Abstract
With the increasing availability of genomics, routine analysis of advanced cancers is now feasible. Treatment selection is frequently guided by the molecular characteristics of a patient’s tumor, and an increasing number of trials are genomically-selected. Furthermore, multiple studies have demonstrated the benefit of therapies that are chosen based upon the molecular profile of a tumor. However, the rapid evolution of genomic testing platforms and emergence of new technologies makes interpreting molecular testing reports more challenging. More sophisticated precision oncology decision support services are essential. This review outlines existing tools available for health care providers and precision oncology teams, and highlights strategies for optimizing decision support. Specific attention is given to the assays currently available for molecular testing, as well as considerations for interpreting alteration information. This article also discusses strategies for identifying and matching patients to clinical trials, current challenges, and proposals for future development of precision oncology decision support.
Keywords: Precision Oncology Decision Support, Bioinformatics
INTRODUCTION
The use of targeted therapies in molecularly-selected patients has led to a paradigm change in cancer medicine. However, although close to 50 targeted therapies have been approved by the Food and Drug Administration (FDA) for solid tumors, only half of these targeted therapies have predictive biomarkers associated with their FDA approval (Table 1).
Table 1.
Food and Drug Administration (FDA) approved targeted therapies for tumors that have an associated biomarker.
| Preferred Name |
Direct drug Target | Company | FDA Approved Indication - Disease(s) |
FDA Approved Indication - Biomarker(s) |
|---|---|---|---|---|
| Abemaciclib | CDK4, CDK6 | Eli Lilly and Company | Breast cancer | ER Positive, PR Positive, HER2 Negative |
| Afatinib | EGFR | Boehringer Ingelheim | Non-small cell lung carcinoma | EGFR Deletion Exon 19, EGFR L858R |
| Alectinib | EML4-ALK, ALK, INSR, RET | Genentech | Non-small cell lung carcinoma | ALK Fusion |
| Anastrozole | Aromatase | AstraZeneca Pharmaceuticals | Breast cancer | ER Positive, PR Positive |
| Bosutinib | ABL1, BCR-ABL1, SRC | Pfizer | Chronic myelogenous leukemia | BCR-ABL1 |
| Brigatinib | ALK, CSF1R, INSR, ABL1, LCK, IGF1R, CAMK2G, FLT4, RET, FGFR1, FGFR2, FGFR3, AURKA, JAK2, FGFR4, FYN, HCK, LYN, SRC, EGFR, EML4-ALK, FER, FES, FLT3, FPS, ROS1, TYK2, YES1, PTK2B, HER4, CAMK2D, CHEK1, CHEK2 | Ariad Pharmaceuticals | Non-small cell lung carcinoma | ALK Fusion |
| Ceritinib | NPM-ALK, ROS1 Fusion, ALK, INSR, IGF1R, TSSK3, FLT3, FGFR2, RET, FGFR3, LCK, JAK2, AURKA, LYN, EGFR, FGFR4 | Novartis Pharmaceuticals | Non-small cell lung carcinoma | ALK Fusion |
| Cetuximab | EGFR | Eli Lilly and Company | Colorectal cancer | KRAS Wildtype, EGFR Positive |
| Cobimetinib | MAP2K1 | F. Hoffmann-La Roche | Melanoma | BRAF V600E, BRAF V600K |
| Crizotinib | ALK, MET, ROS1, NTRK1 | Pfizer | Non-small cell lung carcinoma | ALK Fusion |
| Pfizer | Non-small cell lung carcinoma | ROS1 Positive | ||
| Dabrafenib | BRAF, RAF1 | GlaxoSmithKline | Melanoma | BRAF V600E |
| GlaxoSmithKline | Melanoma | BRAF V600E, BRAF V600K | ||
| GlaxoSmithKline | Non-small cell lung carcinoma | BRAF V600E | ||
| Dasatinib | ABL1, KIT, BRAF, BCR-ABL1, ABL2, PDGFRA, PDGFRB, SRC, DDR1, DDR2, EPHA3 Amplification, EPHA2, FYN, LCK, LYN, YES1 | Bristol-Myers Squibb | Chronic myelogenous leukemia | BCR-ABL1 |
| Acute lymphoblastic leukemia | BCR-ABL1 | |||
| Enasidenib | IDH2 | Agios Pharmaceuticals | Acute myeloid leukemia | IDH2 Mutation |
| Erlotinib | EGFR | Genentech | Non-small cell lung carcinoma | EGFR Deletion Exon 19, EGFR L858R |
| Everolimus | MTOR | Novartis Pharmaceuticals | Breast cancer | ER Positive, PR Positive, HER2 Negative |
| Exemestane | Aromatase | Pfizer | Breast cancer | ER Positive |
| Fulvestrant | ER | AstraZeneca Pharmaceuticals | Breast cancer | ER Positive, PR Positive, HER2 Negative |
| Breast cancer | ER Positive, PR Positive | |||
| Gefitinib | EGFR | AstraZeneca Pharmaceuticals | Non-small cell lung carcinoma | EGFR Deletion Exon 19, EGFR L858R |
| Gemtuzumab Ozogamicin | CD33 | Pfizer | Acute myeloid leukemia | CD33 Positive |
| Ibrutinib | TEC, ABL1, FYN, RIPK2, SRC, LYN, PDGFRA, HER2, BTK, EGFR, BLK, BMX, CSK, FGR, PTK6, HCK, YES1, ITK, JAK3, FRK, LCK, RET, FLT3 | Janssen Biotech|Pharmacyclics | Small lymphocytic lymphoma, Chronic lymphocytic leukemia | c.CHR17p Deletion |
| Imatinib | PDGFRA, KIT, BCR-ABL1, ABL1, PDGFRB | Novartis Pharmaceuticals | Gastrointestinal Stromal Tumors | KIT Positive |
| Chronic myeloid leukemia, Acute lymphoblastic leukemia, | BCR-ABL1 | |||
| Myelodysplastic/myeloproliferative diseases | PDGFRA Fusion | |||
| Chronic eosinophilic leukemia | FIP1L1-PDGFRA | |||
| Lapatinib | EGFR, HER2, HER4 | Novartis Pharmaceuticals | Breast cancer | HER2 Positive |
| Breast cancer | ER Positive, PR Positive, HER2 Positive | |||
| Letrozole | Aromatase | Novartis Pharmaceuticals | Breast cancer | ER Positive, PR Positive |
| Midostaurin | KDR, FLT3, PDGFRA, PDGFRB, SYK, AKT1, FLT1, AKT2, AKT3, KIT, SRC, PRKCA, PRKCB, PRKCG, CDK1, FGR, ETV6-NTRK3 | Novartis Pharmaceuticals | Acute myeloid leukemia | FLT3 Mutation |
| Neratinib | HER2, EGFR, KDR | Puma Biotechnology, Inc. | Breast cancer | HER2 Overexpression, HER2 Amplification |
| Nilotinib | BCR-ABL1 | Novartis Pharmaceuticals | Chronic myelogenous leukemia | BCR-ABL1 |
| Olaparib | PARP1, PARP2 | AstraZeneca Pharmaceuticals | Ovarian cancer | BRCA1 (any deleterious), BRCA2 (any deleterious) |
| Osimertinib | EGFR, EGFR T790M, EGFR Exon 19 deletion | AstraZeneca Pharmaceuticals | Non-small cell lung carcinoma | EGFR T790M |
| Palbociclib | CDK4, CDK6 | Pfizer | Breast cancer | ER Positive, PR Positive, HER2 Negative |
| Panitumumab | EGFR | Amgen | Colorectal cancer | KRAS Wildtype, NRAS Wildtype |
| Pertuzumab | HER2 | Genentech | Breast cancer, Inflammatory breast cancer | HER2 Positive |
| Ponatinib | PDGFRA, KDR, SRC, ABL1, FGFR1, BCR-ABL1, KIT, RET | Ariad Pharmaceuticals | Acute lymphoblastic leukemia /lymphoblastic lymphoma, Chronic myeloid leukemia | BCR-ABL1 T315I |
| Chronic myeloid leukemia, Acute lymphoblastic leukemia | BCR-ABL1 | |||
| Ribociclib | CDK4, CDK6 | Novartis Pharmaceuticals | Breast cancer | ER Positive, PR Positive, HER2 Negative |
| Rituximab | CD20 | Genentech | Non-Hodgkin's lymphoma, Chronic lymphocytic leukemia | CD20 Positive |
| Rucaparib | PARP1 | Clovis Oncology | Ovarian cancer | BRCA1 (any deleterious), BRCA2 (any deleterious) |
| Tamoxifen | ER | AstraZeneca Pharmaceuticals | Breast cancer | ER Positive (may help predict whether therapy will be beneficial) |
| Trametinib | MAP2K1, MAP2K2 | GlaxoSmithKline | Melanoma | BRAF V600E, BRAF V600K |
| Non-small cell lung carcinoma | BRAF V600E | |||
| Trastuzumab | HER2 | Genentech | Breast cancer, Gastric cancer, Gastroesophageal junction | HER2 Positive |
| Trastuzumab Emtansine | HER2, p.Tubulin | Genentech | Breast cancer | HER2 Positive |
| Vemurafenib | BRAF V600E | F. Hoffmann- La Roche | Melanoma | BRAF V600E |
| Venetoclax | BCL2 | AbbVie | Chronic lymphocytic leukemia | c.CHR17p Deletion |
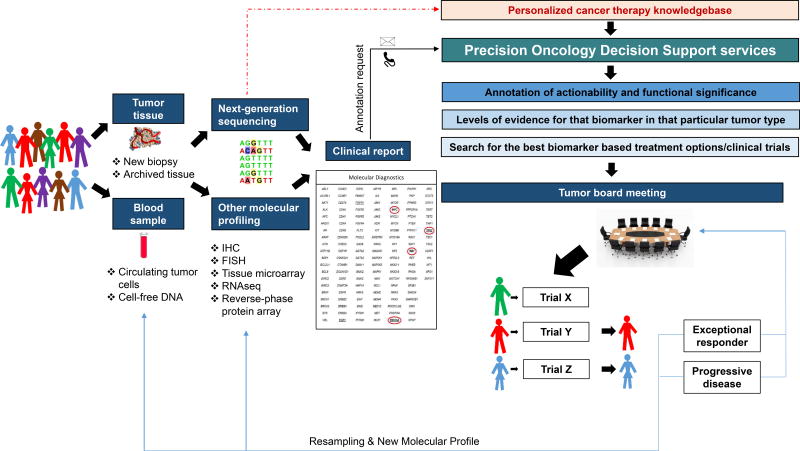
As the breadth of molecular testing and the number of biomarker-selected trials grow, clinicians are less able to rapidly interpret molecular reports during a clinical encounter and determine optimal approved or investigational therapies. The complexities of these processes are highlighted in Figure 1. A survey at a major cancer center demonstrated that many oncologists lack confidence regarding their ability to interpret genomic information(1). As a result, a new field of precision oncology decision support has emerged. The focus of this review is to provide novel insights into the current tools available for precision oncology decision support, as well as to help identify opportunities for future development.
Figure 1.
A flow diagram of precision oncology services that an individual patient may receive while being considered for a targeted agent or investigational therapy.
MOLECULAR TESTING
Genomic Testing
Identifying clinically-relevant characteristics of an individual tumor is critical to precision oncology. Historically, oncologists have chosen therapies and determined prognoses based on site of origin and histology. In select tumor types, oncologists began incorporating biomarkers, such as immunohistochemistry for HER2 and estrogen/progesterone receptor status in breast cancer(2,3). Today, genomic characterization is increasingly being used to guide treatment decisions, especially in patients with advanced disease.
Genomic characterization has been performed using a variety of strategies that range from hotspot exon sequencing of a select panel of genes to whole genome sequencing(4–8). Early genomic evaluations focused primarily on single nucleotide variations (SNVs). Assessments have now expanded to evaluate for other alterations, including indels, translocations, and copy number variations. Although assays to detect these alterations are not as widely available as those for SNVs, several such alterations have been successfully targeted(9–12). Furthermore, some of these successes were for alterations present in rare tumor types that are unlikely to be studied in tumor-specific clinical trials, as in the case of NTRK fusions(12,13). This gave rise to the possibility of tumor-agnostic, molecularly-driven registration strategies.
Panel testing for genomic alterations is becoming more widespread. Many treating physicians utilize commercial testing through companies such as Foundation One (Foundation Medicine, 315 genes) or Caris Molecular Intelligence (Caris Life Sciences, 592 genes). Some institutions have implemented commercial platforms such as Ion Torrent AmpliSeq Cancer Hotspot Panels (ThermoFisher Scientific, 50 genes) or Oncomine Comprehensive Assay (ThermoFisher Scientific, 143 genes V1, 161 genes V3). A small number of institutions have developed their own platforms, such as the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT, 468 genes) assay(14,15). Although local testing requires local molecular diagnostic and bioinformatics support(14,16), it also allows for customization and inclusion of specific rarer alterations within a gene panel. As more actionable alterations are identified, having the ability to alter the available panels to meet local needs may be advantageous.
Whole exome sequencing (WES) is a technique that sequences all the protein-coding genes in a genome (known as exome). This can identify the genetic variants that alter protein sequences at a lower cost than whole genome sequencing (WGS) (the process of sequencing the complete genome). Whole exome and whole genome sequencing have predominantly been research tools, but more recently are being introduced to the clinic(17,18). These strategies have the disadvantage of needing a stronger bioinformatics and decision support pipeline and potentially providing greater information burden on the clinical providers. In contrast, while hot spot sequencing is much cheaper and easier to support from a bioinformatics perspective, these sequencing efforts do not provide as much opportunity for discovery. Panel testing of 100–500 cancer related genes appear to have the greatest utilization at this juncture. Panel tests have the advantages of needed relatively small amounts of DNA, ability to support some discovery, and providing enough coverage and sequencing depth to allow for detection of subclonal variants. WES may especially have a role in patients with no actionable alterations on panel testing, in rare tumors, or in tumors that infrequently have alterations on commonly tested genes. The utility of early routine WES versus selective WES testing is likely to become more apparent over the next few years.
Treating oncologists must be aware of the relevant actionable alterations in the diseases they treat (Table 2). Knowledge of the expected genomic landscape will allow oncologists to order genomic testing early in diseases that have many genomic drivers linked to approved therapies, such as in the case of NSCLC or melanoma(19). This understanding will also ensure that oncologists request assays that evaluate for alterations more frequently found in the patient’s tumor type (e.g., FGFR fusion testing for cholangiocarcinoma, TRK fusions in secretory breast cancer), especially when patients are interested in investigational therapies.
Table 2.
A list of actionable genes, the alteration types, and the alteration frequencies for several common cancer types.
| Tumor type | Actionable genes | Alteration type | Frequency | Comments |
|---|---|---|---|---|
| Non-small cell lung cancer | BRAF | Mutations | 5–10% | |
| DDR2 | Mutations | 1–6% | ||
| EGFR | Mutations | 4–18% | ||
| EML4-ALK | Fusion | 4% | ||
| ERBB2 | Mutations | 2–3% | ||
| FGFR1 | Amplification | 2–17% | ||
| FGFR3 | Fusion | 2% | ||
| KRAS | Mutations | 1–32% | 1% in adenocarcinoma, 32% in squamous cell carcinoma | |
| MAP2K1 | Mutations | 1% | ||
| MET | Amplification | 1–4% | ||
| MET | Mutations | 3–8% | 3% MET exon 14 mutation in lung adenocarcinoma | |
| NF1 | Mutations | 11% | ||
| NTRK1 | Fusion | 2–4% | ||
| PIK3CA | Mutations | 4–16% | ||
| PTEN | Mutations/Deletion | 1–8% | ||
| RET | Fusion | 2–4% | ||
| RICTOR | Amplification | 2–5% | ||
| ROS1 | Fusion | 4–11% | ||
| STK11 | Mutations | 2–17% | ||
| Bladder | AKT1 | Mutations | 3% | |
| CDKN2A | Deletion | 47% | ||
| CDKN2A | Mutations | 5% | ||
| EGFR | Amplification | 11% | ||
| ERBB2 | Amplification | 7% | ||
| ERBB3 | Mutations | 11% | ||
| FGFR3 | Mutations | 45% | 60–80% in non-muscle-invasive; 15–20% in muscle-invasive bladder cancer | |
| FGFR3 | Amplification | 3% | ||
| FGFR3-TACC3 | Fusion | 5% | ||
| KRAS | Mutations | 4% | ||
| MDM2 | Amplification | 9% | ||
| PIK3CA | Mutations | 20% | ||
| PTEN | Mutations | 3% | ||
| PTEN | Deletion | 13% | ||
| TSC1 | Mutations | 9% | ||
| Biliary | BRAF | Mutations | 7% | |
| EGFR | Mutations/Amplification | 5% | ||
| ERBB2 | Mutations/Amplification | 4–18% | 18% in gallbladder carcinoma | |
| FGF19 | Amplification | 3% | ||
| FGFR1 | Mutations/Amplification | 4% | ||
| FGFR2 | Fusion | 5% | 5% in intrahepatic cholangiocarcinoma | |
| IDH1/2 | Mutations | 0–6% | 4–6% in intrahepatic cholangiocarcinoma | |
| KRAS | Mutations | 18% | ||
| MDM2 | Amplification | 5% | ||
| PIK3CA | Mutations | 7% | ||
| PTEN | Deletion | 1–7% | 7% in gallbladder carcinoma | |
| Gastric | EGFR | Mutations | 3–5% | |
| EGFR | Amplification | 6% | ||
| ERBB2 | Mutations | 5–7% | ||
| ERBB2 | Amplification | 13% | ||
| ERBB3 | Mutations | 5–11% | ||
| ERBB3 | Amplification | 4% | ||
| FGFR1 | Mutations | 4% | ||
| FGFR2 | Amplification | 5% | ||
| KRAS | Mutations | 6% | ||
| MET | Mutations | 2% | ||
| MET | Amplification | 4% | ||
| PIK3CA | Mutations/Amplification | 24% | 42% and 72% in MSI-H and EBV+ gastric cancer, respectively | |
| PTEN | Mutations | 4–8% | ||
| PTEN | Deletion | 4% | ||
| Melanoma | BRAF | Mutations | 45% | |
| CDKN2A | Deletion | 13% | ||
| IDH1 | Mutations | 6% | ||
| KDR | Amplification | 3% | ||
| KIT | Amplification | 4% | ||
| MAP2K1 | Mutations | 5% | ||
| NF1 | Mutations | 14% | ||
| NRAS | Mutations | 10–25% | ||
| PDGFRA | Amplification | 3% | ||
| Breast | 11q | Amplification | 15% | |
| AKT1 | Mutations | 2–4% | ||
| CDKN2A | Deletion | 3–4% | ||
| ERBB2 | Mutations/Amplification | 13% | ||
| ESR1 | Mutations | 10% | ER+ breast cancer, metastatic samples and not primary (marker of resistance to antiestrogen therapy) | |
| FGFR1 | Amplification | 10–15% | ||
| FGFR2 | Amplification | 4% | ||
| MAP2K4 | Mutations | 2–7% | ||
| MAP3K1 | Mutations | 4–13% | ||
| NF1 | Mutations | 2–4% | ||
| NTRK3 | Fusion | 92% | Secretory breast cancer | |
| PIK3CA | Mutations | 9–45% | ||
| PIK3CA | Amplification | 4–5% | ||
| PIK3R1 | Mutations | 2% | ||
| PTEN | Mutations/Deletion | 3–8% | ||
| RB1 | Mutations/Deletion | 5–6% | Marker of resistance to CDK 4/6 inhibitors | |
| Colorectal | AKT1 | Mutations | 1–6% | |
| BRAF | Mutations | 3–47% | 47% in MSI-H colorectal cancer | |
| ERBB2 | Mutations/Amplification | 6–13% | ||
| ERBB3 | Mutations | 4–20% | ||
| KRAS | Mutations | 35% | ||
| NRAS | Mutations | 10% | ||
| PIK3CA | Mutations | 15–37% | 37% MSI-H colorectal cancer | |
| PIK3R1 | Mutations | 2–17% | ||
| PTEN | Deletion | 4–20% | 20% MSI-H colorectal cancer | |
| Ovarian | AKT1 | Amplification | 3% | |
| AKT2 | Amplification | 2% | ||
| BRAF | Mutations | 2–6% (low grade serous ovary) | Extremely rare in high grade ovarian cancer; 2–6% low grade serous ovarian cancer (excluding borderline tumors) | |
| BRCA1 (germline or somatic) | Mutations | 9% | ||
| BRCA2 (germline or somatic) | Mutations | 5% | ||
| CCND1 | Amplification | 20% | ||
| CDKN2A | Deletion | 32% | ||
| FGFR1 | Amplification | 5% | ||
| KRAS | Mutations/Amplification | 19–33% (low grade serous ovary) | Extremely rare in high grade; 19–33% low grade serous ovarian cancer (excluding borderline tumors) | |
| NF1 | Mutations/Deletion | 12% | ||
| NOTCH3 | Mutations/Amplification | 11% | ||
| PIK3CA | Mutations/Amplification | 18% | ||
| PTEN | Mutations/Deletion | 7% | ||
| Glioblastoma | BRAF | Mutations | 2% | |
| CDK4 | Amplification | 14% | ||
| CDK6 | Amplification | 2% | ||
| CDKN2A/B | Deletion | 61% | ||
| EGFR | Mutations | 17–21% | ||
| EGFR | Amplification | 41–44% | ||
| FGFR1-TACC1 | Fusion | NA | ||
| FGFR3-TACC3 | Fusion | 3–7% | ||
| IDH1 | Mutations | 5–12% | ||
| MDM2 | Amplification | 7% | ||
| MDM4 | Amplification | 8% | ||
| MET | Amplification | 2% | ||
| NF1 | Mutations | 10% | ||
| NTRK1 | Fusion | 1% | ||
| PDGFRA | Amplification | 10% | ||
| PIK3CA | Mutations/Amplification | 25% | ||
| PTEN | Mutations/Deletion | 41% |
MSI-H = Microsatellite instability high; EBV = Epstein-Barr Virus
Tumors may show genomic evolution over time or may have intra- and intertumor heterogeneity between primary and metastatic sites(20,21); therefore, repeat tumor sampling is being pursued more frequently. Due to greater tissue availability, testing in patients with metastatic cancer is often done on primary tumors. Although one would expect most truncal alterations will be found in the primary tumor, when more than one tumor sample available, the most current sample should be used to account for genomic evolution. Although biopsies can usually be done relatively safely, issues surrounding patient inconvenience, procedure-related pain and complications, and biopsy costs make proximal or serial tissue testing challenging. Insurance coverage and procedural availability may also vary greatly, making broad recommendations difficult to provide. Therefore, the role of repeat biopsies and sequencing remains controversial. Considerations while deciding between analyzing an archival sample and obtaining a new sample include time since sample collection, number of lines of treatment given in the interim, types of treatment lines, as well as logistical considerations including feasibility and safety of a new biopsy. Although there is significant concordance in specific mutations between primary tumors and metastasis, there may be changes in actionable alterations(21). There are also now many established mechanisms of acquired resistance to targeted therapies such as acquired mutations in EGFR and ESR1 with EGFR inhibitors and endocrine therapy respectively, and BRCA mutation reversions with PARP inhibitors, demonstrating the value of repeat sampling and genomic analysis(22,23). For this reason, when possible, genomically-matched trial designs should incorporate “progression biopsies”: biopsy of a progressing lesion in patients who experience progression after initial response or prolonged stable disease. This can give important insights into mechanisms of acquired resistance and ways to overcome them early in drug development.
Liquid Biopsies
The advent of cell-free DNA and circulating tumor cell testing (i.e., “liquid biopsy”) has provided less invasive modalities for genomic testing. Liquid biopsies were initially undertaken using allele-specific polymerase chain reaction and flow cytometry(24). Now, next-generation sequencing panels are increasingly employed(25). The decision to use panel testing versus PCR-based testing depends on the underlying question. When a known mutation is being followed during treatment, it may be more cost effective and sensitive to perform focused testing. However, initial testing or subsequent treatment planning assessments may require a broader panel. Furthermore, serial sampling of plasma with broader sequencing may allow for the detection of acquired resistance mutations in the targeted gene, as shown with EGFR, FGFR and HER2 inhibitors(26–28). Larger panels can also facilitate identification of alternate resistance mechanisms.
Liquid biopsy also has several limitations. It may miss small amounts of mutant DNA, as in the cases of lower mutant allele frequency (subclonal) alterations, patients with limited disease burden, and tumor lineages that release small amounts of DNA into the circulation. It is a strength that liquid biopsy reflects the pool of alterations in a patient, but it may also be more difficult to assess mechanisms of resistance in patients with mixed response. Furthermore, certain alteration types may be more difficult to detect, such as copy number variations(29). With the exciting advances made possible by single cell techniques in the assessment of tumor heterogeneity and subclonal detection of alterations(30,31), it is likely that liquid biopsies will complement rather than replace tumor testing in the foreseeable future.
Multianalyte Testing
Although genomics has been the primary tool in precision oncology recently, there is growing recognition that only a subset of patients has truly compelling genomic alterations and even then only a subset of patients responds to genotype-matched therapy. These is a clear need to move beyond genomics only, and explore the utility of multianalyte testing, independently or in conjunction with genomic testing.
As technology has evolved, the field has also begun incorporating transcriptomics and proteomics. Although microarrays have traditionally been used to evaluate transcriptomics, RNAseq in increasingly being used (6,8,32). RNAseq has already been used to help identify novel alteration types that would be missed at the genomics level, such as fusions and rearrangements(33,34). RNAseq also has the advantage of determining whether the genomic alterations are reflected at the RNA level (e.g., are the oncogenic mutations expressed and are the amplified genes overexpressed?). Newer technologies for proteomics including reverse-phase protein array, mass spectrometry, and cyclic immunofluorescence have great potential. High throughput proteomics techniques will likely provide further novel insights into potential targets and resistance mechanisms once these techniques are optimized for the clinical setting(35,36). Many of these novel characterization strategies will likely transition from the research environment to the clinical setting in the future. Successful clinical utilization of these rapidly evolving technologies requires a decision support framework to help interpret molecular results.
FUNCTIONAL ANNOTATION AND DECISION SUPPORT
Functional impact and therapeutic implications
As genomic testing platforms and both preclinical and clinical data expand, the interpretation of genomic reports becomes increasingly complex. Annotation of molecular alterations is the process of detailing what change has occurred, and the clinical significance of that change. A change in an amino acid can lead to a change in the activity, expression, or stability of the expressed protein, or to no change. Even within the same gene, one alteration might confer sensitivity to treatment, while another may result in resistance (e.g., EGFR mutations and EGFR inhibitors(26,37)). Thus, the functional significance of each alteration must be assessed for clinical decision making. In spite of our growing genomic knowledge, larger panels or whole exome sequencing often reveal variants of unknown significance (VUS), in addition to variants that are well-characterized (such as BRAF V600E). The functional impact of these VUSs is simply not yet known.
There are multiple computational algorithms designed to predict the functional impact of specific aberrations, however, their predictive ability remains limited(38). There is growing interest in functional genomics: generating specific mutations and assessing the effect of introducing these mutations into reporter cells, as well as assaying the effect of introducing the mutant gene versus wildtype gene on cell growth and survival and on pathway activation. However, it remains unclear if these in vitro assays will be uniformly effective in classifying the functional impact of alterations.
A consensus about how to classify therapeutic implications is also lacking. The use of predetermined levels of evidence may aid in data interpretation, but no universally agreed-upon system currently exists(39–41). Recently, the Association for Molecular Pathology, the American Society of Clinical Oncology, and the College of American Pathologists have released a consensus statement. They propose a four-tiered system for clinical significance of somatic sequence variations, ranging from variants with strong clinical significance to variants that are benign or likely benign(42). In practice, however, there are still inconsistencies in terms of predicting therapeutic implications(8,16,39,43–48). Thus, clinical judgment is still required in addition to integration of data from large databases and literature reviews.
Actionability of genomic alterations
“Actionability” of an alteration has been broadly defined to mean having potential clinical utility(8,16,39,43–48). A somatic aberration may be actionable if it alters prognosis or predicts therapy sensitivity/resistance. In some scenarios, prognostic markers may help guide therapeutic choices (e.g., TP53 mutations confer worse prognosis in leukemia and may lead to consideration of transplantation). Germline alterations may also be actionable by increasing the risk of cancer or other hereditary diseases, predicting therapeutic response, or altering drug metabolism and thereby affecting drug efficacy or toxicity. Overall, actionability implies that knowledge of that specific alteration’s presence would change patient management.
In precision oncology, much of the emphasis has been on determining therapeutic actionability. The definitions used at MD Anderson Cancer Center’s Precision Oncology Program are outlined in Supplemental Table 1. However, there remains no consensus on when a genomic alteration should be acted upon. Limiting actionability to FDA-approved therapies for a specific biomarker is restrictive, and fails to allow for use of genomic information to inform enrollment onto clinical trials or for predicting risk. Alternatively, treating patients with suspected benign alterations or variants of unknown significance with genomically-targeted agents would decrease the likelihood of achieving clinical benefit in patients. It would result in patients receiving potentially toxic and costly futile therapy that delays initiation of effective therapeutic options, and would dilute potential efficacy seen in trials. As the field continues to evolve, our understanding of which alterations are actionable will also grow, and selected VUSs will be reclassified as their function and therapeutic impact is understood. We will need to be able to quickly adapt to emerging information.
Use of Available Resources
Most decision support teams report using PubMed or other search engines to review available literature(8,44,47,49–51). Other public databases utilized include COSMIC (cancer.sanger.ac.uk/cosmic), cBio (www.cbioportal.org), ClinGen (www.clinicalgenome.org), UniProt (www.uniprot.org), ClinVar (www.ncbi.nlm.nih.gov/clinvar), 1000 Genomes (www.internationalgenome.org), and dbSNP (www.ncbi.nlm.nih.gov/projects/SNP). Although these resources are important, they are not necessarily designed for point of care clinical use. This underscores the need for specialized processes to review and interpret the existing molecular knowledge-bases. Several groups have therefore developed publically-available resources to help organize and interpret the immense amount of existing molecular data.
Some of the most prominent publically-available precision oncology websites are listed in Table 3. Although most include similar general information, important differences between the websites include content organization, the ability to search for clinical trials, and the level of detail provided. There are also differences in algorithms for deciding which data to include and the definitions of actionability. At present, no direct comparisons of these resources exist, and thus the choice of which website to use depends on users’ individual needs and preferences. While creating and maintaining these publically-available resources is a time-consuming and labor-intensive process, such resources can help bridge the gap between research and practice, particularly when on-site precision oncology support services are not available.
Table 3.
Select publically-available precision oncology decision support websites.
| Website | Maintained by |
Number of genes |
Organized by gene? |
Organized by tumor type? |
Clinical trials? |
|---|---|---|---|---|---|
| Personalized Cancer Therapy www.personalizedcancertherapy.org (Accessed: 26 June 2017) | The University of Texas MD Anderson Cancer Center | 30 | Yes | No: tumor-specific information listed within the gene page | Yes: searchable by gene |
| My Cancer Genome www.mycancergenome.org (Accessed: 26 June 2017) | Vanderbilt-Ingham Medical Center | 823 | No: must search by tumor type (except trials) | Yes | Yes: searchable by gene and tumor type |
| OncoKB oncokb.org (Accessed: 26 June 2017) | Memorial Sloan Kettering Cancer Center, partnership with Quest Diagnostics | 476 | Yes | No: frequency data for tumor types listed within the gene page | No |
| Precision Cancer Medicine www.precisioncancermedicine.org (Accessed: 26 June 2017) | Dana Farber/Brigham and Women's Cancer Center | 17 | No | Yes: clinical trials organized primarily by tumor type | Yes: limited to those offered at Dana Farber |
| Drug Gene Interaction Database dgidb.genome.wustl.edu (Accessed: 26 June 2017) | Washington University in St. Louis | 8,419 | Yes | No: would need to review references for tumor type information | No |
Many providers or institutions have alternatively sought assistance from centralized decision support services. Selected commercial services are listed in Supplemental Table 2.
Molecular Tumor Boards
Many institutions have established Molecular Tumor Boards (MTBs) to help interpret the increasing amount of data available(8,16,43–48,52–54). MTBs are frequently multidisciplinary, and include oncologists, research scientists, bioinformaticians, and genetic counselors(47,53,55).
Most MTBs review a patient’s clinical, pathologic, and molecular information, and ultimately make therapy recommendations that include clinical trial options, use of FDA-approved agents on-label, and occasionally use of investigational agents on a compassionate IND or off-label use of FDA-approved therapies (45–47,49,52,54,56,57). Although clinical trials are potentially the most desirable strategy, studies show that the percentage of patients treated on genomically-driven clinical trials is low (2–18%)(15,46,49,52,56). In recent studies, the proportion of patients whose treatment decision was impacted by genomic testing was 5–40%(5,46,47,52,53,55,56), reflecting many challenges to the routine practice of precision oncology.
As genomic testing becomes more widely available, approaches such as MTBs are unlikely to be scalable. Ideally, available molecular data would be incorporated into existing treatment planning conferences and day-to-day clinical workflow, with point of care decision support.
Institutional Decision Support Services
Several large institutions have created teams to centralize genomic interpretation and provide decision support. At The University of Texas MD Anderson Cancer Center, we have set up a Precision Oncology Decision Support (PODS) team in order to facilitate therapeutic decision-making(39). The PODS team maintains highly curated databases for gene-variants, functional genomics, drugs, and clinical trials, as previously described(58,59). Our current list of actionable genes is available in Supplemental Table 3, but is continuously evolving.
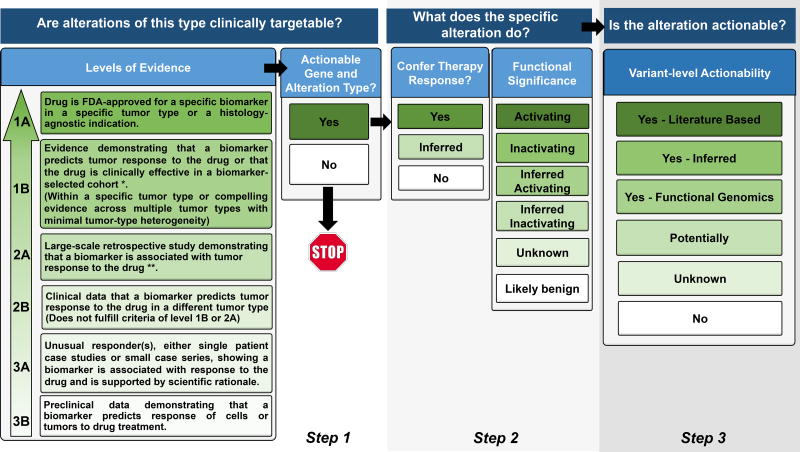
Most decision support requests are for interpretation of genomic results. Clinicians can choose to either interpret the genomic data on their own, or can request an annotation by the PODS team. A conceptual model of the annotation process is shown in Figure 2. Once completed, the annotation is made available via the electronic health record(59). The PODS team also informs the requesting clinician of any actionable alterations and relevant clinical trials(39). Turnaround time for annotation is critical for maintaining clinical utility and we have strived to bring this time down to less than 24 hours, preferably within hours. We expect to transition to routine annotation of testing performed in-house soon, but expect that point-of care services will still be desired for testing performed elsewhere.
Figure 2.
Procedural flow used by the Precision Oncology Decision Support (PODS) team at MD Anderson Cancer Center for annotating an alteration. Our tiers for Levels of Evidence, Functional Significance, and Variant Level of Actionability are included. Terminology is defined in Supplemental Table 1.
* For Level of Evidence 1B, “Evidence” could be:
- An adequately-powered, prospective study with biomarker selection/stratification
- A meta-analysis/overview
- A consensus recommendation for standard of care (as recommended by NCCN guidelines or other consortia)
** For Level of Evidence 2A, “Evidence” could be:
- A prospective trial where biomarker study is the secondary objective
- An adequately-powered retrospective cohort study
- An adequately-powered case-control study
At this time, the PODS team has received over 3,629 annotation requests on 2,741 patients with over 50 tumor types(59). The first 2,444 PODS annotations performed for 669 patients that included an actionable variant call were recently reviewed: 32.5% were actionable, 9.4% potentially actionable, 29.7% unknown, 28.4% non-actionable(59). Of patients with actionable or potentially actionable alterations, 20.6% enrolled on a genomically-matched trial. Trial enrollment was higher for actionable/potentially actionable alterations (27.6%) than those with unknown (11.8%) and non-actionable (3%) alterations (p=0.00004). This demonstrates the interest in and perceived value of decision support, even in academic settings with highly specialized oncologists. Decision support needs in the community may be even higher, with the caveat that therapeutic options may be more limited due to lack of access to trials.
DECISION SUPPORT EFFORTS IN PRECISION ONCOLOGY TRIALS
Decision Support in Precision Oncology Trials
There are now also several large multicenter precision oncology studies. We will highlight four important examples to demonstrate decision support approaches utilized.
NCI-MATCH (Molecular Analysis for Therapy Choice, NCT02465060) is a prospective precision oncology trial in which patients are assigned to receive treatment based on alterations found in their tumors through genomic sequencing (and a few IHC assays)(57,60). Treatment decisions are based upon pre-determined algorithms (MATCHBOX)(57), with review of matches by a multidisciplinary team including oncologists, informaticians, and pathologists.
NCI-MPACT (NCT01827384) is a randomized, biomarker-driven clinical trial. It uses next-generation sequencing to evaluate for alterations impacting RAS/RAF/MEK, PI3K, and DNA damage repair(61). Pre-specified rules designate alterations as being actionable. For the study, the NCI developed a system called GeneMed, which helps to determine appropriate treatment allocation(61). GeneMed automatically identifies suspected actionable alterations, which are then individually reviewed. These data are then used to randomize the patient. Automating part of the process reduces the burden on the decision support team, and standardizes the definition of actionability across sites.
ASCO’s Targeted Agent and Profiling Utilization Registry (TAPUR) is also ongoing. The goal of this registry study is to describe drug activity and toxicity for novel agents used in the setting of an actionable variant(62). This study has the potential to increase access to targeted agents and precision oncology decision support to community oncology providers as well as participating academic centers. In this study, the provider can make the initial determination for patient eligibility and receive central confirmation(62). However, a Molecular Tumor Board which is organized by ASCO is also available for consultation. All drugs are provided by the pharmaceutical companies, and all data are collected and monitored centrally. In addition to providing important toxicity and efficacy information, this study will also assess practitioner knowledge.
Similar trials are also currently being conducted successfully outside of the United States. The WINTHER study is one such study which serves as an interesting model for incorporating transcriptomics into treatment decisions(63). WINTHER also highlights some of the considerations relevant to future international collaborations for precision oncology decision support. Specifically, attention to differences in regulatory testing requirements across countries and barriers to using similar platforms under different regulatory environments will be important for future trials(63).
CHALLENGES TO PRECISION ONCOLOGY DECISION SUPPORT
Expanding Available Biomarkers
Information about transcriptional output, protein expression, epigenetic modifications, and metabolomics may provide a broader understanding of mechanisms of tumor growth, and allow for better treatment decisions(8,32,64). Although difficulties with RNA preservation in formalin-fixed paraffin embedded tissues persist(65), approaches such as RNAseq are now transitioning to the CLIA environment. Further investigation into optimizing techniques for functional proteomics or to operationalize use of multiplex immunohistochemistry will be critical(66). Once these technologies are implemented in the clinical setting, however, the need for scientific and bioinformatics support will only increase.
Patients with multiple alterations remain another challenge. Targets often are prioritized, taking into consideration the allelic frequency of different alterations, relative copy number gain/loss for copy number alterations, and level of evidence of therapeutic implications. Sometimes the presence of a second alteration is thought to be a contraindication for acting on the first alteration (as in the case of activating mutation of EGFR and KRAS). This problem of multiple alterations is likely to grow with broader testing panels. However, multianalyte testing may further assist in decision-making by elucidating which downstream pathway is activated, etc.
Immunotherapy is also a growing field. Although many of the decision support criteria may be different, the precepts underlying the importance of decision support are the same. Biomarkers that predict sensitivity or resistance to immunotherapies are starting to be developed including tumor mutation burden, neoantigen load, neoantigen expression, tumor infiltration lymphocytes, CD8+ cells, PDL1 expression, and immune infiltrate gene expression signatures. However, some are still controversial (e.g., PDL1) and others are not readily available on all tumor types (tumor mutation burden)(67). Although the predictive value of genomics and other multianalyte techniques have shown promise, this work is still in its infancy as compared with biomarker discovery for targeted therapies(68,69). As indications and therapeutic approaches for immunotherapy continue to grow, it is imperative that precision oncology decision support services are equipped to interpret these classes of biomarkers.
Operational Considerations
Cost is an ongoing barrier for precision oncology. Although the cost of next-generation sequencing is decreasing(70), new technologies are continuously being developed. As a field, there is interest in broadening molecular testing for patients in order to address clonal evolution and molecular heterogeneity. However, this will also increase both the amount of data, and the frequency at which data must be analyzed. Furthermore, questions about insurance coverage of testing persist, despite increased use of sequencing in oncologic care(52,71). Additionally, the cost of maintaining decision support infrastructure itself must also be taken into account(72).
Access to investigational therapies through clinical trials is also relevant. The large number of exclusions to eligibility can limit the ability of patients with genomic aberrations to receive matched therapies. Additionally, insurance concerns about enrollment on trials persist. Clinical trial access is also related to geographic limitations, as a large proportion of cancer patients live a burdensome distance from the closest trial site(73). Novel precision oncology initiatives such as ASCO’s TAPUR study will enhance access to off-label drugs while ensuring oncologic outcomes are collected.
The institutional nature of most precision oncology decision support services is both an asset and a weakness. With local review, recommendations are based upon the specific needs and resources of that institution. However, much of the same work is done independently by many institutions. Multiple institutions maintain their own databases for research, including evolving information about actionability and available clinical trials. While this state of decentralized science is not limited to precision oncology, it does limit scalability and reproducibility. However, there are also financial, academic, and logistical incentives for maintaining separate databases and knowledgebases. Regardless, increasing efficiency and decreasing cost of decision support services will be necessary for precision oncology to become the new model for oncology care.
CONCLUSION
Precision oncology is an exciting and rapidly evolving field. With a greater number of therapies and diagnostics, there is a critical need for precision oncology decision support. As our ability to target specific pathways improves, so too must our processes for identifying appropriate patients. “All-comer” trials are increasingly replaced by molecularly-matched clinical trials. While this is likely to improve clinical outcomes, it also means that the knowledge required to practice oncology is becoming increasingly complex. Thus, more sophisticated and organized precision oncology decision support services is critical to implementing precision oncology in routine cancer care.
Supplementary Material
Acknowledgments
Financial support: This work was supported in part by 1U01 CA180964 (FMB, EB), NIH Research Training Grant T32 CA101642 (KCK), The Cancer Prevention and Research Institute of Texas (RP150535) (FMB, EB, AJ, JZ, AB, VH, NSS, KS), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (FMB, GBM, BL, AMJ, YBK, JZ, MAS, AMB, NSS, VH, JM, KMS), NCATS Grant UL1 TR000371 (Center for Clinical and Translational Sciences) (FMB, EB), The Bosarge Family Foundation (AMB, VH, JZ, GBM), and the MD Anderson Cancer Center Support Grant (P30 CA016672) (FMB).
Disclosures: Aileron Therapeutics (FMB – Research Funding), Allostery Inc (GBM – Consulting), AstraZeneca (FMB – Research Funding; GBM – Consulting, Research Funding), Bayer (FMB – Research Funding), Catena Pharmaceuticals (GBM – Consulting, Financial), Celgene (FMB – Consulting), Clearlight Diagnostics (FMB – Consulting), Critical Outcome Technologies (GBM – Consulting, Research Funding), Curis (FMB – Research Funding), CytomX Therapeutics (FMB – Research Funding), Debiopharm (FMB – Research Funding), Effective Pharmaceuticals (FMB – Research Funding), Genetech (FMB – Honoraria, Consulting), Illumina (GBM – Research Funding), ImmunoMet (GBM – Consulting, Financial), Inflection Biosciences (FMB – Consulting), Ionis (GBM – Consulting, Research Funding), Jounce Therapeutics (FMB – Research Funding), Karus (GBM – Research Funding), Medimmune (GBM – Consulting), Merrimack Pharmaceuticals (JM – Board Member), Myriad Genetics (GBM – Licensed Technology [HRD assay]), Nanostring (GBM – Research Funding), Novartis (FMB – Consulting, Research Funding), Nuevolution (GBM – Consulting), Pieris Pharmaceuticals (FMB – Consulting), Precision Medicine (GBM – Consulting), PTV Ventures (GBM – Financial), PUMA Biotechnology (FMB – Research Funding), Roche Diagnostics (FMB – Honoraria, Consulting), Signalchem Lifesciences (GBM – Consulting), Spindletop Ventures (GBM – Financial), Symphogen (GBM – Consulting), Taiho Pharmaceutical (FMB – Research Funding), Takeda/Millenium Pharmaceuticals (GBM – Consulting, Research Funding), Tarveda (GBM – Consulting), Verastem (FMB – Research Funding); Zymeworks (FMB – Research Funding)
The authors would like to thank Ms. Elena Vess, Ms. Amy Simpson, and Dr. Lauren Brusco for their administrative support on this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
Concept development: KCK, FMB
Writing: KCK, EEID, BL, YBK, FMB
Substantial reviewing: KCK, EEID, YBK, AMJ, JZ, MAS, AMB, NSS, VH, JM, KS, EB, GBM, FMB
References
- 1.Gray SW, Hicks-Courant K, Cronin A, Rollins BJ, Weeks JC. Physicians' attitudes about multiplex tumor genomic testing. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(13):1317–23. doi: 10.1200/jco.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwamoto T, Booser D, Valero V, Murray JL, Koenig K, Esteva FJ, et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(7):729–34. doi: 10.1200/jco.2011.36.2574. [DOI] [PubMed] [Google Scholar]
- 3.Toikkanen S, Helin H, Isola J, Joensuu H. Prognostic significance of HER-2 oncoprotein expression in breast cancer: a 30-year follow-up. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1992;10(7):1044–8. doi: 10.1200/jco.1992.10.7.1044. [DOI] [PubMed] [Google Scholar]
- 4.Hasmats J, Green H, Orear C, Validire P, Huss M, Kaller M, et al. Assessment of whole genome amplification for sequence capture and massively parallel sequencing. PloS one. 2014;9(1):e84785. doi: 10.1371/journal.pone.0084785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA oncology. 2015;1(4):466–74. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzilov AV, Ding W, Fink MY, Antipin Y, Brohl AS, Davis C, et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome medicine. 2016;8(1):62. doi: 10.1186/s13073-016-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(25):2753–62. doi: 10.1200/jco.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Science translational medicine. 2011;3(111):111ra21. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. The New England journal of medicine. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. The New England journal of medicine. 2014;371(21):1963–71. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. The New England journal of medicine. 2014;371(23):2167–77. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 12.Patients with NTRK Fusions Respond to Targeted Therapies. Cancer discovery. 2016;6(6):566–7. doi: 10.1158/2159-8290.cd-nb2016-046. [DOI] [Google Scholar]
- 13.Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer discovery. 2015;5(10):1049–57. doi: 10.1158/2159-8290.cd-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17(3):251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zehir A, Benayed R, Shah RH, Syed A, Middha S. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. 2017 doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. The Journal of molecular diagnostics : JMD. 2014;16(1):56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horak P, Frohling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO open. 2016;1(5):e000094. doi: 10.1136/esmoopen-2016-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature reviews Genetics. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 19.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(6):1258–66. doi: 10.1158/1078-0432.ccr-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, Yelensky R, Perez-Fidalgo JA, Wang Y, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Molecular cancer therapeutics. 2014;13(5):1382–9. doi: 10.1158/1535-7163.mct-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757–67. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domchek SM. Reversion Mutations with Clinical Use of PARP Inhibitors: Many Genes, Many Versions. Cancer Discov. 2017;7(9):937–9. doi: 10.1158/2159-8290.CD-17-0734. [DOI] [PubMed] [Google Scholar]
- 24.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 25.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7(9):9707–17. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(8):2240–7. doi: 10.1158/1078-0432.ccr-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer discovery. 2017;7(3):252–63. doi: 10.1158/2159-8290.cd-16-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Bose R, Gao F, Freedman R, Telli M, Kimmick G, et al. Proceedings of the 107th Annual Meeting of the American Association for Cancer Research. Vol. 2017. Washington D.C: AACR; 2017. Circulating tumor DNA (ctDNA) sequencing for HER2 mutation (HER2mut) screening and response monitoring to neratinib in metastatic breast cancer (MBC) [Google Scholar]
- 29.Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clinical chemistry. 2013;59(1):211–24. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 30.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature communications. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LoRusso PM, Boerner SA, Pilat MJ, Forman KM, Zuccaro CY, Kiefer JA, et al. Pilot Trial of Selecting Molecularly Guided Therapy for Patients with Non-V600 BRAF-Mutant Metastatic Melanoma: Experience of the SU2C/MRA Melanoma Dream Team. Molecular cancer therapeutics. 2015;14(8):1962–71. doi: 10.1158/1535-7163.mct-15-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nature medicine. 2012;18(3):375–7. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nature methods. 2010;7(9):681–5. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 36.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(3 Pt 1):839–44. doi: 10.1158/1078-0432.ccr-05-1846. [DOI] [PubMed] [Google Scholar]
- 38.Tarcic G, Kamal M, Edelheit O, Barbash Z, Vidne M, Miron B, et al. Functional mutational analysis to assess the oncogenic activity of variant of uncertain significance (VUS) detected in patients included in the SHIVA trial. European Journal of Cancer. 69:S6–S7. doi: 10.1016/S0959-8049(16)32618-1. [DOI] [Google Scholar]
- 39.Johnson A, Zeng J, Bailey AM, Holla V, Litzenburger B, Lara-Guerra H, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug discovery today. 2015;20(12):1433–8. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature medicine. 2014;20(6):682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andre F, Mardis E, Salm M, Soria JC, Siu LL, Swanton C. Prioritizing targets for precision cancer medicine. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2014;25(12):2295–303. doi: 10.1093/annonc/mdu478. [DOI] [PubMed] [Google Scholar]
- 42.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. The Journal of molecular diagnostics : JMD. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bardia A, Iafrate JA, Sundaresan T, Younger J, Nardi V. Metastatic Breast Cancer With ESR1 Mutation: Clinical Management Considerations From the Molecular and Precision Medicine (MAP) Tumor Board at Massachusetts General Hospital. The oncologist. 2016;21(9):1035–40. doi: 10.1634/theoncologist.2016-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knepper TC, Bell GC, Kevin Hicks J, Padron E, Teer JK, Vo TT, et al. Key Lessons Learned from Moffitt's Molecular Tumor Board: The Clinical Genomics Action Committee Experience. The oncologist. 2017;22(2):144–51. doi: 10.1634/theoncologist.2016-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons HA, Beaver JA, Cimino-Mathews A, Ali SM, Axilbund J, Chu D, et al. Individualized Molecular Analyses Guide Efforts (IMAGE): A Prospective Study of Molecular Profiling of Tissue and Blood in Metastatic Triple-Negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(2):379–86. doi: 10.1158/1078-0432.ccr-16-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaderbhai CG, Boidot R, Beltjens F, Chevrier S, Arnould L, Favier L, et al. Use of dedicated gene panel sequencing using next generation sequencing to improve the personalized care of lung cancer. Oncotarget. 2016;7(17):24860–70. doi: 10.18632/oncotarget.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tafe LJ, Gorlov IP, de Abreu FB, Lefferts JA, Liu X, Pettus JR, et al. Implementation of a Molecular Tumor Board: The Impact on Treatment Decisions for 35 Patients Evaluated at Dartmouth-Hitchcock Medical Center. The oncologist. 2015;20(9):1011–8. doi: 10.1634/theoncologist.2015-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;2(1):82–93. doi: 10.1158/2159-8290.cd-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantripragada KC, Olszewski AJ, Schumacher A, Perez K, Birnbaum A, Reagan JL, et al. Clinical Trial Accrual Targeting Genomic Alterations After Next-Generation Sequencing at a Non-National Cancer Institute-Designated Cancer Program. Journal of oncology practice. 2016;12(4):e396–404. doi: 10.1200/jop.2015.008433. [DOI] [PubMed] [Google Scholar]
- 50.Bailey AM, Mao Y, Zeng J, Holla V, Johnson A, Brusco L, et al. Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps. Discovery medicine. 2014;17(92):101–14. [PMC free article] [PubMed] [Google Scholar]
- 51.Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(15):1849–57. doi: 10.1200/jco.2012.45.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGowan ML, Ponsaran RS, Silverman P, Harris LN, Marshall PA. "A rising tide lifts all boats": establishing a multidisciplinary genomic tumor board for breast cancer patients with advanced disease. BMC medical genomics. 2016;9(1):71. doi: 10.1186/s12920-016-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gunderson CC, Rowland MR, Wright DL, Andrade KL, Mannel RS, McMeekin DS, et al. Initiation of a formalized precision medicine program in gynecologic oncology. Gynecologic oncology. 2016;141(1):24–8. doi: 10.1016/j.ygyno.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 54.Hirshfield KM, Tolkunov D, Zhong H, Ali SM, Stein MN, Murphy S, et al. Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers. The oncologist. 2016;21(11):1315–25. doi: 10.1634/theoncologist.2016-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker BA, Schwaederle M, Scur MD, Boles SG, Helsten T, Subramanian R, et al. Breast Cancer Experience of the Molecular Tumor Board at the University of California, San Diego Moores Cancer Center. Journal of oncology practice. 2015;11(6):442–9. doi: 10.1200/jop.2015.004127. [DOI] [PubMed] [Google Scholar]
- 56.Sohal DP, Rini BI, Khorana AA, Dreicer R, Abraham J, Procop GW, et al. Prospective Clinical Study of Precision Oncology in Solid Tumors. Journal of the National Cancer Institute. 2015;108(3) doi: 10.1093/jnci/djv332. [DOI] [PubMed] [Google Scholar]
- 57.Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Seminars in oncology. 2014;41(3):297–9. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Kurnit KC, Bailey AM, Zeng J, Johnson AM, Shufean MA, Brusco L, et al. "Personalized Cancer Therapy": A Publically Available Precision Oncology Resource. Cancer research. doi: 10.1158/0008-5472.CAN-17-0341. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson A, Khotskaya Y, Brusco L, Zeng J, Holla V, Bailey A, et al. Clinical Utilization of Precision Oncology Decision Support. JCO Precis Oncol. doi: 10.1200/PO.17.00036. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNeil C. NCI-MATCH launch highlights new trial design in precision-medicine era. Journal of the National Cancer Institute. 2015;107(7) doi: 10.1093/jnci/djv193. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, Polley EC, Li MC, Lih CJ, Palmisano A, Sims DJ, et al. GeneMed: An Informatics Hub for the Coordination of Next-Generation Sequencing Studies that Support Precision Oncology Clinical Trials. Cancer informatics. 2015;14(Suppl 2):45–55. doi: 10.4137/cin.s17282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oncology ASoC. [Accessed 2017];2017 Targeted Agent and Profiling Utilization Registry Study. < www.tapur.org>.
- 63.Rodon J, Soria JC, Berger R, Batist G, Tsimberidou A, Bresson C, et al. Challenges in initiating and conducting personalized cancer therapy trials: perspectives from WINTHER, a Worldwide Innovative Network (WIN) Consortium trial. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2015;26(8):1791–8. doi: 10.1093/annonc/mdv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA: a cancer journal for clinicians. 2016;66(1):75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staff S, Kujala P, Karhu R, Rokman A, Ilvesaro J, Kares S, et al. Preservation of nucleic acids and tissue morphology in paraffin-embedded clinical samples: comparison of five molecular fixatives. Journal of clinical pathology. 2013;66(9):807–10. doi: 10.1136/jclinpath-2012-201283. [DOI] [PubMed] [Google Scholar]
- 66.Assadi M, Lamerz J, Jarutat T, Farfsing A, Paul H, Gierke B, et al. Multiple protein analysis of formalin-fixed and paraffin-embedded tissue samples with reverse phase protein arrays. Molecular & cellular proteomics : MCP. 2013;12(9):2615–22. doi: 10.1074/mcp.M112.023051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY) 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(15):4242–50. doi: 10.1158/1078-0432.ccr-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV, et al. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity. 2016;45(5):1122–34. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Nimwegen KJ, van Soest RA, Veltman JA, Nelen MR, van der Wilt GJ, Vissers LE, et al. Is the $1000 Genome as Near as We Think? A Cost Analysis of Next-Generation Sequencing. Clinical chemistry. 2016;62(11):1458–64. doi: 10.1373/clinchem.2016.258632. [DOI] [PubMed] [Google Scholar]
- 71.Trosman JR, Weldon CB, Kelley RK, Phillips KA. Challenges of coverage policy development for next-generation tumor sequencing panels: experts and payers weigh. Journal of the National Comprehensive Cancer Network : JNCCN. 2015;13(3):311–8. doi: 10.6004/jnccn.2015.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think. Genome biology. 2011;12(8):125. doi: 10.1186/gb-2011-12-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galsky MD, Stensland KD, McBride RB, Latif A, Moshier E, Oh WK, et al. Geographic accessibility to clinical trials for advanced cancer in the United States. JAMA internal medicine. 2015;175(2):293–5. doi: 10.1001/jamainternmed.2014.6300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.