Abstract
Background
The cellulolytic fungus Neurospora crassa is considered a potential host for enzyme and bioethanol production. However, large scale applications are hindered by its filamentous growth. Although previous investigations have shown that mycelial morphology in submerged culture can be controlled by altering physical factors, there is little knowledge available about the potential for morphology control by genetic modification.
Results
In this study, we screened morphological mutants in the filamentous fungus N. crassa. Of the 90 morphological mutants screened, 14 mutants exhibited considerably higher viscosity compared with that of the wild type strain, and only two mutants showed low-viscosity morphologies in submerged culture. We observed that disruption of gul-1 (NCU01197), which encodes an mRNA binding protein involved in cell wall remodeling, caused pellet formation as the fermentation progressed, and resulted in the most significant decrease in viscosity of culture broth. Moreover, over-expression of gul-1 caused dramatically increased viscosity, suggesting that the gul-1 had an important function in mycelial morphology during submerged cultivation. Transcriptional profiling showed that expression of genes encoding eight GPI-anchored cell wall proteins was lowered in Δgul-1 while expression of genes associated with two non-anchored cell wall proteins was elevated. Meanwhile, the expression levels of two hydrophobin genes were also significantly altered. These results suggested that GUL-1 affected the transcription of cell wall-related genes, thereby influencing cell wall structure and mycelial morphology. Additionally, the deletion of gul-1 caused increased protein secretion, probably due to a defect in cell wall integrity, suggesting this as an alternative strategy of strain improvement for enzyme production. To confirm practical applications, deletion of gul-1 in the hyper-cellulase producing strain (∆ncw-1∆Ncap3m) significantly reduced the viscosity of culture broth.
Conclusions
Using the model filamentous fungus N. crassa, genes that affect mycelial morphology in submerged culture were explored through systematic screening of morphological mutants. Disrupting several candidate genes altered viscosities in submerged culture. This work provides an example for controlling fungal morphology in submerged fermentation by genetic engineering, and will be beneficial for industrial fungal strain improvement.
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0944-5) contains supplementary material, which is available to authorized users.
Keywords: Neurospora crassa, Mycelial morphology, Pellet, Viscosity, Protein secretion
Background
Filamentous fungi are widely used for industrial scale production of antibiotics, proteins and many other useful chemicals [1–3]. Unlike bacteria and yeast, filamentous fungi are morphologically complex microorganisms, which often leads to lots of process engineering problems during the fermentation process. When grown in submerged culture, fungi display different morphological forms, such as freely dispersed mycelia, clumps and pellets [4]. In general, the clump form predominates in fungal fermentations, resulting in high viscosity and a fermentation broth that shows several non-Newtonian characteristics. Although these disadvantages may not significant impact preliminary Erlenmeyer flask batch fermentations, when scaled up there are negative impacts on nutrient consumption and oxygen uptake, and a decrease in productivity. The simplest strategy to overcome this problem is to increase agitation speed. However, high impeller speed increases power consumption, and produces high shear stress that often damages fungal mycelia reducing product yield [5]. In contrast, the pellet growth form leads to Newtonian rheological behavior and low viscosities. As expected, lower power inputs are required for achieving sufficient agitation and mass transfer. Thus, the pellet morphology is preferred for many industrial processes, and it would greatly facilitate cost-effective production if one could design a way of controlling mycelial morphology during the fermentation process.
Many studies have reported on the effects of cultivation conditions and physical factors on fungal morphology, including media constituents, inoculum size, pH, temperature, agitation systems and the type of fermenter [5–8]. Since the genetics of morphogenesis is very complicated in filamentous fungi, there is little knowledge available about morphology control through genetic modification. In recent years, chemical and UV mutagenesis strategies have been widely used to create mutants that show low-viscosity morphology in submerged culture [9]. However, random mutagenesis is quite time-consuming, and it is also difficult to determine which gene is responsible for a desired phenotype.
In the past century, the filamentous fungus Neurospora crassa has been used as a model organism for genetic and molecular studies [10], leading to a range of genetic techniques and tools [11]. Compared with other filamentous fungi, only N. crassa possesses a near full genome deletion strain collection. Recently, N. crassa was deployed as a model to unravel mechanisms of lignocellulase expression and regulation due to its capacity to secrete lots of enzymes involved in lignocellulose utilization [12]. In addition, N. crassa has been reported to be a potential alternative candidate for heterologous protein expression and bioethanol production [13]. Several approaches have been employed to improve cellulase production, including regulation of the cellulase induction pathway, regulation of the unfolded protein response pathway, and by enhancing protein secretion [14–16]. Furthermore, N. crassa can be easily sexually crossed to generate multiple-gene mutants, which facilitates construction of hyper-cellulase producers. However, to our knowledge, N. crassa has not been used in industrial scale processes. One of the most limiting factors is its mycelial morphology in submerged culture. The N. crassa wild type strain exhibits a clump type of morphology, which results in high viscosity and mass transfer limitations. Although it is reported that N. crassa can grow as dispersed pellets following the addition of Triton N-101 or anionic polymer carboxypolymethylene [17, 18], investigations into the molecular controls of morphology could have great significance in industrial application.
Therefore, we screened 90 N. crassa morphological mutants to identify putative genes affecting mycelial morphology during submerged cultivation. Several candidate genes affected the viscosity of media when disrupted in N. crassa. Intriguingly, we found that disruption of gul-1 (NCU01197), the homolog of Saccharomyces cerevisiae ssd1, led to a significant change in viscosity. In S. cerevisiae, SSD1 plays critical roles in maintaining cell wall integrity [19], and has been found to specifically associate with the mRNAs of proteins that are involved in cell wall remodeling [20]. SSD1 is also negatively regulated by the NDR family kinase Cbk1 through phosphorylation [21]. Deletion of ssd1 alters the composition and cell wall architecture of the yeast cell surface [22]. Similar to the functions described for SSD1, GUL-1 is an important component of the NDR kinase COT-1 pathway, which is involved in the regulation of polarized growth in N. crassa [23, 24]. The NDR family kinase COT-1, which is a homolog of the S. cerevisiae Cbk1, is required for normal hyphal elongation [25]. The temperature-sensitive cot-1 mutant exhibits cessation of tip extension and hyperbranching after being shifted to restrictive temperature. Disruption of gul-1 can partially suppress the growth defects of cot-1. Thus, gul-1 has been initially identified as a dominant modifier of cot-1 [26]. Further investigation demonstrated that the Ste20 kinase POD-6 is involve in polar tip extension and also acts in the NDR kinase COT-1 pathway. The growth defects in pod-6 and cot-1 can be suppressed by the common extragenic suppressor, gul-1, suggesting that gul-1 may function as a downstream mediator in this pathway [23]. Recently, Herold and Yarden provided strong evidence to support this hypothesis. Their results illuminated that the NDR family kinase COT-1 affects hyphal growth by influencing the transcript levels of cell wall remodeling genes, which is mediated by gul-1 [24]. Despite the previous studies described above, the mechanism of how gul-1 affects mycelial morphology in submerged culture remains unclear. In this study, we demonstrated that gul-1 alters cell wall structure by influencing the expression of cell wall protein genes, which leading to changes in mycelial morphology. In addition, we also illustrated that morphology engineering enables enhanced protein secretion. Our findings enhance our understanding of the genetics of morphogenesis in filamentous fungi and provided a novel strategy for morphological engineering of filamentous fungi by genetic modification.
Results
A low-viscosity mutant from a morphological mutant library
Colot et al. [27] constructed a nearly complete genome disruption mutant collection. There were two 96-well plates in this collection relevant to our study. One plate was annotated as “Morphologicals”, the other was identified as “Hyphal Growth Set” [28]. To identify the key genes that play important roles in mycelial morphology during submerged cultivation, we screened these 90 N. crassa morphological mutants. The viscosities of the fermentation broths are shown in Fig. 1 and Table 1. According to microscope observations, the majority of mutants resembled the wild type (WT) in growth characteristics. Compared to WT, 14 mutants showed dramatically increased viscosity (> 50%) and two mutants showed decreased viscosity (> 50%). Deletion of NCU03938 (encoding an alternative oxidase-5) reduced viscosity by approximately 55%, but this mutant still grew as clumps. Conversely, loss of NCU01197, which was annotated as gul-1 previously [23], resulted in the pellet growth form and reduced viscosity by more than 80% compared with WT. Thus, gul-1 was selected for further investigation.
Fig. 1.
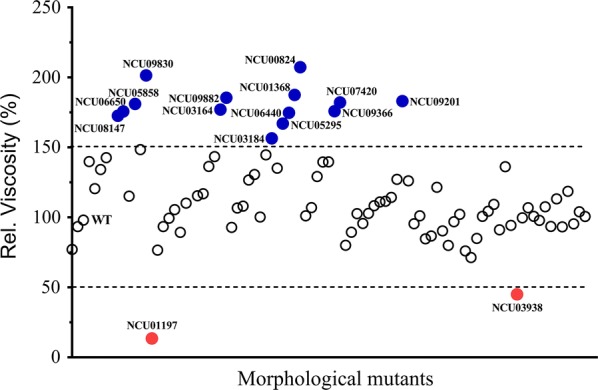
Screening of 90 morphological mutants in Neurospora crassa. Conidia from the wild type (WT) and morphological mutants were separately inoculated into Avicel medium and batch cultured for 7 days. The viscosities of culture broths altered by more than 50% compared with the WT are indicated as follows: blue dots, high-viscosity mutants; red dots, low-viscosity mutants
Table 1.
List of morphological mutants in Neurospora crassa
| No. | NCU no. | Annotation | Viscosity (cP) | Increased vs WT (%) |
|---|---|---|---|---|
| 1 | NCU07075 | Calcium exchanger | 267.5 ± 3.5 | − 22.9 |
| 2 | NCU09866 | Thyroid hormone receptor interactor 12 | 324.5 ± 34.6 | − 6.5 |
| 3 | NCU09423 | Secreted protein | 340.5 ± 12.0 | − 1.9 |
| 4 | NCU05854 | Hypothetical protein | 430.0 ± 127.3 | 23.9 |
| 5 | NCU05790 | Phytochrome-2 | 418.5 ± 9.2 | 20.6 |
| 6 | NCU05956 | Glycosylhydrolase family 2–2 | 466.0 ± 36.8 | 34.3 |
| 7 | NCU07617 | Aconidiate-3 | 407.5 ± 88.5 | 17.4 |
| 8 | NCU08147 | P-type ATPase | 599.0 ± 58.0 | 72.6 |
| 9 | NCU06650 | Secretory phospholipase A2 | 610.0 ± 56.6 | 75.8 |
| 10 | NCU06531 | Hypothetical protein | 458.0 ± 116.0 | 32.0 |
| 11 | NCU05858 | Fatty acid oxygenase | 630.0 ± 42.4 | 81.6 |
| 12 | NCU06419 | MAPK/ERK kinase | 495.0 ± 41.4 | 42.6 |
| 13 | NCU09830 | Menadione-induced gene-12 | 700.0 ± 28.3 | 101.7 |
| 14 | NCU01197 | Gulliver-1 | 42.0 ± 2.8 | − 87.9 |
| 15 | NCU09364 | Heat shock protein 30 | 261.0 ± 26.9 | − 24.8 |
| 16 | NCU06265 | Hypothetical protein | 324.5 ± 54.4 | − 6.5 |
| 17 | NCU01213 | Superoxide dismutase-2 | 345.0 ± 83.4 | − 0.6 |
| 18 | NCU09494 | Hypothetical protein | 366.5 ± 75.7 | 5.6 |
| 19 | NCU08741 | Hyphal anastomosis-3 | 380.0 ± 127.3 | 9.5 |
| 20 | NCU03894 | Serine/threonine protein kinase-4 | 382.5 ± 95.5 | 10.2 |
| 21 | NCU01833 | Nonidentical kinase-2 | 401.0 ± 79.2 | 15.6 |
| 22 | NCU02542 | Embden–meyerhof pathway-1 | 406.0 ± 19.8 | 17.0 |
| 23 | NCU01181 | Acyl-CoA dehydrogenase-3 | 473.5 ± 79.9 | 36.5 |
| 24 | NCU08225 | High affinity nickel transporter nic1 | 498.0 ± 46.7 | 43.5 |
| 25 | NCU03164 | Two-component system response regulator | 615.0 ± 62.2 | 77.2 |
| 26 | NCU09882 | Metacaspase-1A | 644.5 ± 21.9 | 85.7 |
| 27 | NCU09450 | Regulatory particle, non-ATPase-like-2 | 322.5 ± 64.3 | − 7.1 |
| 28 | NCU02260 | Regulatory particle, ATPase-like-3 | 370.5 ± 43.1 | 6.8 |
| 29 | NCU00634 | Ribosomal protein L14 | 375.0 ± 77.8 | 8.1 |
| 30 | NCU03702 | rRNA 2′-O-methyltransferase fibrillarin | 440.0 ± 67.9 | 26.8 |
| 31 | NCU08050 | Hypothetical protein | 453.5 ± 50.2 | 30.7 |
| 32 | NCU06764 | Proteasome catalytic alpha-2 | 490.0 ± 155.6 | 41.2 |
| 33 | NCU00396 | Pre-mRNA-splicing factor rse-1 | 502.5 ± 51.6 | 44.8 |
| 34 | NCU03184 | C2H2 conidiation transcription factor FlbC | 543.0 ± 28.3 | 56.5 |
| 35 | NCU02604 | Hypothetical protein | 456.0 ± 33.9 | 31.4 |
| 36 | NCU05295 | Proteasome catalytic alpha-5 | 580.5 ± 26.2 | 67.3 |
| 37 | NCU06440 | Proteasome catalytic alpha-4 | 607.5 ± 41.7 | 75.1 |
| 38 | NCU01368 | Proteasome catalytic beta-4 | 651.5 ± 68.6 | 87.8 |
| 39 | NCU00824 | Histone deacetylase-3 | 720.0 ± 14.1 | 107.5 |
| 40 | NCU06429 | Alpha-actinin | 351.0 ± 38.2 | 1.2 |
| 41 | NCU00554 | Homoserine-1 | 371.5 ± 64.3 | 7.1 |
| 42 | NCU08093 | Hypothetical protein | 448.5 ± 78.5 | 29.3 |
| 43 | NCU00105 | 60S ribosome subunit biogenesis protein NIP7 | 484.5 ± 17.7 | 39.6 |
| 44 | NCU03479 | Endoribonuclease ysh-1 | 485.0 ± 77.8 | 39.8 |
| 45 | NCU09366 | Proteasome catalytic beta-6 | 611.0 ± 55.2 | 76.1 |
| 46 | NCU07420 | eIF4A | 633.0 ± 53.7 | 82.4 |
| 47 | NCU00467 | COP9 signalosome-5 | 267.5 ± 17.7 | − 22.9 |
| 48 | NCU01408 | COP9 signalosome-3 | 310.5 ± 0.7 | − 10.5 |
| 49 | NCU00923 | Topogenesis of outer membrane beta barrel protein 37 | 336.5 ± 19.1 | − 3.0 |
| 50 | NCU04669 | Hypothetical protein | 332.0 ± 72.1 | − 4.3 |
| 51 | NCU04242 | Period-6 | 357.0 ± 42.4 | 2.9 |
| 52 | NCU02057 | Autoinducer 2 sensor kinase/phosphatase luxQ | 376.0 ± 1.4 | 8.4 |
| 53 | NCU09842 | Mitogen activated protein kinase-1 | 385.5 ± 33.2 | 11.1 |
| 54 | NCU01033 | Hypothetical protein | 387.0 ± 83.4 | 11.5 |
| 55 | NCU08875 | Cullin binding protein CanA | 397.0 ± 49.5 | 14.4 |
| 56 | NCU00810 | Glycosylhydrolase family 2–3 | 436.5 ± 19.1 | 25.8 |
| 57 | NCU09201 | Hypothetical protein | 636.0 ± 58.0 | 83.3 |
| 58 | NCU04096 | Protein kinase-9 | 462.9 ± 18.3 | 33.4 |
| 59 | NCU00204 | Hypothetical protein | 306.3 ± 33.6 | − 11.7 |
| 60 | NCU00355 | Catalase-3 | 331.1 ± 26.7 | − 4.6 |
| 61 | NCU01225 | Ubiquitin conjugating enzyme | 259.0 ± 29.8 | − 25.4 |
| 62 | NCU01312 | Regulator of conidiation in Aspergillus-1 | 336.1 ± 33.8 | − 3.1 |
| 63 | NCU01613 | Protoperithecia-2 | 372.0 ± 31.1 | 7.2 |
| 64 | NCU01642 | Hypothetical protein | 283.8 ± 22.9 | − 18.2 |
| 65 | NCU02133 | Superoxide dismutase-1 | 262.7 ± 24.5 | − 24.3 |
| 66 | NCU02387 | Nuclear import and export protein Msn5 | 306.3 ± 33.6 | − 11.7 |
| 67 | NCU02498 | Cullin-3 | 334.9 ± 21.4 | − 3.5 |
| 68 | NCU02794 | Soft | 269.0 ± 43.9 | − 22.5 |
| 69 | NCU03013 | Anchored cell wall protein-10 | 277.7 ± 45.7 | − 20.0 |
| 70 | NCU03076 | Delta-1-pyrroline-5-carboxylate dehydrogenase | 305.0 ± 21.2 | − 12.1 |
| 71 | NCU03125 | NIMA-interacting protein TinC | 304.9 ± 21.0 | − 12.1 |
| 72 | NCU03281 | Transport of copper-2 | 347.3 ± 18.0 | 0.1 |
| 73 | NCU03314 | mob2-like-a | 364.6 ± 20.6 | 5.1 |
| 74 | NCU03623 | Ubiquitin-conjugating enzyme E | 291.3 ± 12.4 | − 16.1 |
| 75 | NCU06175 | Peroxin 3 | 473.0 ± 38.1 | 36.3 |
| 76 | NCU06255 | Hypothetical protein | 317.4 ± 17.8 | − 8.5 |
| 77 | NCU03938 | Alternative oxidase-5 | 156.9 ± 32.6 | − 54.8 |
| 78 | NCU04264 | Extracellular developmental signal biosynthesis protein FluG | 331.1 ± 26.7 | − 4.6 |
| 79 | NCU04302 | Ubiquitin-conjugating enzyme E | 331.0 ± 15.6 | − 4.6 |
| 80 | NCU04513 | Ubiquitin conjugating enzyme Ubc14 | 364.6 ± 20.6 | 5.1 |
| 81 | NCU04533 | Abundant perithecial protein | 339.9 ± 28.5 | − 2.1 |
| 82 | NCU04834 | Phytochrome-1 | 373.4 ± 23.4 | 7.6 |
| 83 | NCU05046 | E1–E2 ATPase-1 | 329.7 ± 27.9 | − 5.0 |
| 84 | NCU05591 | ABC transporter CDR4 | 353.3 ± 47.1 | 1.8 |
| 85 | NCU07378 | Serine/threonine protein kinase-12 | 323.7 ± 37.2 | − 6.7 |
| 86 | NCU07728 | Siderophore regulation | 416.9 ± 9.8 | 20.1 |
| 87 | NCU08055 | b-ZIP transcription factor IDI4 | 331.1 ± 26.7 | − 4.6 |
| 88 | NCU08791 | Catalase-1 | 361.0 ± 26.9 | 4.0 |
| 89 | NCU03725 | Vegetative incompatibility blocked-1 | 415.7 ± 20.2 | 19.8 |
| 90 | NCU03043 | C2H2 finger domain-containing protein FlbC | 356.0 ± 19.8 | 2.6 |
GUL-1 is involved in mycelial morphology in N. crassa
In order to determine the extent of the morphological changes conferred by gul-1 disruption, we conducted a microscopic analysis of Δgul-1 grown under permissive temperature. When grown on agar plates at 25 °C, there were significant differences in hyphal elongation and branching frequency between Δgul-1 and WT. The Δgul-1 exhibited slow growth rate and hyperbranching (Additional file 1: Figure S1), which is consistent with previous observations [23]. These results indicated that disruption of gul-1 substantially impact hyphal development in N. crassa.
A detailed analysis of the effects of disrupting gul-1 on the rheological properties of mycelial broth was also performed. Conidia of WT and Δgul-1 were inoculated into Avicel medium and batch cultured for 7 days. The viscosity of WT increased to 557 ± 44 cp in the first 72 h before stabilizing until 120 h when the viscosity decreased significantly (Fig. 2). However, the rheological properties of Δgul-1 cultures were quite different from that of the WT. Firstly, the viscosity of Δgul-1 was much higher than that of WT 48 h post-inoculation, with its viscosity then decreasing dramatically during the time course of batch fermentation. After 7 days fermentation, the viscosity of Δgul-1 was only one-fifth that of WT (Fig. 2). According to microscopic observations, the WT showed a clump type of morphology. However, Δgul-1 grew as pellets that became more compact as fermentation progressed (Fig. 2). The diameter of pellets was about 350 ± 62 μm. Furthermore, scanning electron microscopy demonstrated that Δgul-1 exhibited much smoother surface than the WT (Additional file 2: Figure S2).
Fig. 2.
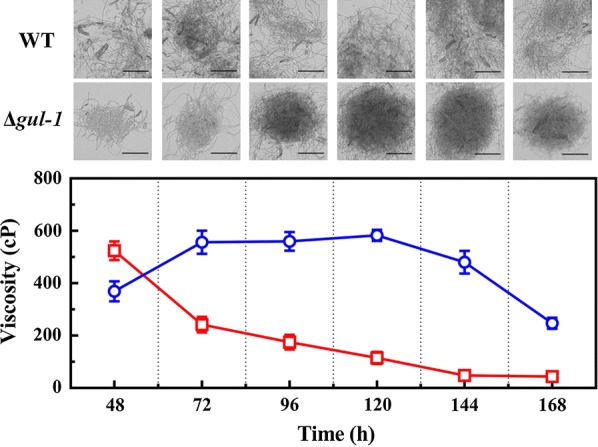
Mycelial morphologies of wild type and Δgul-1 mutant during submerged cultivation. Conidia from the wild type (WT) and gul-1 mutant (Δgul-1) were separately inoculated into Avicel medium and batch cultured for 7 days, and the viscosities of the broths were measured at 24 h intervals. The Δgul-1 mutant grew in pellet form, whereas the wild type exhibited a clump type morphology. Blue line indicates WT; Red line indicates the Δgul-1 mutant. Scale bar is 200 μm. Values represent the means of at least three replicates, error bars show standard deviation
It is well established that mycelial biomass significantly affects the apparent viscosity of fermentation broth. Thus, the biomass of Δgul-1 and WT were determined as described [14]. The results revealed that the dry weight of Δgul-1 was similar to that of the WT (Additional file 3: Figure S3). Thus, the change in rheological properties was due to the morphological change in mycelia, and not to a reduction in biomass.
In addition, fungal morphology in submerged cultures is affected by the type of carbon source [29]. The Δgul-1 grew as pellets, and exhibited dramatically reduced viscosity (> 50%) compared with that of WT when grown on glucose, xylose and fructose (Additional file 4: Figure S4) indicated that growth form is independent of the type of carbon source. This suggests that gul-1 might be considered as a universal target for morphology control under different cultivation conditions.
To the best of our knowledge, this is the first report indicating that N. crassa can change morphological forms in submerged culture (from clump type to pellet type) through genetic modification.
Expression level of gul-1 affected culture viscosity
To determine whether gul-1 expression levels affect the rheological properties of N. crassa fermentation, we engineered the Δgul-1 strain to express a C-terminal GFP-tagged GUL-1 under the regulation of either the native gul-1 promoter (strain Pn-gul-1, complemented strain) or the constitutive ccg-1 promoter (strain Pc-gul-1, gul-1 over-expression strain). Complemented Pn-gul-1 was indistinguishable from WT in any of the phenotypic assays performed in this study (data not shown). The Pc-gul-1 strain showed WT growth rates on MM agar (Additional file 5: Table S1) and WT clump type morphology in submerged culture, but the viscosity was approximately 50% greater than that of WT (Fig. 3). These results indicate that gul-1 plays important roles in controlling mycelial morphology during submerged cultivation.
Fig. 3.
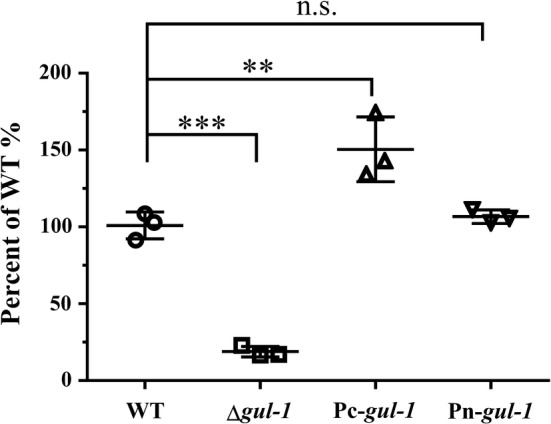
Comparison of the viscosity of culture broths from WT, Δgul-1, Pc-gul-1 and Pn-gul-1 strains grown on Avicel medium for 7 days. The following strains were grown in 2% (w/v) Avicel media: the WT, the gul-1 gene knockout mutant (Δgul-1) and the complemented strain under either the control of the ccg-1 promoter (Pc-gul-1) or the native promoter (Pn-gul-1). The viscosity was measured and displayed after normalization to the WT control according to percentage. Values represent the means of at least three biological replicates, error bars show standard deviation. Statistical significance was performed using a two-tailed Student’s t test (**P < 0.01; ***P < 0.001; n.s., not significant)
Subcellular localization of the GUL-1 protein in N. crassa
To assess the subcellular localization of GUL-1 protein, we tracked the C-terminal GFP-tagged GUL-1 in the Δgul-1 strain. Under its native promoter, GFP-tagged GUL-1 fluorescence was hard to observe in mycelia, but detectable in conidia (data not shown), indicating that gul-1 had a lower expression level in mycelium than conidia. When the GUL-1-GFP fusion encoding gene was under the control of the ccg-1 constitutive promoter, GFP fluorescence was uniformly distributed in the cytoplasm of both hyphae and conidia (Fig. 4), similar to that reported for the Colletotrichum lagenarium Classd1p [30]. However, recent studies have shown that S. cerevisiae Ssd1, the homolog of N. crassa GUL-1, transiently localizes to the nucleus [31]. To address whether GUL-1 also enters the nucleus, we surveyed GUL-1 for a signal sequence using NLStradamus which predicted a putative lysine-rich nuclear localization signal (amino acids 497–507: KREKEEKKKRK) [32]. Thus, GUL-1 is probable a nucleocytoplasmic protein, which usually localizes to the cytoplasm. The detailed mechanism of nucleocytoplasmic shuttling of GUL-1 remains to be elucidated.
Fig. 4.
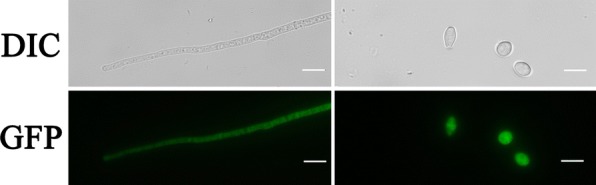
The subcellular localization of GUL-1 in Neurospora crassa. Locations of GUL-1 proteins were monitored by recording enhanced green fluorescent protein signal. Microscopic observation was performed with an OLYMPUS BX51 microscope. Scale bar is 10 μm
Sensitivity to cell wall inhibitors and oxidative agents
Our results indicate that gul-1 influences the mycelial morphology of N. crassa, so we determined whether Δgul-1 suffers defects in cell wall integrity. We therefore examined its sensitivity to cell wall perturbation, oxidative stress and high osmolarity. The results showed that Δgul-1 had increased sensitivity to Congo Red, which binds to β-1,3-glucans and interferes with cell wall construction [33]. Growth of Δgul-1 on MM containing 1 mg/mL Congo Red was reduced by 29.7 ± 0.3%, whereas growth of WT was only reduced by 17.7 ± 0.5%. Likewise, Δgul-1 showed increased sensitivity to Calcofluor White, which disrupts cell wall synthesis by binding to chitin [34]. Growth of Δgul-1 on MM supplemented with 200 μg/mL Calcofluor White was reduced by over 29.3 ± 1.3%, whereas growth of WT was only reduced by 19.1 ± 2.1% (Fig. 5), suggesting that the mutant had altered cell wall integrity. These data are consistent with other studies, where deletion of the gul-1 homolog in the plant pathogen Magnaporthe grisea also resulted in hypersensitivity to Calcofluor White [30].
Fig. 5.
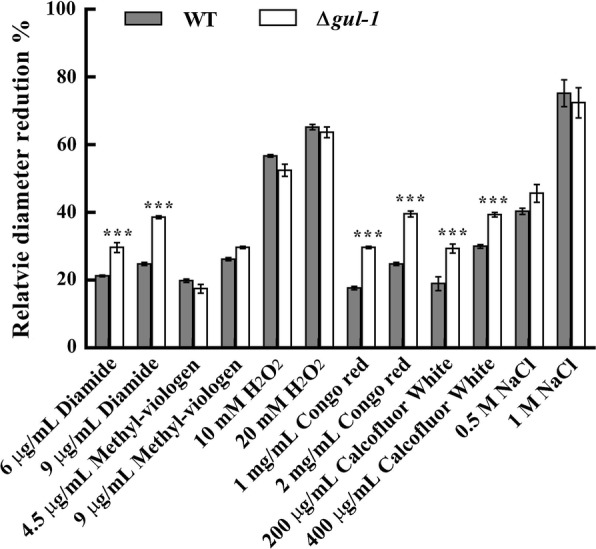
Effect of different concentrations of chemicals on hyphal growth in WT and Δgul-1. Aliquots of 5 μL 1 × 107 mL−1 spore suspensions of WT and Δgul-1 were incubated at 28 °C for 18 h on MM plates incorporating H2O2 (10, 20 mM), diamide (6, 9 μg/mL), methyl-viologen (4.5, 9 μg/mL), NaCl (0.5, 1.0 M), Congo Red (1, 2 mg/mL) or Calcofluor White (200, 400 μg/mL), and then the diameter of each colony was measured. Relative diameter reduction means the reduction of growth rate on MM containing chemicals compared with the growth on MM only. Values represent the means of at least three biological replicates, error bars show standard deviation. Statistical significance was performed using a two-tailed Student’s t test (***P < 0.001)
Oxidative stress was provided by hydrogen peroxide (H2O2), diamide or methyl-viologen. These oxidants elicit oxidative damage to fungal cells in different ways. H2O2 is a common oxidative agent able to react with many biological molecules including proteins and DNA [35]. Diamide triggers oxidative stress by oxidizing intracellular glutathione [36]. Methyl-viologen is a redox cycling reagent that produces superoxide anions under aerobic condition [37]. All three oxidative agents partially inhibited the elongation of hyphae in both Δgul-1 and WT strains; only diamide was more effective on Δgul-1 than the WT (Fig. 5). The growth of Δgul-1 on MM agar containing 9 μg/mL diamide was reduced by 38.6 ± 0.4% as compared to 24.8 ± 0.5% for the WT. These results indicate that gul-1 has a role in glutathione-mediated antioxidant processes, but not in general oxidative stress protection of N. crassa.
Δgul-1 grown on MM medium supplemented with NaCl exhibited no greater sensitivity to osmotic stress than the wild type strain (Fig. 5), indicating that gul-1 is not required for osmotic stress tolerance.
Deletion of gul-1 improved protein secretion
As a cellulolytic filamentous fungus, N. crassa possesses an extraordinary capacity to secret a wide variety of lignocellulolytic enzymes, making it a potential workhorse for production of industrial enzymes and high-value vaccines [38]. Based on our findings, we interrogated whether the defect in cell wall integrity would enhance protein secretion in N. crassa. The ability of WT and Δgul-1 to secrete cellulases was compared in Avicel medium. The Δgul-1 mutant secreted 25% more extracellular proteins 7 days post-inoculation. Although endoglucanase activity was unaffected, β-glucosidase activity in Δgul-1 was 56% higher than that of WT (Fig. 6). We also assessed whether disrupting gul-1 would enhance protein secretion on other carbon sources. Compared with WT, the secreted protein titers in the Δgul-1 strain were 181, 57 and 43% higher than those of WT strain on sucrose, fructose or xylose, respectively (Additional file 4: Figure S4), suggesting that the enhancement of protein secretion in the Δgul-1 strain is independent of the carbon source. These results also indicate that cell wall integrity could play a critical role in protein secretion with implications for other filamentous fungi.
Fig. 6.
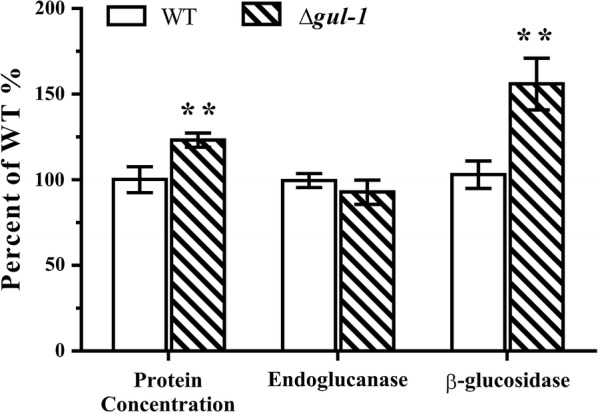
Phenotype of WT and Δgul-1 strains when grown on Avicel medium. Conidia from the wild type (WT) and the gul-1 knockout mutant (Δgul-1) were separately inoculated into Avicel medium and batch cultured. After 7 days, total extracellular protein concentration, endoglucanase activity and β-glucosidase activity were measured. Data were normalized to the WT control according to percentage. Values represent the means of at least three biological replicates, error bars show standard deviation. Statistical significance was performed using a two-tailed Student’s t test (**P < 0.01)
Comparative analysis of transcriptomes of WT and the Δgul-1 strain
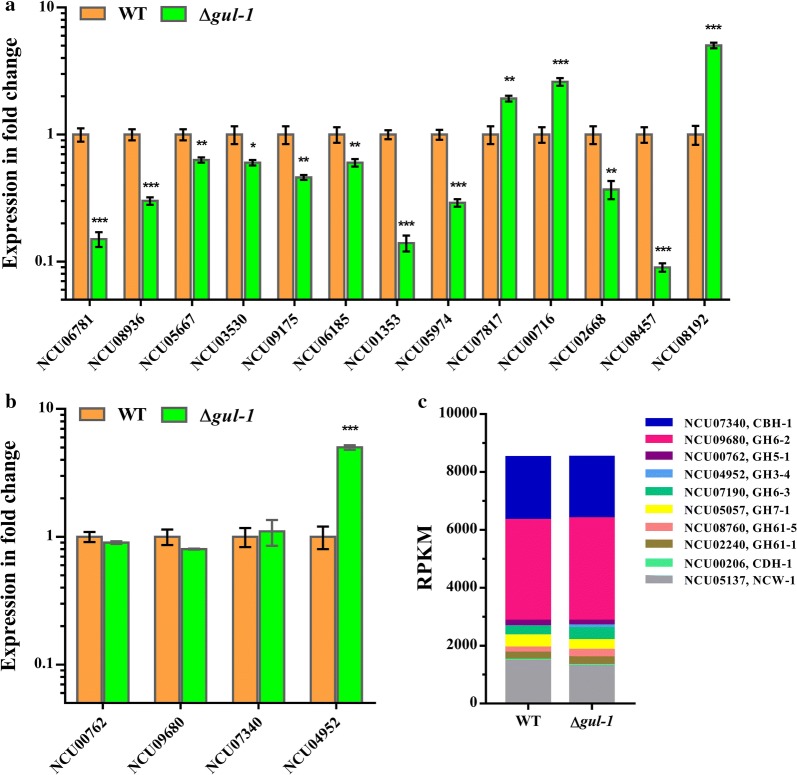
Disruption of gul-1 in N. crassa caused pellet growth in submerged cultivation (Fig. 2). It is well established that fungal pellet formation is due to the combination of electrostatic interactions, hydrophobicity and specific interactions from cell wall components [39]. Based on the analysis above, we therefore hypothesized that the mechanism of pellet formation in Δgul-1 might feasibly be explained by changes in cell wall organization. To test this hypothesis, we evaluated transcriptional changes in the Δgul-1 mutant when grown on Avicel medium for 3 days using the WT as a control. RNA-seq data showed that the expression level of 101 genes was significantly lower in the Δgul-1 mutant, while the expression level of 145 genes increased (Additional file 6: Table S2). It is worth noting that the expression levels of several cell wall-related genes were significantly altered in Δgul-1. These cell wall proteins are critical components of the fungal cell wall. Based on the manner in which they are attached to the cell wall polysaccharides, the cell wall proteins have been classified into two classes in N. crassa: GPI-anchored cell wall protein (ACW) and non-anchored cell wall protein (NCW) [40]. As previously reported, 22 cell wall proteins have been identified in N. crassa vegetative hyphae cell wall proteomic analysis [40]. Of the 22 identified genes, we detected 10 (45%) genes with altered expression (Fig. 7a and Additional file 7: Table S3). Eight GPI-anchored cell wall protein genes showed a decrease in expression level > twofold. Among these genes, four GPI-anchored glycoside hydrolase genes (NCU09175, NCU01353, NCU05974 and NCU6781), which were probably involved in cell wall remodeling, were down-regulated 2.1-, 7.1-, 3.4- and 6.7-fold in gul-1 mutant, respectively, suggesting a reorganized arrangement of the cell wall. Moreover, anchored cell wall genes acw-1 (NCU08936), acw-3 (NCU05667), acw-6 (NCU03530), and acw-9 (NCU06185) were also significantly down-regulated in Δgul-1. In S. cerevisiae, acw-1 ortholog (ecm33) played a critical role in cell wall organization. Deletion of ecm33 results in a weakened and disorganized cell wall [41]. On the contrary, non-anchored cell wall genes ncw-3 (NCU07817) and ncw-5 (NCU00716) were up-regulated 2.7- and 3.2-fold, respectively. Moreover, in S. cerevisiae, SUN4 is involved in cell wall remodeling, and regulated by the yeast GUL-1 homologue Ssd1 [21]. As expected, the cell wall synthesis gene (NCU02668), a homolog of S. cerevisiae sun4, was down-regulated 2.9-fold in Δgul-1, suggesting a possible direct regulation of NCU02668 by GUL-1 in N. crassa. Furthermore, a previous study has demonstrated that hydrophobins play important roles in pellet formation. In Aspergillus nidulans, disruption of hydrophobin gene rodA led to a decrease in pellet biomass and size [42]. According to our results, the Class I hydrophobin gene eas (NCU08457) was down-regulated 15-fold in the Δgul-1 strain, whereas the Class II hydrophobin gene NCU08192 was up-regulated 6.2-fold, indicating that hyphal hydrophobicity might be changed in Δgul-1. Taken together, the significant changes in expression of cell wall-related proteins suggested that the Δgul-1 mutant has an abnormal cell wall structure, which might be the reason for pellet formation.
Fig. 7.
Transcriptome analysis of the Δgul-1 strain on Avicel medium. a Expression levels of genes encoding cell wall proteins in Δgul-1 mutant relative to wild-type (WT) strain on Avicel. b Expression levels of major cellulase genes in Δgul-1 mutant relative to the wild-type (WT) strain on Avicel. c Expression levels from RNA-seq data of genes encoding major secreted proteins from WT and Δgul-1 when grown on Avicel medium. After Δgul-1 and WT conidia were grown on Avicel for 3 days, the transcriptional abundance of major cellulase genes and cell wall protein genes was evaluated by RNA-seq and quantitative real-time PCR. Values represent the means of at least three biological replicates, error bars show standard deviation. Statistical significance was performed using a two-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001)
In addition, the expression levels of major cellulase genes were not affected by the gul-1 deletion, except that the extracellular β-glucosidase gene (NCU04952) had significantly higher expression level in Δgul-1 (Fig. 7b and c). These results are consistent with the enzyme activity assay shown in Fig. 6.
Genetic engineering of N. crassa for hyper-cellulase production and lower viscosity
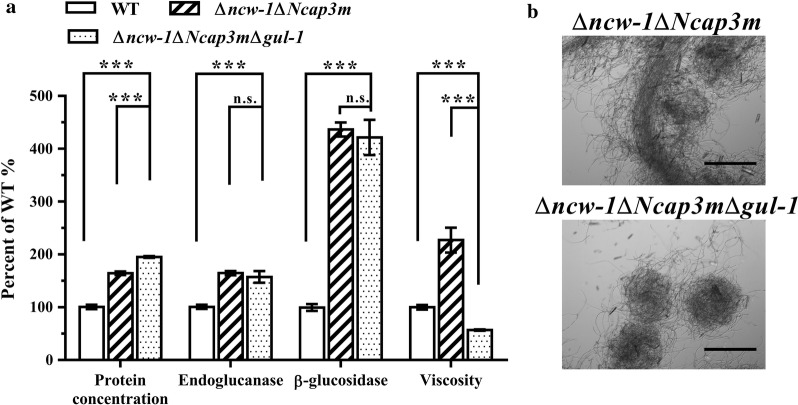
Recent investigations have revealed that N. crassa is a good model system for unraveling the mechanisms of lignocellulose degradation [43, 44]. Meanwhile, several key genes involved in cellulase expression and secretion have been identified and characterized in N. crassa. NCU05137, which encodes a non-anchored cell wall protein (NCW-1), could be detected in the secreted proteome of cultures containing cellulose as sole carbon [45]. The disruption of ncw-1 resulted in significantly increased cellulase production and cellulolytic activity [12]. In addition, we previously reported that disrupting Ncap3m (NCU03998, which encodes the μ subunit of the adaptor protein 3 complex) also dramatically increases lignocellulase secretion [16]. Based on these findings, a hyper cellulase producing strain ∆ncw-1∆Ncap3m was generated by sexual crosses. Compared to WT, the extracellular protein concentration of the ∆ncw-1∆Ncap3m strain was improved by approximately 65% (~ 0.8 g/L) and endoglucanase activity increased by about 73% (Fig. 8a). However, ∆ncw-1∆Ncap3m exhibited poor rheological properties, and the viscosity of ∆ncw-1∆Ncap3m was almost twofold higher than that of the WT. We therefore determined whether the rheological properties of the broth could be improved by deleting the gul-1 gene in ∆ncw-1∆Ncap3m. The triple deletion strain (∆ncw-1∆Ncap3m∆gul-1) was obtained by crossing Δgul-1 with ∆ncw-1∆Ncap3m, and the progeny were verified by PCR. Not surprisingly, the triple deletion strain grew as pellets (Fig. 8b), and the viscosity of ∆ncw-1∆Ncap3m∆gul-1 was only 23% that of the double deletion strain (∆ncw-1∆Ncap3m), and 53% that of the WT strain (Fig. 8a). Production of extracellular protein by ∆ncw-1∆Ncap3m∆gul-1 was significantly improved relative to ∆ncw-1∆Ncap3m. However, no significant differences in endoglucanase and β-glucosidase activities were apparent.
Fig. 8.
Phenotype of ∆ncw-1∆Ncap3m∆gul-1 triple mutant when grown on Avicel medium. Conidia of the wild type (WT), the double deletion strain (∆ncw-1∆Ncap3m) and the triple deletion strain (∆ncw-1∆Ncap3m∆gul-1) were separately inoculated into Avicel medium, and then cultured for 7 days. a Total extracellular protein concentration, endoglucanase activity, β-glucosidase activity and the viscosity of culture broth were measured and normalized to the WT according to percentage. Values represent the means of at least three biological replicates, error bars show standard deviation. Statistical significance was performed using a two-tailed Student’s t test (***P < 0.001; n.s., not significant). b Mycelial morphologies of the triple deletion strain (∆ncw-1∆Ncap3m∆gul-1) and its parental strain (∆ncw-1∆Ncap3m) after 7 days cultivation. The images were acquired by an Olympus SZX-7 stereomicroscopy with a digital camera attached. Scale bar is 300 μm
Discussion
In fungal fermentations, high apparent viscosity and non-Newtonian behavior of broths are critical problems in industrial application. Thus, controlling the morphology of filamentous fungus in submerged fermentations is a major challenge. Many strategies have been applied to solve these problems, principally by optimization of fermentation process parameters [5–8]. However, morphology control by altering chemical and physical factors is not sufficient. Moreover, these optimized parameters are not always suited for achieving maximum productivity. Recently, several attempts have been employed to control mycelial morphology by genetic modification. As previously reported, the motor proteins played critical roles in polarized growth in Aspergillus spp. Disruption of kinesin motor KipA in Aspergillus glaucus caused pellet growth in submerged cultivation and the aspergiolide A production was 82% higher than that of wild type strain [46]. Furthermore, disturbance of cell wall synthesis had significant influences on mycelial morphology [47]. In Aspergillus orzyae, disruption of chsB had no effect on enzyme production, but the viscosity was lower than that of parental strain [48]. In Penicillium chrysogenum, down-regulation of chitin synthase gene resulted in pellet growth in submerged culture, and also caused enhanced penicillin production [49]. Thus, rationally shaping mycelial morphology through genetic modification seems to be an attractive strategy. However, large-scale generation and screening of morphological mutants is difficult to achieve. Fortunately, N. crassa possesses a near full genome deletion strain collection [27]. In this work, we systematically screened the 90 N. crassa morphological mutants to identify the critical genes that could influence mycelial morphology during submerged cultivation. The viscosity of fermentation broth was used to reflect mycelial morphology change. Fourteen mutants showed increased viscosity compared to the WT strain. Among these mutants, we found several genes involved in the proteasome pathway, e.g., NCU01368, NCU05295, NCU09366 and NCU06440. All these four genes encode different subunits of the proteasome, implying that interruption of the protein degradation pathway might influence mycelial morphology. Moreover, NCU00824 encodes a histone deacetylase which possibly contributes to chromatin structure modification. Previous work in the yeast C. albicans has demonstrated that histone deacetylase interacting with transcription factors plays a critical role in morphological transition [50]. The high-viscosity phenotype induced by a defect in NCU00824 implies a close link between chromatin structure modification and morphology control in filamentous fungi.
Of most interest, we found that disruption of gul-1 (NCU01197) in N. crassa led to favorable morphological changes in submerged culture. The Δgul-1 mutant grew as pellets and showed a low viscosity phenotype. Furthermore, scanning electron microscopy observations revealed that the cell wall structure was altered in the gul-1 mutant. Our results with Congo Red and Calcofluor White also illustrated that gul-1 is required for cell wall integrity. Based on these results, we hypothesized that pellet formation is closely associated with changes in cell wall structure. In filamentous fungi, β-(1,3)-glucan and chitin are two major structural components of the fungal cell wall. However, according to our results, the transcript levels of genes encoding chitin and glucan synthases (chs1-7 and gls-1), which are involved in cell wall synthesis, were not changed by the gul-1 deletion (Additional file 6: Table S2). Moreover, the expression levels of chit-1 (NCU02184) and gh18-5 (NCU04554), which are involved in cell wall maintenance and remodeling [51], were also not altered in Δgul-1 (Additional file 6: Table S2). It suggested that these genes are probably not involved in conferring the gul-1 phenotype. On the other hand, cell wall proteins are the important components of the fungal cell wall, and also play important roles in cell wall biosynthesis and interactions with external environment [40]. Intriguingly, the transcript abundance of several cell wall protein genes was found to be significantly altered in Δgul-1. Among these genes, the GPI-anchored cell wall β-1,3-endoglucanase gene bgt-2 (NCU09175), which is seemingly involved in cell wall remodeling, was down-regulated 2.1-fold in Δgul-1. A recent study also showed the cell wall architecture is altered in the Δbgt-2 mutant [52]. Likewise, the transcript abundances of GPI-anchored glycoside hydrolases NCU05974 and NCU06781, the homologs of S. cerevisiae crh1 and Aspergillus fumigatus gel2, respectively, were also lower in Δgul-1. It has been reported that both S. cerevisiae crh1 and A. fumigatus gel2 are required for normal cell wall structure. Loss of S. cerevisiae crh1 causes hypersensitivity to Congo Red and Calcofluor White, suggesting defects in cell wall structure [53]. In A. fumigatus, the gel2 disruption strain exhibits slower growth and an altered cell wall composition [54]. These implicated that down-regulation of both NCU05974 and NCU06781 may results in an abnormal cell wall structure, leading to influencing mycelial morphology during submerged cultivation. Besides that, other GPI-anchored cell wall proteins, which are of unknown function, are annotated as ACW (GPI-anchored cell wall protein) [40]. They may serve as important structural components of the wall, and play critical roles in maintaining cell wall stability and function [55]. For example, AfuEcm33, a homolog of the N. crassa GPI-anchored cell wall protein-1 (acw-1), has been shown to be involved in maintaining cell wall integrity and virulence in A. fumigatus [56]. It is worth noting that disruption of AfuEcm33 results in increased cell–cell adhesion. The disruption strain forms large ‘star-shaped’ clusters of germinating conidia during submerged cultivation [56]. In Candida albicans, a similar aggregate phenotype has been also observed in the Caecm33 deletion strain [57]. In this study, the GPI-anchored cell wall protein ACW-1 encoded by NCU08936 was found to be down-regulated in Δgul-1, suggesting that ACW-1 might, at least in part, contribute to pellet formation by increased cell–cell adhesion. Taken together, our results indicated that gul-1 affects mycelial morphology during submerged cultivation by influencing the expression of cell wall-related protein. However, a detailed mechanistic description of how the gul-1 influences these genes expression requires further investigation.
According to our results, the deletion of gul-1 led to a hypersecretion phenotype in N. crassa. When grown on 2% (w/v) Avicel, the extracellular protein secreted by the Δgul-1 mutant was about 25% higher than that in the WT, while only β-glucosidase activity was found to be improved. These results were also confirmed by our transcriptomic analysis (Fig. 7b and c). However, the increase in activity was disproportionate to the increase in total protein, suggesting that the effect of gul-1 on protein secretion is not due to increased transcription levels. Thus, we hypothesized that the defects in cell wall integrity benefit protein secretion. According to the ‘bulk flow’ hypothesis, growing hyphal tips are filled with vesicles that contain extracellular proteins, and these proteins get carried to the outer region of the wall for secretion [58]. However, some extracellular proteins might become trapped in the rigid cell wall. These ‘trapped’ enzymes could be released into an external medium through a weakening of cell wall integrity [59], which may explain the Δgul-1 mutant having a higher protein secretion capacity. Similar results could be observed in the N. crassa slime mutant which has serious defects in cell wall synthesis [60].
Our previous study demonstrated that the disruption of ncw-1 led to enhanced cellulase production in N. crassa, but unfortunately the viscosity of ∆ncw-1 was increased two fold [14]. A similar phenotype was also observed in the ∆ncw-1∆Ncap3m strain. Having identified the impact of gul-1 disruption on mycelial morphology during submerged cultivation, we applied this approach to the hyper-cellulase producing strain ∆ncw-1∆Ncap3m. The triple deletion strain (∆ncw-1∆Ncap3m∆gul-1) grew in pellet form and exhibited a low viscosity phenotype, indicating that gul-1 is a promising target gene for morphological improvement of hyper-cellulase-producing strains. In addition, phylogenetic analyses demonstrated that GUL-1 is highly conserved in other industrial filamentous fungi, including Aspergillus niger (XP_001392470, identity 66%), Myceliophthora thermophila (XP_003661716, 75% identity) and Trichoderma reesei (XP_006964501, 78% identity) (Additional file 8: Figure S5). Thus, manipulation of gul-1 homologs could be a highly useful strategy for morphology engineering of industrial filamentous fungi.
Conclusions
In this work, we systematically screened morphological mutants of N. crassa and identified several potential genes that affect mycelial morphology in submerged cultivation. Deletion of one of these genes, gul-1, resulted in the pellet form and enhanced protein secretion in N. crassa. Transcriptional profiling revealed that a deletion of gul-1 led to an alteration in transcription of genes encoding cell wall proteins, which may be the reason for changes in mycelial morphology during submerged cultivation. Furthermore, a significant reduction in viscosity of broth culture was obtained when gul-1 was deleted from the hyper cellulolytic strain ∆ncw-1∆Ncap3m. These results indicated that gul-1 might be a potential target to control mycelial morphology in submerged cultivation. This work provides a novel strategy for morphological engineering of filamentous fungi through genetic modification, especially for industrially relevant species.
Methods
Strains, growth conditions and microscopy
Neurospora crassa strains were obtained from Fungal Genetics Stock Centre (FGSC), including wild type (WT, FGSC 2489, A), a his-3 mutant strain (FGSC 9716, his-3, a) and the morphological mutants. The his-3; Δgul-1, a strain was obtained by crossing FGSC 9716 with FGSC 11288 (NCU01197, A). Multiple deletion strains were made by performing sequential crosses. All of the constructed strains were verified by PCR.
To obtain conidia, N. crassa was grown on slant tubes containing Vogel’s minimal media (1 × Vogel’s salts, 2% w/v sucrose and 1.5% w/v agar) for 10 days at 28 °C. For flask culture, 10-day-old conidia of N. crassa strains were suspended in sterile water and inoculated using a final concentration of 1 × 105 per mL into a 250-mL Erlenmeyer flask containing 100 mL Avicel medium (1 × Vogel’s salts, 2% w/v Avicel, 0.75% w/v yeast extract and 0.2% v/v Tween 80), then cultured for 7 days at 25 °C, with shaking at 200 rpm under constant light. The observations were carried out on 7 consecutive days at 24 h intervals.
Microscopic observations were carried out using an OLYMPUS BX51 microscope equipped with a QImagingRetiga 2000R camera (QImaging, Surrey, Canada), and analyzed with the Image-Pro Express 6.3 software. The images were also acquired by an Olympus SZX-7 stereomicroscopy with a digital camera attached.
To compare the differences in cell surface structure between the WT and Δgul-1, fungal conidia were incubated in Avicel medium at 25 °C for 5 days. The harvested mycelia were fixed in 0.1 M phosphate buffer (pH 7.2) containing a final concentration of 2.5% (w/v) glutaraldehyde. The samples were rinsed in thrice-distilled PBS, and dehydrated with a series of ethanol solutions (30–100%, v/v). After which the samples were dried by critical point drying (Leica CPD300) and coated with platinum (Hitachi E-1045). Observations were conducted using a scanning electron microscope (Hitachi SU8010) operating at 1 kV.
Plasmid construction and transformation
Genomic DNA from the WT strain was extracted as described previously [61]. To complement Δgul-1, the open reading frame (ORF) of gul-1 was cloned by PCR with primers ORF-F and ORF-R, and the fragment was inserted into the XbaI and PacI sites of pMF272 to form pMF272-Pc-gul-1-gfp. The promoter of gul-1 was amplified with primers PF and PR, and then cloned into the NotI and XbaI sites of pMF272-Pc-gul-1-gfp to form pMF272-Pn-gul-1-gfp. The primers used in this study are listed in Additional file 9: Table S4. All PCR products in this study were cloned into the pGEM-T vector (Promega) and sequenced for confirmation.
Plasmid DNA (10 μg) was transformed into a (his-3; Δgul-1, a) strain as described previously [16]. The constructs were targeted to the his-3 locus by homologous recombination, and correct integration was confirmed by GFP fluorescence and PCR. To recover homokaryotic strains, His+GFP+ transformants were crossed with FGSC 11288 (NCU01197, A). Progeny were selected for histidine prototrophy and GFP fluorescence. The resulting complemented strains were termed Pc-gul-1 and Pn-gul-1.
Enzyme activity assay
For protein assays, 5 mL of culture supernatants were collected at each time point, centrifuged at 12,000 rpm for 10 min to remove mycelia and stored at 4 °C for further analysis. Total extracellular proteins were quantified using a Bio-Rad Protein Assay kit based on absorbance at 595 nm, with bovine serum albumin as the standard. The endoglucanase activity was determined using the azo-CMC assay kit according to manufacturer’s instruction (Megazyme). We used p-nitrophenyl-β-d-glucopyranoside (pNPG) for the measurement of β-glucosidase activity as described previously [62]. Briefly, a 250 μL culture supernatant that was diluted with 50 mM sodium citrate (pH 4.8) was added to 250 μL of 1 mg/mL pNPG, and then incubated immediately for 10 min at 50 °C. The reaction mixture was terminated by adding 500 μL of 1 M Na2CO3 and determined at an absorbance of 420 nm. Inactive enzyme, which was boiled at 100 °C for 10 min, was used as a control. pNP was used to generate a standard curve. One unit (U) of β-glucosidase activity was defined as the amount of pNP released from the substrate per minute using 1 mL enzyme under the standard assay conditions.
Fungal biomass assay and rheological measurement
The dry weight of fungal biomass was indirectly measured as previously described [14]. Briefly, a 5-mL culture broth was centrifuged at 4000 rpm for 10 min, the supernatant was discarded, and the pellet was dried and weighed. The dried residue was resuspended in 3 mL of acetic nitric reagent (80:20, v/v), and bathed with boiling water for 2 h to remove mycelia. The residual Avicel was washed thoroughly, then dried, and reweighed. Mycelial dry weight was defined as the dry weight of the original 5-mL culture minus that of residual Avicel.
Rheological properties of fermentation broth were measured with a Brookfield digital HBDV-III Ultra rheometer according to the manufacturer’s instructions. Viscosity was measured as the torque exerted by the mycelial sample on the rheometer impeller at 50 rpm. Torque was recorded after the reading had stabilized (45–60 s).
Oxidative stress, high osmotic stress and cell wall integrity test
For plate assays, 5 μL aliquots of conidial suspensions (107 mL−1) were applied onto Vogel’s solid medium (MM: 1 × Vogel’s salts, 2% w/v sucrose and 1.5% w/v agar) supplemented with H2O2 (10, 20 mM), diamide (6, 9 μg/mL), methyl-viologen (4.5, 9 μg/mL), NaCl (0.5, 1.0 M), Congo Red (1, 2 mg/mL) or Calcofluor White (200, 400 μg/mL). Plates were incubated at 28 °C for 18 h. All colonies were cross-measured to determine their growth indices under each stress.
RNA extraction, sequencing and data analysis
For gene expression analysis, conidia of ∆gul-1 and WT strains were separately inoculated into Avicel medium (1 × Vogel’s salt, 2% w/v crystalline cellulose, 0.75% w/v yeast extract and 0.2% v/v Tween 80) and batch cultured for 3 days at 25 °C with constant light. Mycelia were harvested by vacuum filtration and frozen immediately in liquid nitrogen. Total RNA from frozen samples was isolated using Trizol reagent (Invitrogen) in accordance with the manufacturer’s protocol, and further treated with DNase I (RNeasy Mini Kit, Qiagen). The RNA integrity was checked by agarose gel electrophoresis. RNA-seq was performed on the Illumina HiSeqTM 2000 platform of Beijing Genomics Institute (BGI, Shenzhen, China). Filtered clean reads were mapped against predicted transcripts from the N. crassa OR74R genome (version 12) [63] using TopHat (version 2.0.8b) [64]. The alignment results were stored in SAM format files for subsequent analysis. The counts of reads that uniquely mapped to only one gene were calculated for each gene by Htseq-count (http://www-huber.embl.de/users/anders/HTSeq) using SAM files and genome annotation as input. Abundance for each transcript was calculated using the reads per kilobase per million (RPKM) [65]. Genes with altered expression were performed by using R package NOISeq (version 2.6.0) [66] (Q value ≥ 0.95 used as threshold, which approximately corresponds to a |log2 ratio| ≥ 1). To discover significantly upregulated and downregulated genes between WT and Δgul-1, only the genes with relative high transcriptional abundance (RPKM values above 20 in at least one strain) went into further analysis [67]. Expression differences detected by RNA-seq were also verified by RT-PCR. Profiling data are listed in Additional file 6: Table S2, and the sequence data produced in this study can be accessed (GEO: GSE113321).
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed using a CFX96 real-time PCR detection system (Bio-Rad) with reagents from TOYOBO (One-step qPCR Kit). All assays were performed in triplicate with actin (NCU04173) as the endogenous standard, according to the manufacturer’s instruction. All primers used in this study were listed in Additional file 9: Table S4.
Phylogenetic analysis
Putative orthologs of gul-1 in selected fungal species were identified as best reciprocal BLAST hits from the National Center for Biotechnology Institute (NCBI) protein database. The phylogenetic analysis was carried out using the Neighbor-Joining in MEGA4 software. Bootstrap values were adjacent to each internal node, representing the percentage of 1000 bootstrap replicates.
Statistical analyses
Unless otherwise noted, all experiments were performed in triplicate and statistical tests for significance were determined by two-tailed Student’s t test.
Additional files
Additional file 1: Figure S1. Mycelial morphology of Δgul-1 and wild type strains. Cultures were grown on Vogel’s minimal medium for 18 h at 28 °C. Scale bar is 500 μm.
Additional file 2: Figure S2. Scanning electron micrographs of hyphal morphology of wild type and Δgul-1 strains. Cultures were grown on Avicel medium for 5 days at 25 °C. Scale bar is 10 μm.
Additional file 3: Figure S3. Biomass accumulation of WT and Δgul-1 mutant when grown on Avicel medium. Conidia from Δgul-1 and wild type (WT) strains were separately inoculated into Avicel medium and batch cultured for 7 days. The biomass accumulation was measured. Values represent the means of at least three replicates, error bars show standard deviation.
Additional file 4: Figure S4. Phenotype of WT and Δgul-1 strains when grown on different carbon sources. (A) Viscosity; (B) Fungal morphology; (C) Protein secretion. Conidia were inoculated into 100 mL liquid media [1×Vogel’s salts, 0.75% w/v yeast extract, 0.2% v/v Tween 80 and 2% w/v carbon source (sucrose, xylose or fructose)] at 105 conidia/mL and grown at 25 °C in constant light and shaking (200 rpm). Statistical significance was performed using a two-tailed Student’s t-test (*P<0.05) Scale bar is 1500 μm.
Additional file 5: Table S1. Growth rates of wild type and gul-1 mutant. Aliquots of 5 μL 1×107 mL−1 spore suspensions of WT, Δgul-1, Pn-gul-1 and Pc-gul-1 were incubated at 28 °C for 18 h on MM plates.
Additional file 6: Table S2. Gene list of the 246 genes differentially expressed in Δgul-1 as compared to WT on Avicel.
Additional file 7: Table S3. Comparative analysis of cell wall-related gene expression between Δgul-1 and WT by RNA-seq.
Additional file 8: Figure S5. Phylogenetic analysis of GUL-1 and its homologs. MEGA 4 software was used to carry out the analysis. Bootstrap values are adjacent to each internal node, representing the percentage of 1,000 bootstrap replicates.
Additional file 9: Table S4. Primers used in this study.
Authors’ contributions
LL and CT conceived and designed the project. LL, ZS, YC, QL and WS performed the experiments. LL, JL and CT analyzed the data. LL and CT wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Raymond J St. Leger from University of Maryland, College Park for revising the manuscript. We also thank Dr. Timothy C. Cairns for the critical discussions and Huanhuan Zhai for the technical assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this manuscript (and its Additional files).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (31501007 and 31471186).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Liangcai Lin and Zhiyong Sun contributed equally to this work
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0944-5) contains supplementary material, which is available to authorized users.
Contributor Information
Liangcai Lin, Email: lin_lc@tib.cas.cn.
Zhiyong Sun, Email: sun_zy@tib.cas.cn.
Jingen Li, Email: li_jg@tib.cas.cn.
Yong Chen, Email: chen_y@tib.cas.cn.
Qian Liu, Email: liu_q1@tib.cas.cn.
Wenliang Sun, Email: sun_wl@tib.cas.cn.
Chaoguang Tian, Phone: 86-22-84861947, Email: tian_cg@tib.cas.cn.
References
- 1.Nevalainen KM, Te’o VS, Bergquist PL. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 2005;23:468–474. doi: 10.1016/j.tibtech.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Meyer V. Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol Adv. 2008;26:177–185. doi: 10.1016/j.biotechadv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Ward OP. Production of recombinant proteins by filamentous fungi. Biotechnol Adv. 2012;30:1119–1139. doi: 10.1016/j.biotechadv.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Li ZJ, Shukla V, Fordyce AP, Pedersen AG, Wenger KS, Marten MR. Fungal morphology and fragmentation behavior in a fed-batch Aspergillus oryzae fermentation at the production scale. Biotechnol Bioeng. 2000;70:300–312. doi: 10.1002/1097-0290(20001105)70:3<300::AID-BIT7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Amanullah A, Christensen LH, Hansen K, Nienow AW, Thomas CR. Dependence of morphology on agitation intensity in fed-batch cultures of Aspergillus oryzae and its implications for recombinant protein production. Biotechnol Bioeng. 2002;77:815–826. doi: 10.1002/bit.10181. [DOI] [PubMed] [Google Scholar]
- 6.Chao Y, Singh D, Yu L, Li Z, Chi Z, Chen S. Secretome characteristics of pelletized Trichoderma reesei and cellulase production. World J Microbiol Biotechnol. 2012;28:2635–2641. doi: 10.1007/s11274-012-1073-7. [DOI] [PubMed] [Google Scholar]
- 7.Nair RB, Lennartsson PR, Taherzadeh MJ. Mycelial pellet formation by edible ascomycete filamentous fungi, Neurospora intermedia. AMB Express. 2016;6:31. doi: 10.1186/s13568-016-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abd-Aziz S, Fernandez CC, Salleh MM, Illias RM, Hassan MA. Effect of agitation and aeration rates on chitinase production using Trichoderma virens UKM1 in 2-l stirred tank reactor. Appl Biochem Biotechnol. 2008;150:193–204. doi: 10.1007/s12010-008-8140-4. [DOI] [PubMed] [Google Scholar]
- 9.Bocking SP, Wiebe MG, Robson GD, Hansen K, Christiansen LH, Trinci AP. Effect of branch frequency in Aspergillus oryzae on protein secretion and culture viscosity. Biotechnol Bioeng. 1999;65:638–648. doi: 10.1002/(SICI)1097-0290(19991220)65:6<638::AID-BIT4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 10.Roche CM, Loros JJ, McCluskey K, Glass NL. Neurospora crassa: looking back and looking forward at a model microbe. Am J Bot. 2014;101:2022–2035. doi: 10.3732/ajb.1400377. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass NL, McCluskey K, Plamann M, Galagan JE, Birren BW, et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dogaris I, Mamma D, Kekos D. Biotechnological production of ethanol from renewable resources by Neurospora crassa: an alternative to conventional yeast fermentations? Appl Microbiol Biotechnol. 2013;97:1457–1473. doi: 10.1007/s00253-012-4655-2. [DOI] [PubMed] [Google Scholar]
- 14.Lin L, Chen Y, Li J, Wang S, Sun W, Tian C. Disruption of non-anchored cell wall protein NCW-1 promotes cellulase production by increasing cellobiose uptake in Neurospora crassa. Biotechnol Lett. 2016;39:545–551. doi: 10.1007/s10529-016-2274-1. [DOI] [PubMed] [Google Scholar]
- 15.Fan F, Ma G, Li J, Liu Q, Benz JP, Tian C, Ma Y. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels. 2015;8:66. doi: 10.1186/s13068-015-0248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei X, Fan F, Lin L, Chen Y, Sun W, Zhang S, Tian C. Involvement of the adaptor protein 3 complex in lignocellulase secretion in Neurospora crassa revealed by comparative genomic screening. Biotechnol Biofuels. 2015;8:124. doi: 10.1186/s13068-015-0302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajan JS, Virkar PD. Induced pelletized growth of Neurospora crassa for tyrosinase biosynthesis in airlift fermenters. Biotechnol Bioeng. 1987;29:770–772. doi: 10.1002/bit.260290617. [DOI] [PubMed] [Google Scholar]
- 18.Wen SuW, Jun He B. Secreted enzyme production by fungal pellets in a perfusion bioreactor. J Biotechnol. 1997;54:43–52. doi: 10.1016/S0168-1656(97)01690-8. [DOI] [Google Scholar]
- 19.Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen JM, Wanless AG, Seidel CW, Weiss EL. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr Biol. 2009;19:2114–2120. doi: 10.1016/j.cub.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler RT, Kupiec M, Magnelli P, Abeijon C, Fink GR. A Saccharomyces cerevisiae mutant with increased virulence. Proc Natl Acad Sci USA. 2003;100:2766–2770. doi: 10.1073/pnas.0437995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seiler S, Vogt N, Ziv C, Gorovits R, Yarden O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol Biol Cell. 2006;17:4080–4092. doi: 10.1091/mbc.e06-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold I, Yarden O. Regulation of Neurospora crassa cell wall remodeling via the cot-1 pathway is mediated by gul-1. Curr Genet. 2017;63:145–159. doi: 10.1007/s00294-016-0625-z. [DOI] [PubMed] [Google Scholar]
- 25.Yarden O, Plamann M, Ebbole DJ, Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terenzi HF, Reissig JL. Modifiers of the cot gene in Neurospora: the gulliver mutants. Genetics. 1967;56:321–329. doi: 10.1093/genetics/56.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watters MK, Boersma M, Johnson M, Reyes C, Westrick E, Lindamood E. A screen for Neurospora knockout mutants displaying growth rate dependent branch density. Fungal Biol. 2011;115:296–301. doi: 10.1016/j.funbio.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Papagianni M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol Adv. 2004;22:189–259. doi: 10.1016/j.biotechadv.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka S, Yamada K, Yabumoto K, Fujii S, Huser A, Tsuji G, Koga H, Dohi K, Mori M, Shiraishi T, et al. Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol Microbiol. 2007;64:1332–1349. doi: 10.1111/j.1365-2958.2007.05742.x. [DOI] [PubMed] [Google Scholar]
- 31.Kurischko C, Kuravi VK, Herbert CJ, Luca FC. Nucleocytoplasmic shuttling of Ssd1 defines the destiny of its bound mRNAs. Mol Microbiol. 2011;81:831–849. doi: 10.1111/j.1365-2958.2011.07731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ba ANN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopecka M, Gabriel M. The influence of congo red on the cell wall and (1→3)-beta-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol. 1992;158:115–126. doi: 10.1007/BF00245214. [DOI] [PubMed] [Google Scholar]
- 34.Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 35.Collinson LP, Dawes IW. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- 36.Wemmie JA, Steggerda SM, Moye-Rowley WS. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J Biol Chem. 1997;272:7908–7914. doi: 10.1074/jbc.272.12.7908. [DOI] [PubMed] [Google Scholar]
- 37.Srijaruskul K, Charoenlap N, Namchaiw P, Chattrakarn S, Giengkam S, Mongkolsuk S, Vattanaviboon P. Regulation by SoxR of mfsA, which encodes a major facilitator protein involved in paraquat resistance in Stenotrophomonas maltophilia. PLoS ONE. 2015;10:e0123699. doi: 10.1371/journal.pone.0123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allgaier S, Taylor RD, Brudnaya Y, Jacobson DJ, Cambareri E, Stuart WD. Vaccine production in Neurospora crassa. Biologicals. 2009;37:128–132. doi: 10.1016/j.biologicals.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Zhang J. The filamentous fungal pellet and forces driving its formation. Crit Rev Biotechnol. 2016;36:1066–1077. doi: 10.3109/07388551.2015.1084262. [DOI] [PubMed] [Google Scholar]
- 40.Maddi A, Bowman SM, Free SJ. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet Biol. 2009;46:768–781. doi: 10.1016/j.fgb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardo M, Monteoliva L, Vazquez P, Martinez R, Molero G, Nombela C, Gil C. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology. 2004;150:4157–4170. doi: 10.1099/mic.0.26924-0. [DOI] [PubMed] [Google Scholar]
- 42.Dynesen J, Nielsen J. Surface hydrophobicity of Aspergillus nidulans conidiospores and its role in pellet formation. Biotechnol Prog. 2003;19:1049–1052. doi: 10.1021/bp0340032. [DOI] [PubMed] [Google Scholar]
- 43.Znameroski EA, Glass NL. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol Biofuels. 2013;6:6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips CM, Iavarone AT, Marletta MA. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J Proteome Res. 2011;10:4177–4185. doi: 10.1021/pr200329b. [DOI] [PubMed] [Google Scholar]
- 46.Cai M, Zhang Y, Hu W, Shen W, Yu Z, Zhou W, Jiang T, Zhou X, Zhang Y. Genetically shaping morphology of the filamentous fungus Aspergillus glaucus for production of antitumor polyketide aspergiolide A. Microb Cell Fact. 2014;13:73. doi: 10.1186/1475-2859-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. BioEssays. 2006;28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 48.Muller C, Hansen K, Szabo P, Nielsen J. Effect of deletion of chitin synthase genes on mycelial morphology and culture viscosity in Aspergillus oryzae. Biotechnol Bioeng. 2003;81:525–534. doi: 10.1002/bit.10491. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Zheng Z, Wang P, Gong G, Wang L, Zhao G. Morphological changes induced by class III chitin synthase gene silencing could enhance penicillin production of Penicillium chrysogenum. Appl Microbiol Biotechnol. 2013;97:3363–3372. doi: 10.1007/s00253-012-4581-3. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Lee JE, Lee JS. Histone deacetylase-mediated morphological transition in Candida albicans. J Microbiol. 2015;53:805–811. doi: 10.1007/s12275-015-5488-3. [DOI] [PubMed] [Google Scholar]
- 51.Kovacs Z, Szarka M, Kovacs S, Boczonadi I, Emri T, Abe K, Pocsi I, Pusztahelyi T. Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet Biol. 2013;54:1–14. doi: 10.1016/j.fgb.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Nunez L, Riquelme M. Role of BGT-1 and BGT-2, two predicted GPI-anchored glycoside hydrolases/glycosyltransferases, in cell wall remodeling in Neurospora crassa. Fungal Genet Biol. 2015;85:58–70. doi: 10.1016/j.fgb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Cabib E, Blanco N, Grau C, Rodriguez-Pena JM, Arroyo J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol Microbiol. 2007;63:921–935. doi: 10.1111/j.1365-2958.2006.05565.x. [DOI] [PubMed] [Google Scholar]
- 54.Mouyna I, Morelle W, Vai M, Monod M, Lechenne B, Fontaine T, Beauvais A, Sarfati J, Prevost MC, Henry C, Latge JP. Deletion of GEL2 encoding for a beta(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol Microbiol. 2005;56:1675–1688. doi: 10.1111/j.1365-2958.2005.04654.x. [DOI] [PubMed] [Google Scholar]
- 55.Bowman SM, Piwowar A, Al Dabbous M, Vierula J, Free SJ. Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa. Eukaryot Cell. 2006;5:587–600. doi: 10.1128/EC.5.3.587-600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romano J, Nimrod G, Ben-Tal N, Shadkchan Y, Baruch K, Sharon H, Osherov N. Disruption of the Aspergillus fumigatus ECM33 homologue results in rapid conidial germination, antifungal resistance and hypervirulence. Microbiology. 2006;152:1919–1928. doi: 10.1099/mic.0.28936-0. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology. 2004;150:3341–3354. doi: 10.1099/mic.0.27320-0. [DOI] [PubMed] [Google Scholar]
- 58.Wessels JGH. Tansley Review No. 45 Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 1993;123:397–413. doi: 10.1111/j.1469-8137.1993.tb03751.x. [DOI] [PubMed] [Google Scholar]
- 59.Peberdy JF. Protein secretion in filamentous fungi—trying to understand a highly productive black box. Trends Biotechnol. 1994;12:50–57. doi: 10.1016/0167-7799(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 60.Bigger CH, White MR, Braymer H. Ultrastructure and invertase secretion of the slime mutant of Neurospora crassa. J Gen Microbiol. 1972;71:159–166. doi: 10.1099/00221287-71-1-159. [DOI] [Google Scholar]
- 61.Lin L, Fang W, Liao X, Wang F, Wei D, St Leger RJ. The MrCYP52 cytochrome P450 monoxygenase gene of Metarhizium robertsii is important for utilizing insect epicuticular hydrocarbons. PLoS ONE. 2011;6:e28984. doi: 10.1371/journal.pone.0028984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang B, Cai P, Sun W, Li J, Tian C, Ma Y. A transcriptomic analysis of Neurospora crassa using five major crop residues and the novel role of the sporulation regulator rca-1 in lignocellulase production. Biotechnol Biofuels. 2015;8:21. doi: 10.1186/s13068-015-0208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 64.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 66.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McIntyre LM, Lopiano KK, Morse AM, Amin V, Oberg AL, Young LJ, Nuzhdin SV. RNA-seq: technical variability and sampling. BMC Genomics. 2011;12:293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Mycelial morphology of Δgul-1 and wild type strains. Cultures were grown on Vogel’s minimal medium for 18 h at 28 °C. Scale bar is 500 μm.
Additional file 2: Figure S2. Scanning electron micrographs of hyphal morphology of wild type and Δgul-1 strains. Cultures were grown on Avicel medium for 5 days at 25 °C. Scale bar is 10 μm.
Additional file 3: Figure S3. Biomass accumulation of WT and Δgul-1 mutant when grown on Avicel medium. Conidia from Δgul-1 and wild type (WT) strains were separately inoculated into Avicel medium and batch cultured for 7 days. The biomass accumulation was measured. Values represent the means of at least three replicates, error bars show standard deviation.
Additional file 4: Figure S4. Phenotype of WT and Δgul-1 strains when grown on different carbon sources. (A) Viscosity; (B) Fungal morphology; (C) Protein secretion. Conidia were inoculated into 100 mL liquid media [1×Vogel’s salts, 0.75% w/v yeast extract, 0.2% v/v Tween 80 and 2% w/v carbon source (sucrose, xylose or fructose)] at 105 conidia/mL and grown at 25 °C in constant light and shaking (200 rpm). Statistical significance was performed using a two-tailed Student’s t-test (*P<0.05) Scale bar is 1500 μm.
Additional file 5: Table S1. Growth rates of wild type and gul-1 mutant. Aliquots of 5 μL 1×107 mL−1 spore suspensions of WT, Δgul-1, Pn-gul-1 and Pc-gul-1 were incubated at 28 °C for 18 h on MM plates.
Additional file 6: Table S2. Gene list of the 246 genes differentially expressed in Δgul-1 as compared to WT on Avicel.
Additional file 7: Table S3. Comparative analysis of cell wall-related gene expression between Δgul-1 and WT by RNA-seq.
Additional file 8: Figure S5. Phylogenetic analysis of GUL-1 and its homologs. MEGA 4 software was used to carry out the analysis. Bootstrap values are adjacent to each internal node, representing the percentage of 1,000 bootstrap replicates.
Additional file 9: Table S4. Primers used in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript (and its Additional files).