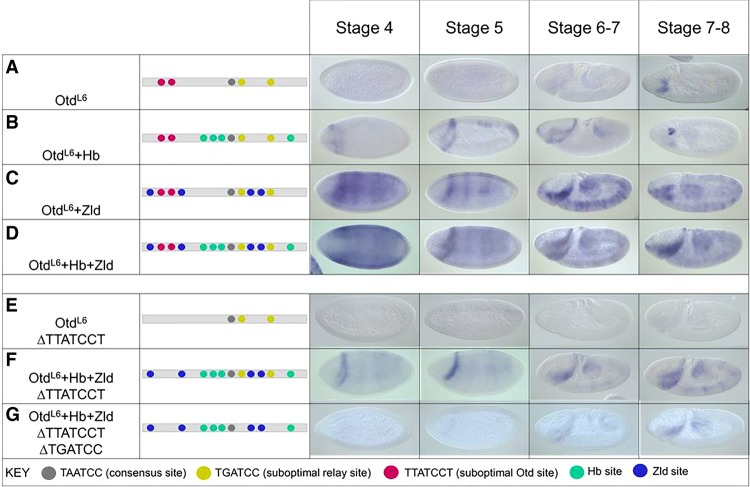
We hypothesized that early activation of enhancer activity by Bcd is controlled in part by cofactor interactions and that the addition of Hb and/or Zld sites might convert a late acting enhancer into a relay enhancer that is expressed earlier. We tested this hypothesis on OtdL6, which is bound by Otd at S6–S8 and active only at this later stage. This enhancer is not active early and is not bound by Bcd in vivo despite having two copies of the relay motif (TGATCC) (Fig. 7A). We added four high-affinity Hb sites to the OtdL6 enhancer, which resulted in early activation of expression as an anterior stripe (Fig. 7B). Adding four canonical Zld sites into this element also caused it to be activated early but in a complex pattern along the length of the embryo (Fig. 7C), and adding both Hb and Zld sites had an additive effect on the enhancer's activities (Fig. 7D). OtdL6 enhancers containing extra Hb and Zld sites were crossed into bcd mutants, which caused a complete loss of expression (data not shown), consistent with the hypothesis that the addition of Hb and Zld sites successfully converted the OtdL6 enhancer into an early acting Bcd-dependent enhancer.
Figure 7.
Binding site manipulations that change the timing of enhancer activity. (A–D) lacZ RNA expression patterns at four different time points driven by the wild-type OtdL6 enhancer (A) and the OtdL6 enhancer with four added Hb sites (B), four added Zld sites (C), and four extra Hb sites plus four extra Zld sites (D). (E) lacZ expression driven by the OtdL6 enhancer carrying mutations in three suboptimal Otd (TTATCCT) sites (compare with the patterns in A). (F) lacZ expression driven by the OtdL6 enhancer carrying mutations in three TTATCCT sites and four additional sites each for Zld and Hb. (G) lacZ expression driven by the OtdL6 enhancer carrying mutations in three TTATCCT sites, two TGATCC sites, and four additional sites each for Zld and Hb. See the text for a description of results and interpretations.
Our experiments above suggest that the specific sequence variants TGATCC and TTATCCT are required for the activities of relay and Otd-activated enhancers, respectively (Figs. 3, 5, 6). The OtdL6 enhancer contains both sequence variants and thus provides an opportunity to test how each motif might interact with the Zld and Hb cofactors. Mutating the TTATCCT motifs in the OtdL6 enhancer causes a loss of expression even though there are two intact copies of the TGATCC sequence (Figs. 6E, 7E). We hypothesized that adding Hb and Zld sites to this inactivated enhancer might “rescue” regulatory activity. Adding Hb and Zld sites to OtdL6ΔTTATCCT resulted in a stripe of expression that was expressed early and maintained in S6–S8 (Fig. 7F), a temporal pattern similar to that observed for other relay enhancers. This result suggests that the combination of only two TGATCC sites can mediate early activation if augmented with binding sites for Zld and Hb. A further mutation of the two TGATCC sites in the OtdL6 + Hb + Zld ΔTTATCCT enhancer resulted in a loss of expression, indicating the importance of those sites for early activation despite the presence of strong binding sites for Zld and Hb (Fig. 7G).
Taken together, our experiments suggest that the specific responses of enhancers to Bcd- and Otd-mediated activation are controlled in part by suboptimal motifs preferred by each protein and that early activation by Bcd requires interactions with the cofactors Hb and Zld.
Discussion
In this study, we compared the in vivo functions of two K50HD proteins (Bcd and Otd) within a framework of evolution. We showed that Otd and Bcd have evolved independent functions in vivo, and HD swaps between the two proteins indicated that the major structural differences mediating their distinct in vivo activities can be traced to their HDs. The two proteins bind to unique enhancers, and Otd also binds enhancers previously bound by Bcd via a FFR mechanism that extends the timing of the gene expression patterns that they regulate. We presented evidence that Otd binding does not lead to enhancer activation in the early embryo but is an effective activator after the first wave of zygotic target gene activation. Finally, we showed that enhancers respond in specific ways to Bcd and Otd through suboptimal binding sites for K50HD protein and binding sites for cofactors.
The Bcd–Otd FFR in Drosophila
Bcd has evolved rapidly to become a powerful morphogen in Drosophila but is not well conserved even within Diptera (Stauber et al. 1999). In Drosophila, otd has evolved to become a zygotic target gene of Bcd (Finkelstein and Perrimon 1990). otd RNA appears at S4, and its protein is detectable at early S5—at the same time as most other Bcd target genes. Despite having a lethal mutant phenotype, little is known about Otd's molecular functions in Drosophila embryogenesis. Otd binds to many enhancers that were initially activated by Bcd at S4 (Fig. 3). The reduced expression driven by these enhancers in otd mutants at S5 suggests that Otd protein binding is functional, and the loss of expression at S6–S8 demonstrates that Otd is required for maintaining their activity (Fig. 4). These results suggest strongly that Bcd and Otd participate in a relay in which Bcd initiates transcriptional activation of target genes, including otd, and Otd maintains its own expression and the expression of other target genes after the Bcd gradient decays. This relay is similar to the C1 feed-forward loop (FFL) with an “OR”-like input function described by Alon (2007) but is distinct in two respects. First, all target enhancers regulated by the Bcd–Otd FFR are initially activated by Bcd before Otd is present. Second, the relay involves the sequential binding of Bcd and Otd to the same binding motifs in the same target gene enhancers.
Because Bcd and Otd recognize similar sequence motifs in vitro and are involved in an FFR in Drosophila, one would predict that a maternal gradient of Otd (Mat:Otd) would activate many Bcd-dependent target genes when Bcd is genetically removed. On the contrary, we found that Mat:Otd cannot activate the great majority (more than 90%) of the tested Bcd-dependent transcriptional targets, including many that are regulated by the Bcd–Otd FFL. Also, our ChIP-seq experiments identified hundreds of peaks that bind Otd but not Bcd in early embryos. We tested 10 such fragments from euchromatic regions, but none showed any enhancer-like activity. Taken together, these results suggest that Otd is unable to efficiently activate transcription on its own in the early embryo.
The most likely explanation for Otd's inability to replace or function well without Bcd is that it is unable to interact efficiently with the Bcd cofactors Zld or Hb. Bcd activates the very first zygotic target genes as part of the maternal-to-zygotic transition (MZT), and a key factor in the MZT is Zld, which is thought to act as a pioneer that opens chromatin (Li et al. 2014; Sun et al. 2015). We showed previously that Zld facilitates Bcd binding to target gene enhancers (Xu et al. 2014). Similarly, most Bcd target genes require the activity of Hb to increase target gene sensitivity to Bcd-dependent activation (Simpson-Brose et al. 1994; Porcher et al. 2010). Interestingly, because insertion of the Bcd HD into the Otd protein confers on it many of Bcd's normal functions, these critical interactions may involve direct contacts with peptide motifs within Bcd's HD. We propose that similar motifs are not present in Otd's HD, which accounts for its inactivity at the early time point.
Mechanisms controlling different enhancer activities
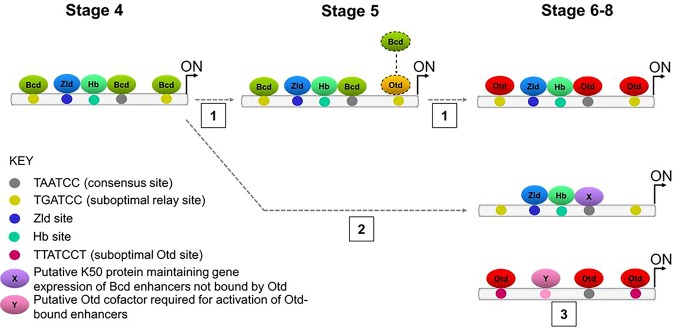
The ChIP-seq experiments enabled us to classify genomic fragments based on their ability to bind Bcd and/or Otd and their temporal expression patterns (Fig. 8). Fragments in class 1 bind Bcd early and Otd late and mediate the FFR described above. Fifty-three of the 66 known Bcd-dependent enhancers belong to this class. Fragments in class 2 are bound early by Bcd and never bound by Otd. Only 13 of the 66 known Bcd-dependent enhancers are in this class. These enhancers are also active after the Bcd gradient degrades but do not bind Otd, and it is not clear how their expression is maintained. Fragments in class 3 (OtdL) are never bound by Bcd but are bound by Otd at S6–S8. We tested nine OtdL candidate enhancers, and all showed enhancer activity.
Figure 8.
Three classes of Bcd- and Otd-dependent enhancers. Class 1: Bcd–Otd FFR enhancers are activated at S4 by Bcd with the cofactors Hb and Zld. Bcd protein binds to consensus (TAATCC) and suboptimal (TGATCC) sites required for gene activity. At S5, Otd protein binds to suboptimal sites, replacing Bcd. At S6–S8, Otd protein binds to all consensus and suboptimal sites to maintain gene activity. Class 2: These enhancers are activated by Bcd and maintained after the Bcd gradient decays but are never bound by Otd. These enhancers may be regulated by a FFL involving an unknown factor (X). Class 3: Other Otd activates gene targets in the late stage through binding to both consensus and suboptimal (TTATCCT) sites. Otd enhancer activation at S6–S8 might require another cofactor (Y).
We have begun to understand the molecular mechanisms that distinguish the activities of the three enhancer classes. Motif searches identified two suboptimal motifs, TGATCC and TTATCCT, in class 1 enhancers and class 3 enhancers, respectively (Fig. 3G,H). Mutating these motifs abolished enhancer function, which is consistent with recent studies showing that suboptimal sites play critical roles in other systems (Lebrecht et al. 2005; Rowan et al. 2010; Ramos and Barolo 2013; Crocker et al. 2015; Farley et al. 2016). However, it is important to point out that almost all genomic regions bound by Bcd and Otd and all enhancers tested here also contain optimal TAATCC motifs. Previous studies have shown that optimal sites are critical for enhancer activation (Driever et al. 1989; Struhl et al. 1989; Stanojevic et al. 1991; Arnosti et al. 1996) and sufficient for in vivo activation when clustered (Driever et al. 1989; Schier and Gehring 1992; Hanes et al. 1994; Crauk and Dostatni 2005; Lebrecht et al. 2005; Barr et al. 2017). These studies and our results suggest that both optimal and suboptimal sites contribute binding events required for productive activation of the basal promoter. However, we propose that the presence or absence of a suboptimal site can bias an enhancer toward activation by a specific family member.
Sequence motifs for Zld and Hb were also overrepresented in ChIP-seq peaks bound by Bcd (Fig. 3), consistent with the combinatorial activation model mentioned above. Adding Zld- and Hb-binding sites to a class 3 enhancer normally activated later by Otd results in earlier activation and conversion of that enhancer into a Bcd-dependent class 1 enhancer (Fig. 7B–D,F). We hypothesize that class 3 enhancers contain binding sites for unknown Otd-specific cofactors, and identifying these sites and the proteins that bind them will be the focus of future studies. Taken together, our results indicate that both intrinsic DNA-binding preferences and interactions with cofactors control the distinct temporal and spatial patterns of expression driven by individual enhancers in vivo.
Convergent evolution and a robust core anterior patterning network
bcd and its paralog, zen, arose through duplication of an ancestral maternally expressed Hox3-like gene (Stauber et al. 1999, 2002). In insects lacking Bcd, Otd has Bcd-like properties (maternal expression and anterior mRNA localization and patterning) (Schroder 2003; Lynch et al. 2006). Since the two proteins do not share common ancestry (Otd is a paired class homeoprotein that diverged from the Hox cluster >800 million years ago), some functional convergence of these proteins must have occurred during insect evolution.
The evolution of Bcd likely involved the retention of the maternal promoter and the acquisition of three characteristics required for anterior embryonic patterning: (1) UTR sequences that control anterior mRNA localization, (2) protein motifs that mediate translational repression, and (3) amino acid substitutions that alter its DNA-binding preferences; namely, a Q50-to-K50 mutation in its HD. Since otd is zygotic in Drosophila, some of these characteristics described above must have been lost in otd in the lineage that led to Drosophila. Such dramatic changes in these two genes may be attributed to reduced selective pressure on maternal genes (Barker et al. 2005; Demuth and Wade 2007), which permits the exploration of the evolutionary landscape and the acquisition of new functional roles.
What is striking is that Otd binds to at least half of the Bcd-bound target genes after they are activated by Bcd. Perhaps this set of target genes represents an ancestral core network that is well conserved in evolution. Many feed-forward enhancers are associated with the gap genes, which cross-regulate each other via repressive interactions. All of these enhancers contain DNA motifs that are recognized by a K50HD TF such as Bcd or Otd. Thus, it is possible that the cis-regulatory motifs (and consequently the enhancers) are functionally robust in the evolution of anterior embryo patterning, while trans-acting factors can accumulate mutations. This allows for a conserved set of targets that makes up a canalized anterior patterning network that allows the regulating TFs to evolve.
Materials and methods
D. melanogaster stocks
The following stocks were used in these experiments: yw (wild type), ±/Cyo bcd+;bcdE1/bcdE1, Cyo bcd+/Sco;bcdE1/bcdE1, and ΦC31 (y+);38F1 (w+).
Maternal gene chimeras
The bcd- and otd-coding regions were amplified by PCR from pBS-SK+ cDNA clones. We cloned an injection plasmid (piattB40-Bcd) using traditional techniques from pBS-SK+ (Asp718/SacI) and industrially synthesized oligonucleotides. This plasmid contained inverted ΦC31-specific recombination sequences (AgeI/HindIII), the fluorescent green eye marker Gmr-Gfp (HindIII/AscI), and a polylinker flanked by the bcd promoter and 3′ UTR (AgeI/AscI). The amplification product was digested with RsrII and AscI and ligated into the plasmid fragment. HD swaps and residue changes were generated using standard cloning techniques and nested PCR. All transgenic lines were generated using the ΦC31 integration system, and constructs were integrated into the 38F1 landing site on the third chromosome (Bateman et al. 2006).
Embryo collections, cuticle preparations, and immunohistochemistry
Embryos were collected 2–3 and 3–5 h AEL. Embryos were dechorionated for 2 min in bleach, and a 2:1 mixture of methanol and heptane was used to remove the vitelline membrane. ISH and FISH were performed as described previously (Kosman et al. 2004) using digoxigen-, fluorescein-, and biotin-labeled probes. Cuticle preparations were performed on embryos aged 20–24 h. For cuticle preparations, larvae were fixed overnight at 65°C in a 1:4 mixture of glycerol and acetic acid and mounted in a 1:1 mixture of Hoyer's medium and lactic acid. Rabbit anti-Bcd (1:400) and guinea pig anti-Otd (1:1000) were used for immunostaining (with rabbit FITC and 647 guinea pig as secondary antibodies at 1:400 each; Invitrogen). Guinea pig anti-Cad (1:400) and Alexa flour-conjugated 488 donkey anti-guinea pig (1:500; Invitrogen) were used to examine Cad protein expression. All antibodies were diluted in PBT. Data for immunostaining images were collected on a Leica TCS SP5 confocal microscope using the Leica confocal analysis software. Gradient quantifications were performed as described previously (Ochoa-Espinosa et al. 2009).
ChIP-seq
ChIP was performed using Bcd antibody (rabbit) and two Otd antibodies (guinea pig; GP-5 and GP-6, both provided by Tiffany Cook) on two biological replicates (two technical replicates per sample) of wild-type chromatin collected at 2–3 and 3–5 h AEL. Antibodies were protein A-purified using the protein A antibody purification kit (Sigma). The embryos from each collection were DAPI-stained to confirm embryonic stages, and 85% of the embryos were the correct age in each collection. Embryos were treated and fixed in 1.8% formaldehyde as described previously (Zeitlinger et al. 2007; Xu et al. 2014). Samples were sonicated for 10 min at setting 3 (30 sec on and 30 sec off) followed by 2.5 min at setting 4 (30 sec on and 30 sec off) using a Sonic Dismembrator model 550. Two-hundred microliters of embryonic lysate was used for each immunoprecipitation reaction. Two-hundred-forty microliters of buffer FA + PI (50 mM HEPES/KOH at pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl, protease inhibitor) was added to each sample to bring the total volume to 440 µL. Five percent of each extract was removed as input. Ten microliters of each antibody was added to the sample (minus input) and incubated overnight at 4°C. Forty microliters of protein A sepharose bead slurry (Amersham Biosciences) was added per ChIP sample with 1 mL of FA buffer. The sample was centrifuged at 2500g for 1 min, and supernatant was discarded. One milliliter of FA buffer was added to each tube. Beads were suspended by inverting the tubes a few times and then centrifuging them again. This wash was repeated three times. After the final wash, the beads were suspended in 40 µL of FA buffer. The ChIP sample was added to the bead slurry and rotated for 2 h at 4°C. Two microliters of 20 mg/mL RNase A was added to the inputs. The beads were washed at room temperature by adding 1 mL of each of the following buffers and incubating on a nutator (or a rotator): FA (twice for 5 min), FA–1 M NaCL (once for 5 min; 50 mM HEPES/KOH at pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 M NaCl), FA–500 mM NaCl (once for 10 min; 50 mM HEPES/KOH at pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 500 mM NaCl), TEL (once for 10 min; 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl at pH 8.0), and Tris-EDTA (twice for 5 min). The beads were collected after each wash by centrifugation at 2500g for 1 min, and the supernatant was removed. To elute the immunocomplexes, we added 125 µL of ChIP elution buffer and placed the tube for 15 min in a 65°C heat block. We spun down the beads at 6000g for 1 min and transferred the supernatant to a new tube. The elution was repeated, and supernatants were combined.
ChIP-seq library preparation and data processing
NEXTflex ChIP-seq kits from BIOO Scientific (5143-01) were used to prepare libraries, which were then barcoded using NEXTflex ChIP-seq barcodes (BIOO Scientific, 514120). We performed single-end HiSeq 2000 sequencing (Illumina) at the New York University Genome Center. Sequencing reads were mapped to the D. melanogaster genome release 5.3 using Bowtie, and duplicate reads were removed after combining data from each biological replicate. The MACS program was used to call peaks over input (Zhang et al. 2008; Liu et al. 2011). We removed all heterochromatic and uncharacterized chromatin from the analysis and further narrowed down relevant peaks by using only those that appeared in both antibodies (for Otd) and across all replicates. All ChIP-seq data processing was done on the Galaxy Cistrome platform. A false discovery rate cutoff of 5% was used for all further analysis on ChIP-seq peaks.
Motif enrichment analyses
Matlab was used for all motif analyses unless stated otherwise. Bcd and Otd peaks were compared, and peaks were considered shared if they overlapped by at least 200 base pairs. We compared all of our ChIP-seq data sets with prior DNase I, Hb ChIP–chip, and Zld ChIP-seq experiments using the same overlapping criterion. We used MEME-ChIP (Machanick and Bailey 2011) in the MEME suite (Bailey et al. 2009) to search for overrepresented motifs in each ChIP data set. We then used a discriminative de novo motif search to look for overrepresented 6-mers, 7-mers, 8-mers, and 9-mers in ChIP-seq peaks. Cofactor position–weight matrices used to search Bcd- and Otd-bound genomic regions were derived as follows: Bcd and Otd from PBMs done in this study and Zld and Hb from Fly Factor Survey (http://mccb.umassmed.edu/ffs).
GST-tagged proteins
HD-coding sequences (as well as 15-amino-acid flanking regions) for Bcd and Otd were PCR-amplified and cloned into a N-terminal GST fusion Gateway expression vector (pDEST15, Invitrogen), and the correct clones were confirmed by sequencing. Rosetta (DE3)-competent cells (BL21 derivatives from Novagen) were transformed with the GST-BcdHD or GST-OtdHD plasmids. For each transformation, a single colony was used to inoculate 0.5 L of LB + Amp100 and shaken at 200 rpm at 37°C until the OD600 reached 0.5–0.6. Proteins were induced for 3 h at 37°C by the addition of 0.5 mM IPTG. After induction, bacteria were harvested by centrifugation at 5000g for 15 min, and the cell pellets were stored at −80°C. Each frozen pellet was resuspended in 40 mL of lysis buffer (150 mM NaCl, 10 mM HEPES at pH 7.9, 5 mM DTT, 10% glycerol, 0.5% Triton X-100, 0.5 mg/mL lysozyme, 1 mM MgCl2, 1 mM benzamidine, 3 µM pepstatin A, 2 mM leupeptin, 1 mM PMSF) and sonicated using a Branson Sonifier 450 homogenizer equipped with a midi tip. The sonication parameters were six to eight strokes of 1 min with an output of 6, duty cycle of 50%, and a resting time of 2 min on ice between strokes. The crude lysate was centrifuged at 27,000g for 30 min at 4°C, and the supernatant was transferred to a new tube containing 0.5 mL of settled, prewashed, and pre-equilibrated glutathione sepharose 4B beads (GE Healthcare, 17075601) and rotated for 6 h. Elutions were performed with lysis buffer containing 5 mM (E1 and E2), 7.5 mM (E3), 10 mM (E4 and E5), 20 mM (E6 and E7) and 50 mM (E8 and E9) imidazole. A total of 10 mL of the peak eluates was dialyzed twice for 12 h each time against 1 L of storage buffer (150 mM NaCl, 5 mM HEPES at pH 7.9, 1 mM DTT, 10% glycerol, 1 mM PMSF). After dialysis, the samples were centrifuged at 21,000g for 5 min, aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C.
PBMs
PBMs were performed as described previously (Berger et al. 2006) using a custom-designed “all 10-mer” universal array (Berger et al. 2008) in the 8x60K array format (Agilent Technologies, Inc., Agilent microarray design identifier [AMADID] 030236) (Nakagawa et al. 2013). Both proteins were tested at a concentration of 87 nM. Duplicate PBMs were performed for each protein. The array data were quantified as described previously (Berger et al. 2006), and 8-mer data were averaged over duplicate PBM experiments using the Universal PBM analysis suite (Berger and Bulyk 2009). Motifs were derived by the Seed-and-Wobble algorithm (Berger et al. 2006; Berger and Bulyk 2009), modified to use 90% of the foreground and background features (Busser et al. 2012). As described previously (Busser et al. 2012), each 9-mer was then assigned the E-score of the lower scoring of its two constituent 8-mers. In previous assays using PBM data for other Drosophila HDs, 9-mers with an E-score >0.31 were used with confidence to predict real binding events (Busser et al. 2012). 9-mer sequences bound preferentially by Bcd versus Otd were identified as described previously (Busser et al. 2012).
Generation of otd HD deletion by CRISPR
The CRISPR flies were generated as described (Gratz et al. 2013, 2014). gRNAs were inserted into the pU6-BbsI-chiRNA vector (gRNA1, CTTCGAAAAAAAAAAACGAGTTAGC; gRNA2, CTTCGCATATATAAACATTATGTAC). Two homology arms flanking the cleavage sites were inserted into the multiple cloning sites of the pHD-DsRed-attP vector as donor template. The mixture of 500 ng/µL donor vector and two gRNA vectors (100 ng/µL each) was injected into embryos of y[1] M(vas-Cas9).RFP-ZH-2A w[1118]/FM7a. The F1 flies were crossed to YW flies, and F2s were screened for the dsRed+ transformants.
Synthetic enhancer constructs, transgenesis, and K50-dependent enhancer analysis
All genomic fragments were cloned into the piB-HC-lacZ vector described previously (Chen et al. 2012) using BglII and Asc1. All constructs were inserted using ΦC31 integrase-mediated cassette exchange at 38F1 on chromosome II. We converted low-affinity Zld and Hb sites into high-affinity sites in the OtdLA enhancer. Synthetic enhancer fragments for BcdOtdEL4 (HC_01), BcdOtdEL25 (HC_49), and OtdL6 were synthesized by Integrated DNA Technologies. OtdEL and OtdL enhancers were cloned from genomic DNA. The dependency of enhancers on Otd, Bcd, and Mat > Otd was determined by crossing the enhancers to otd and bcd mutants and Mat > Otd virgin females, collecting progeny embryos, and checking to see how lacZ expression was affected by ISH.
Supplementary Material
Acknowledgments
We are grateful to Tiffany Cook for the Otd antibody, and the Bloomington Stock Center for fly stocks. We thank the New York University Center for Genomics and Systems Biology for all of their help and support with next-generation sequencing. We thank Jinshuai Cao for technical assistance, and Steve Gisselbrecht for help with PBM K-mer analysis. We also thank Anastasia Vendenko for help with the initial stages of PBM analyses. We are grateful to Claude Desplan, Lionel Christiaen, and Pinar Onal for invaluable feedback on the manuscript. M.L.B. was supported by National Institutes of Health (NIH) RO1 HG005287. R.L. was supported by a New York University Dean's Undergraduate Research Fellowship. S.S. and R.J.J. were supported by NIH GM 106090.
Author contributions: R.R.D., G.Y., R.J.J., and S.S. conceived the study. R.R.D., P.S., M.L.B., and Z.X. developed the methodology. R.R.D., J.L., I.B., and J.K. provided the software. R.R.D., J.L., and J.K. performed the formal analysis. R.R.D., J.M., X.R., T.B., J.K., and R.L. performed the investigations. R.R.D. and S.S. wrote the original draft of the manuscript. R.R.D., S.S., and R.J.J. reviewed and edited the manuscript. R.R.D. visualized the study. R.R.D., S.S., and M.L.B. supervised the work. R.R.D., R.J.J., and S.S. were the project administrators. S.S., M.L.B., and R.J.M. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.311985.118.
References
- Abascal F, Corpet A, Gurard-Levin ZA, Juan D, Ochsenbein F, Rico D, Valencia A, Almouzni G. 2013. Subfunctionalization via adaptive evolution influenced by genomic context: the case of histone chaperones ASF1a and ASF1b. Mol Biol Evol 30: 1853–1866. [DOI] [PubMed] [Google Scholar]
- Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, Small S. 1996. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122: 205–214. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Demuth JP, Wade MJ. 2005. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet 1: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr KA, Martinez C, Moran JR, Kim AR, Ramos AF, Reinitz J. 2017. Synthetic enhancer design by in silico compensatory evolution reveals flexibility and constraint in cis-regulation. BMC Syst Biol 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. 2006. Site-specific transformation of Drosophila via ΦC31 integrase-mediated cassette exchange. Genetics 173: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. 2006. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol Biol 338: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. 2009. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc 4: 393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW III, Bulyk ML. 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J 7: 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser BW, Shokri L, Jaeger SA, Gisselbrecht SS, Singhania A, Berger MF, Zhou B, Bulyk ML, Michelson AM. 2012. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development 139: 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Casillas S, Negre B, Barbadilla A, Ruiz A. 2006. Fast sequence evolution of Hox and Hox-derived genes in the genus Drosophila. BMC Evol Biol 6: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu Z, Mei C, Yu D, Small S. 2012. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149: 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crauk O, Dostatni N. 2005. Bicoid determines sharp and precise target gene expression in the Drosophila embryo. Curr Biol 15: 1888–1898. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Wade MJ. 2007. Maternal expression increases the rate of bicoid evolution by relaxing selective constraint. Genetica 129: 37–43. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1989. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337: 138–143. [DOI] [PubMed] [Google Scholar]
- Driever W, Thoma G, Nusslein-Volhard C. 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340: 363–367. [DOI] [PubMed] [Google Scholar]
- Driever W, Siegel V, Nusslein-Volhard C. 1990. Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development 109: 811–820. [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. 2016. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc Natl Acad Sci 113: 6508–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Boncinelli E. 1994. From fly head to mammalian forebrain: the story of otd and Otx. Trends Genet 10: 310–315. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Perrimon N. 1990. The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature 346: 485–488. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Smouse D, Capaci TM, Spradling AC, Perrimon N. 1990. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev 4: 1516–1527. [DOI] [PubMed] [Google Scholar]
- Frohnhofer HG, Nusslein-Volhard C. 1986. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324: 120–124. [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD, Riddihough G, Ish-Horowicz D, Brent R. 1994. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol 14: 3364–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TY, Cook CE, Davis GK, Shigenobu S, Chen RP, Chang CC. 2010. Anterior development in the parthenogenetic and viviparous form of the pea aphid, Acyrthosiphon pisum: hunchback and orthodenticle expression. Insect Mol Biol 19: 75–85. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. 2004. Multiplex detection of RNA expression in Drosophila embryos. Science 305: 846. [DOI] [PubMed] [Google Scholar]
- Lebrecht D, Foehr M, Smith E, Lopes FJ, Vanario-Alonso CE, Reinitz J, Burz DS, Hanes SD. 2005. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc Natl Acad Sci 102: 13176–13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3: e03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S, Tkacik G, Kneeland T, Wieschaus E, Gregor T. 2011. The formation of the bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol 9: e1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. 2005. Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol Dev 7: 629–646. [DOI] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J, Desplan C. 2003. ‘De-evolution’ of Drosophila toward a more generic mode of axis patterning. Int J Dev Biol 47: 497–503. [PubMed] [Google Scholar]
- Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. 2006. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439: 728–732. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. 2013. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci 110: 12349–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7: e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008a. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. 2008b. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res 36: 2547–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. 2005. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci 102: 4960–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. 2009. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci 106: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AD. 2008. The evolution of developmental gene networks: lessons from comparative studies on holometabolous insects. Philos Trans R Soc Lond B Biol Sci 363: 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. 2015. Genomic control process: development and evolution. Academic Press, London. [Google Scholar]
- Porcher A, Abu-Arish A, Huart S, Roelens B, Fradin C, Dostatni N. 2010. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development 137: 2795–2804. [DOI] [PubMed] [Google Scholar]
- Ramos AI, Barolo S. 2013. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos Trans R Soc Lond B Biol Sci 368: 20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. 2010. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev 24: 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. 1992. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature 356: 804–807. [DOI] [PubMed] [Google Scholar]
- Schroder R. 2003. The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422: 621–625. [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. 2004. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol 2: E271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Brose M, Treisman J, Desplan C. 1994. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell 78: 855–865. [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J 11: 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254: 1385–1387. [DOI] [PubMed] [Google Scholar]
- Stauber M, Jackle H, Schmidt-Ott U. 1999. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci 96: 3786–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Prell A, Schmidt-Ott U. 2002. A single Hox3 gene with composite bicoid and zerknullt expression characteristics in non-Cyclorrhaphan flies. Proc Natl Acad Sci 99: 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57: 1259–1273. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nien CY, Chen K, Liu HY, Johnston J, Zeitlinger J, Rushlow C. 2015. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res 25: 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet 12: 692–702. [DOI] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. 2003. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 301: 1714–1717. [DOI] [PubMed] [Google Scholar]
- Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. 1989. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell 59: 553–562. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, Small S. 2014. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev 28: 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. 2007. Whole-genome ChIP–chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.