Abstract
Accurate chromosome segregation is critical to ensure the faithful inheritance of the genome during cell division. Human chromosomes distinguish the location of the centromere from general chromatin by the selective assembly of CENP-A containing nucleosomes at the active centromere. The location of centromeres in most higher eukaryotes is determined epigenetically, independent of DNA sequence. CENP-A containing centromeric chromatin provides the foundation for assembly of the kinetochore that mediates chromosome attachment to the microtubule spindle and controls cell cycle progression in mitosis. Here we review recent work demonstrating the role of posttranslational modifications on centromere function and CENP-A inheritance via the direct modification of the CENP-A nucleosome and pre-nucleosomal complexes, the modification of the CENP-A deposition machinery and the modification of histones within existing centromeres.
Epigenetic specification of centromeres
Chromosome segregation during mitosis and meiosis is orchestrated by the centromere. Centromeres are unique chromosome domains that direct the assembly of the kinetochore during mitosis, which mediates microtubule attachment, checkpoint signaling and chromosome movement [1]. Centromeres are defined by the presence of the histone H3 variant Centromere Protein A (CENP-A).
DNA sequence is sufficient to determine centromere location in budding yeast; however, centromere identity in most higher eukaryotes is not dependent on DNA sequence. In most higher eukaryotes, centromere specification relies on the incorporation of CENP-A nucleosomes, and the location of CENP-A nucleosomes is sufficient to specify the site of centromere formation [2]. Many neocentromeres have been identified, where centromere proteins and function have been relocalized to a new site within the chromosome devoid of alpha-satellite sequence [3]. Centromeres are therefore defined by the epigenetic mechanisms that facilitate CENP-A deposition and stable inheritance across the cell cycle, and across multiple generations, all of which are critical for maintaining centromere identity.
Covalent posttranslational modification (PTM) of histones is a major mechanism to regulate chromatin function. Here we review recent advances in our understanding of how PTMs of CENP-A, canonical histones within the centromere, and proteins involved in CENP-A deposition regulate centromere specification. We have focused our review on the human centromere; however, several examples of PTMs in important model systems are also included.
Intrinsic features of CENP-A modifications and CCAN recruitment
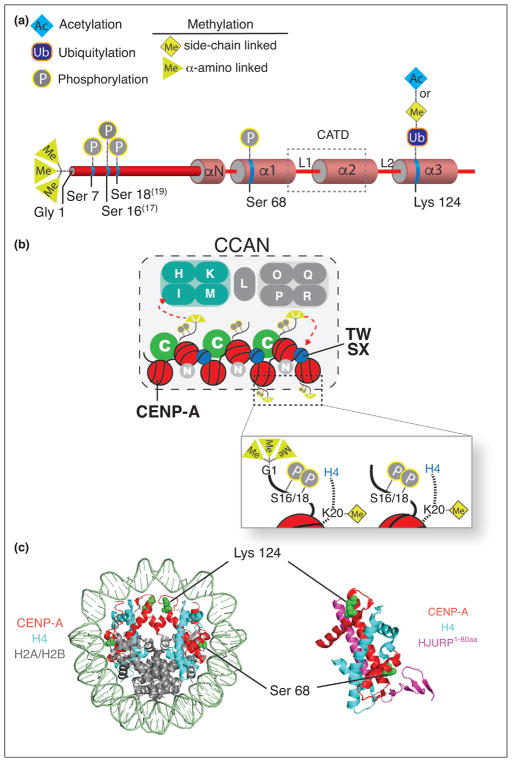
Posttranslational modifications of histones control chromatin function through intrinsic effects on chromatin structure, as well as the selective recruitment of proteins. CENP-A undergoes different types of PTMs in the amino terminal tail and within the histone fold domain (Figure 1a). Several modifications of CENP-A contribute to the recruitment of the CCAN (constitutive centromere associated network) (Figure 1a,b). The first PTM of human CENP-A identified was phosphorylation of Ser7, based on its similar location with H3 amino terminal tail phosphorylation at Ser10 [4]. Mutation of this site leads to errors in chromosome segregation and cytokinesis [5,6]. Ser7 phosphorylation is mediated by the Aurora kinases, and has been shown to indirectly recruit CENP-C through phospho-binding protein 14-3-3 [4–7]; although, the primary direct binding site for CENP-C is the carboxy terminus of CENP-A [8,9].
Figure 1.
Posttranslational modifications of CENP-A and CCAN recruitment. (a) A schematic of CENP-A secondary structure showing the locations of posttranslational modifications. Note that Ser16, Ser18 and Gly1 are numbered based on the removal of the initiating methionine, in convention with other histone proteins; however, these modifications have also been referred to by Ser17, Ser19 and Gly2, respectively. (b) Centromeric histone H4 and CENP-A posttranslational modifications found in centromeric chromatin. CCAN proteins (CENP-H/I/K/M, CENP-T/W/S/X and CENP-C) whose localization are influenced by CENP-A modifications are colored. (c) Location of phosphorylated Ser68 and modified Lys124 (green) within the CENP-A nucleosome (left) and the CENP-A/H4/HJURPScm3 pre-nucleosomal complex (right). Based on crystal structures, 3AN2 (human CENP-A nucleosome) and 3R45 (human CENP-A and histone H4 heterodimer bound to HJURP amino acids 1–80).
More recently, CENP-A was shown to be phosphorylated at two highly conserved residues, Ser16 and Ser18, both prior to its deposition and in the CENP-A nucleosome [10] (Figure 1a). CENP-A Ser18 is a substrate for cyclinE1/CDK2 phosphorylation, and is cell cycle regulated [11]. Loss of phosphorylation or hyper-phosphorylation at these sites, lead to chromosome missegregation [10,11]. In vitro studies suggest that CENP-A has the ability to form a salt-bridged secondary structure through intramolecular association, which is dependent on phosphorylation of Ser16/18, and therefore this dual phosphorylation may impact the higher order chromatin organization of the centromere [10].
The initiating methionine of CENP-A is removed and the exposed glycine (Gly1) residue is trimethylated on the alpha-amino group by the enzyme NRMT1 [10,12••] (Figure 1a). Amino-terminal trimethylation of CENP-A is required for full recruitment of the CENP-T and CENP-I proteins of the CCAN, but does not affect CENP-C binding [12••] (Figure 1b). S. pombe centromeres show a similar specific requirement for the SpCENP-A amino terminus in CENP-I and CENP-T recruitment [13]. Loss of CENP-A alpha-amino-terminal methylation leads to chromosome missegregation and abnormal microtubule spindles.
Modification of CENP-A also occurs within the histone fold domain. Lys124 in the α3 helix of CENP-A has been shown to undergo methylation and acetylation, as well as ubiquitylation (discussed below) [14–16] (Figure 1a,c). Molecular dynamics simulations suggest that acetylation of the Lys124 may induce conformation changes in the CENP-A nucleosomes resulting in limited access of the C-terminal tail and thus inhibition of CENP-C binding.
Proteolytic degradation of non-centromeric CENP-A is a potential mechanism to restrict CENP-A to centromeres and eliminate incorrectly or ectopically deposited CENP-A nucleosomes. The ubiquitylation-dependent proteolytic degradation of CENP-ACse4/CID is clearly established in yeast and Drosophila; although the pathways in human cells are still unknown. So far, four E3 ubiquitin ligases have been reported to mediate CENP-ACse4 ubiquitylation. These include Psh1, Rcy1, Slx5 and Ubr1 [17–19,20•,21•,22,23]. CENP-ACse4 is protected from degradation at the centromere, while CENP-ACse4 in chromosome arms is degraded. Notably, Slx5 mediated ubiquitylation is dependent on SUMOylation of CENP-ACse4 [21•], suggesting that CENP-ACse4 degradation may be under complex regulatory control. Drosophila dCENP-ACID protein levels are regulated by the F-Box Protein Partner of Paired (Ppa), a component of the SCF E3-ubiquitin ligase complex [24]. E3 ligases regulating the stability of human CENP-A have not yet been identified.
Modifications of the CENP-A nucleosome that are present within the histone H4 tail also contribute to CENP-A nucleosome function. Monomethylation of Lys20 on the histone H4 tail within the CENP-A nucleosome has been observed in chicken cells [25] (Figure 1b). The removal of this mark by targeting an H4 Lys20 demethylase to the centromere leads to errors in centromere formation and chromosome segregation.
Pre-nucleosomal posttranslational modification of centromeric histones
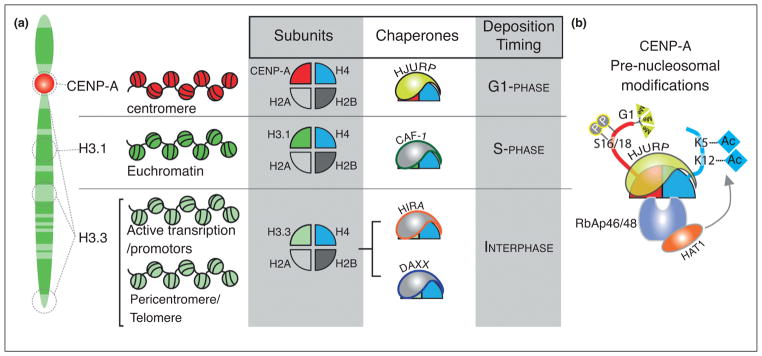
Histones associate with a series of chaperone proteins to facilitate their assembly into nucleosomes [26,27]. The histone H3.1 and H3.3 variants differ by only a few amino acids (96% identity), whereas CENP-A is more divergent (~50% identity). Based on these differences, the histone H3 variants interact with distinct chaperones that determine the timing and the location of their deposition in chromatin (Figure 2a). HJURP (Holliday junction recognition protein) is the CENP-A specific histone chaperone. The CENP-A binding domain of HJURP is conserved from budding yeast to humans which is essential for the deposition of new CENP-A [28–30]. Drosophila has evolved a non-homologous CENP-A chaperone, CAL1 [31,32], and some organisms, including C. elegans lack a clear HJURP or CAL1 homolog, suggesting these organisms have evolved different, but potentially related mechanisms to establish centromeric chromatin.
Figure 2.
Histone variant chaperones and pre-nucleosomal posttranslational modifications of CENP-A. (a) The interaction of histone H3 variants with distinct chaperone proteins determines the timing and site of variant nucleosome deposition. (b) The amino-terminus of prenucleosomal CENP-A is trimethylated and phosphorylated at Ser16/18. Recruitment of the HAT1 acetyltransferase through RbAp46/48 binding to CENP-A and histone H4 leads to acetylation of the histone H4 tail within the prenucleosomal complex.
HJURP and the yeast homolog Scm3 interact with the heterodimer that includes a single CENP-A and histone H4 [33,34] (Figures 1c and 2b). Posttranslational modification of the histone H4 subunit is required for proper deposition of the CENP-A nucleosome. RbAp46 and RbAp48 subunits found in many chromatin assembly complexes are associated with pre-nucleosomal CENP-A/H4 and HJURP [29,35,36••] (Figure 2b). Mis16, the S. pombe homolog of RbAp46/48, recognizes both CENP-A and histone H4 in the heterodimer [37]. RbAp46/48 recruits HAT1 (histone acetyltransferase 1) to the hetero-dimer and acetylates Lys5 and Lys12 on histone H4. In this respect, the modification of the preassembled CENP-A/H4 dimer is similar to histone H3/H4, which is also acetylated on the same residues of histone H4 [26]. Depletion of RbAp46/48/Mis16 in several organisms, or the mutation of histone H4 Lys5 or Lys12 residues to non-acetylated amino acids, reduces new CENP-A deposition [36••,38,39]; implying that the modification of the H4 subunit of the pre-nucleosomal heterodimer is essential for CENP-A deposition.
Posttranslational modification of CENP-A is emerging as a possible mechanism to regulate HJURP binding and thus CENP-A deposition. However, contradictory experimental evidence for the roles of CENP-A PTMs in this process means that the importance of the regulatory step remains somewhat murky. The ubiquitin ligase CUL4 in complex with RDX1-COPS8 or DDB1 has been shown to be important for CENP-A nucleosome deposition, without leading to CENP-A degradation [14,40,41]. Ubiquitylation of Lys124 of CENP-A is proposed to be required for CENP-A binding to HJURP. Biochemical evidence suggests that the addition of monoubiquitin to CENP-A Lys124 promotes HJURP binding, and Lys124 mutants fail to bind HJURP and reduce new CENP-A deposition [14,41]. While Lys124 does not lie within the CATD, which is sufficient for CENP-A deposition [42], the size of the ubiquitin moiety could bridge the distance and contribute to HJURP binding. Despite the intriguing biochemical data, gene replacement experiments show that mutant CENP-A that cannot be ubiquitylated, can completely replace the endogenous CENP-A and support cell viability [43]. Therefore, direct ubiquitylation of CENP-A by CUL4 does not play an essential role in CENP-A deposition; although, we cannot preclude that Lys124 modification may play important roles under certain cellular conditions. The Drosophila dCENP-ACID is also subjected to ubiquitylation, which facilitates its centromeric deposition [44]. CAL1 also serves as an adaptor for CENP-ACID ubiquitylation by directly associating with the ubiquitin ligase Cul3/RDX1 complex.
CENP-A undergoes several other modifications prior to deposition into chromatin. The amino terminus of pre-nucleosomal CENP-A in humans is already methylated on the alpha-amino group of Gly1 by NRMT1 and phosphorylated on Ser16/18 (Figure 2b). As discussed above, both of these modifications persist after CENP-A is deposited into chromatin [10]. CENP-A is also phos-phorylated on Ser68, within the histone fold domain in a cell cycle specific manner, and is discussed below. Drosophila dCENP-ACID is phosphorylated at Ser75/77, a site that may be analogous to Ser16/18 in human CENP-A and was enriched in the nuclear fraction [39]. In contrast, dCENP-ACID is acetylated on Lys105 selectively in the prenucleosomal complex. This suggests a potential role for differential modifications in controlling distinct steps in processing prenucleosomal CENP-A.
Cell cycle control of centromere inheritance
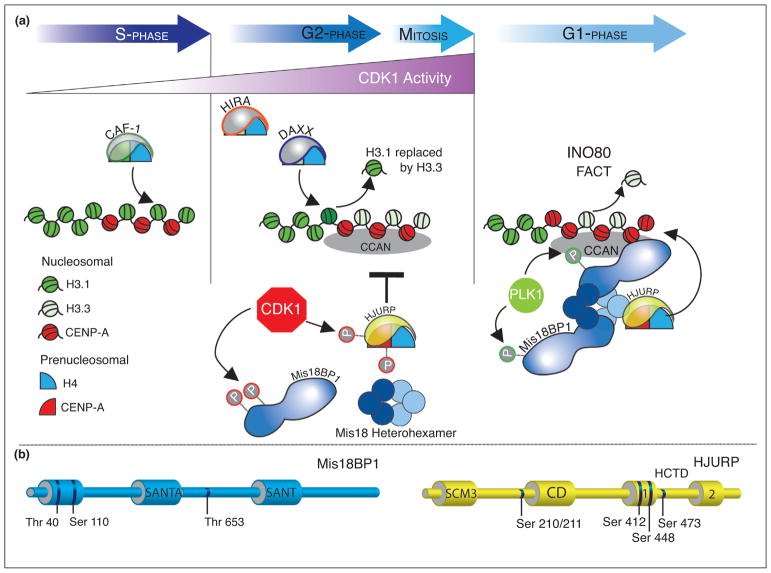
The location of the centromere is highly fixed on the chromosome, with new CENP-A nucleosomes assembled near the domain of existing CENP-A [45]. The selective deposition of new CENP-A at the existing centromere is achieved by coupling the proteins required for CENP-A deposition to CCAN proteins, including CENP-A itself. The Mis18 complex binds HJURP directly and is required for the recruitment of HJURP and new CENP-A to the centromere in G1-phase [38,46–50] (Figure 3a). The human Mis18 complex forms an oligomeric structure that includes Mis18α and Mis18β paralogs, and Mis18BP1 [51••,52••]. CENP-C has been shown to participate in recruiting the Mis18 complex proteins to the existing centromere; although, it is unclear whether this interaction fully accounts for Mis18 recruitment [53–55]. More recently, a direct interaction between Mis18BP1 and CENP-A has been identified in Xenopus and chickens [56,57]. Whether direct binding of the CENP-A nucleosome by the Mis18BP1 plays a role in human centromere assembly is not yet clear.
Figure 3.
Cell cycle control of vertebrate CENP-A deposition. (a) CDK1 activity increases in G2/M phase and inhibits the deposition of CENP-A nucleosomes by directly phosphorylating Mis18BP1 and the CENP-A chaperone HJURP. Mis18BP1 phosphorylation disrupts the binding to the Mis18α/β oligomer and thus inhibits recruitment of the complex to centromeres. Reduction of CDK1 activity in response to activation of the APC/C following mitotic checkpoint satisfaction leads to CENP-A deposition in G1. PLK1 activity positively regulates CENP-A deposition through phosphorylation of Mis18BP1. FACT mediated transcription in Drosophila and INO80 remodeling complex in budding yeast promote histone exchange within centromeric chromatin (reviewed by [70]). (b) CDK1 targeted phosphorylation sites within Mis18BP1 and HJURP that are known to inhibit CENP-A deposition are depicted.
Deposition of new CENP-A nucleosomes in human cells is restricted to early G1, immediately following mitosis, and the restricted timing involves both negative and positive regulation of the deposition pathway [58]. CDK1/2 negatively regulates CENP-A deposition in G2 and mitosis [59] (Figure 3a). Pharmacological inhibition of CDKs or the genetic knockout of CDK1 and CDK2 leads to the mis-timed loading of CENP-A in G2 phase.
The targets of CDK1 phosphorylation include CENP-A and proteins within the deposition pathway, including HJURP and Mis18BP1. CDK1 phosphorylates CENP-A at Ser68, and the degree of Ser68 phosphorylation peaks in mitosis, coincident with maximal CDK1 activity [60]. Phosphomimetic mutants result in a reduction of new CENP-A deposition, and show a decreased association with HJURP [34,60]. These data suggest that CDK1 mediated phosphorylation of CENP-A during G2-phase and mitosis contributes to the inhibition of new CENP-A deposition until G1, when CDK1 activity is lost. However, non-phosphorylatable CENP-A (S68A) does not result in CENP-A loading outside of G1-phase; therefore, additional mechanisms further inhibit CENP-A deposition. There are conflicting data about the importance of Ser68 phosphorylation in cells. Fachinetti et al. [43] show no effect of a phosphomimetic mutant of Ser68 (S68D) on cell viability in gene replacement assays; whereas, Wang et al. [61] show that cell viability is reduced to 50% when the non-phosphorylated mutant (S68A) is introduced into the endogenous CENP-A locus by CRISPR. Ser18 within the amino terminal tail of CENP-A has also been proposed to negatively regulate CENP-A deposition; however, the mechanism is unknown [11].
Mis18BP1 and HJURP are the major targets of CDK phosphorylation in the CENP-A deposition pathway [51••,52••,59,62,63••] (Figure 3a,b). The expression of Mis18BP1 containing mutations within the CDK phosphorylation sites leads to CENP-A loading in G2; however, the level of CENP-A deposited is much lower than that observed in G1 [51••,59,63••]. Phosphorylation of sites within the Mis18α/β binding domain in the amino terminus of Mis18BP1, specifically Thr40 and Ser110, inhibit the binding of the Mis18α/β heterohexamer [51••,52••] (Figure 3b), providing a molecular basis of the inhibition of CENP-A deposition by CDK1 phosphorylation of Mis18BP1. Phosphorylation of Thr653 also inhibits recruitment of the Mis18 complex to centromeres, but lies outside of the Mis18α/β interaction domain [63••], suggesting there is more to be learned about how phosphorylation by CDK1 inhibits this complex.
Likewise, the alanine mutations of CDK-dependent phosphorylation sites in HJURP (Ser210, Ser211 and Ser412) lead to HJURP recruitment to centromeres and loading of CENP-A in G2, but not to the levels observed when cells enter G1 (Figure 3b) [62,63••]. The mechanism of how HJURP phosphorylation inhibits recruitment and CENP-A deposition remains unclear. Regardless, combining phospho-mutants of HJURP and Mis18BP leads to accumulation of new CENP-A in G2 that is comparable to the level of CENP-A normally deposited in G1. These data suggest that the combinatorial modification of HJURP and Mis18BP1 by CDK1 accounts for the major regulation of CENP-A deposition. Although mutations in HJURP and Mis18BP1 lead to unscheduled loading in G2-phase, S-phase seems to be refractory to new CENP-A deposition even under these conditions [62].
The PLK1 mediated phosphorylation of the human Mis18 complex also plays a regulatory role in new CENP-A deposition pathway. Inhibition of the PLK1 kinase activity results in new CENP-A deposition defects and mitotic errors [64]. The timing of CENP-A nucleosome deposition varies between organisms, and so the mechanisms that control CENP-A deposition are also likely to diverge from those discussed.
Influence of chromatin context on CENP-A deposition
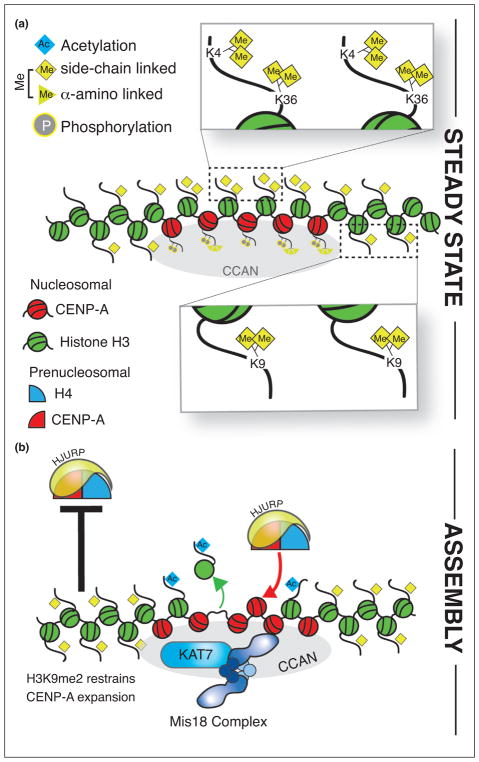
Only a subset of the alpha-satellite DNA present at the centromere is occupied by CENP-A nucleosomes. Estimates based on in vivo fluorescence measurements suggest that there are approximately 200 CENP-A nucleosomes per centromere [65]. Histone H3 nucleosomes flank regions of CENP-A containing nucleosomes and are also interspersed between CENP-A nucleosomes within the endogenous centromere [66–68]. H3.3-variant nucleosomes predominate within the CENP-A domain [69]. The presence of H3.3 may reflect the active transcription of alpha-satellite DNA at the centromere [70]. Interspersed centromeric H3 nucleosomes are often marked by histone H3 Lys4 and Lys36 dimethylation [66,67]; while nucleosomes flanking the CENP-A nucleosomes contain histone H3 Lys9 dimethylation (Figure 4a). This suggests that the CENP-A containing alpha-satellite DNA are associated with both active and repressive marks. Although endogenous centromeres show H3 Lys4 and Lys36 methylation within the CENP-A domain, a strong correlation between centro-mere function and H3 Lys4 methylation has not been observed at neocentromeres, leaving open the question of whether these modifications are essential for centromere function outside of alpha-satellite repetitive regions [71].
Figure 4.
Role of the chromatin context in CENP-A deposition. (a) Histone H3 nucleosomes are interspersed within and surround CENP-A containing chromatin throughout the cell cycle (steady-state). H3 nucleosomes within CENP-A containing chromatin are enriched for H3 Lys4 and Lys36 dimethylation, and H3 Lys9 dimethylation marks are found in nucleosomes flanking centromeres. (b) During G1-phase, recruitment of the KAT7 acetyltransferase complex by the Mis18 complex facilitates histone H3 removal and the assembly of new CENP-A nucleosomes.
The modification state of the histone H3 within chromatin is critical for the assembly of the centromere during G1. The efficiency with which centromeres form on exogenously provided alpha-satellite repeats of a human artificial chromosome (HAC) is influenced by the histone H3 Lys9 methylation/acetylation status. H3 Lys9 tri-methylation inhibits HAC stability and CENP-A accumulation on the alpha-satellite repeats of the HAC, and the presence of H3 Lys9 dimethylated histones limits the spread of CENP-A nucleosomes into flanking heterochromatin [72•,73] (Figure 4b). Notably, the removal of histone H3 Lys4 dimethylation by the artificial tethering of LSD1 (lysine-specific demethylase 1) also inhibits HJURP recruitment and new CENP-A deposition [67]. In contrast, the acetylation of H3 Lys9 within the alpha-satellite of the HAC, enhances new CENP-A deposition and HAC stability [72•]. The active alteration of H3 histone modification through the recruitment of histone modifying enzymes occurs to facilitate new CENP-A deposition. Mis18BP1 interacts with the KAT7 acetyltransferase complex and is necessary for new CENP-A deposition [74] (Figure 4b). Recruitment of KAT7 to the centromere antagonizes H3 Lys9 trimethylation, which leads to increased histone H3 turnover and enhanced new CENP-A deposition.
Conclusion
CENP-A overexpression and misincorporation at non-centromeric loci is observed in many cancers, and serves as a potent prognostic marker in conjunction with other centromere genes [75–78]. Inhibition of CENP-A amino terminal methylation and hyperphosphorylation of Ser18 by cyclin E1/CDK2 contributes to chromosome instability [11,12••], suggesting that perturbations in posttranslational modifications of CENP-A may contribute to cancer phenotypes. Although several E3 ligases have been identified that lead to degradation of non-centromeric CENP-ACse4 in yeast, almost nothing is known about the pathways that mark and degrade non-centromeric CENP-A in humans. These processes may be at play in CENP-A overexpressing tumors and provide a potential way to target non-centromeric CENP-A for degradation.
The epigenetic specification of centromere location by the unique CENP-A containing nucleosome means that assembling new CENP-A nucleosomes as cells replicate their genome and divide, is a critical process to ensure the inheritance of the centromere. The recent studies reviewed here show that posttranslational modifications have emerged as important regulators of CENP-A deposition and function within the centromere.
Acknowledgments
Our sincere apologies to those we were unable to reference due to space limitations. D.R.F. was supported by NIH R01GM111907 and by a Zell Scholar award from the Robert H. Lurie Comprehensive Cancer Center.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Musacchio A, Desai A. A molecular view of kinetochore assembly and function. Biology (Basel) 2017:6. doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekulic N, Black BE. Molecular underpinnings of centromere identity and maintenance. Trends Biochem Sci. 2012;37:220–229. doi: 10.1016/j.tibs.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeitlin SG, Barber CM, Allis CD, Sullivan K. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- 5.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 7.Goutte-Gattat D, Shuaib M, Ouararhni K, Gautier T, Skoufias DA, Hamiche A, Dimitrov S. Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function. Proc Natl Acad Sci U S A. 2013;110:8579–8584. doi: 10.1073/pnas.1302955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci U S A. 2013;110:11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takada M, Zhang W, Suzuki A, Kuroda TS, Yu Z, Inuzuka H, Gao D, Wan L, Zhuang M, Hu L, et al. FBW7 loss promotes chromosomal instability and tumorigenesis via cyclin E1/CDK2-mediated phosphorylation of CENP-A. Cancer Res. 2017;77:4881–4893. doi: 10.1158/0008-5472.CAN-17-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Sathyan KM, Fachinetti D, Foltz DR. alpha-Amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat Commun. 2017;8:14678. doi: 10.1038/ncomms14678. This paper, along with previous work from Bailey et al. (2011), demonstrates that CENP-A is subjected to the alpha-amino terminal methylation, and demonstrates the importance of this uncommon modification in assembling the CCAN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folco HD, Campbell CS, May KM, Espinoza CA, Oegema K, Hardwick KG, Grewal SI, Desai A. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr Biol. 2015;25:348–356. doi: 10.1016/j.cub.2014.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell. 2015;32:589–603. doi: 10.1016/j.devcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bui M, Pitman M, Nuccio A, Roque S, Donlin-Asp PG, Nita-Lazar A, Papoian GA, Dalal Y. Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenet Chromatin. 2017;10:17. doi: 10.1186/s13072-017-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 20•.Cheng H, Bao X, Gan X, Luo S, Rao H. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci Rep. 2017;7:8565. doi: 10.1038/s41598-017-08923-w. This work demonstrates that Cse4/CENP-A protein levels in S. cerevisiae are controlled by multiple independent E3-ligases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Ohkuni K, Takahashi Y, Fulp A, Lawrimore J, Au WC, Pasupala N, Levy-Myers R, Warren J, Strunnikov A, Baker RE, et al. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol Biol Cell. 2016 doi: 10.1091/mbc.E15-12-0827. This work demonstrates Cse4/CENP-A protein levels in S. cerevisiae are controlled through SUMO-dependent ubiquitylation to prevent ectopic assembly of CENP-A nucleosomes in chromosome arms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H, Bao X, Rao H. The F-box protein Rcy1 is involved in the degradation of histone H3 variant Cse4 and genome maintenance. J Biol Chem. 2016;291:10372–10377. doi: 10.1074/jbc.M115.701813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, Basrai MA. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013;194:513–518. doi: 10.1534/genetics.113.149898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno-Moreno O, Medina-Giro S, Torras-Llort M, Azorin F. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID) Curr Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 25.Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev Cell. 2014;29:740–749. doi: 10.1016/j.devcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almouzni G, Cedar H. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol. 2016:8. doi: 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 30.Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phansalkar R, Lapierre P, Mellone BG. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 2012;20:493–504. doi: 10.1007/s10577-012-9299-7. [DOI] [PubMed] [Google Scholar]
- 32.Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, et al. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun. 2016;7:13465. doi: 10.1038/ncomms13465. This work demonstrates the function of the RbAp46/48 (the yeast of homolog of Mis16) in the CENP-A prenucleosomal is to recruit HAT1 acetylate histone h4 on Lys 5 and Ly12, and these modifications are important for nucleosome deposition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An S, Kim H, Cho US. Mis16 independently recognizes histone H4 and the CENP-ACnp1-specific chaperone Scm3sp. J Mol Biol. 2015;427:3230–3240. doi: 10.1016/j.jmb.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Boltengagen M, Huang A, Boltengagen A, Trixl L, Lindner H, Kremser L, Offterdinger M, Lusser A. A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. Nucleic Acids Res. 2016;44:2145–2159. doi: 10.1093/nar/gkv1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouysset J, Gilberto S, Meier MG, Lampert F, Belwal M, Meraldi P, Peter M. CRL4(RBBP7) is required for efficient CENP-A deposition at centromeres. J Cell Sci. 2015;128:1732–1745. doi: 10.1242/jcs.162305. [DOI] [PubMed] [Google Scholar]
- 41.Niikura Y, Kitagawa R, Kitagawa K. CENP-A ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell. 2017;40:7–8. doi: 10.1016/j.devcel.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 43.Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, Black BE. CENP-A modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev Cell. 2017;40:104–113. doi: 10.1016/j.devcel.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bade D, Pauleau AL, Wendler A, Erhardt S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell. 2014;28:508–519. doi: 10.1016/j.devcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Ross JE, Woodlief KS, Sullivan BA. Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenet Chromatin. 2016;9:20. doi: 10.1186/s13072-016-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR. Licensing of centromeric chromatin assembly through the Mis18alpha-Mis18beta heterotetramer. Mol Cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone Holliday junction recognition protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A. CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. Elife. 2017:6. doi: 10.7554/eLife.23352. Together with Spiller et al. this paper demonstrates that CDK phosphorylation of Mis18BP1 inhibits new CENP-A deposition by inhibiting the ability of Mis18BP1 to complex with Mis18α and Mis18β. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Spiller F, Medina-Pritchard B, Abad MA, Wear MA, Molina O, Earnshaw WC, Jeyaprakash AA. Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition. EMBO Rep. 2017;18:894–905. doi: 10.15252/embr.201643564. Together with Pan et al. this paper demonstrates that CDK phosphorylation of Mis18BP1 inhibits new CENP-A deposition by inhibiting the ability of Mis18BP1 to complex with Mis18α and Mis18β. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stellfox ME, Nardi IK, Knippler CM, Foltz DR. Differential binding partners of the Mis18alpha/beta YIPPEE domains regulate Mis18 complex recruitment to centromeres. Cell Rep. 2016;15:2127–2135. doi: 10.1016/j.celrep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hori T, Shang WH, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, Fukagawa T. Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev Cell. 2017;42:181–189. e183. doi: 10.1016/j.devcel.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 57.French BT, Westhorpe FG, Limouse C, Straight AF. Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev Cell. 2017;42:190–199. e110. doi: 10.1016/j.devcel.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 60.Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32:68–81. doi: 10.1016/j.devcel.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 61.Wang K, Yu Z, Liu Y, Li G. Ser68 phosphorylation ensures accurate cell-cycle-dependent CENP-A deposition at centromeres. Dev Cell. 2017;40:5–6. doi: 10.1016/j.devcel.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 62.Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3 (CENP-A) loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 63••.Stankovic A, Guo LY, Mata JF, Bodor DL, Cao XJ, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE, et al. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol Cell. 2017;65:231–246. doi: 10.1016/j.molcel.2016.11.021. This study demonstrates that independent regulation of both Mis18BP1 and HJURP by CDK1 phosphorylation accounts for the majority of CDK regulation of CENP-A deposition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. Elife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunleavy EM, Almouzni G, Karpen GH. H3. 3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duda Z, Trusiak S, O’Neill R. Centromere transcription: means and motive. Prog Mol Subcell Biol. 2017;56:257–281. doi: 10.1007/978-3-319-58592-5_11. [DOI] [PubMed] [Google Scholar]
- 71.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenet Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Ohzeki JI, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012 doi: 10.1038/emboj.2012.82. This study demonstrates that the Mis18BP1 protein recruits the KAT7 acetyltransferase to centromeres to facilitate histone h3 removal and new CENP-A deposition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H. KAT7/HBO1/MYST2 regulates CENP-A chromatin assembly by antagonizing Suv39h1-mediated centromere inactivation. Dev Cell. 2016;37:413–427. doi: 10.1016/j.devcel.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung MH, Dalal Y. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet Chromatin. 2015;8:2. doi: 10.1186/1756-8935-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, Karpen GH. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 78.Wu Q, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012 doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]