Abstract
Purpose
Thoracic aortic aneurysm/aortic dissection (TAAD) is a disorder with highly variable age of onset and phenotype. We sought to determine the prevalence of pathogenic variants in TAAD-associated genes in a mixed cohort of sporadic and familial TAAD patients and identify relevant genotype-phenotype relationships.
Methods
We used a targeted PCR and next generation sequencing-based panel for genetic analysis of 15 TAAD associated genes in 1025 unrelated TAAD cases.
Results
We identified 49 pathogenic or likely pathogenic (P/LP) variants in 47 cases (4.9% of those successfully sequenced). Almost half of the variants were in non-syndromic cases with no known family history of aortic disease. Twenty-five variants were within FBN1 and two patients were found to harbour two P/LP variants. Presence of a related syndrome, younger age at presentation, family history of aortic disease and involvement of the ascending aorta increased the risk of carrying a P/LP variant.
Conclusions
Given the poor prognosis of TAAD that is undiagnosed prior to acute rupture or dissection, genetic analysis of both familial and sporadic cases of TAAD will lead to new diagnoses, more informed management and possibly reduced mortality through earlier, preclinical diagnosis in genetically determined cases and their family members.
Keywords: Thoracic Aortic Aneurysm/Aortic Dissection (TAAD), TAAD Genetics, FBN1, Sporadic TAAD, High-Throughput DNA Sequencing
Introduction
Thoracic aortic aneurysm/aortic dissection (TAAD) manifests a devastating clinical course if undiagnosed and untreated 1. Recent studies of post-mortem computerised tomographic autopsy indicate that 7% of out-of-hospital sudden deaths are due to Type A aortic dissection 2.
The major mortality and morbidity associated with TAAD lies with acute dissection or rupture, both of which are difficult to predict 1,3. Early perioperative mortality of a ruptured aneurysm remains high (28-46%) despite recent advances in surgical technology compared with the safety of elective treatment of unruptured thoracic aneurysms where surgical results have improved significantly (6-7% mortality) 4. Aneurysm size (5-5.5cm) is currently the major determinant of the timing of surgery5. In syndromic aortic disease (Marfan (MFS), Loeys-Dietz (LDS), Ehlers-Danlos Syndrome (EDS)), earlier surgical intervention is recommended as dissection can occur at diameters less than 5.0cm 6 although these associated syndromes may be difficult to diagnose 7,8. Whilst genetic testing is recommended in familial and syndromic TAAD, it is not widely available in sporadic TAAD (STAAD) and is generally restricted to those with a strong suspicion of a genetic aetiology5,9.
The study of large pedigrees with multiply affected members has led to the identification of causal genes in both syndromic and non-syndromic TAAD 6,10. Causative mutations have been identified in non-syndromic FTAAD pedigrees within the smooth muscle contractile (SMC) genes ACTA2, MYH11, PRKG1 and MYLK in non-syndromic patients 11–13. Syndromic TAAD is associated with numerous connective tissue disorders and their corresponding genes; Marfan syndrome (FBN1), Loeys-Dietz syndrome (TGFBR1, TGFBR2, SMAD3 & TGFB2), Ehlers-Danlos syndrome (COL1A1, COL1A2, COL3A1, COL5A1 and COL5A2), Arterial tortuosity syndrome (SLC2A10) and Shprintzen-Goldberg syndrome (SKI) 10,14.
Many of the genes in syndromic TAAD are associated with dissections occurring in patients with an aortic size below the 5.5cm threshold recommended for reparative surgery 6. The identification of causal variants in probands can lead to additional diagnoses by family screening in family members who have yet to develop clinical manifestations of the disorder 9. In addition, endovascular repair is contraindicated in cases diagnosed with a connective tissue syndrome, when open surgery is recommended 15. Genetic testing can therefore identify causal variants leading to a definitive preclinical diagnosis that can better determine the optimal timing and technique of prophylactic surgery.
Next generation sequencing (NGS) is increasingly used for mutation testing and clinical diagnosis 16. Using a cost-effective PCR and NGS-based targeted sequencing approach 17,18, we sought to identify the prevalence of causative genetic variants in known TAAD genes and to elucidate genotype-phenotype relationship in a mixed cohort of 1,025 cases of familial and sporadic TAAD from the UK and USA.
Methods
Patient Cohorts
Surgical intervention in 927 consecutive unrelated cases was undertaken over a 10 year period at the Aortic Institute at Yale-New Haven (Yale University, New Haven, USA): 785 surgical samples from these cases were used for DNA analysis within this study (Yale cohort) and 240 unrelated cases, treated and followed up at three UK centres from 2000-2013 (St Mary’s hospital, London; Royal Brompton & Harefield hospitals, London; Liverpool Heart & Chest Hospital, Liverpool) (UK cohort) were recruited to the study. Twenty-five cases within the Yale cohort were also present in a previous whole-exome sequencing study 9. The Yale cohort was approved by the Human Investigation Committee of Yale University (IRB protocol 12617) and the UK cohort was approved by the West London Research Ethics Committee (REC reference 11/LO/0883). Both cohorts complied with the Declaration of Helsinki and written informed consent was obtained from all participants. MFS and LDS diagnoses in the UK cohort were identified by clinical case note review in which diagnoses were made by consultant clinical geneticists following a standard referral pathway. MFS diagnoses in the Yale cohort were made according to the Revised Ghent Nosology19. There were no LDS cases in the Yale cohort.
DNA Extraction
For the UK cohort, saliva samples were collected using the Oragene DNA kit (Genotek, Ontario, Canada) and QIAamp DNA Blood Midi kit (Qiagen, Venlo, Netherlands) was used to extract DNA from whole-blood samples. For the Yale cohort, DNA from fresh frozen aortic specimens collected at surgery were extracted using the DNeasy Blood & Tissue kit (Qiagen, Venlo, Netherlands). All DNA samples were subsequently normalized to 25-50ng/ul.
Targeted Exon Sequencing
Targeted exon sequencing was carried out on two Fluidigm assays named Aortopathy panel 1 (TAAD-X), which included 363 primer-pairs for FBN1, TGFBR1, TGFBR2, MYH11, ACTA2, SMAD3 and MYLK and Aortopathy panel 2 (TAAD-Z) containing 493 primer pairs for SKI, TGFB2, SLC2A10, COL1A1, COL1A2, COL3A1, COL5A1 and COL5A2 (Table S1 & S2). Multiplex PCR was performed using the Access Array System (Fluidigm, South San Francisco, CA) and the MiSeq sequencing platform (Illumina, San Diego, CA) as previously described 17,18.
Read Mapping, Variant Calling & Annotation
FastQC was used to assess the sequencing read quality. Primer sequences were trimmed from FASTQ files using cutadapt (v 1.9.1) 20 prior to read mapping to GRCh37/hg19 human reference sequence using BWA-MEM V0.7.12 21. Realignment of reads around indels and base quality score recalibration was performed using GATK v3.4 22. The GATK UnifiedGenotyper was used for calling variants 23. Variants were annotated using Ensembl Variant Effect Predictor v84 24.
Variant and Sample Filtering
Synonymous variants and intronic variants (excluding splice sites) were omitted from downstream analysis unless annotated as pathogenic or disease-causing in ClinVar 25 or HGMD 26 databases. Variants with a minor allele frequency greater than 0.1% in either ExAC release 0.3.1 27 or dbSNP146 28 datasets were excluded. We found a high frequency of false positives in variants within SKI exon 1 and those with an allele balance below 0.3. These variants were therefore excluded from the datasets. Samples with less than 80% of target bases covered by more than 49 reads were not considered for downstream analysis.
Pathogenicity Assignment
Variants passing filters were assigned to one of three categories: either ‘pathogenic’, ‘likely pathogenic’, ‘variant of uncertain significance (VUS)’ or ‘likely benign’ based on the presence of one or more pieces of evidence (Table S3 & S4). Variants were assigned to the ‘pathogenic’ or ‘likely pathogenic’ category if they met any of the following criteria: if the variant is already reported as disease causing in HGMD or ClinVar unless sufficient evidence could not be found for categorising it as ‘pathogenic/likely pathogenic’ (P/LP) (Table S5); if the variant results in the same amino acid substitution as a variant already reported as disease causing in HGMD or ClinVar; premature termination of translation; a substitution of a glycine residue within a GlyXY repeat in collagen triple helical domains; an insertion of amino acids disrupting the GlyXY repeat sequence; alteration of a key residue in a protein feature (e.g. active site, disulphide bond) in keeping with previously ascribed molecular mechanisms for a given gene (see Table S6). Variants predicted to be damaging by PolyPhen and SIFT and with CADD scores above 10 were assigned to the VUS category, as were variants resulting in a substitution at an amino acid position at which a different substitution is assigned disease-causing status in ClinVar or HGMD 25,26,29–31. Absence of PolyPhen or SIFT predictions for missense variants resulted in classification as VUS. Remaining variants were assigned ‘likely benign’ status. All variants assigned P/LP status were validated by Sanger sequencing.
Statistical Analysis
Significant differences in categorical variables between individuals in different genotype or phenotype groups were estimated by Fisher’s exact test. The unpaired Wilcoxon rank-sum test was used to assess non-parametric phenotypic continuous measurements.
Results
Patient Demographics and Clinical Characteristics
Of the total of 785 cases recruited to the Yale cohort and 240 to the UK cohort, sequence depth reached the assay threshold of 50 reads in at least 80% of target bases in 93% of the Yale cohort and 98% of the UK cohort. A total of 732 patients in the Yale cohort and 235 patients in the UK cohort were therefore taken forward for further analyses: their demographic and clinical characteristics are given in Table 1.
Table 1.
Patient demographics and clinical characteristics of the cohort.
| Yale Cohort | UK Cohort | Whole Cohort | |
|---|---|---|---|
| Demographics | |||
| Number (%) | 732 (75.7) | 235 (24.3) | 967 (100) |
| Age at Diagnosis, Median | 60 | 60 | 60 |
| Min | 10 | 11 | 10 |
| Max | 86 | 84 | 86 |
| Male (%) a | 503 (68.7) | 154 (65.5) | 657 (67.9) |
| Female (%) | 223 (30.4) | 80 (34.2) | 303 (31.3) |
| Caucasian (%) | 634 (86.7) | 200 (85.2) | 833 (86.1) |
| Probable/Proven Family History (%) | 214 (29.2) | 31 (13.2) | 245 (25.3) |
| No Family History (%) b | 444 (60.7) | NA* | NA* |
| Undergone Aortic Surgery (%) | 732 (100) | 199 (84.7) | 931 (96.3) |
| Primary Aortic Pathology | |||
| Aneurysm (%) | 650 (88.8) | 151 (64.3) | 801 (82.8) |
| Dissection (%) | 72 (9.8) | 79 (33.6) | 151 (15.6) |
| PAU/IMH (%) | 10 (1.4) | 4 (1.7) | 14 (1.4) |
| Rupture (%) | 3 (0.4) | 2 (0.9) | 5 (0.5) |
| Primary Anatomical Presentation | |||
| Ascending/Arch (%) | 683 (93.3) | 152 (64.7) | 835 (86.3) |
| Descending/Thoracoabdominal (%) | 48 (6.6) | 83 (35.3) | 131 (13.5) |
| Aortic Size | |||
| Maximum Aortic Diameter (cm), Median | 5.1 | 5.5 | 5.1 |
| Min | 3.4 | 2.7 | 2.7 |
| Max | 11 | 13 | 13 |
| Maximum Aortic Diameter < 5.5cm (%) | 488 (66.7) | 103 (43.8) | 591 (61.1) |
| Known Syndrome c | |||
| MFS (%) | 4 (0.5) | 20 (8.5) | 24 (2.5) |
| LDS (%) | 0 (0) | 3 (1.3) | 3 (0.3) |
| EDS (%) | 0 (0) | 0 (0) | 0 (0) |
| Other (%) d | 1 (0.1) | 2 (0.9) | 3 (0.3) |
PAU, penetrating aortic ulcer. IMH, intramural haematoma. MFS, Marfan Syndrome. LDS, Loeys-Dietz syndrome. EDS, Ehlers-Danlos syndrome. NA, not available.
Absence of family history was only recorded for the Yale cohort.
Gender information un available for 7 patients.
Family history unavailable for 207 patients in UK cohort and 78 in Yale cohort.
The majority of known Marfan patients in the Yale cohort were operated on as emergencies without research consent, explaining the low number of Marfan cases within the Yale cohort.
One individual from the Yale cohort had several features suggestive of a connective tissue disorder but did not fit any classic syndrome presentations while two individuals from the UK cohort had scoliosis in addition to TAAD.
Variants Identified by NGS
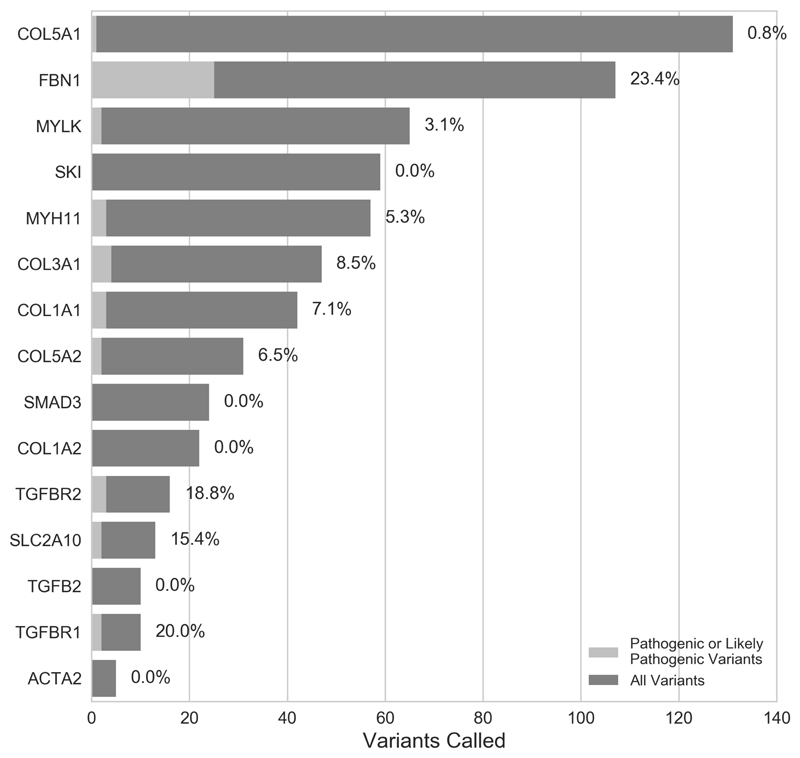
We identified 17 pathogenic variants, 32 likely pathogenic variants and 68 VUS within the whole cohort. Two patients, Y_91_1 and Y_17_1, each carried two P/LP variants, giving a total of 49 P/LP variants in 47 patients, constituting 4.9% of the 967 samples sequenced to our target threshold (Tables 2 and S7). The majority of the identified P/LP variants were in the FBN1 gene (n = 25) (Figure 1, Table S7). A total of 68 VUS were found within 67 patients (Table S8).
Table 2.
Pathogenic and likely pathogenic variants identified by the NGS panels
| Gene Affected | Variant | Functional Category | Classification | Previously Reported? | ID | Primary Diagnosis | Clinical Diagnosis | Family History |
|---|---|---|---|---|---|---|---|---|
| COL1A1 | c.1042G>A:p.A348T | missense | Likely Pathogenic | yes | Y_109_21 | Aneurysm | N/A | no |
| COL1A1 | c.1042G>A:p.A348T | missense | Likely Pathogenic | yes | Y_17_1 | Dissection | N/A | no |
| COL1A1 | c.2932C>T:p.Pro978Ser | missense | Likely Pathogenic | yes | Y_12_61 | Aneurysm | N/A | no |
| COL1A1 | c.2932C>T:p.Pro978Ser | missense | Likely Pathogenic | yes | Y_50_38 | Aneurysm | N/A | yes |
| COL3A1 | c.1178G>A:p.Gly393Asp | missense | Likely Pathogenic | no | Y_5_23 | Aneurysm | Other* | yes |
| COL3A1 | c.1204G>A:p.Gly402Ser | missense | Likely Pathogenic | no | Y_112_51 | Aneurysm | N/A | no |
| COL3A1 | c.1744G>A:p.Gly582Ser | missense | Pathogenic | yes | UK_24_0727 | Aneurysm | N/A | unknown |
| COL3A1 | c.536delC:p.Pro179GlnfsTer43 | frameshift | Pathogenic | no | Y_130_31 | Aneurysm | N/A | yes |
| COL5A1 | c.2504G>C:p.Gly835Ala | missense | Likely Pathogenic | no | Y_68_20 | Aneurysm | N/A | no |
| COL5A2 | c.3275G>A:p.Gly1092Asp | missense | Likely Pathogenic | no | UK_21_0261 | Aneurysm | N/A | unknown |
| COL5A2 | c.808G>A:p.Gly270Ser | missense | Likely Pathogenic | no | Y_1_1 | Aneurysm | N/A | yes |
| FBN1 | c.59A>G:p.Tyr20Cys | missense | Likely Pathogenic | yes | Y_128_61 | Aneurysm | N/A | no |
| FBN1 | c.626G>A:p.Cys209Tyr | missense | Likely Pathogenic | yes | Y_95_7 | Dissection | N/A | unknown |
| FBN1 | c.1090C>T:p.Arg364Ter | stop gained | Pathogenic | yes | UK_24_0907 | Aneurysm | Marfan | yes |
| FBN1 | c.1090C>T:p.Arg364Ter | stop gained | Pathogenic | yes | UK_24_0916 | Aneurysm | Marfan | yes |
| FBN1 | c.1422T>G:p.Cys474Trp | Missense | Likely Pathogenic | yes | UK_24_0712 | Dissection | Marfan | yes |
| FBN1 | c.1468+5G>A | splice region | Likely Pathogenic | yes | UK_24_0719 | Aneurysm | N/A | unknown |
| FBN1 | c.1468+5G>A | splice region | Likely Pathogenic | yes | UK_24_0720 | Aneurysm | N/A | unknown |
| FBN1 | c.2168-1G>T | splice acceptor | Likely Pathogenic | no | Y_82_41 | Aneurysm | N/A | unknown |
| FBN1 | c.2306G>A:p.Cys769Tyr | missense | Likely Pathogenic | yes | UK_24_0904 | Aneurysm | Marfan | yes |
| FBN1 | c.2554_2555dupAC:p.Cys853LeufsTer20 | frameshift | Pathogenic | no | UK_21_0250 | Dissection | Marfan | yes |
| FBN1 | c.2581C>T:p.Arg861Ter | stop gained | Pathogenic | yes | Y_17_1 | Dissection | N/A | no |
| FBN1 | c.2645C>T:p.Ala882Val | missense | Likely Pathogenic | yes | UK_21_0003 | Aneurysm | N/A | unknown |
| FBN1 | c.2896G>T:p.Glu966Ter | stop gained | Pathogenic | yes | UK_21_0281 | Dissection | N/A | unknown |
| FBN1 | c.3012C>G:p.Tyr1004Ter | stop gained | Pathogenic | yes | UK_21_0083 | Dissection | N/A | unknown |
| FBN1 | c.3193delG:p.Glu1065LysfsTer23 | frameshift | Pathogenic | yes | Y_21_18 | Aneurysm | N/A | yes |
| FBN1 | c.4406G>C:p.Arg1469Pro | missense | Likely Pathogenic | yes | Y_133_86 | Aneurysm | Marfan | yes |
| FBN1 | c.5235_5236dupTA:p.Thr1746IlefsTer148 | frameshift | Pathogenic | no | UK_21_0242 | Aneurysm | Marfan | yes |
| FBN1 | c.5917+6T>C | splice region | Likely Pathogenic | yes | UK_24_0796 | Aneurysm | Marfan | unknown |
| FBN1 | c.5917+6T>C | splice region | Likely Pathogenic | yes | UK_24_0842 | Aneurysm | Marfan | yes |
| FBN1 | c.6402dupC:p.Asp2135ArgfsTer4 | frameshift | Pathogenic | no | UK_21_0355 | Aneurysm | Marfan | yes |
| FBN1 | c.7039_7040delAT:p.Met2347ValfsTer19 | frameshift | Pathogenic | yes | Y_26_51 | Aneurysm | N/A | no |
| FBN1 | c.7788C>A:p.Tyr2596Ter | stop gained | Likely Pathogenic | no | Y_94_61 | Aneurysm | N/A | yes |
| FBN1 | c.7956T>A:p.Cys2652Ter | stop gained | Pathogenic | no | Y_47_31 | Aneurysm | N/A | yes |
| FBN1 | c.8504dupC:p.Leu2836ThrfsTer3 | frameshift | Likely Pathogenic | no | Y_56_99 | Aneurysm | N/A | yes |
| FBN1 | c.8504dupC:p.Leu2836ThrfsTer3 | frameshift | Likely Pathogenic | no | Y_59_47 | Aneurysm | N/A | yes |
| MYH11 | c.1A>G:p.Met1? | start loss | Likely Pathogenic | no | Y_91_1 | Aneurysm | N/A | unknown |
| MYH11 | c.4861A>C:p.Lys1621Gln | missense | Likely Pathogenic | yes | Y_20_10 | Aneurysm | N/A | no |
| MYH11 | c.5273G>A:p.Arg1758Gln | missense | Likely Pathogenic | yes | Y_21_41 | Aneurysm | N/A | no |
| MYH11 | c.5273G>A:p.Arg1758Gln | missense | Likely Pathogenic | yes | Y_35_28 | Aneurysm | Marfan | no |
| MYLK | c.2390+2T>C | splice donor | Likely Pathogenic | no | Y_19_18 | Aneurysm | N/A | yes |
| MYLK | c.5275T>C:p.Ser1759Pro | missense | Likely Pathogenic | yes | Y_84_18 | Dissection | N/A | yes |
| SLC2A10 | c.394C>T:p.Arg132Trp | missense | Likely Pathogenic | yes | Y_51_21 | Aneurysm | N/A | yes |
| SLC2A10 | c.648C>G:p.Tyr216Ter | stop gained | Pathogenic | no | Y_91_1 | Aneurysm | N/A | unknown |
| TGFBR1 | c.974-2A>G | splice acceptor | Likely Pathogenic | no | Y_105_61 | Dissection | N/A | no |
| TGFBR1 | c.1255+1G>A | splice donor | Pathogenic | no | UK_21_1025 | Dissection | N/A | unknown |
| TGFBR2 | c.1489C>T:p.Arg497Ter | stop gained | Pathogenic | yes | Y_18_71 | Aneurysm | N/A | yes |
| TGFBR2 | c.1524+1G>T | splice donor | Pathogenic | no | UK_24_0795 | Aneurysm | LDS | unknown |
| TGFBR2 | c.1609C>T:p.Arg537Cys | missense | Likely Pathogenic | no | Y_112_30 | Aneurysm | N/A | yes |
LDS, Loeys-Dietz Syndrome.
This individual had a suspected but unconfirmed connective tissue disorder
Figure 1.
Numbers of pathogenic or likely pathogenic (P/LP) variants identified by gene across both the Yale and UK cohorts. Percentages shown are the overall proportion of P/LP variants for each gene.
We also tested our pipeline with a more lenient allele frequency threshold of 1%, which yielded three more P/LP variants. However, each of these additional variants were reclassified to VUS upon inspection of the literature (Table S5), so a maximum allele frequency of 0.1% was used for all variant filtering.
Genotype-phenotype correlation with P/LP variants
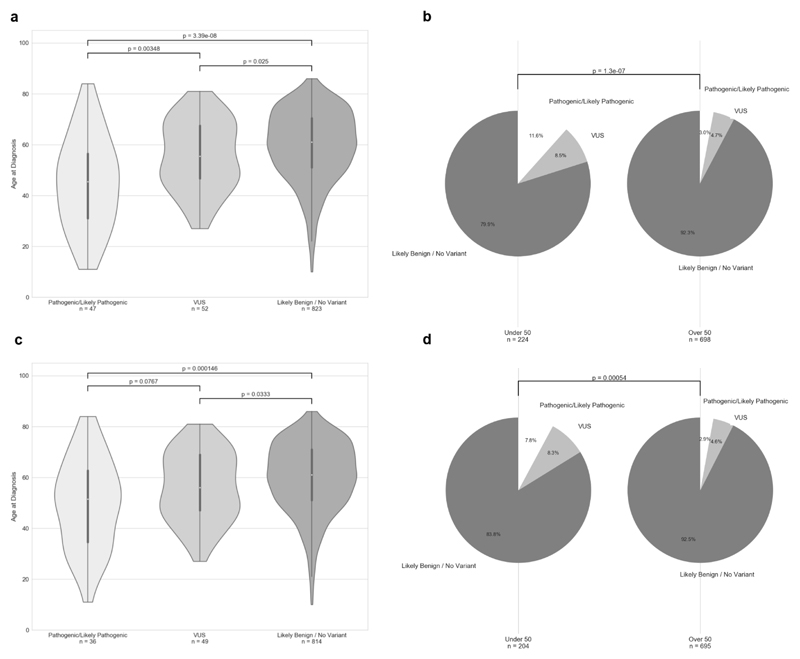
A lower age at diagnosis was found to significantly increase the likelihood of identifying a P/LP variant. The median age at diagnosis was 46.0 years for patients carrying a P/LP variant compared to 55.5 years for those carrying a VUS (p=3.5e-3), and 61.0 for those in the likely benign and no variant group (p=3.4e-8) (Figure 2a). Patients who were diagnosed under 50 years of age were far more likely to carry a P/LP variant with 11.6% of those diagnosed before 50 carrying a P/LP variant compared to 3.0% in the over 50 category (p=1.3e-7) (Figure 2b). Removing Marfan cases from these datasets increases the median age at diagnosis for patients harbouring a P/LP variant to 51.5 (Figure 2c) and decreases the percentage of patients under 50 harbouring a P/LP variant to 7.8% (Figure 2d). The median age of TAAD diagnosis in clinically suspected MFS cases was 33.5 years in our cohort.
Figure 2.
Influence of variant type and diagnosis of Marfan syndrome on age of diagnosis. In cases with more than one P/LP variant or VUS, only the most damaging was included in this analysis. Age distribution and impact of age on variant type in whole cohort (a, b), and in non-Marfan patients (c, d).
TAAD patients with a family history were four times more likely to carry a P/LP variant than those without a family history (p=1.05e-4; Figure S1a). Gender did not significantly influence the likelihood of harbouring a P/LP variant within our cohorts (p=0.29; Figure S1b). A median maximum aortic size of 5.25cm was found in patients harboring P/LP variants compared to 5.1cm in those not harboring a P/LP variant (p = 0.45). Although aortic size is the main determinant for surgery, no association was found in our cohorts between maximum aortic size and variant categories (unpublished data). Similarly, splitting the data by pathology type and location yielded nothing significant (unpublished data).
To estimate the influence of clinical characteristics in determining the likelihood of obtaining a genetic diagnosis, we calculated the relative risk of detecting a P/LP variant by clinical phenotype (Table 3). Likelihood of genetic diagnosis was associated with presence of a related syndrome, a younger age at diagnosis, family history of aortic disease and ascending aortic aneurysm compared to other locations. All but 7 of patients harbouring a P/LP variant possessed at least one of these phenotypes. Other clinical features yielded small, non-significant risk.
Table 3.
Probability of harbouring a pathogenic or likely pathogenic variant according to phenotype
| Total | Number (Percentage) with a Pathogenic or Likely Pathogenic Variant Validated by Sanger | RR (95% CI) | P-Value | |
|---|---|---|---|---|
| Syndromic | 30 | 13 (43) | 11.94 (7.06-20.20) | 9.89e-11 |
| Young Age, < 50 | 237 | 26 (11) | 3.83 (2.20-6.67) | 4.28e-6 |
| Known or Probable Family History | 257 | 23 (9) | 3.80 (1.88-7.66) | 1.39e-4 |
| Ascending Aorta | 477 | 29 (6) | 1.84 (1.04-3.27) | 0.036 |
| Male | 694 | 28 (4) | 0.68 (0.39-1.20) | 0.20 |
| Presence of Dissection | 158 | 9 (6) | 1.28 (0.63-2.59) | 0.53 |
| Short-Term Mortality* | 575 | 24 (5) | 1.18 (0.42-3.32) | 1.00 |
| Large Aortic Diameter (>5cm) | 612 | 29 (5) | 1.05 (0.60-1.84) | 0.88 |
Mortality data was only available from the Yale cohort
A median age at diagnosis of 39 and 29 was observed in patients who carried a P/LP variant within FBN1 (n=25) and TGFBR2 (n=3) respectively (Figure S2). Of the 24 cases of clinically diagnosed MFS, 10 cases were identified as harbouring a P/LP variant in FBN1; the remaining case (Y_35_28) in whom a presumptive genetic diagnosis was made had a likely pathogenic variant in MYH11 (Table 2). Although no pathogenic FBN1 variants were found in this patient by our panel assay, subsequent clinical exome analysis identified a probable copy number variant in FBN1 in this patient (unpublished data), suggesting two possible genetic causes for this patient’s TAAD.
Discussion
To date, the majority of genetic abnormalities in TAAD have been identified in syndromic or familial cases 6. The aim of our study was to determine the prevalence of likely disease-causing variants in a mixed, clinically relevant cohort of patients with sporadic and familial TAAD. We sequenced 967 of 1025 unselected TAAD patients from the UK and USA above our target coverage threshold and identified a total of 49 P/LP variants in 47 patients that are the likely cause of their disease. To our knowledge, this is the largest genetic analysis of familial and sporadic cases of TAAD to date and defines a lower limit of 4.9% for the prevalence of TAAD in our cohorts with a pathogenic abnormality amongst most of the currently known TAAD genes. This frequency is similar to that of deleterious mutations (3.9%) reported previously in a series of 102 TAAD patients analysed by exome sequencing 9.
The majority (87.8%) of identified P/LP variants were found within known syndromic genes. Mutations in FBN1 accounted for more than half of all identified P/LP variants, with the majority affecting functionally significant domains of the gene. P/LP variants in FBN1 were discovered in 2.6% of the sequenced cohort and 5.8% of patients who had a probable or proven family history of TAAD. This is somewhat higher than was reported in a previous study of FTAAD, in which 2.7% of familial cases carried a pathogenic FBN1 variant 32.
FBN1 is clearly an important contributor to Mendelian cases of TAAD and MFS. A recent genome-wide association study showed that common variants in FBN1 are associated with STAAD, suggesting a common pathogenesis of thoracic aortic disease in MFS and STAAD 33. We found that only 40% of those with a P/LP variant in FBN1 had a known or suspected clinical diagnosis of MFS. Similarly, only 48.9% of those with a P/LP variant in any gene had a known or probable family history of TAAD. Although these results may be partly due to an incomplete clinical record (UK records did not definitively record the absence of a family history), they are also consistent with observations over the last 20 years that FBN1 mutations are a cause of TAAD in patients who do not have clinical MFS 32,34. In keeping with this, a recent study of probands with FBN1 mutations found that, only 56-79% met formal clinical criteria for MFS by Ghent systemic scores 35.
Patients Y_91_1 and Y_17_1 were each found to harbour two P/LP variants (in SLC2A10 and MYH11, and FBN1 and COL1A1 respectively). No common phenotypic features (young age-of-onset, family history) were seen in these two patients, although a much larger maximum aortic size was found in both patients (7cm and 6.5cm respectively) compared to the median maximum aortic size identified in patients harbouring a P/LP variant (5.25cm).
The age at diagnosis within our patients that have a P/LP variant (46.0 years) is much lower than has been observed previously in FTAAD (56.8 years) and STAAD (64.3 years) patients 36. This is likely due to the fact that 11 of the 47 patients harbouring a P/LP variant had clinically suspected MFS, 10 of whom had a P/LP variants in FBN1. The diagnosis of TAAD in MFS has been reported to have a lower age at diagnosis (24.8 years) than non-syndromic TAAD 36; our cohort had a similarly lower age at diagnosis (33.5 years). If the patients in our cohort with suspected MFS are removed, the median age of TAAD diagnosis in our cohort rises from 46.0 to 51.5 years. Overall, the percentage of patients with a P/LP variant was 3-4x higher within the under 50 age group than in those over the age of 50.
Although aortic size is the main determinant for surgery, we found no association between maximum aortic size and variant categories. This may in part be explained by early evaluation and surgical intervention in cases with syndromic presentations or family history of TAAD.
Comparing all 47 patients with a P/LP variant with the rest of the cohort, we were able to identify statistically significant risks of carrying a P/LP variant associated with developing the disorder: a syndromic component (RR 11.94; 95% CI 7.06-20.20), a younger age at presentation (RR 3.83; 95% CI 2.20-6.67), a probable or known family history of aortic disease (RR 3.80; 95% CI 1.88-7.66) and an aneurysm or dissection occurring partially or wholly within the ascending aorta (RR 1.84; 95% CI 1.04-3.27). The first three factors are clearly suggestive of Mendelian disease and may be suitable criteria for prioritizing TAAD patients for genetic testing if genetic testing is not applied to all cases. The fourth factor, disease location in the ascending aorta, is reflective of the known stronger genetic aetiology of TAAD in the ascending aorta compared to the descending aorta 9.
Previous studies have suggested that genetic testing should be undertaken in patients who present at a young age without any additional risk factors 37, although it is unclear to what extent this is implemented in routine clinical practice. Non-syndromic TAAD can have a similarly severe clinical course to those with syndromic TAAD 38, highlighting the importance of identifying genetically predisposed, non-syndromic TAAD patients prior to the development of symptoms. Seven (14.9%) patients harbouring a P/LP variant did not possess any of the above phenotypes (syndromic features, young age-of-onset, family history, involvement of the ascending aorta) associated with greater risk of a genetic aetiology. Therefore, whilst these risk factors increase the likelihood of identifying P/LP variants in known TAAD genes, restricting genetic testing solely to cases with these risk factors would miss a significant, albeit low percentage (14.9%) of cases in whom a genetic diagnosis could be made.
Our study has a number of limitations that include possibly having underestimated the number of truly pathogenic variants. We employed intentionally stringent criteria for defining P/LP variants, but this may have led to a number of truly pathogenic variants being classified as VUS’s. The difficulty in unequivocally designating variants as either P/LP or benign highlights the need for concerted efforts to systematically classify mutations in these disease genes, for example with prospective functional assays39. As more information becomes available we anticipate that many of the VUS identified in this study, particularly those with strong supporting in silico data, (Tables S8 and S9) may be unambiguously reclassified as causative variants. A further limitation to our study is that our sequencing assay is limited to the coding sequences and intron/exon boundaries of 15 TAAD genes that represent most of the common causes of TAAD - our study would not detect variants in as yet undiscovered genes or in non-coding sequence. In time, with greater understanding of genotype-phenotype correlation, whole genome sequencing could ultimately detect a higher frequency of pathogenic variants than detected herein. Indeed, we were unable to identify a P/LP variant in 44 of 62 individuals with three or more of the risk factors we identified as increasing the likelihood of harbouring a P/LP variant, and only eight of these individuals were found to carry a VUS. These individuals with a high relative probability of carrying a P/LP variant but no genetic diagnosis after panel testing are likely to be worth prioritising for future whole genome/exome sequencing studies.
In summary, we found 4.9% of patients carried a P/LP variant as the underlying cause of their TAAD, predominantly within FBN1 but with substantial contributions from TGFBR and COL genes. Consistent with previous reports, amongst these cases were patients with non-syndromic TAAD in whom we found P/LP variants in genes normally associated with connective tissue disease. A higher likelihood of harbouring a P/LP variant was found to be associated with a syndromic component to the disease, early age at presentation, positive family history and aneurysm location in the ascending aorta. However, restricting genetic testing only to TAAD patients with these features is likely to miss a small but significant number of cases in whom a definitive genetic diagnosis could be made.
Supplementary Material
Acknowledgements
We thank the patients for providing their informed consent to this research. The research was supported by a Medical Research Council transitional award to TJA, a Welcome Trust Clinical Fellowship grant to RW and by the National Institute for Health Research (NIHR) Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London. We acknowledge support from Laurence Game and the London Institute of Medical Sciences Genomics Facility.
Footnotes
Conflict of Interest
Tim Aitman reports grants from Wellcome, grants from National Institute for Health Research, grants from UK Medical Council, personal fees from Illumina, during the conduct of the study; personal fees from AstraZeneca, outside the submitted work.
Colin Bicknell reports personal fees from Medtronic, personal fees from Bolton Medical, non-financial support from Vascutek, non-financial support from Gore, outside the submitted work.
Data Access
All P/LP variants have been submitted to ClinVar database (SUB2992095). Detailed phenotype and genotype data is available on request. Source code for data cleaning and statistical analysis can be found at https://github.com/superDross/TAAD_analysis
References
- 1.Goldfinger JZ, Halperin JL, Marin ML, et al. Thoracic aortic aneurysm and dissection. J Am Coll Cardiol. 2014;64:1725–1739. doi: 10.1016/j.jacc.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka Y, Sakata K, Sakurai Y, et al. Prevalence of Type A Acute Aortic Dissection in Patients With Out-Of-Hospital Cardiopulmonary Arrest. Am J Cardiol. 2016;117:1826–1830. doi: 10.1016/j.amjcard.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Booher AM, Eagle KA. Diagnosis and management issues in thoracic aortic aneurysm. Am Heart J. 2011;162:38–46. doi: 10.1016/j.ahj.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Goodney PP, Travis L, Lucas FL, et al. Survival after open versus endovascular thoracic aortic aneurysm repair in an observational study of the medicare population. Circulation. 2011;124:2661–2669. doi: 10.1161/CIRCULATIONAHA.111.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erbel R, Aboyans V, Boileau C, et al. 2014 ESC guidelines on the diagnosis and treatment of aortic diseases. Eur Heart J. 2014;35:2873–2926. doi: 10.1093/eurheartj/ehu281. [DOI] [PubMed] [Google Scholar]
- 6.Milewicz DM, Regalado ES. Use of genetics for personalized management of heritable thoracic aortic disease: How do we get there? J Thorac Cardiovasc Surg. 2015;149:S3–S5. doi: 10.1016/j.jtcvs.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faivre L, Collod-Beroud G, Adès L, et al. The new Ghent criteria for Marfan syndrome: What do they change? Clin Genet. 2012;81:433–442. doi: 10.1111/j.1399-0004.2011.01703.x. [DOI] [PubMed] [Google Scholar]
- 8.Williams JA, Loeys BL, Nwakanma LU, et al. Early Surgical Experience With Loeys-Dietz: A New Syndrome of Aggressive Thoracic Aortic Aneurysm Disease. Ann Thorac Surg. 2007;83:S757–S763. doi: 10.1016/j.athoracsur.2006.10.091. [DOI] [PubMed] [Google Scholar]
- 9.Ziganshin BA, Bailey AE, Coons C, et al. Routine genetic testing for thoracic aortic aneurysm and dissection in a clinical setting. Ann Thorac Surg. 2015;100:1604–1612. doi: 10.1016/j.athoracsur.2015.04.106. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay ME, Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. doi: 10.1038/nature10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Guo DC, Cao J, et al. Mutations in myosin light chain kinase cause familial aortic dissections. Am J Hum Genet. 2010;87:701–707. doi: 10.1016/j.ajhg.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo D-C, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 13.Guo DC, Regalado E, Casteel DE, et al. Recurrent gain-of-function mutation in PRKG1 causes thoracic aortic aneurysms and acute aortic dissections. Am J Hum Genet. 2013;93:398–404. doi: 10.1016/j.ajhg.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle AJ, Doyle JJ, Bessling SL, et al. Mutations in the TGF-β repressor SKI cause Shprintzen-Goldberg syndrome with aortic aneurysm. Nat Genet. 2012;44:1249–1254. doi: 10.1038/ng.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabenwöger M, Alfonso F, Bachet J, et al. Thoracic endovascular aortic repair (TEVAR) for the treatment of aortic diseases: A position statement from the european association for cardio-thoracic surgery (EACTS) and the european society of cardiology (ESC) Eur J Cardio-thoracic Surg. 2012;42:17–24. doi: 10.1093/ejcts/ezs107. [DOI] [PubMed] [Google Scholar]
- 16.Baker MW, Atkins AE, Cordovado SK, et al. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: a technical feasibility study. Genet Med. 2015;18:231–238. doi: 10.1038/gim.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weerakkody RA, Vandrovcova J, Kanonidou C, et al. Targeted next-generation sequencing makes new molecular diagnoses and expands genotype-phenotype relationship in Ehlers-Danlos syndrome. Genet Med. 2016;18:1119–1127. doi: 10.1038/gim.2016.14. [DOI] [PubMed] [Google Scholar]
- 18.Vandrovcova J, Thomas ERa, Atanur SS, et al. The use of next-generation sequencing in clinical diagnosis of familial hypercholesterolemia. Genet Med. 2013;15:948–957. doi: 10.1038/gim.2013.55. [DOI] [PubMed] [Google Scholar]
- 19.Loeys BL, Dietz HC, Braverman AC, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47:476–485. doi: 10.1136/jmg.2009.072785. [DOI] [PubMed] [Google Scholar]
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. [Google Scholar]
- 21.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [Accessed October 20, 2017];2013 Mar; http://arxiv.org/abs/1303.3997.
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ, Lee JM, Benson M, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Weisburd B, Thomas B, et al. The ExAC browser: Displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45:D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei I, Jordan DM, Sunyaev SR. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. 2013 doi: 10.1002/0471142905.hg0720s76. Vol Chapter 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regalado ES, Guo DC, Santos-Cortez RLP, et al. Pathogenic FBN1 variants in familial thoracic aortic aneurysms and dissections. Clin Genet. 2016;89:719–723. doi: 10.1111/cge.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeMaire SA, McDonald M-LN, Guo D-C, et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat Genet. 2011;43:996–1000. doi: 10.1038/ng.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milewicz DM, Michael K, Fisher N, et al. Fibrillin-1 (FBN1) mutations in patients with thoracic aortic aneurysms. Circulation. 1996;94:2708–2711. doi: 10.1161/01.cir.94.11.2708. [DOI] [PubMed] [Google Scholar]
- 35.Faivre L, Collod-Beroud G, Child A, et al. Contribution of molecular analyses in diagnosing Marfan syndrome and type I fibrillinopathies: an international study of 1009 probands. J Med Genet. 2008;45:384–390. doi: 10.1136/jmg.2007.056382. [DOI] [PubMed] [Google Scholar]
- 36.Luyckx I, Loeys BL. The genetic architecture of non-syndromic thoracic aortic aneurysm. Heart. 2015;101:1678–1684. doi: 10.1136/heartjnl-2014-306381. [DOI] [PubMed] [Google Scholar]
- 37.De Backer J, Renard M, Campens L, et al. Genes in Thoracic Aortic Aneurysms and Dissections – Do they Matter? Translation and Integration of Research and Modern Genetic Techniques into Daily Clinical Practice. Aorta. 2013;1:135–145. doi: 10.12945/j.aorta.2013.13-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keramati AR, Sadeghpour A, Farahani MM, Chandok G, Mani A. The non-syndromic familial thoracic aortic aneurysms and dissections maps to 15q21 locus. BMC Med Genet. 2010;11:143. doi: 10.1186/1471-2350-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majithia AR, Tsuda B, Agostini M, et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.