Abstract
Persistent pain, which is poorly treated and estimated to afflict one third of the world’s population, is largely mediated by the sensitization of nociceptive neurons. This sensitization involves de novo gene expression to support biochemical and structural changes required to maintain amplified pain signaling that frequently persists even after injury to tissue resolves. While transcription-dependent changes in gene expression are important, recent work demonstrates that activity-dependent regulation of mRNA translation is key to controlling the cellular proteome and the development and maintenance of persistent pain. In this review, we highlight recent advances in translational regulation of gene expression in nociceptive circuits, with a focus on key signaling pathways and mRNA targets that may be tractable for the creation of next-generation pain therapeutics.
Why Is Translational Control Important for Persistent Pain?
Dorsal root ganglion (DRG) and trigeminal ganglion (TG) neurons are remarkable structures, with a cell body located up to a meter away from the transduction mechanisms in nerve endings that innervate skin, muscles, and visceral organs. Approximately 50% of these neurons are nociceptive, meaning that they detect damaging or potentially damaging stimuli and transmit signals to the brain that are eventually perceived as pain. The traditional view of these neurons as static transmitters of information has been overturned in recent decades because numerous studies have demonstrated that injury and/or endogenous mediators are capable of fundamentally changing the neurochemical and electrophysiological phenotypes of these neurons, making them extraordinarily hyperexcitable as pain becomes persistent. This is a key feature of clinical pain disorders and has emerged as a major intervention point for pain therapeutics.
Why translational control? Changes in gene expression are key to nociceptor (see Glossary) plasticity and phenotypic alterations, but many of these changes evidently do not start at the cell body, because transcriptional changes and protein sorting to nerve endings are not rapid enough to explain this physiology. A solution that has emerged to solve this biological riddle is localized translation at nociceptor endings and/or along injured axons to control changes in gene expression. This solution is important because: (i) it explains how many endogenous pain-promoting molecules induce long-lasting plasticity [1–10]; (ii) it is supported by findings from experimental pain models in humans that are best explained by localized translation regulation [11,12]; and (iii) it provides insight into the generation of ectopic activity after peripheral nerve injury [6,13–16], which is a major driver of neuropathic pain. In this review, we summarize rapidly growing, recent literature that elucidates the intricacies of translational control in persistent pain and highlight new therapeutic opportunities.
The Basics of Translational Control
Local protein synthesis (e.g., in dendrites and axonal terminals) provides a mechanism by which neurons rapidly respond to their changing environment to modify the protein repertoire in specific compartments. This mechanism requires the presence of mRNAs, translation machinery (ribosomes and translation factors), and activation of signaling pathways upstream of translation [17]. Studies in the central and peripheral nervous systems (CNS and PNS, respectively) demonstrate that these components are present in axonal (predominantly in the PNS) and dendritic compartments (predominantly in the CNS), where they support activity-dependent protein synthesis to modify the local proteome in response to a multitude of extracellular and intracellular stimuli (reviewed in [18]). Translation is divided into three steps: initiation, elongation, and termination. Initiation is the rate-limiting step and, therefore, is a major target for activity-dependent regulation in neuronal plasticity and an emerging drug target.
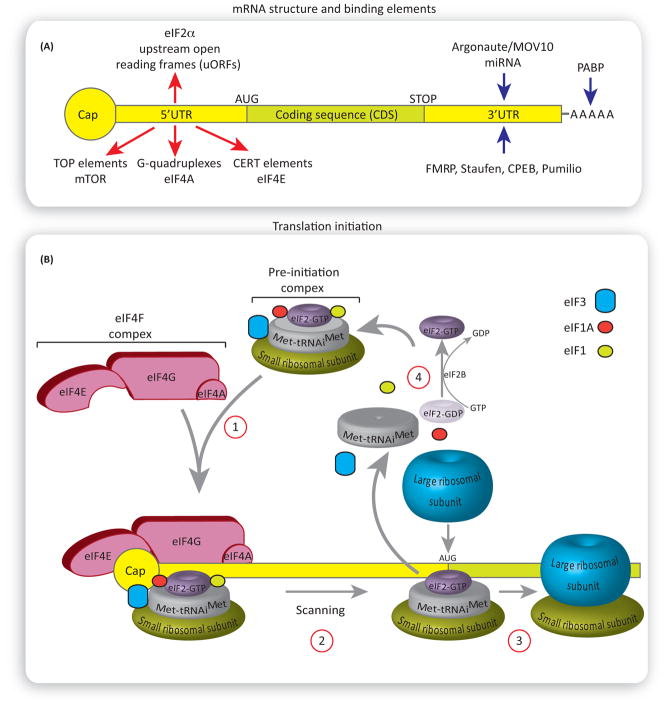
All nuclear transcribed eukaryotic mRNAs contain at their 5′ end a structure termed the ‘cap’, which is a 7-methylguanosine linked to the first nucleotide [19]. The cap is located at the beginning of the mRNA 5′ untranslated region (5′UTR), which is followed by a coding sequence, a 3′UTR and a poly(A) tail at the end of the mRNA (Figure 1A). During the initiation step of translation, a small (40S) ribosomal subunit joins the initiator Met-tRNAiMet and the GTP-bound eukaryotic initiation factor 2 (eIF2-GTP), forming the pre-initiation 43S complex (Figure 1B). This pre-initiation complex is then recruited to the mRNA cap structure by a group of translation initiation factors, termed eukaryotic initiation factor 4 (eIF4) to scan the mRNA 5′UTR for the AUG start codon. Upon encountering the start codon, the large ribosomal subunit (60S) joins the pre-initiation complex to form an active ribosome (80S) that then proceeds to the elongation step. The formation of the preinitiation complex, the recruitment to the ribosome, and the joining of the large ribosomal subunit are controlled by translation initiation factors, whose activity is regulated by phosphorylation, and potentially other post-translational modifications, to allow for rapid and reversible control of translation initiation rates [20].
Figure 1. Regulation of Translation Initiation.
(A) General structure of the mammalian mRNA and its binding elements. Red arrows show the 5′ untranslated region (UTR) elements rendering selective translation of specific mRNAs. mRNAs harboring G-quadruplexes in their 5′UTR are dependent on the activity of eukaryotic initiation factor 4F (eIF4A) helicase for their translation. Cytosine-enriched regulator of translation (CERT) renders eIF4E selectivity. Translation of mRNAs with 5′ terminal oligopyrimidine tract (TOP) is mammalian of rapamycin complex 1 (mTORC1) sensitive. Blue arrows show binding elements that regulate the rates of mRNA translation via binding the 3′UTR or poly(A) tail. (B) An overview of translation initiation. The eIF4F complex recruits the pre-initiation complex to the mRNA (1). The pre-initiation complex scans the 3′UTR in a 5′ to 3′ direction for the presence of the start codon, AUG. Once it encounters the start codon, the eIF2-GDP and other factors (eIF1, eIF3, eIF4F, eIF4B, and eIF5) are displaced, followed by binding of the large ribosomal subunit 60S (2) to form the 80S ribosomal initiation complex, which proceeds to the elongation step (3). eIF2B catalyzes the recycling of GDP-eIF2 to GTP-eIF2, allowing for a new round of translation initiation (4).
Regulation of mRNA Translation via eIF4F Complex Formation
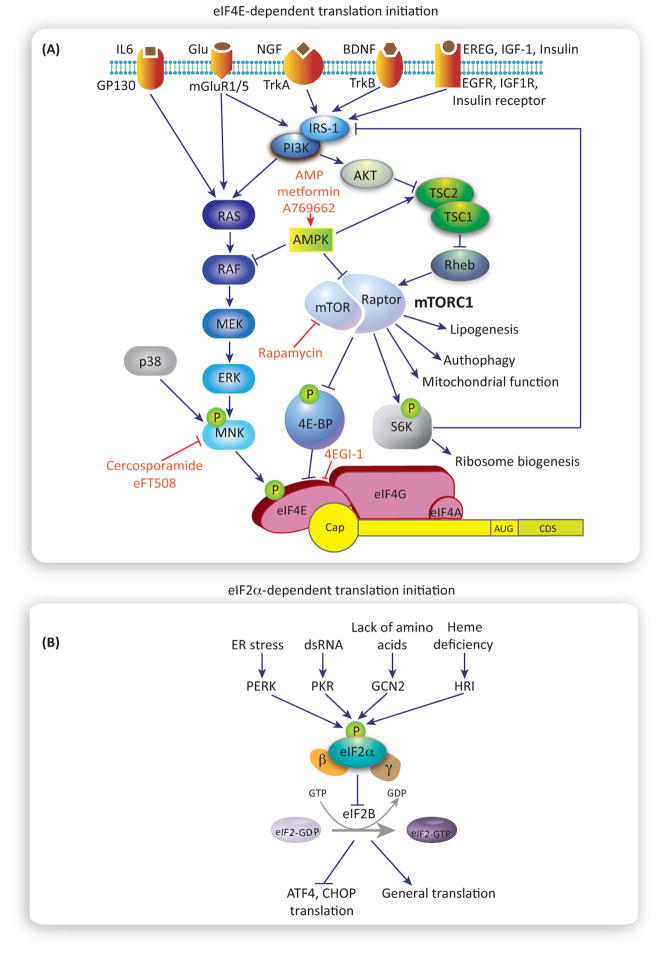
mRNA translation can be triggered by activation of various membrane receptors, including tyrosine receptor kinases (trkA and trkB), insulin-like receptors [IR, IGF1R, and epidermal growth factor receptor (EGFR)], NMDA receptors, and metabotropic glutamate receptors. Activating these receptors stimulates mRNA translation via both the mammalian/mechanistic target of rapamycin complex 1 (mTORC1) and the extracellular-signal-regulated kinase (ERK) and p38 signaling pathways (Figure 2A). mTORC1, a complex comprising mTOR, raptor, and several other proteins, integrates a variety of extracellular signals (via membrane receptors) as well as intracellular signals (via energy status sensors and inputs from other pathways) to control key cellular processes, including autophagy, lipogenesis, mitochondria function, and mRNA translation [21]. mTORC1 regulates the rate of mRNA translation via phosphorylation of two primary downstream effectors, 4E-binding protein (4E-BP) and p70S6 ribosomal kinase (S6K1/2). 4E-BPs are a family of small translational inhibitors (4E-BP1, 2, and 3 in mammals) that suppress the assembly of a trisubunit complex called eIF4F, which is critical for the recruitment of the pre-initiation complex to the mRNA cap. eIF4F comprises eIF4E, which specifically interacts with the cap; eIF4A, an RNA helicase that unwinds the 5′UTR secondary structure to allow for scanning of the start codon; and eIF4G, a large scaffolding protein that binds to both eIF4E and eIF4A. In its hypophosphorylated form, 4E-BP binds to eIF4E, preventing the eIF4E–eIF4G interaction, thus inhibiting eIF4F complex formation. The phosphorylation of 4E-BP by mTORC1 leads to the dissociation of 4E-BPs from eIF4E, thus increasing the amount of free eIF4E, and promoting the assembly of the eIF4F complex, ribosome recruitment to the mRNA, and translation initiation. eIF4E activity is also regulated by phosphorylation of a single residue, serine 209 [22]. This phosphorylation event is associated with increased rates of translation initiation, although the underlying mechanism remains unknown. Activation of Ras-Raf signaling leads to ERK-dependent activation of mitogen-activated protein kinase-interacting kinase 1 (MNK1), which phosphorylates eIF4E. p38, another important mitogen-activated protein kinase (MAPK) in the pain pathway, also phosphorylates MNK1. A subset of ‘eIF4E-sensitive’ mRNAs critically depends on eIF4E activity. These mRNAs encode proteins that stimulate cell proliferation and survival. In neurons, they are often involved in the regulation of synaptic function and plasticity. Unique features of these mRNAs remain controversial, because some groups have suggested that they have long and highly structured 5′UTR, whereas others found the presence of 5′ terminal oligopyrimidine tracts (5′TOPs) [23] and cis-regulatory elements [20,24–26]. There is now strong evidence linking mTORC1 and MAPK activity to translation regulation and the sensitization of nociceptors and/or dorsal horn neurons that receive nociceptive input. Therefore, these pathways have emerged as important pharmacological targets for new pain medicines, and identification of mRNAs that are translated downstream of these pathways in the context of pain plasticity may lead to the elucidation of additional novel targets.
Figure 2. Activity-Dependent Translation Control.
(A) Eukaryotic initiation factor 4E (eIF4E) -dependent translation initiation. The eIF4E complex is illustrated at the bottom, recruited to the mRNA. Activation of several membrane receptors (top) stimulates cap-dependent translation via both mammalian of rapamycin complex 1 (mTORC1) (right) and extra-cellular-signal-regulated kinase (ERK) (left) pathways, which converge on regulation of eIF4E activity. (B) eIF2α-dependent translation initiation. Distinct eIF2α-kinases increase eIF2αphosphorylation (middle of the diagram) in response to cellular stress. Increased eIF2αphosphorylation inhibits, in turn, guanine nucleotide exchange factor (GEF) eIF2B, resulting in reduction of general translation and enhanced translation of mRNAs with uORFs, such as ATF4 and CHOP.
Regulation of mRNA Translation via eIF2α Phosphorylation
Another major mode of regulating translation initiation is via the phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2α) [27]. eIF2α is a key integrator of diverse cellular stress responses and an important regulator of mRNA translation. eIF2α binds GTP, the initiator (Met-tRNAiMet), and the small ribosomal subunit to form the pre-initiation complex. Upon encountering the start codon, the GTP is hydrolyzed to GDP. The recycling of the GDP to GTP on eIF2α is mediated by the guanine nucleotide exchange factor, eIF2B (Figures B and B). Phosphorylation of eIF2α converts eIF2α from a substrate to an inhibitor of eIF2B, resulting in the reduced availability of GTP-bound eIF2α, decreased amounts of pre-initiation complex, and inhibition of translation. Different stress conditions promote eIF2α phosphorylation via activation of four eIF2α kinases, which are activated by distinct stressors, including endoplasmic reticulum stress [via PKR-like ER kinase (PERK)], double-stranded RNA during viral infection [via double-stranded RNA-dependent protein kinase (PKR)], nutrient deprivation [via general control non-derepressible-2 (GCN2)], and heme deficiency [via heme-regulated inhibitor (HRI)] (Figure 2B). This signaling network, convergent on eIF2α, is called the ‘integrated stress response’ (ISR). An increase in p-eIF2α decreases general translation, but paradoxically stimulates the translation of mRNAs with upstream open reading frames (uORFs) in their 5′UTRs, such as ATF4 and CHOP. A subset of these mRNAs encodes transcription factors, which activate gene expression programs to promote cell adaptation to stress. Other uORFs, which are found pervasively in 5′UTRs, as revealed by ribosome footprinting experiments, may encode other short peptides or act as translational repressors of canonical ORFs [28].
Regulation of mRNA Translation by Other Mechanisms
mRNA translation initiation is regulated via several additional mechanisms, including regulation of the length of the poly(A) tail, which protects mRNA from degradation and stimulates the translation of mRNA by promoting mRNA circularization. mRNA circularization is mediated by poly(A)-binding protein (PABP), which simultaneously binds the 3′ poly(A) tail and eIF4G [29]. Binding of the cytoplasmic polyadenylation element (CPE)-binding protein (CPEB) to specific mRNAs harboring CPE sequences at their 3′UTR elongates the poly(A) tail, and likewise stimulates translation [30]. miRNA-dependent mechanisms affect translation initiation, mRNA stability, the length of the poly(A) tail, and mRNA circularization [31]. Finally, internal ribosome entry sites (IRESs) mediate cap-independent translation initiation [32]. IRESs are present in many mammalian mRNAs [32–34], but their functional role remains elusive.
Local mRNA Translation
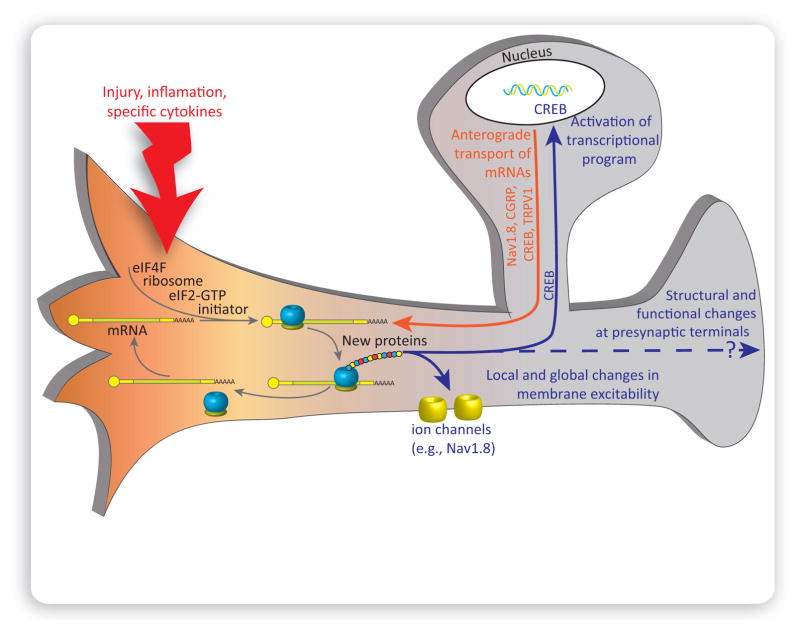
mRNAs, ribosomes, and other components of translational machinery are present in PNS axonal and CNS dendritic compartments [17,35,36]. Release of endogenous molecules associated with tissue damage increases nascent protein synthesis in DRG neurons and their axons via activation of signaling pathways upstream of translation (Figure 3). There is now abundant evidence that this process leads to sensitization of sensory neurons (Table 1). For instance, inhibition of mTORC1, MNK1/2, cap-dependent translation, or general translation by local administration of rapamycin, cercosporamide, 4EGI-1, or anisomycin, respectively, reduces injury-induced changes in nociceptor excitability and mechanical hypersensitivity [5,10,37]. However, the identity of the full repertoire of mRNAs that are translated to lead to this change in excitability is only starting to emerge. While much of work has been done to identify mRNAs that localize to DRG axons in vitro [35], there are substantial technical challenges to confirm that these mRNAs localize to DRG axons in vivo. Nevertheless, several mRNAs have been localized to DRG axons in mice and rats, and some of these have been linked to pain hypersensitivity, such as the mRNAs for CREB, CPEB, and Nav1.8 [38,39] (Table 2). Recent findings have also demonstrated that many other ion channel mRNAs localize to DRG axons and that the machinery needed to traffic these complex transmembrane proteins is also found in axons [13,16]. The development of genetic techniques for tagging ribosomes translating ribosome affinity purification (TRAP)] [40] in specific cellular populations will likely lead to important breakthroughs in our understanding of which mRNAs are translated locally in nociceptor axons to modulate excitability in response to injury.
Figure 3. Local Translation in Nociceptors Promotes Persistent Pain.
Tissue injury, inflammation, and specific cytokines (red) activate local cap-dependent translation of distinct mRNAs. The synthesized proteins act either locally, for example by altering membrane excitability, or retrogradely, where they are transported to the cell body (solid-blue line) to modulate transcriptional gene expression program. Transcriptional upregulation in the cell body leads to the production of new mRNAs, which can then be translocated (orange line) to the periphery in a suppressed state, derepressed, and translated into proteins by local translation machinery. Activation of translation machinery can also lead to changes in central branches of sensory neurons and affect the structure and activity of presynaptic terminals. However, the underlying mechanisms of this process have not been studied extensively.
Table 1.
Key References Demonstrating an Important Role of Translation Regulation in Sensitization of DRG Neurons in Pain Models
| Refs | Mechanism/signaling pathway | Pain model/target | Key findings |
|---|---|---|---|
| Willis et al. 2005 [85] | mRNA transport into axons, regulation by NGF | Preconditioning nerve crush | First paper to detail mRNAs that localize to DRG neuron axons after a preconditioning nerve crush; also showed that NGF treatment enhanced axonal localization of subset of mRNAs |
| Price et al. 2007 [37] | FMRP, mTORC1, ERK | Formalin, peripheral nerve injury, mGLuR1/5 activation | Deficits in formalin phase 2 and mGluR1/5-mediated pain in Fmr1- knockout mice. Formalin and mGluR1/5-induced pain hypersensitivity are dependent on peripheral mTORC1/ERK signaling |
| Jimenez-Diaz et al. 2008 [15], Geranton et al. 2009 [41] | mTORC1 | Peripheral nerve injury/Neuropathic pain | Continuous mTORC1 activity is required for maintaining excitability in A-fiber nociceptors and that blockade of mTORC1 attenuates nerve injury-induced changes in excitability |
| Huang et al. 2008 [16] | Local translation in neuromas | Peripheral nerve injury/Neuropathic pain | Used proteomics from experimental neuromas to demonstrate that local translation of neuron-specific cytoskeletal, oxidative stress, and other enzymes in neuromas likely contribute to enhanced excitability at this known site of ectopic activity generation |
| Thakor et al. 2009 [14], Ruangsri et al. 2012 [39] | Axonal transport of Nav1.8 mRNA and local translation | Peripheral nerve injury/neuropathic pain | Strong evidence that Nav1.8 mRNA is trafficked into axons after peripheral nerve injury and that its local translation leads to enhanced excitability promoting neuropathic pain |
| Melemedjian et al. 2010 [5] | mTORC1, ERK | NGF and IL6, hyperalgesic priming | NGF and IL6 induce mTORC1 and ERK signaling, respectively, to enhance eIF4F complex formation in DRG neurons and their axons; also that local inhibition of translation blocks behavioral response to NGF and IL6 and hyperalgesic priming |
| Rukwied et al. 2010 [11], Obreja et al. 2017 [12] | NGF-induced pain in humans | NGF injection combined with microneurography in human skin | Used NGF injections into skin in human volunteers to show that hypersensitivity generated is locally regulated and occurs without inflammation; concluded that mechanism must be driven by local translation. [12] showed that, when NGF is injected into one of several axonal branches of a single DRG nociceptor, only the injected branch is sensitized, whereas the others show signs of ‘priming’ |
| Melemedjian et al. 2011 [6] | AMPK, mTORC1, ERK | Peripheral nerve injury | Increase in injured DRG neuron axons for mTORC1, ERK, and other downstream effectors, as well as mRNA transport proteins in both mouse and rat models. Targeting AMPK reduced DRG neuron excitability, inhibited mTORC1 and MAPK signaling and alleviated neuropathic pain |
| Bogen et al. 2012 [2], Ferrari et al. 2013 [9] | CPEB, CaMKIIα | Hyperalgesic priming | First to show role for CPEB and poly(A) tail modulation in peripheral response that generates hyperalgesic priming. CPEB effect is neuron specific because knockdown was done by targeting DRG. [9] linked CPEB to local translation of CaMKIIα which drives signaling changes that promote hyperalgesic priming |
| Inceoglu et al. 2015 [3], Khoutorsky et al. 2016 [4] | eIF2α, integrated stress response | Diabetic neuropathy and inflammatory pain | Inceoglu et al. demonstrated that diabetic neuropathy causes a strong increase in ISR induction in sciatic nerve. Khoutorsky et al. used a genetic approach to show that eIF2α regulates thermal nociception through a mechanism that involves TRPV1. Inflammation enhances eIF2α phosphorylation in DRG neurons and this is linked to thermal hyperalgesia |
| Moy et al. 2017 [10] | MNK, eIF4E | Mechanical and affective pain response to NGF, IL6 and PAR2 agonist and hyperalgesic priming | Used eIF4E phosphorylation mutant mice and MNK1/2 double- knockout mice to show that this signaling pathway regulates mechanical and ongoing pain induced by NGF, IL6, and PAR2 agonists as well as by inflammation. Changes in intrinsic excitability in DRG nociceptors in response to these factors are also lost when MNK- eIF4E signaling is disrupted |
| Martin et al. 2017 [8] | EGFR, mTORC1 | Chronic inflammatory pain, human genetics | Epiregulin signaling to EGFR is linked to human pain disorders (using genetics) and has a key role in pain in mice and flies. EGFR signals via mTORC1 to enhance pain and nociceptor sensitivity |
Table 2.
mRNAs That Are Transported in DRG Neuron Axons with Details of Evidence for Local Translation in Pain States
| mRNA transported in DRG neuron axons with relevance for pain | Evidence | Refs |
|---|---|---|
| Nav1.8 | qPCR on adult axons from sciatic nerve with and without nerve injury and RNA sequencing | [13,14,39] |
| Nav1.6 | RT-PCR on adult sciatic nerve with immunolocalization and RNA sequencing on adult sciatic nerve from nerve-injured animals | [13,86] |
| TRPV1 | Carrageenan-induced increases in sciatic nerve TRPV1 mRNA by RT-PCR. TRPV1 protein is increased in axons after inflammation in a p38-dependent fashion that occurs independently of transcriptional changes | [87,88] |
| CREB | RT-PCR on developing DRG axons and modified methionine labeling of nascently synthesized CREB and retrograde axonal transport | [38,89] |
| CGRP | In situ hybridization on developing olfactory sensory neuron axons and localized increases in regenerating axons after nerve crush that was not explained by increased expression in the soma | [90,91] |
| Kappa opioid receptor (KOR) | Activity-dependent transport in DRG axons measured by RT-PCR and reporter constructs | [92] |
| CaMKIV | Localization of protein and mRNA in DRG axons in the IB4 subset of nociceptors controlled by expression of a novel 3′UTR | [93] |
| Papers with large-scale resources for DRG axonal mRNAs | Used microarray- or RNA sequencing-based resources for potential DRG axonal RNAs in different models. Most of these studies relied on in vitro methods to isolate axons | [13,16,85,94] |
Tissue Injury-Induced Modulation of mRNA Translation Promotes Pain Hypersensitivity
Nerve injury and tissue inflammation evoke robust changes in signaling pathways upstream of translation in nociceptors and spinal neurons that receive inputs from PNS neurons, suggesting that translation regulation has a key role in pain amplification along the pain pathway. The phosphorylation of PI3K, AKT, MAPKs, as well as mTOR, eIF4E, and S6, is increased in sensory neurons in models of nerve injury, such as spared nerve injury (SNI) and spinal nerve ligation (SNL), and in models of chronic inflammation, such as complete Freund’s adjuvant (CFA) and carrageenan injection [6,10,15,41]. In the dorsal horn of the spinal cord, the activation of mTOR and ERK pathways has been well documented following peripheral inflammation and nerve injury [15,42–49]. Peripheral (subcutaneous) and central (intrathecal) administration of rapamycin, an allosteric mTORC1 inhibitor, and its paralogs, effectively reduces mechanical hypersensitivity in models of nerve injury, inflammation, cancer pain, and postsurgical pain [1,5,9,15,37,41,44,50–52]. The activation of mTORC1 has also been detected in the spinal cord dorsal horn following repeated opioid administration [7]. Rapamycin alleviated opioid-induced hyperalgesia, supporting the notion that the activation of mRNA translation in the spinal cord following chronic opioid administration contributes to plastic changes leading to this drug-induced pain state. Altogether, these results indicate that activity-dependent translation regulation in the PNS and CNS significantly contributes to maladaptive plasticity that promotes persistent pain. A series of recent studies have begun to dissect how specific aspects of these signaling pathways contribute to plasticity in distinct locations along the neuraxis.
eIF4E Integrates Inputs from the MAPK and mTOR Pathways to Maintain Persistent Pain
Recent studies provided evidence that the ERK/MNK1 and mTORC1/4E-BP1 pathways converge on a key step in translational control, eIF4E activation, to regulate cap-dependent translation and promote the sensitization of sensory neurons [5,10]. The ERK pathway regulates translation mainly via MNK1-dependent phosphorylation of eIF4E, but it also affects the mTORC1 pathway via the phosphorylation of TSC2 and Raptor. Inhibition of eIF4E phosphorylation, via either MNK1 inhibition using cercosporamide or mutation of eIF4E serine 209 to alanine, attenuates increases in the excitability of nociceptors in response to pain-promoting endogenous mediators, such as nerve growth factor (NGF) and interleukin 6 (IL-6) [10]. In vivo experiments with these pharmacological and genetic tools demonstrated that eIF4E phosphorylation is a key mediator of persistent nociceptive sensitization induced by NGF, IL6, and inflammation. eIF4E phosphorylation also has a key role in cold hypersensitivity induced by peripheral nerve injury [10].
mTORC1 activates eIF4E and promotes cap-dependent translation via the phosphorylation and suppression of the eIF4E inhibitor 4E-BP. 4E-BP1 is the predominant isoform in the DRG and spinal cord, whereas its expression in the forebrain is lower than that of 4E-BP2 [53]. Eif4ebp1− knockout mice mimic hyperactivation of the mTORC1/4E-BP1 pathway because eIF4E activity cannot be suppressed in the absence of 4E-BP1, leading to enhanced eIF4F complex formation [53]. Interestingly, Eif4ebp1-knockout mice exhibit enhanced long-term potentiation (LTP)-like potentiation of synaptic transmission in spinal neurons, increased excitatory and inhibitory synaptic activity in lamina II neurons, and mechanical hypersensitivity (see Table 3 for translation regulation mechanisms governing CNS pain plasticity).
Table 3.
Key References Demonstrating an Important Role of Translation Regulation in Dorsal Horn Plasticity in Pain Models
| Refs | Signaling pathway | Pain model/target | Key findings |
|---|---|---|---|
| Zhuo et al. 1998 [95] | General translation regulation | Formalin | First report to show that spinal inhibition of protein synthesis inhibits second phase of formalin test |
| Price et al. 2007 [37] | mTORC1 | Formalin, peripheral nerve injury, mGLuR1/5 activation | Phase II formalin- and mGluR1/5-mediated behaviors are decreased in Fmr1-knockout mice and that spinal inhibition of mTORC1 inhibits pain behaviors in both of these models at late time points |
| Asante et al. 2009 [52], Asante et al. 2010 [96] | mTORC1 | Formalin, peripheral nerve injury, electrophysiology | Used spinal electrophysiology to show that inhibition of mTORC1 with rapamycin causes a decrease in sensitization of deep dorsal horn neurons in response to formalin or peripheral nerve injury |
| Asiedu et al. 2011 [97], Melemedjian et al. 2013 [98] | BDNF signaling to mTORC1 leading to atypical PKC translation | Hyperalgesic priming | Demonstrated that mTORC1-mediated translation regulates atypical PKC translation in dorsal horn, contributing to initiation of hyperalgesic priming; [98] also showed that this process is regulated by BDNF signaling |
| Xu et al. 2014 [7] | mTORC1 | Opioid tolerance, opioid hyperalgesia | Chronic opioid treatment increases spinal mTORC1 activity; inhibition of this signaling pathway blocks and reverses opioid tolerance and opioid-induced hyperalgesia |
| Bonin and De Koninck 2014 [74], Kim et al. 2015 [77] | Spinal reconsolidation | Capsaicin-induced pain hypersensitivity | [74] showed that a reconsolidation window can be opened in spinal dorsal horn that allows for reversal of pain hypersensitivity when that reconsolidation event is paired with a protein synthesis inhibitor. [77] showed that this paradigm can be generalized to hyperalgesic priming models where dopamine D1/D5 agonists open a reconsolidation window that is also sensitive to protein synthesis inhibition |
| Khoutorsky et al. 2015 [53] | 4E-BPs | Enhanced mechanical pain with 4E-BP1 deletion | Genetic removal of 4E-BP1 causes mechanical hypersensitivity (but no changes in thermal pain) that is accounted for by increased neuroligin 1 expression and increased excitatory neurotransmission in dorsal horn |
| Maixner et al. 2015 [99] | AMPK | Peripheral nerve injury | Neuropathic pain leads to downregulation of AMPK activity in dorsal horn. Activating spinal AMPK alleviates neuropathic pain and restores deficits in glial glutamate transporter expression |
An important question is how these signaling mechanisms are engaged in response to inflammation and/or nerve injury. While there is clear evidence that NGF and IL-6, among other pain-promoting molecules, can activate signaling pathways upstream of translation, a new mechanism by which mTORC1 is stimulated in nociceptors in acute and chronic pain conditions has been recently suggested: the EGFR [8]. This transmembrane receptor from the tyrosine kinase family is activated in neuropathic and chronic inflammation pain models. mTORC1 is one of the major downstream targets of EGFR. Consistent with this, the activation of EGFR leads to an AKT- and mTORC1-dependent stimulation of eIF4F complex formation and an activation of MMP9 mRNA translation in the DRG. MMP9 was previously shown to be locally translated in neurons [54] and involved in the early stages of neuropathic pain in mice [55]. Work in Drosophila, where downregulation of EGFR expression impaired thermal sensitivity, supports a role for this signaling pathway as an evolutionarily conserved regulator of nociception. The potential role of EGFR gene polymorphism in humans has been assessed in several cohorts of patients with temporomandibular disorder (TMD), where a correlation between single nucleotide polymorphisms (SNPs) in EGFR and the incidence of TMD has been identified. Finally, studies in both humans and mice demonstrated that expression levels of epiregulin (EREG), an endogenous agonist of EGFR, are significantly increased in persistent pain conditions [8] and several clinical case reports suggest a role of EGFR in cancer pain [56,57]. Therefore, these studies collectively support a model wherein elevated levels of EREG lead to the stimulation of EGFR and its major downstream effector, mTORC1, to promote the translation of MMP9 and potentially other mRNAs that are involved in promoting pain hypersensitivity.
eIF2α-Dependent Translational Control in Pain
As discussed above, the regulation of eIF2α phosphorylation is an important mechanism to control the rate of protein synthesis. eIF2α phosphorylation is increased in the DRG and sciatic nerve in models of chronic inflammation, nerve injury, and diabetes [3,4,6]. A study using transgenic mice with reduced phosphorylation of eIF2α showed that the increase in eIF2α phosphorylation in the CFA model of inflammation promotes thermal but not mechanical hypersensitivity [4]. This effect on thermal sensitivity is mediated by sensitization of the transient receptor potential vanilloid subfamily, member 1 (TRPV1). Phosphorylation of eIF2α in the DRG and sciatic nerve is also increased in animal models of diabetes and peripheral nerve injury. This suggests induction of the ISR in response to nerve injury, but the idea has not been thoroughly vetted. Since elevated eIF2α phosphorylation attenuates general protein synthesis, while stimulating the translation of mRNAs with uORFs, it is tempting to speculate that translation of mRNAs harboring functional uORFs leads to pain hypersensitivity. In the immune system, small peptides translated from uORFs can be presented on the cell surface by MHC class I ligands [58], suggesting a functional role for uORF translation in neuro-immune interactions.
Targeting Translation for Therapeutic Development of Next-Generation Pain Medications
Based on the evidence provided above, an appealing therapeutic approach is to suppress mTORC1 to achieve pain relief. While mTORC1 and S6K1 inhibitors show efficacy in acute treatment paradigms in rodent pain models, their long-term administration leads to diminished effects on pain, resulting in feedback signaling in the MAPK pathway and promoting the development of mechanical hypersensitivity [59]. Recently, several novel approaches have emerged to target dysregulated translation in pathological pain conditions. One potential target is the MNK1 kinase, which is the only known kinase for eIF4E phosphorylation. Inhibiting eIF4E phosphorylation with the MNK1 inhibitor, cercosporamide, alleviated tissue injury-induced increases in neuronal excitability and blocked the transition of acute to chronic pain state in hyperalgesic priming models [10,60]. This approach also alleviated cold hypersensitivity in neuropathic pain. New MNK1 inhibitors with better selectivity and pharmacokinetic profiles are in clinical development, creating an opportunity to test this mechanism in human trials in the future.
An additional promising strategy to normalize aberrant mRNA translation in persistent pain involves the activation of a cellular energy sensor, AMP-activated protein kinase (AMPK), using metformin or other compounds [61,62]. Metformin is a US Food and Drug Administration (FDA)-approved first-line anti-type 2 diabetic compound. It has been shown to have numerous positive health effects beyond its original therapeutic use, including lowered incidence of cancer, and correction of some of the abnormalities in Fragile X syndrome (FXS) [63,64]. Metformin is thought to exert its effects by activating AMPK and inhibiting mRNA translation via TSC2, Raptor, and BRAF; however, several AMPK-independent actions of metformin have also been documented [63]. Metformin showed dramatic effects in mouse and rat models of neuropathic and inflammatory pain, leading to an alleviation of thermal and mechanical hypersensitivity, and promoting neuroprotection in models of chemotherapy-induced peripheral neuropathic pain [6,65]. A key feature of metformin is its disease-modifying properties in neuropathic pain models, wherein metformin treatment leads to sustained pain relief several months after cessation of treatment. Intriguingly, retrospective [66] and prospective human [67] trials have shown beneficial pain effects of metformin. In animal models, several other AMPK activators have shown similar effects to metformin, supporting the notion that AMPK activation is critical for the effect of metformin on pain [68–70]. Therefore, further metformin trials are warranted, as is the continued development of more-specific AMPK activators that may be able to produce disease-modifying properties in patients with pain.
Hyperalgesic priming, mentioned briefly above, is a model system for studying the development of persistent pain plasticity [60,71]. In these models, an initial insult is given and then animals are allowed to fully recover, such that they no longer show any overt signs of ongoing or evoked pain hypersensitivity. Then, a normally subthreshold dose of an inflammatory mediator is given. In naïve animals, this dose of inflammatory mediator does not produce a response, whereas, in ‘primed’ animals, it causes a robust pain response that can last for more than a week. This suggests a transition to a chronic form of pain plasticity [60,71]. AMPK activators, mTORC1 inhibitors, and disruption of MNK1-eIF4E signaling all block the development of hyperalgesic priming if these manipulations are made near the time of the priming injury [1,5,10,38]. Moreover, disrupting CPEB and/or poly(A) tail-mediated regulation of translation block the development of hyperalgesic priming [1,2,9]. Experiments in this model system support the hypothesis that the mechanism of priming initiation requires local translation in the nociceptor axons that are proximal to the initial insult but not in the nociceptor cell bodies [72]. Human experimental data using NGF injection also support the view that local translation is required for nociceptor sensitization [11,12]. Collectively, these findings suggest that interfering with local translation at the time of injury, such as surgical incision, could alleviate the acute generation of pain hypersensitivity and block the transition to a chronic pain state.
A new form of hyperalgesic priming, type II priming, was recently discovered. This form of priming involves activation of GPCRs that are linked to Gαi signaling proteins, such as opioid receptors. This form of priming is notable because it is engaged by inhibitory GPCRs and proceeds independently of local translation regulation mechanisms [73].
Targeting another protein synthesis-dependent process, called spinal reconsolidation, reverses established spinal LTP and pain hypersensitivity [74]. Reconsolidation, which was first discovered in the amygdala as a mechanism to update fear memory, has also been found to occur in many brain areas and species. When a memory trace is formed, it is consolidated into a neuronal circuitry in a protein synthesis-dependent manner. Following exposure to a stimulus that reactivates this circuit, the memory trace becomes labile again, allowing it to be manipulated pharmacologically. Reconsolidation has been proposed as a potential strategy to erase unwanted memories in conditions associated with adverse events, such as in post-traumatic stress disorder (PTSD) [75,76]. Remarkably, reconsolidation also occurs in the spinal cord [74,77,78], raising the potential for therapeutic strategies based on this approach. Bonin and De Koninck demonstrated that, after establishing an initial sensitization of nociceptive synapses in the dorsal horn using peripheral administration of capsaicin, repeated capsaicin administration in the presence of the general protein synthesis inhibitor anisomycin completely reversed the sensitization in an NMDA receptor-dependent fashion. Kim et al. expanded on this finding to show that dopamine, acting on D1 or D5 receptors in the spinal cord, is able to open a reconsolidation window in mice that have transitioned to a chronic pain state [77]. When these mice were given a D1/D5 agonist in combination with spinal anisomycin, their chronic pain state was reversed. It is currently not clear whether this form of reconsolidation requires pre- or postsynaptic protein synthesis. These findings demonstrate a striking similarity to therapeutic strategies being developed for PTSD.
Concluding Remarks
Despite progress in deciphering the signaling mechanisms upstream of translation in different pathological pain conditions, we still do not understand the precise mechanisms by which altered mRNA translation promotes neuronal hyperexcitability, changes synaptic transmission in the spinal cord, and causes persistent pain (see Outstanding Questions). What are the mRNAs whose translation efficiency is altered (increased or decreased) in pathological pain conditions, and what is the function of the corresponding proteins? Several groups are working toward resolving these questions by measuring the translation efficiency of mRNAs on a genome-wide scale in different pathological pain conditions, along the continuum of pain and in a cell type-specific fashion. Another key question is whether specific translation pathways promote the translation of distinct subsets of mRNAs. Evidence from other cell or tissue types suggests that this is indeed the case, but little work has been done in this area in neurons, where the transcriptional landscape is unique.
Outstanding Questions.
What are the mRNAs whose activity-dependent translation leads to neuronal hyperexcitability and pain? New technologies, such as ribosome footprinting, to characterize the translation landscape and ribosome tagging to identify transcripts in specific cell types will facilitate discovery in this area.
Most recent work has focused on the identification of translational control mechanisms in neurons. What is the role of translational control in non-neuronal cells (microglia, astrocytes, and satellite glial cells) in the development of persistent pain?
Synaptic plasticity in different systems relies on de novo protein synthesis to tag activated synapses and contribute to their structural and functional remodeling. How does mRNA translation regulation promote LTP-like potentiation of synaptic transmission in the spinal cord?
Lastly, ERK/mTOR and eIF2α pathways in the brain are distinctly involved in protein synthesis-dependent forms of synaptic plasticity, such as LTP and long-term depression (LTD) [79–84]. 4E-BP1 represses spinal cord synaptic potentiation [53], and mTORC1-mediated inhibition of 4E-BP1 activity appears to be a prominent mechanism to induce central sensitization. The function of other translational control pathways in the regulation of spinal synaptic plasticity remains unknown. Given the requirement for protein synthesis in the sensitization of peripheral and central nociceptive circuits, and in the transition of acute to chronic pain, deciphering the molecular and cellular mechanisms by which protein synthesis regulation promotes persistent pain provides an opportunity to identify novel pain targets and develop more-targeted and efficient pain therapeutics.
Highlights.
Persistent pain is largely mediated by the sensitization of nociceptive neurons. This sensitization relies on de novo gene expression to support biochemical and structural changes that maintain amplified pain signaling.
Nerve injury and peripheral inflammation stimulate local translation from pre-existing mRNAs in nociceptive sensory neurons. The newly synthesized proteins act locally to increase neuronal excitability or are transported to the cell body to activate transcriptional gene expression programs.
Eukaryotic initiation factor 4F (eIF4F) complex formation is a convergent point downstream of mammalian target of rapamycin (mTOR) and extracellular-signal-regulated kinase (ERK) signaling pathways to regulate cap-dependent translation and promote neuronal excitability, synaptic transmission, and pain. A cellular energy status sensor AMP-activated protein kinase (AMPK) emerges as an attractive target to normalize aberrant mRNA translation in pathological pain conditions. Metformin, an AMPK activator and US Food and Drug Administration (FDA)-approved antidiabetic drug, corrects abnormal mRNA translation and reverses enhanced neuronal excitability and pain hypersensitivity in a variety of animal models of chronic pain.
Glossary
- 5′ untranslated region (5′UTR) and 3′ untranslated region (3′ UTR)
sections of mRNA that are not translated into a protein. 5′UTR precedes the sequence encoding the protein (coding sequence), whereas 3′UTR follows it. These regions are involved in the regulation of mRNA translation and stability.
- AMP-activated protein kinase (AMPK)
a central cellular energy sensor and a master regulator of energy homeostasis. It senses the energy level by monitoring AMP:ATP and ADP:ATP ratios, and regulates a variety of cellular processes, including gluconeogenesis, protein synthesis, autophagy, and lipid metabolism.
- Cap
a modified guanine nucleotide located at the 5′ end of all nuclear transcribed mRNAs. The cap facilitates ribosome recruitment to the mRNA to initiate protein synthesis. It also participates in mRNA splicing, stability, and transport.
- Eukaryotic initiation factor 4F (eIF4F)
a three-subunit complex that recruits a small ribosomal subunit to the cap structure at the 5′ end of mRNA to promote translation initiation. eIF4F comprises: (i) eIF4A (an RNA helicase); (ii) eIF4E, which specifically interacts with the cap structure; and (iii) eIF4G, a large scaffolding protein that binds to both eIF4E and eIF4A. The expression level of eIF4E is the lowest of all eukaryotic initiation factors; therefore, the cap recognition is a rate-limiting step for translation initiation.
- ‘eIF4E-sensitive’ mRNAs
a subset of mRNAs that are sensitive to the activity of eIF4E. These mRNAs harbor highly structured 5′UTRs and generally encode proteins involved in growth, proliferation, and survival.
- Mammalian/mechanistic target of rapamycin complex 1 (mTORC1)
a protein kinase complex that integrates a variety of extracellular and intracellular cues to regulate key cellular processes, such as mRNA translation, lipid synthesis, mitochondrial functions, and autophagy. By regulating these processes, mTORC1 controls cell metabolism, growth, and proliferation. In the nervous system, mTORC1 has critical roles in the regulation of neuronal functions and synaptic plasticity.
- mRNA translation
a process describing the production of a protein from a sequence of mRNA; also referred to as protein synthesis.
- Neuropathic pain
pain caused by an injury or disease affecting the somatosensory system. In most cases, neuropathic pain is caused by damage to the peripheral nerves, but can also be produced by an injury or disease affecting the spinal cord or brain.
- Nociceptors
a subset of peripheral sensory neurons that transmit information about noxious stimuli, such as extreme heat, cold, and pressure, as well as toxic molecules and inflammatory mediators.
- Ribosome footprinting
a technique used for genome-wide quantitative analysis of mRNA translation. In ribosome footprinting, ribosome/RNA complexes are isolated from cell lysates and digested with an endoribonuclease, which degrades all RNAs that are not protected by bound ribosomes. This generates approximately 30-nucleotide-long fragments of ribosome-protected mRNAs, or ‘footprints’. The footprints are reverse-transcribed and cloned into a cDNA library for RNA sequencing. This approach provides information on: (i) the position of ribosomes on mRNAs (mapping); and (ii) the number of ribosomes associated with a given mRNA (translation efficiency).
- Sensitization
a process caused by a tissue injury or disease, resulting in reduced threshold and/or enhanced responsiveness of the somatosensory system to stimuli.
- Translating ribosome affinity purification (TRAP)
methodology for characterization of translated transcripts in a genetically defined cell population. Cell type-specific tagging of ribosomes (using eGFP-tagged ribosomal subunit L10a), followed by affinity purification of tagged ribosomes and subsequent sequencing of the ribosome-associated mRNAs allows the identification of translated mRNAs in specific cell types.
References
- 1.Ferrari LF, et al. Peripheral administration of translation inhibitors reverses increased hyperalgesia in a model of chronic pain in the rat. J Pain. 2013;14:731–738. doi: 10.1016/j.jpain.2013.01.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari LF, et al. Role of nociceptor alphaCaMKII in transition from acute to chronic pain (hyperalgesic priming) in male and female rats. J Neurosci. 2013;33:11002–11011. doi: 10.1523/JNEUROSCI.1785-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inceoglu B, et al. Endoplasmic reticulum stress in the peripheral nervous system is a significant driver of neuropathic pain. Proc Natl Acad Sci U S A. 2015;112:9082–9087. doi: 10.1073/pnas.1510137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoutorsky A, et al. eIF2alpha phosphorylation controls thermal nociception. Proc Natl Acad Sci U S A. 2016;113:11949–11954. doi: 10.1073/pnas.1614047113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melemedjian OK, et al. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J Neurosci. 2010;30:15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melemedjian OK, et al. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain. 2011;7:70. doi: 10.1186/1744-8069-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu JT, et al. Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J Clin Invest. 2014;124:592–603. doi: 10.1172/JCI70236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin LJ, et al. Epiregulin and EGFR interactions are involved in pain processing. J Clin Invest. 2017;127:3353–3366. doi: 10.1172/JCI87406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogen O, et al. Generation of a pain memory in the primary afferent nociceptor triggered by PKCepsilon activation of CPEB. J Neurosci. 2012;32:2018–2026. doi: 10.1523/JNEUROSCI.5138-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moy JK, et al. The MNK - eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. J Neurosci. 2017;37:7481–7499. doi: 10.1523/JNEUROSCI.0220-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obreja O, et al. NGF locally sensitizes nociceptors in human skin. Pain. 2017 doi: 10.1097/j.pain.0000000000001108. Published online November 13, 2017. http://dx.doi.org/10.1097/j.pain.0000000000001108. [DOI] [PubMed]
- 12.Rukwied R, et al. NGF induces non-inflammatory localized and lasting mechanical and thermal hypersensitivity in human skin. Pain. 2010;148:407–413. doi: 10.1016/j.pain.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Hirai T, et al. Aberrant plasticity of peripheral sensory axons in a painful neuropathy. Sci Rep. 2017;7:3407. doi: 10.1038/s41598-017-03390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruangsri S, et al. Relationship of axonal voltage-gated sodium channel 1.8 (NaV1.8) mRNA accumulation to sciatic nerve injury-induced painful neuropathy in rats. J Biol Chem. 2011;286:39836–39847. doi: 10.1074/jbc.M111.261701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geranton SM, et al. A rapamycin-sensitive signaling pathway is essential for the full expression of persistent pain states. J Neurosci. 2009;29:15017–15027. doi: 10.1523/JNEUROSCI.3451-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang HL, et al. Proteomic profiling of neuromas reveals alterations in protein composition and local protein synthesis in hyper-excitable nerves. Mol Pain. 2008;4:33. doi: 10.1186/1744-8069-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glock C, et al. mRNA transport & local translation in neurons. Curr Opin Neurobiol. 2017;45:169–177. doi: 10.1016/j.conb.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Jung H, et al. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinnebusch AG, et al. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 22.Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 23.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truitt ML, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 28.Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derry MC, et al. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 30.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miR-ISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 33.Weingarten-Gabbay S, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 34.Kolekar P, et al. IRESPred: Web server for prediction of cellular and viral internal ribosome entry site (IRES) Sci Rep. 2016;6:27436. doi: 10.1038/srep27436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kar AN, et al. Expanding axonal transcriptome brings new functions for axonally synthesized proteins in health and disease. Neuroscientist. 2017 doi: 10.1177/1073858417712668. Published online June 8, 2017. http://dx.doi.org/10.1177/1073858417712668. [DOI] [PMC free article] [PubMed]
- 36.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price TJ, et al. Decreased nociceptive sensitization in mice lacking the fragile X mental retardation protein: role of mGluR1/5 and mTOR. J Neurosci. 2007;27:13958–13967. doi: 10.1523/JNEUROSCI.4383-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melemedjian OK, et al. Local translation and retrograde axonal transport of CREB regulates IL-6-induced nociceptive plasticity. Mol Pain. 2014;10:45. doi: 10.1186/1744-8069-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakor DK, et al. Increased peripheral nerve excitability and local NaV1.8 mRNA up-regulation in painful neuropathy. Mol Pain. 2009;5:14. doi: 10.1186/1744-8069-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heiman M, et al. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jimenez-Diaz L, et al. Local translation in primary afferent fibers regulates nociception. PLoS One. 2008;3:e1961. doi: 10.1371/journal.pone.0001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih MH, et al. Spinal cord NMDA receptor-mediated activation of mammalian target of rapamycin is required for the development and maintenance of bone cancer-induced pain hypersensitivities in rats. J Pain. 2012;13:338–349. doi: 10.1016/j.jpain.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norsted Gregory E, et al. Mammalian target of rapamycin in spinal cord neurons mediates hypersensitivity induced by peripheral inflammation. Neuroscience. 2010;169:1392–1402. doi: 10.1016/j.neuroscience.2010.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Q, et al. Spinal phosphinositide 3-kinase-Akt-mammalian target of rapamycin signaling cascades in inflammation-induced hyperalgesia. J Neurosci. 2011;31:2113–2124. doi: 10.1523/JNEUROSCI.2139-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang L, et al. mTOR and its downstream pathway are activated in the dorsal root ganglion and spinal cord after peripheral inflammation, but not after nerve injury. Brain Res. 2013;1513:17–25. doi: 10.1016/j.brainres.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang F, et al. Activation of mammalian target of rapamycin mediates rat pain-related responses induced by BmK I, a sodium channel-specific modulator. Mol Pain. 2013;9:50. doi: 10.1186/1744-8069-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, et al. Activation of mTOR in the spinal cord is required for pain hypersensitivity induced by chronic constriction injury in mice. Pharmacol Biochem Behav. 2013;111:64–70. doi: 10.1016/j.pbb.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 48.Karim F, et al. Metabotropic glutamate receptor subtypes 1 and 5 are activators of extracellular signal-regulated kinase signaling required for inflammatory pain in mice. J Neurosci. 2001;21:3771–3779. doi: 10.1523/JNEUROSCI.21-11-03771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji RR, et al. ERK MAP kinase activation in superficial spinal cord neurons induces prodynorphin and NK-1 upregulation and contributes to persistent inflammatory pain hypersensitivity. J Neurosci. 2002;22:478–485. doi: 10.1523/JNEUROSCI.22-02-00478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obara I, Hunt SP. Axonal protein synthesis and the regulation of primary afferent function. Dev Neurobiol. 2014;74:269–278. doi: 10.1002/dneu.22133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price TJ, Geranton SM. Translating nociceptor sensitivity: the role of axonal protein synthesis in nociceptor physiology. Eur J Neurosci. 2009;29:2253–2263. doi: 10.1111/j.1460-9568.2009.06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asante CO, et al. Formalin-induced behavioural hypersensitivity and neuronal hyperexcitability are mediated by rapid protein synthesis at the spinal level. Mol Pain. 2009;5:27. doi: 10.1186/1744-8069-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khoutorsky A, et al. Translational control of nociception via 4E-binding protein 1. Elife. 2015;4:e12002. doi: 10.7554/eLife.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dziembowska M, et al. Activity-dependent local translation of matrix metalloproteinase-9. J Neurosci. 2012;32:14538–14547. doi: 10.1523/JNEUROSCI.6028-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawasaki Y, et al. Distinct roles of matrix metalloproteases in the early- and late-phase development of neuropathic pain. Nat Med. 2008;14:331–336. doi: 10.1038/nm1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kersten C, Cameron MG. Cetuximab alleviates neuropathic pain despite tumour progression. BMJ Case Rep. 2012;2012:22707700. doi: 10.1136/bcr.12.2011.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kersten C, et al. Epidermal growth factor receptor-inhibition (EGFR-I) in the treatment of neuropathic pain. Br J Anaesth. 2015;115:761–767. doi: 10.1093/bja/aev326. [DOI] [PubMed] [Google Scholar]
- 58.Starck SR, et al. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016;351:aad3867. doi: 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melemedjian OK, et al. mTORC1 inhibition induces pain via IRS-1-dependent feedback activation of ERK. Pain. 2013;154:1080–1091. doi: 10.1016/j.pain.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichling DB, Levine JD. Critical role of nociceptor plasticity in chronic pain. Trends Neurosci. 2009;32:611–618. doi: 10.1016/j.tins.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asiedu MN, et al. Targeting AMPK for the alleviation of pathological pain. EXS. 2016;107:257–285. doi: 10.1007/978-3-319-43589-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price TJ, Dussor G. AMPK: an emerging target for modification of injury-induced pain plasticity. Neurosci Lett. 2013;557(Pt A):9–18. doi: 10.1016/j.neulet.2013.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pernicova I, Korbonits M. Metformin–mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 64.Gantois I, et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat Med. 2017;23:674–677. doi: 10.1038/nm.4335. [DOI] [PubMed] [Google Scholar]
- 65.Mao-Ying QL, et al. The anti-diabetic drug metformin protects against chemotherapy-induced peripheral neuropathy in a mouse model. PLoS One. 2014;9:e100701. doi: 10.1371/journal.pone.0100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor A, et al. The use of metformin is associated with decreased lumbar radiculopathy pain. J Pain Res. 2013;6:755–763. doi: 10.2147/JPR.S52205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kialka M, et al. Metformin increases pressure pain threshold in lean women with polycystic ovary syndrome. Drug Design Dev Ther. 2016;10:2483–2490. doi: 10.2147/DDDT.S109086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asiedu MN, et al. The AMPK activator A769662 blocks voltage-gated sodium channels: discovery of a novel pharmacophore with potential utility for analgesic development. PLoS One. 2017;12:e0169882. doi: 10.1371/journal.pone.0169882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burton MD, et al. Pharmacological activation of AMPK inhibits incision-evoked mechanical hypersensitivity and the development of hyperalgesic priming in mice. Neuroscience. 2017;359:119–129. doi: 10.1016/j.neuroscience.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tillu DV, et al. Resveratrol engages AMPK to attenuate ERK and mTOR signaling in sensory neurons and inhibits incision-induced acute and chronic pain. Mol Pain. 2012;8:5. doi: 10.1186/1744-8069-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price TJ, Inyang KE. Commonalities between pain and memory mechanisms and their meaning for understanding chronic pain. Prog Mol Biol Transl Sci. 2015;131:409–434. doi: 10.1016/bs.pmbts.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferrari LF, et al. Distinct terminal and cell body mechanisms in the nociceptor mediate hyperalgesic priming. J Neurosci. 2015;35:6107–6116. doi: 10.1523/JNEUROSCI.5085-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Araldi D, et al. Repeated mu-opioid exposure induces a novel form of the hyperalgesic priming model for transition to chronic pain. J Neurosci. 2015;35:12502–12517. doi: 10.1523/JNEUROSCI.1673-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonin RP, De Koninck Y. A spinal analog of memory reconsolidation enables reversal of hyperalgesia. Nat Neurosci. 2014;17:1043–1045. doi: 10.1038/nn.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nader K, et al. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- 76.Lee JLC, et al. An update on memory reconsolidation updating. Trends Cogn Sci. 2017;21:531–545. doi: 10.1016/j.tics.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JY, et al. Spinal dopaminergic projections control the transition to pathological pain plasticity via a D1/D5-mediated mechanism. J Neurosci. 2015;35:6307–6317. doi: 10.1523/JNEUROSCI.3481-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonin RP, De Koninck Y. Reconsolidation and the regulation of plasticity: moving beyond memory. Trends Neurosci. 2015;38:336–344. doi: 10.1016/j.tins.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Costa-Mattioli M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa-Mattioli M, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costa-Mattioli M, et al. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Di Prisco GV, et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2al-pha. Nat Neurosci. 2014;17:1073–1082. doi: 10.1038/nn.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelleher RJ, 3rd, et al. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 84.Banko JL, et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25:9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Willis D, et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gonzalez C, et al. Axons provide the secretory machinery for trafficking of voltage-gated sodium channels in peripheral nerve. Proc Natl Acad Sci U S A. 2016;113:1823–1828. doi: 10.1073/pnas.1514943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tohda C, et al. Axonal transport of VR1 capsaicin receptor mRNA in primary afferents and its participation in inflammation-induced increase in capsaicin sensitivity. J Neurochem. 2001;76:1628–1635. doi: 10.1046/j.1471-4159.2001.00193.x. [DOI] [PubMed] [Google Scholar]
- 88.Ji RR, et al. p38 MAPK activation by NGF in primary sensory neurons after inflammation increases TRPV1 levels and maintains heat hyperalgesia. Neuron. 2002;36:57–68. doi: 10.1016/s0896-6273(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 89.Cox LJ, et al. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Denis-Donini S, et al. Localization of calcitonin gene-related peptide mRNA in developing olfactory axons. Cell Tissue Res. 1998;294:81–91. doi: 10.1007/s004410051158. [DOI] [PubMed] [Google Scholar]
- 91.Li XQ, et al. CGRP peptide and regenerating sensory axons. J Neuropathol Exp Neurol. 2004;63:1092–1103. doi: 10.1093/jnen/63.10.1092. [DOI] [PubMed] [Google Scholar]
- 92.Bi J, et al. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci U S A. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harrison BJ, et al. IB4-binding sensory neurons in the adult rat express a novel 3′UTR-extended isoform of CaMK4 that is associated with its localization to axons. J Comp Neurol. 2014;522:308–336. doi: 10.1002/cne.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minis A, et al. Subcellular transcriptomics – dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev Neurobiol. 2014;74:365–381. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- 95.Kim SJ, et al. Macromolecular synthesis contributes to nociceptive response to subcutaneous formalin injection in mice. Neuropharmacology. 1998;37:1091–1093. doi: 10.1016/s0028-3908(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 96.Asante CO, et al. Mammalian target of rapamycin signaling in the spinal cord is required for neuronal plasticity and behavioral hypersensitivity associated with neuropathy in the rat. J Pain. 2010;11:1356–1367. doi: 10.1016/j.jpain.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Asiedu MN, et al. Spinal protein kinase M zeta underlies the maintenance mechanism of persistent nociceptive sensitization. J Neurosci. 2011;31:6646–6653. doi: 10.1523/JNEUROSCI.6286-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melemedjian OK, et al. BDNF regulates atypical PKC at spinal synapses to initiate and maintain a centralized chronic pain state. Mol Pain. 2013;9:12. doi: 10.1186/1744-8069-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maixner DW, et al. Adenosine monophosphate-activated protein kinase regulates interleukin-1beta expression and glial glutamate transporter function in rodents with neuropathic pain. Anesthesiology. 2015;122:1401–1413. doi: 10.1097/ALN.0000000000000619. [DOI] [PMC free article] [PubMed] [Google Scholar]